Abstract
Background:
The global prevalence of non-alcoholic fatty liver disease (NAFLD) continues to rise. Non-invasive diagnostic modalities including ultrasonography and clinical scoring systems have been proposed as alternatives to liver biopsy but with limited performance. Artificial intelligence (AI) is currently being integrated with conventional diagnostic methods in the hopes of performance improvements. We aimed to estimate the performance of AI-assisted systems for diagnosing NAFLD, non-alcoholic steatohepatitis (NASH), and liver fibrosis.
Methods:
A systematic review was performed to identify studies integrating AI in the diagnosis of NAFLD, NASH, and liver fibrosis. Pooled sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and summary receiver operating characteristic curves were calculated.
Results:
Twenty-five studies were included in the systematic review. Meta-analysis of 13 studies showed that AI significantly improved the diagnosis of NAFLD, NASH and liver fibrosis. AI-assisted ultrasonography had excellent performance for diagnosing NAFLD, with a sensitivity, specificity, PPV, NPV of 0.97 (95% confidence interval (CI): 0.91–0.99), 0.98 (95% CI: 0.89–1.00), 0.98 (95% CI: 0.93–1.00), and 0.95 (95% CI: 0.88–0.98), respectively. The performance of AI-assisted ultrasonography was better than AI-assisted clinical data sets for the identification of NAFLD, which provided a sensitivity, specificity, PPV, NPV of 0.75 (95% CI: 0.66–0.82), 0.82 (95% CI: 0.74–0.88), 0.75 (95% CI: 0.60–0.86), and 0.82 (0.74–0.87), respectively. The area under the curves were 0.98 and 0.85 for AI-assisted ultrasonography and AI-assisted clinical data sets, respectively. AI-integrated clinical data sets had a pooled sensitivity, specificity of 0.80 (95%CI: 0.75–0.85), 0.69 (95%CI: 0.53–0.82) for identifying NASH, as well as 0.99–1.00 and 0.76–1.00 for diagnosing liver fibrosis stage F1–F4, respectively.
Conclusion:
AI-supported systems provide promising performance improvements for diagnosing NAFLD, NASH, and identifying liver fibrosis among NAFLD patients. Prospective trials with direct comparisons between AI-assisted modalities and conventional methods are warranted before real-world implementation.
Protocol registration:
PROSPERO (CRD42021230391)
Keywords: AI-assisted system, artificial intelligence, diagnostic tool, fatty liver, liver fibrosis, machine-learning, NAFLD, NASH, non-invasive tests
Introduction
Chronic liver disease (CLD) and cirrhosis have a high burden on global health. CLD is the 11th leading cause of death globally, attributing to 1.1 million deaths annually. 1 In previous decades, the major causes of cirrhosis were chronic hepatitis B (HBV) and hepatitis C (HBC) infection. More recently, the main causes of cirrhosis have shifted to non-alcoholic steatohepatitis (NASH). 2 The global prevalence of non-alcoholic fatty liver disease (NAFLD) is estimated at 25% and is predicted to increase up to 30% in 2030.3,4 Moreover, liver-specific deaths are also significantly increasing in patients with NAFLD, especially patients with NASH. 4 An updated term ‘metabolic associated fatty liver disease (MAFLD)’ has been proposed to replace NAFLD which establishes the disease as a metabolic disorder.5,6 This revision highlights the importance of early detection and risk factor modification to slow steatosis and fibrosis progression.
The gold standard for the diagnosis of NAFLD, NASH, and cirrhosis is liver biopsy. It provides an assessment of hepatic steatosis, inflammation, and fibrosis. However, liver biopsy is relatively invasive with complications, such as hemoperitoneum and hemothorax. 7 Due to its invasive nature, liver biopsy is also not pragmatic as a follow-up tool. Alternative diagnostic methods for NAFLD, such as clinical/laboratory scores and imaging modalities have been proposed, but with limited performance. For example, NAFLD Liver Fat Score has a sensitivity of 86% and specificity of 71%, 8 whereas ultrasonography have a reasonable performance for the diagnosis of moderate steatosis (>33% of hepatocytes contain steatosis) but is less reliable for mild steatosis (⩽33% steatosis). 9 Magnetic resonance imaging proton density fat fraction (MRI-PDFF) has greater accuracy but comes with a high cost and limited availability. 10 Moreover, limitations also extend to the detection of NASH and significant fibrosis among NAFLD patients. For example, the previously reported area under the receiver operating characteristic curves (AUROCs) for the diagnosis of NASH among NAFLD were up to 0.82 for ultrasonography scores (e.g. ultrasonography fatty liver indicator and ultrasonography fatty score) and 0.82 for transient elastography (TE). 11 On the contrary, the AUROCs for detecting significant fibrosis among NAFLD were 0.83 for TE, 0.88 for MRE and 0.64–0.75 for clinical scoring systems, for example, BARD score (0.64) and FIB-4 (0.75). 12 Artificial intelligence (AI) has begun to be incorporated into these clinical scoring systems and imaging modalities in order to improve diagnostic performance.
Over the past decade, AI has been used to identify and predict patterns or connections within large data sets in various fields of medicine, demonstrating particular usefulness in the diagnostic process. Previous systematic review of AI in hepatology reported on the utilization of machine-learning for assessing liver fibrosis, predicting liver decompensation, screening eligible liver transplant recipients as well as predicting post-transplant survival and complications.13,14 Another recent systematic review summarized the integration of AI in imaging modalities, digital pathology, and electronic health records for the diagnosis and staging of NAFLD. 15 The review emphasized on the high accuracy of AI-based system for NAFLD diagnosis and staging. However, very few meta-analyses have been conducted to summarize the overall diagnostic performance of AI-assisted diagnosis of liver diseases. 16 In this systematic review and meta-analysis, we aimed to determine the performance of AI-assisted systems for the diagnosis of NAFLD, NASH, and liver fibrosis.
Methods
The study was conducted based on the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) checklist. 17 The protocol was registered with PROSPERO (CRD42021230391).
Search strategy
The objective of the search was to identify studies utilizing AI in the diagnosis and classification of NAFLD, NASH and liver fibrosis among NAFLD patients. A literature search was conducted on MEDLINE, Scopus, Web of Science, and Google Scholar databases. The search was conducted from January 2000 through September 2021. We excluded studies published prior to the year 2000 to avoid obsolete computer-based algorithms which are not consistent with the modern AI classification. The keywords for the search included: ‘artificial intelligence’, ‘computer-assisted’, ‘computer-aided’, ‘neural network’, ‘machine learning’, ‘deep learning’, ‘liver’, ‘hepatic’, ‘steatosis’, ‘fatty’, ‘NAFLD’, ‘NASH’, ‘steatohepatitis’, ‘fibrosis,’ and ‘cirrhosis’. Due to the previously mentioned updated nomenclature, the search term ‘metabolic associated fatty liver disease’ or ‘MAFLD’ was also included. However, at the time of literature search, no studies with MAFLD and AI were identified. The search strategies for all databases are present in the Supplemental method.
Inclusion and exclusion criteria
We included articles using AI to assist in the diagnosis and grading of NAFLD. The inclusion criteria consisted of studies with sufficient data to generate a 2 × 2 table of true positive (TP), true negative (TN), false positive (FP), and false negative (FN). The articles also had to specify the reference standard (diagnostic method) and class(es) of AI. The exclusion criteria were studies which did not report the desired outcomes or did not have sufficient data to complete the 2 × 2 table. We also excluded studies that did not clearly describe validation methods or characteristics of training and validation cohorts. Studies in languages other than English as well as reviews, editorials, conference proceedings, and abstracts with incomplete information on the study population or characteristics of source image data sets were also excluded.
Data extraction
Two authors (PD and TT) independently screened the abstracts and titles to select the studies for full-text review. After screening, data extraction and quality assessment were also independently performed and cross-checked by the two authors (PD and TT). Any disagreements were discussed and decided by the third author (RC). Extracted data included author’s last name, publication year, study location, study design (prospective or retrospective cohort), validation methods (k-fold cross-validation and independent validation cohort), characteristics of training and validation cohorts (general population or at-risk population with specific diseases), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) as well as TP, TN, FP, and FN values. For studies with multiple AI classifiers, we selected the AI classifier with the best performance indicated by the best accuracy or greatest area under the curve (AUC).
Quality assessment
The quality of the studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool which is comprised of 12 questions assessing risk of bias and applicability in four domains (patient selection, appropriate index test, reference standard, and flow and timing). 18 As mentioned in our previous work, some questions were slightly modified to better assess the quality of AI-related studies. 16 For instance, the interpretation of the index test in clinical diagnostic studies should be conducted with an optimal pre-specified threshold in order to avoid overfitting. In AI-related research, separate validation or testing cohorts should be conducted in order to prevent the overfitting issue. Therefore, we assessed whether the included studies provided clear validation methods. Other questions for the assessment of human-oriented bias were also modified including whether knowing the reference standard results influenced the index test results. This was interpreted as a risk of bias caused by human manipulation in the AI protocol which could affect the AI output.
Statistical analysis
We used Covidence (Veritas Health Innovation, Melbourne, and Australia) for the screening, data extraction, and quality assessment process. After data extraction, TP, FP, TN, and FN values were exported from Covidence. If not available, the values were calculated from sensitivity, specificity, and prevalence using Review Manager version 5.3.5. 19 All statistical analysis was conducted using R software, version 3.6.3, Vienna, Austria. 20 Pooled sensitivity, specificity, PPV, NPV, and diagnostic odds ratio (DOR) with 95% confidence intervals (95% CI) were calculated using random effects model. Summary receiver operating characteristics (SROC) with AUCs were also generated. AUC values of 0.5–0.7, 0.7–0.9, and 0.9–1 indicated low, moderate, and high accuracy, respectively. Heterogeneity was assessed using I2 and Cochrane’s Q statistics. Publication bias was assessed with Deeks’ funnel plot. Subgroup analysis and meta-regression were pre-specified according to population, AI classifiers and diagnostic methods. P values of <0.05 were considered statistically significant. Sensitivity analysis was also performed by excluding studies with uncertain risk or high risk for bias and applicability assessed by the QUADAS-2 criteria.
Results
Literature search
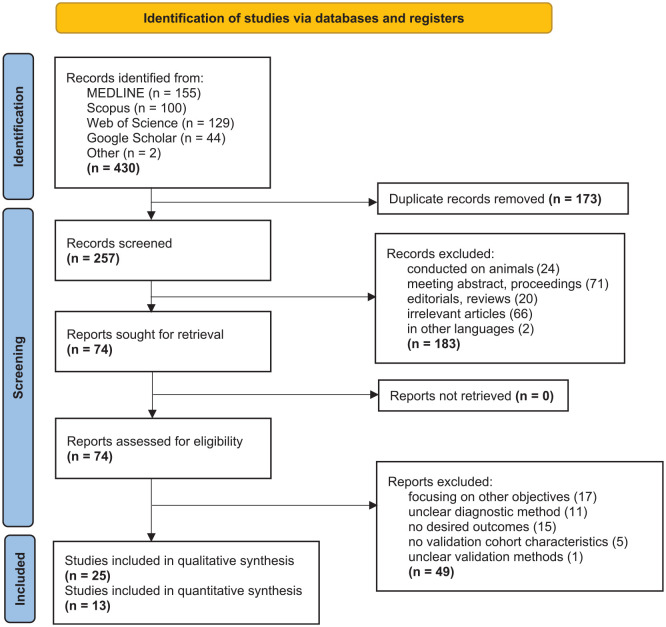
The searching process and results are shown in Figure 1. After literature search, a total of 430 articles were identified. After removing 173 duplicates, 257 abstracts were screened and 183 articles were excluded due to the following reasons: conducted on animals (n = 24), meeting abstracts or proceedings (n = 71), editorials or reviews (n = 20), irrelevant articles (n = 66), and written in languages other than English, that is, Chinese (n = 1) and Arabic (n = 1). Next, 74 full-text articles were assessed for eligibility and 49 were excluded due to the following reasons: focusing on other objectives (n = 17), unclear diagnostic method (n = 11), no desired outcomes (n = 15), no validation cohort characteristics (n = 5), and unclear validation methods (n = 1). A final total of 25 studies were included in the systematic review with 13 of the studies identified for the meta-analysis.
Figure 1.
Flow diagram of search methodology and literature selection process.
The studies in the systematic review were divided into 5 categories: (1) AI-assisted ultrasonography to diagnose NAFLD (n = 6), (2) AI-assisted analysis of clinical data sets to diagnose NAFLD (n = 6), (3) AI-assisted system for the diagnosis of NASH (n = 5), (4) AI-assisted system for the diagnosis of liver fibrosis in NAFLD (n = 5), and (5) AI-assisted system for steatosis quantification in pathological specimen (n = 4). One study evaluated both AI-integrated diagnosis of NASH and fibrosis among NAFLD patients, the result for each category was therefore extracted and included in the respective categories. 21 Seven studies contained multiple AI classifiers. For the studies with a single AI model (n = 18), nine studies used neural network models including artificial neural network (ANN) and convolutional neural network (CNN). Nine studies utilized non-neural network models such as regression tree (RT), rule extraction algorithm, and lasso regression. The diagnostic methods for NAFLD, NASH, and liver fibrosis applied in the included articles were liver biopsy, MRI, elastography, ultrasonography, and ultrasonography in combination with elevated liver chemistries. Details of the extracted data on the studies’ information, developmental and validation cohort characteristics, validation methods, diagnostic method, AI classifiers, and performance are shown in Table 1.
Table 1.
Characteristics of included studies in systematic review (13 studies included in meta-analysis are in bold).
| Study | Country | Study cohort | Diagnostic method | AI classifier | Development cohort (n) | Validation cohort (n) | Validation methods | Sensitivity | Specificity | TP | FP | TN | FN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AI-assisted ultrasonography to diagnose NAFLD | NAFLD/total, % Steatosis | NAFLD/total, % Steatosis | |||||||||||
| Kuppili et al.b22 | Portugal | Retrospective | Liver biopsy (not defined) | ELM a , SVM | 36/63 N/A |
N/A | k-fold cross-validation | 0.913 | 0.921 | 33 | 2 | 25 | 3 |
| Byra, et al.c23 | Poland | Prospective | Liver biopsy (>5% hepatocyte steatosis) | CNN | 38/55 50% had steatosis <30% |
N/A | 5-fold cross-validation | 1.00 | 0.882 | 38 | 3 | 15 | 0 |
| Biswas et al. c24 | Portugal | Retrospective | Liver biopsy (not defined) | CNN a , SVM, ELM | 36/63 N/A |
N/A | 10-fold cross-validation | 1.00 | 1.00 | 36 | 0 | 27 | 0 |
| Shi et al. 25 | China | Prospective | MRI (>5% hepatic fat content) | RT | 34/60 92% had steatosis < 20% |
N/A | 10-fold cross-validation | 0.875 | 0.9286 | 30 | 2 | 24 | 4 |
| Han et al. 26 | US | Prospective | MRI (>5% hepatic fat content) | CNN | 70/102 Average 11 ± 9% |
70/102 Average 11 ± 8% |
Validation cohort | 0.97 | 0.94 | 68 | 2 | 30 | 2 |
| Zamanian et al. c27 | Poland | Prospective | Liver biopsy (>5% hepatocyte steatosis) | CNN + SVM | 38/55 50% had steatosis < 30% |
N/A | 10-fold cross-validation | 0.972 | 1.00 | 70 | 0 | 74 | 2 |
| AI-assisted clinical data sets to diagnose NAFLD | NAFLD/total | NAFLD/total | |||||||||||
| Ma et al. 28 | China | Prospective | Ultrasonography | BN a , kNN, SVM, LR, NB, RF, BN, AdaBoost, HNB, Bagging, AODE | 2522/10,508 | N/A | 10-fold cross-validation | 0.675 | 0.878 | 1702 | 974 | 7012 | 820 |
| Islam et al. 29 | Taiwan | Retrospective | Ultrasonography | LR a , RF, SVM, ANN | 593/994 | N/A | 10-fold cross-validation | 0.741 | 0.649 | 439 | 141 | 260 | 154 |
| Wu et al. 30 | Taiwan | Retrospective | Ultrasonography | RF a , LR, ANN, NB | 377/577 | N/A | 10-fold cross-validation | 0.872 | 0.859 | 329 | 28 | 172 | 48 |
| Atabaki-Pasdar et al. 31 | United Kingdom | Retrospective | MRI (⩾5% hepatic fat content) | RF | 640/1514 | 1011/4617 | Validation cohort | 0.67 | 0.74 | 677 | 838 | 2668 | 334 |
| Chen et al. 32 | China | Retrospective | Ultrasonography | ANN | Total 10,354 | 2218/4436 | Validation cohort | 0.837 | 0.804 | 1857 | 435 | 1783 | 361 |
| Liu et al. 33 | China | Retrospective | Ultrasonography | XGBoost a , LR, SVM, SGD, CNN, MLP, LSTM | 4018/10,373 | 1860/4942 | Validation cohort | 0.611 | 0.909 | 1136 | 280 | 2802 | 724 |
| AI-assisted diagnosis of NASH in patients at-risk for NASH | |||||||||||||
| Gallego-Duran et al. 21 | Spain | Prospective | Liver biopsy | LR | NASH/NAFLD 21/39 |
NASH/NAFLD 44/87 |
Validation cohort | 0.87 | 0.60 | 38 | 17 | 26 | 6 |
| Naganawa et al. 34 | Japan | Retrospective | Liver biopsy | LR | Total 53 | NASH/non-NASH 7/28 |
Validation cohort | No suspicion of fibrosis: 1.00 Suspicion of fibrosis: 1.00 |
No suspicion of fibrosis: 0.92 Suspicion of fibrosis: 0.31 |
4 3 |
1 11 |
11 5 |
0 0 |
| Uehara et al. 35 | Japan | Retrospective | Liver biopsy | Rule extraction algorithm | NASH/non-NASH 79/23 |
NASH/non-NASH 65/12 |
Validation cohort | 0.862 | 0.417 | 56 | 7 | 5 | 9 |
| Garcia-Carretero et al. 36 | Spain | Retrospective | Ultrasonography with LFTs | Lasso regression | NASH/non-NASH 204/1587 |
NASH/non-NASH 51/397 |
Validation cohort | 0.70 | 0.79 | 36 | 83 | 314 | 15 |
| Docherty et al. 37 | United States | Retrospective | Liver biopsy | kNN, RF, XGBoost a | NASH/NAFLD 270/152 |
NASH/NAFLD 180/102 |
Validation cohort | 0.81 | 0.66 | 146 | 34 | 68 | 34 |
| AI-assisted diagnosis of liver fibrosis in NAFLD | |||||||||||||
| Pournik et al. 38 | Iran | Retrospective | Liver biopsy | ANN | Cirrhotic/non-cirrhotic 52/248 |
Cirrhotic/non-cirrhotic 15/65 |
Validation cohort | 0.657 | 0.987 | 44 | 4 | 309 | 23 |
| Gallego-Duran et al. 21 | Spain | Prospective | Liver biopsy | LR | F0-1/F2-4 20/19 |
F0-1/F2-4 56/31 |
Validation cohort | F2-4 0.77 | F2-4 0.80 | 24 | 11 | 45 | 7 |
| Shahabi et al. 39 | Iran | Retrospective | Elastography | ANN | F0/F1/F2/F3/F4 415/151/132/23/5 |
15% of data set (same proportion) |
Validation cohort | F1 0.993 F2 0.939 F3 1.000 F4 1.000 |
F1 0.757 F2 0.938 F3 0.993 F4 1.000 |
- | - | - | - |
| Okanoue et al.d40 | Japan | Retrospective | Liver biopsy and ultrasonography | ANN | Normal/F0/F1/F2/F3/F4 48/106/74/56/65/23 |
F0/F1/F2/F3-F4 17/18/15/24 |
Validation cohort | NAFLD (F0) vs. NASH (F1-4) 0.877 |
NAFLD (F0) vs. NASH (F1-4) 0.941 |
50 | 7 | 1 | 16 |
| AI-assisted diagnosis of liver fibrosis in NAFLD | |||||||||||||
| Okanoue et al.d41 | Japan | Retrospective | Liver biopsy | ANN | F0/F1/F2/F3/F4 106/74/56/65/23 |
F0/F1/F2/F3-F4 30/27/24/29 |
Validation cohort | F0 vs. F1-4: 0.85 F0-1 vs. F2-4: 0.755 F0-2 vs. F3-4: 0.828 |
F0 vs. F1-4: 0.867 F0-1 vs. F2-4: 0.877 F0-2 vs. F3-4: 0.877 |
68 40 24 |
4 7 10 |
26 50 71 |
12 13 5 |
| AI-assisted steatosis quantification of pathological specimen | |||||||||||||
| Vanderbeck et al. 42 | United States | Retrospective | Pathologist | SVM | Macrosteatosis/other features 1100/859 |
N/A | 10-fold cross-validation | 0.98 | 0.94 | 1072 | 48 | 859 | 28 |
| Liu et al. 43 | China | Prospective | Pathologist | Linear regression | Steatosis grade 0: 0 Grade 1: 77 Grade 2: 45 Grade 3: 24 |
Steatosis grade 0: 1 Grade 1: 41 Grade 2: 22 Grade 3: 9 |
Validation cohort | Steatosis grade 0 vs. ⩾ 1: 0.99 grade ⩽ 1 vs. ⩾ 2: 0.91 grade ⩽ 2 vs. 3: 0.67 |
Steatosis grade 0 vs. ⩾ 1: 1.00 grade ⩽ 1 vs. ⩾ 2: 0.85 grade ⩽ 2 vs. 3: 0.98 |
71 28 6 |
0 6 1 |
1 36 63 |
0 3 3 |
| Sun et al. 44 | United States | Prospective | Pathologist | CNN | 30 | 66 | Validation cohort | ⩾ 30% steatosis 0.714 |
⩾ 30% steatosis 0.973 |
15 | 2 | 73 | 6 |
| Teramoto et al. 45 | Japan | Retrospective | Pathologist | Logistic regression | Matteoni classification
46
Type 1/type 2/type 3-4 (NASH) 33/33/33 |
Matteoni classification Type 1/type 2/type 3-4 (NASH) 33/33/33 |
Validation cohort | type 1 vs. NASH: 0.879 type 2 vs. NASH: 0.909 |
type 1 vs. NASH: 1.00 type 2 vs. NASH: 0.909 |
29 30 |
0 6 |
66 60 |
4 3 |
ANN, artificial neural network; AODE, aggregating one-dependence estimators; BN, Bayesian network; CNN, convolutional neural networks; ELM, extreme learning machine; F0-4, METAVIR fibrosis staging; FLD, fatty liver disease; HNB, hidden naïve Bayes; kNN, k-nearest network; LFTs, liver function tests; LR, logistic regression; LSTM, long short-term memory; MLP, multilayer perceptron; MRI, magnetic resonance imaging; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; NB, naïve Bayes; RF, random forest; RT, regression tree; SGD, stochastic gradient descent; SVM, support vector machine; XGBoost, extreme gradient boosting.
Selected AI in the analysis.
Studies conducted on the same population cohorts.
Quality assessment by QUADAS-2 showed that most studies contained low risk of bias and had no applicability concerns, except for the four studies which contained uncertain risk of bias, one study with high risk of bias, and one study with high risk for applicability concerns. Studies with uncertain risk for bias were studies referring to both alcoholic and non-alcoholic fatty liver disease (n = 3) of which two of the three studies did not provide detailed distribution of the patient’s degree of liver steatosis which could affect the performance of AI-assisted methods. Another study with uncertain risk of bias used non-standard reference diagnostic methods, that is, ultrasonography with elevated liver function for the diagnosis of NASH (n = 1). The study with high risk of bias had multiple diagnostic methods, that is, using ultrasonography for control group and liver biopsy for NAFLD group. Regarding applicability concerns, one high-risk study included genomic data as AI inputs which could be difficult to obtain in clinical setting (n = 1). Detailed assessments for each study are summarized in Supplemental Table 1.
Performance of AI-assisted ultrasonography for the diagnosis of NAFLD
Systematic review included six studies incorporating AI into ultrasonography for NAFLD diagnosis.22–27 Three studies relied on multiple AI classifiers22,24,27 and three studies utilized a single AI classifier (2 CNN23,26 and 1 RT 25 ). Liver biopsy was employed as the diagnostic method for NAFLD in four studies,22–24,27 whereas the other two studies chose MRI-PDFF.25,26 Two studies included 50% of patients with less than 30% steatosis.23,27 One study consisted of 92% of patients with less than 20% steatosis 25 and the other study had a mean steatosis of 11%. 26 Two pairs of studies (Kuppili et al. 22 and Biswas et al., 24 Byra et al. 23 and Zamanian et al. 27 ) were conducted in the same patient cohorts. In the meta-analysis, we included one study from each population cohort which was more recent and reported the better performance of the AI system.24,27 Eventually, a total of four studies were included in the meta-analysis.24–27
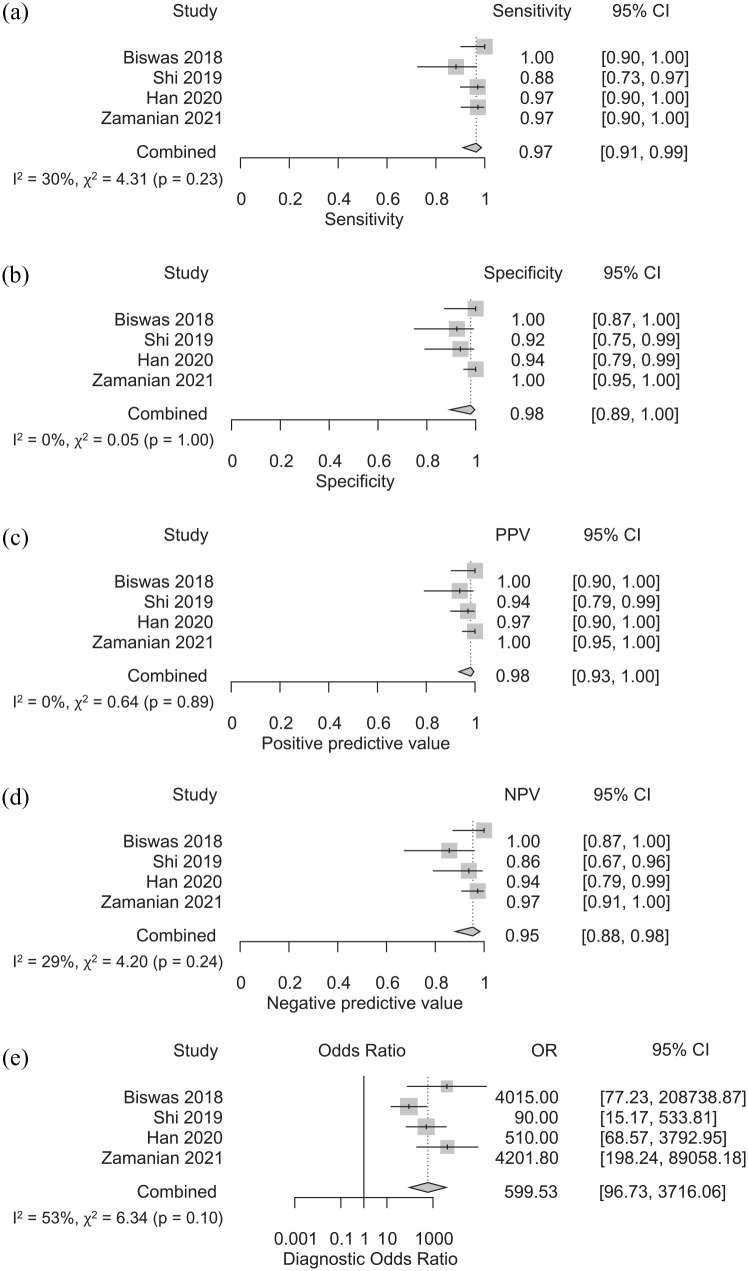
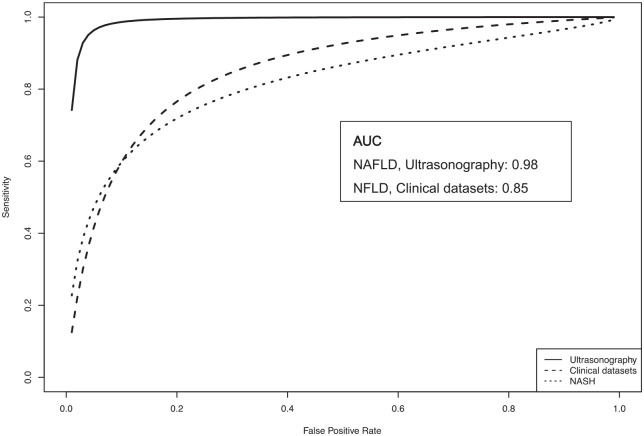
The pooled sensitivity, specificity, PPV, NPV, and DOR for the four studies was computed as 0.97 (95% CI: 0.91–0.99), 0.98 (95% CI: 0.89–1.00), 0.98 (95% CI: 0.93–1.00), 0.95 (95% CI: 0.88–0.98), and 599.53 (95% CI: 96.73–3716.06), respectively (Figure 2(a)–(e)). Heterogeneity was relatively low with I2 of 0 for pooled specificity and PPV, I2 of 30, 29, and 53 for sensitivity, PPV, and DOR, respectively. Cochrane’s Q results were also not significant (p ⩾ 0.1) for all analyses. SROC curve with AUC of 0.98 is shown in Figure 3.
Figure 2.
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of AI-assisted ultrasonography for the diagnosis of NAFLD.
Figure 3.
SROC curves demonstrating performance of AI-assisted diagnosis of NAFLD (AI-assisted ultrasonography and AI-assisted clinical data sets) and AI-assisted diagnosis of NASH).
We further performed meta-regression with the diagnostic method (liver biopsy vs MRI), AI classifier (neural network vs non-neural network) and study population (general population vs specific at-risk population) as covariates in order to determine whether these factors affected the overall results of the meta-analysis. The p values for the AI classifier, diagnostic method, and study population as covariates were 0.04, 0.04, and 0.23, respectively. This finding suggested that different AI classifiers and diagnostic methods significantly affected the overall performance of AI-assisted diagnosis of NAFLD.
Subgroup analysis by AI classifiers revealed that neural network AI had slightly higher sensitivity, NPV, and DOR than non-neural network AI, with sensitivity of 0.98 (95% CI: 0.94–0.99) vs 0.88 (95% CI: 0.73–0.97), NPV of 0.97 (95% CI: 0.92–0.99) vs 0.86 (95% CI: 0.67–0.96) and DOR of 1197.75 (95% CI: 255.84–5607.32) vs 90.00 (95% CI: 15.17–533.81), respectively (Supplemental Table 2). Nevertheless, interpretation of the subgroup analysis should be approached with caution because there was only one study that utilized non-neural network. Moreover, subgroup analysis by diagnostic method showed that studies using liver biopsy had higher NPV and DOR compared to studies using MRI with NPV of 0.98 (95% CI: 0.93–1.00) vs 0.90 (95% CI: 0.79–0.95) and DOR of 4130.95 (95% CI: 368.73–46279.93) vs 200.90 (95% CI: 36.88–1094.46), respectively. Interestingly, heterogeneity was also significantly lower in liver biopsy subgroup with I2 of 0 for all pooled analysis (Supplemental Table 2). However, no significant difference was found in the subgroup analysis based on the population. Sensitivity analysis excluding studies with uncertain or high risk for bias according to the QUADAS-2 showed consistent results with sensitivity, specificity, PPV, NPV, and DOR of 0.96 (95% CI: 0.87–0.99), 0.95 (95% CI: 0.88–0.98), 0.97 (0.93–0.99), 0.94 (95% CI: 0.82–0.98), and 336.58 (95% CI: 53.80–2105.69), respectively (Supplemental Table 3).
Performance of AI-assisted clinical data sets for the diagnosis of NAFLD
We performed a meta-analysis of six studies incorporating AI into clinical data sets for NAFLD diagnosis.28–33 Examples of clinical data sets primarily included demographic data (age, sex, weight, and height) and laboratory values (liver and renal function tests, lipid profile, and plasma glucose). Multiple AI classifiers were used in four studies,28–30,33 while the other two studies used a single AI classifier (1 ANN 32 and 1 random forest 31 ). Five articles selected ultrasonography as the diagnostic method,28–30,32,33 while one study relied on MRI. 31
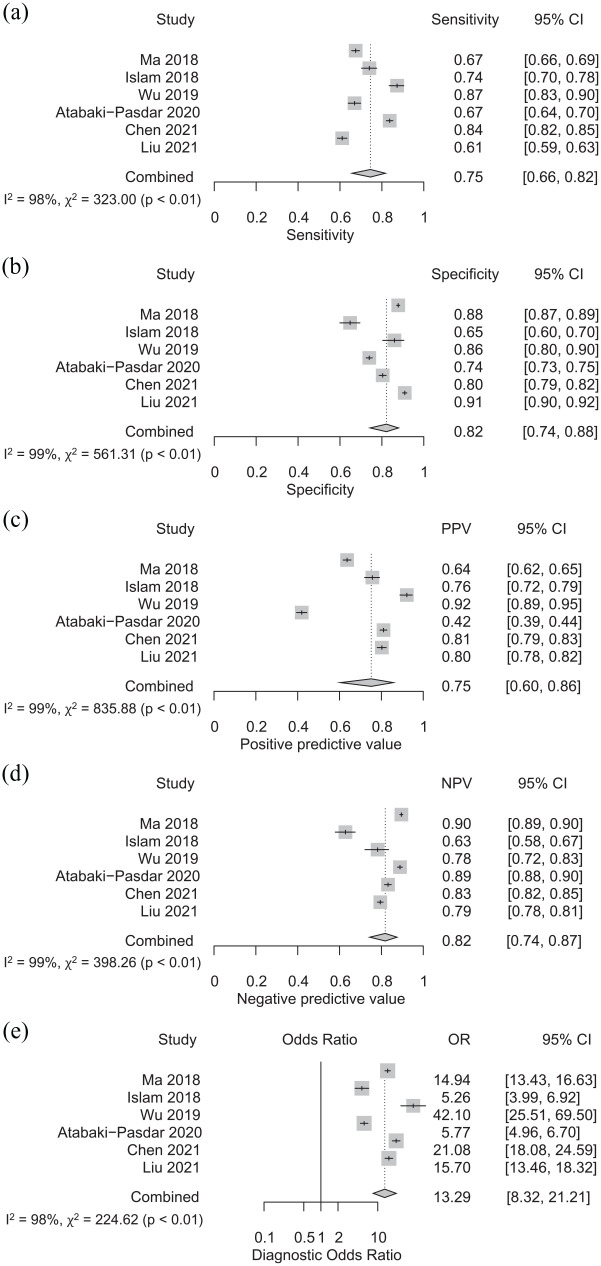
The pooled sensitivity, specificity, PPV, NPV, and DOR were 0.75 (95% CI: 0.66–0.82), 0.82 (95% CI: 0.74–0.88), 0.75 (95% CI: 0.60–0.86), 0.82 (0.74–0.87), and 13.29 (95% CI: 8.32–21.21), respectively (Figure 4(a)–(e)). Figure 3 shows the SROC with an AUC of 0.85. We observed a high degree of heterogeneity with I2 of 98%, 99%, 99%, 99%, and 98% for pooled sensitivity, specificity, PPV, NPV, and DOR, respectively.
Figure 4.
Sensitivity (a), specificity (b), positive predictive value (c), negative predictive value (d), and diagnostic odds ratio (e) of AI-assisted clinical data sets for the diagnosis of NAFLD.
Meta-regression performed with diagnostic method and AI classifier as covariates resulted in p values of 0.20 and 0.55, respectively. Subgroup analysis by AI classifiers revealed that neural network AI had slightly higher sensitivity and DOR than non-neural network AI, with sensitivity of 0.84 (95% CI: 0.82–0.85) vs 0.72 (95% CI: 0.63–0.80) and DOR of 21.08 (95% CI: 18.08–24.59) vs 12.09 (95% CI: 7.13–20.50), respectively. Subgroup analysis according to the diagnostic method yielded a higher specificity, PPV, NPV, and DOR of 0.84 (95% CI: 0.75–0.89), 0.80 (95%CI: 0.70–0.87), 0.80 (95%CI: 0.72–0.86), and 15.60 (95% CI: 10.62–22.92) for studies using ultrasonography (n = 5), compared to 0.74 (95% CI: 0.73–0.75), 0.42 (95% CI: 0.40–0.44), 0.89 (95% CI: 0.88–0.90), and 5.77 (95% CI: 4.96–6.70) for the study using MRI (n = 1). Since only one study utilized neural network as AI classifier and one study used MRI as the diagnostic method, interpretation of the subgroup analysis should be approached cautiously. Results for the subgroup analyses are presented in Supplemental Table 2. Sensitivity analysis, excluding articles with uncertain or high risk for bias revealed similar results with sensitivity, specificity, PPV, NPV, and DOR of 0.72 (95% CI: 0.63–0.80), 0.83 (95% CI: 0.72–0.90), 0.76 (95% CI: 0.69–0.82), 0.80 (95% CI: 0.70–0.88), and 12.94 (95% CI: 8.74–19.15), respectively (Supplemental Table 3).
Performance of AI-assisted diagnosis of NASH in patients at-risk for NASH
We identified five studies focusing on the diagnosis of NASH among patients with NAFLD or with at-risk for NAFLD (i.e. obese and hypertensive).21,34–37 In this category, two studies integrated AI with imaging modalities21,34 and three studies incorporated AI with clinical data sets.35–37 Almost all studies selected liver biopsy as the diagnostic methods, except for one study which used ultrasonography findings in combination with elevated liver enzymes. 36 The pooled sensitivity, specificity, PPV, NPV, and DOR for the diagnosis of NASH were 0.80 (95% CI: 0.75–0.85), 0.69 (95% CI: 0.53–0.82), 0.71 (95% CI: 0.36–0.91), 0.75 (95% CI: 0.35–0.94), and 8.27 (95% CI: 5.53–12.37), respectively. The heterogeneity was relatively high with I2 ranging from 0–98% (Supplemental Figure 1A–1E). SROC curve showed an AUC of 0.81 (Figure 3).
Performance of AI-assisted diagnosis of liver fibrosis in NAFLD
Systematic review included a total of five studies integrating AI for the diagnosis of liver fibrosis among NAFLD patients.21,38–41 However, the meta-analysis was not feasible due to differences in diagnostic modalities and outcomes of the included studies. Three studies integrated AI with clinical data38,39,41 and one study incorporated AI with imaging biomarkers 21 to evaluate liver fibrosis in NAFLD patients. The other study investigated AI-assisted clinical data sets for evaluating both the diagnosis of NASH and fibrosis. 40 Two studies conducted by the same investigator group contained overlapping study population.40,41 Regarding diagnostic methods in each study, three study relied on liver biopsy,21,38,41 one study used elastography 39 and one study selected liver biopsy and ultrasonography as diagnostic method for the NAFLD group and control group, respectively. 40 Overall, the reported sensitivity and specificity varied by different stages of fibrosis. For example, one study found that the performance for identifying METAVIR F1-F4 ranged from a sensitivity of 0.993 for F1 to 1.00 for F4 and a specificity of 0.757 for F1 to 1.00 for F4. 39
Performance of AI-assisted steatosis quantification in pathological specimen
Our systematic review identified four studies integrating AI with pathological imaging analysis for steatosis quantification and diagnosis of NAFLD.42–45 The outcome of each study was different from each other, including steatosis grading, differentiating macrosteatosis from other structures, identify significant steatosis or macrosteatosis and diagnosing NASH among NAFLD samples. Therefore, meta-analysis was not performed. All studies relied on pathologist as the reference standard. The diagnostic performance varied by outcomes of the study. For example, the AI-assisted identification of macrosteatosis showed a sensitivity and specificity of 0.98 and 0.94, respectively, 42 while the sensitivity and specificity for diagnosing ⩾30% steatosis were 0.714 and 0.973, respectively. 44 The performance of AI-assisted system for steatosis grading according to the NASH Clinical Research Network histological scoring system ranged from a sensitivity of 0.99 for grade 1 to 0.67 for grade 3 and a specificity of 1.00 for grade 1 to 0.98 for grade 3 steatosis. 43 Furthermore, the AI-assisted pathological identification of NASH among NAFLD had a sensitivity and specificity of 0.879–0.909 and 0.909–1.00, respectively. 45
Publication bias
In the Deeks funnel plot, the slope coefficients were relatively symmetrical with a p- value of 0.40 for AI-assisted ultrasonography for the diagnosis of NAFLD, 0.78 for AI-assisted clinical data sets for the diagnosis of NAFLD and 0.23 for AI-assisted clinical data sets for the diagnosis of NASH, indicating that no publication bias was detected for the selected studies (Supplemental Figure 2A–C).
Discussion
This systematic review and meta-analysis have identified many types of AI-assisted methods to diagnose NAFLD, NASH, and fibrosis among NAFLD patients and quantify liver steatosis in pathological specimens. Meta-analysis results showed excellent performance of AI-assisted ultrasonography for the diagnosis of NAFLD, with an AUC of 0.98 and relatively low heterogeneity. Combining AI with clinical data sets also demonstrated an acceptable performance level for the diagnosis of NAFLD, with an AUC of 0.85, with a higher degree of heterogeneity, which was likely due to variations in clinical input data.
Integrating AI into ultrasonography can improve the performance of NAFLD diagnosis. Ultrasonography is widely available in most hospitals and healthcare facilities. The equipment is also relatively inexpensive and the procedure is non-invasive. However, since the image analysis is user-dependent, it is also subject to inter- and intra-observer variations. The performance of conventional ultrasonography is often less reliable for the diagnosis of early-stage NAFLD. Therefore, incorporating AI with ultrasonography image analysis can minimize both human-related errors as well as improve overall performance. Our meta-analysis found that three out of the four studies had enrolled patients with mild steatosis (50–92% of patient cohorts had less than 30% steatosis) emphasizing the ability of AI-integrated methods to identify early-stage steatosis. The meta-analysis results show promising performance of AI-assisted ultrasonography with excellent sensitivity, specificity, PPV, and NPV of 0.95 and above as well as high accuracy with an AUC of 0.98. Heterogeneity assessment for AI-assisted ultrasonography was also relatively low with I2 of <40 for all pooled analyses except for the I2 of 53 for DOR. Subgroup analysis by AI classifier indicated the superior performance of neural network AI over non-neural network AI. Interestingly, subgroup also showed that studies using liver biopsy as diagnostic method has significantly lower degree of heterogeneity with I2 of 0 for all pooled analysis, implying that different diagnostic method could be the cause of heterogeneity for AI-assisted ultrasonography. Moreover, compared to currently available imaging modalities, the performance of an AI-assisted system for the diagnosis of NAFLD exceeded the performance of conventional ultrasonography, TE, and dual-gradient echo magnetic resonance imaging (DGE-MRI) reported in previous studies (Table 2).47,48 Our results support the benefits and robustness of using AI-assisted ultrasonography. Nevertheless, randomized controlled trials with head-to-head comparisons between AI-assisted system and conventional imaging modalities are warranted to validate the performance differences.
Table 2.
Comparisons between the performance of AI-assisted systems in this meta-analysis and the performance of conventional methods reported in previous studies for the diagnosis of NAFLD.
| Analysis | AI-assisted ultrasonography | AI-assisted clinical datasets | Conventional ultrasonography 48 (⩾5% steatosis) | Transient elastography 47 S0 vs S1–3 | DGE-MRI 48 (⩾5% steatosis) |
|---|---|---|---|---|---|
| Sensitivity | 0.97 (0.91–0.99) | 0.75 (0.66–0.82) | 0.62 (0.49–0.73) | 0.69 (0.60–0.75) | 0.77 (0.65–0.86) |
| Specificity | 0.98 (0.89–1.00) | 0.82 (0.74–0.88) | 0.81 (0.72–0.88) | 0.82 (0.76–0.90) | 0.87 (0.79–0.92) |
| Positive predictive value | 0.98 (0.93–1.00) | 0.75 (0.60–0.86) | 0.66 (0.53–0.77) | – | 0.78 (0.66–0.87) |
| Negative predictive value | 0.95 (0.88–0.98) | 0.82 (0.74–0.87) | 0.78 (0.69–0.85) | – | 0.86 (0.78–0.92) |
| AUC | 0.98 | 0.85 | – | 0.82 | 0.88 |
DGE-MRI, dual-gradient echo magnetic resonance imaging.
AI has also been employed to analyze large clinical data sets with various inputs, such as demographic data, physical findings, and laboratory results. The performance of AI in this category is promising but less satisfactory with findings showing only moderate accuracy compared to AI-assisted ultrasonography (AUC: 0.85 vs 0.98) and large heterogeneity (I2: 98–99%). To identify the source of heterogeneity, we performed a meta-regression which suggested that heterogeneity was not driven by various AI classifiers and diagnostic method. We hypothesized that the differences in performance are likely due to the differences in type and quantity of the AI inputs. The inputs for AI-assisted ultrasonography are usually images of the liver which contain diverse and potentially relevant features to be extracted by AI. The inputs for AI-assisted clinical data sets are limited to clinical parameters pre-selected by the investigators. The numbers of selected clinical parameters inputted in the AI were relatively small with great variation among the different studies. This may explain the lower performance and higher degree of heterogeneity. Nonetheless, the overall performance of AI-assisted clinical data sets was still comparable to those of TE and slightly lower than DGE-MRI as shown in Table 2. Incorporating AI into patient information already available in routine clinical practice could provide a preliminary screening method to identify patients at risk for NAFLD, especially in resource-limited settings where TE or MRI machines are unavailable or cost-prohibitive.
Other applications of AI in NAFLD are the identification of NASH and fibrosis which could offer tremendous clinical benefits as the degree of hepatic inflammation or fibrosis is associated with liver-related mortality. 4 Regarding the AI-assisted diagnosis of NASH, our meta-analysis showed an acceptable sensitivity of 80% and AUC of 0.8 but with relatively high heterogeneity. We hypothesized that the different diagnostic methods and different population might in part contribute to the high heterogeneity. Due to the limited number of studies included in the meta-analysis (n = 3), interpretation of the results needs to be done with caution. More studies in this topic are required for a more comprehensive analysis. Moreover, various scoring systems and imaging modalities have been proposed as screening tools for early detection of fibrosis, including aspartate aminotransferase-to-platelet ratio index (APRI) and Fibrosis-4 score (FIB-4). A previous meta-analysis reported a relatively low diagnostic performance of these conventional scoring systems, with a pooled sensitivity and specificity of 60% and 77%, respectively, for diagnosing significant fibrosis, and 67% and 77%, respectively, for diagnosing advanced fibrosis. 12 We found that when AI was integrated into the clinical datasets, it provided a better tool for screening fibrosis. For example, AI-integrated clinical data sets had a pooled sensitivity and specificity of 0.99–1.00 and 0.76–1.00 for diagnosing liver fibrosis stage F1–F4, respectively. Nevertheless, more studies focusing on using AI to improve diagnostic capabilities of clinical scoring systems are critically needed.
The last application of AI in NAFLD is to quantify liver steatosis in pathological specimens. Previous studies have shown that conventional identification of pathological specimen is susceptible to inter- and intra-observer variations and also considered to be a time-consuming process.49,50 AI-supported analysis has shown that it can provide reliable results with acceptable performance levels including a sensitivity and specificity of 0.71 and 0.97 for the diagnosis of more than 30% steatosis 44 as well as 0.67 – 0.99 and 0.85 – 1.00 for steatosis grading. 43
This manuscript represents one of the very first meta-analyses focusing on the application of AI in the diagnosis of NAFLD. In the production of this effort, we conducted a comprehensive literature search, including articles from medical journals, computer science, and engineering journals. Our selection criteria also only included articles with clear validation methods which is crucial for evaluating performance of AI technology. We do recognize some limitations remain present in this study. No AI algorithms were completely identical among the included articles. Since AI inputs were slightly different among the studies despite being classified as similar, interpretation of the pooled diagnostic performance must proceed with caution. More studies in each subgroup are required for comprehensive subgroup analysis. Another limitation is the difference in the diagnostic method among the included studies. The gold standard for the diagnosis of NAFLD and steatosis quantification is liver biopsy or MRI-PDFF as the best alternative. However, some studies integrating AI with clinical data sets instead relied on ultrasonography which may affect performance results. In order to accurately evaluate the performance of the AI-assisted diagnostic system, liver biopsy or MRI-PDFF should be employed as the diagnostic method for NAFLD. Finally, prospective or randomized controlled studies comparing AI-supported analysis with conventional methods would be beneficial in assessing the potential utility of AI in clinical practice.
Conclusion
AI-assisted ultrasonography and clinical data sets delivered satisfactory performance as a diagnostic tool for NAFLD. AI-assisted systems used in the identification of fibrosis and NASH as well as the quantification of steatosis of a pathological specimen also yielded promising results albeit the limited number of the studies available for review. Randomized controlled studies or prospective studies are warranted to validate the benefit of AI use in clinical setting.
Supplemental Material
Supplemental material, sj-docx-1-tag-10.1177_17562848211062807 for Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: a systematic review and meta-analysis by Pakanat Decharatanachart, Roongruedee Chaiteerakij, Thodsawit Tiyarattanachai and Sombat Treeprasertsuk in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-2-tag-10.1177_17562848211062807 for Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: a systematic review and meta-analysis by Pakanat Decharatanachart, Roongruedee Chaiteerakij, Thodsawit Tiyarattanachai and Sombat Treeprasertsuk in Therapeutic Advances in Gastroenterology
Footnotes
Author contributions: All authors were involved in the conception of the research. PD created the search strategy. Abstract screening and full-text assessment were individually performed by PD and TT. Data extraction and quality assessment were done by PD and TT. Data interpretation was performed by all authors. PD drafted the manuscript. Critical revision was done by RC and ST. All authors reviewed and provided final approval of the manuscript.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Ratchadapisek Sompoch Endowment Fund (2019) under Telehealth Cluster, Chulalongkorn University, Bangkok, Thailand and Chulalongkorn University CU-GRS-62-02-30-01.
ORCID iD: Roongruedee Chaiteerakij  https://orcid.org/0000-0002-7191-3881
https://orcid.org/0000-0002-7191-3881
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Pakanat Decharatanachart, Department of Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
Roongruedee Chaiteerakij, Division of Gastroenterology, Department of Medicine, Faculty of Medicine, Chulalongkorn University and King Chulalongkorn Memorial Hospital, Thai Red Cross Society, 1873 Rama IV Rd., Pathum Wan, Bangkok 10330, ThailandCenter of Excellence for Innovation and Endoscopy in Gastrointestinal Oncology, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
Thodsawit Tiyarattanachai, Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand.
Sombat Treeprasertsuk, Division of Gastroenterology, Department of Medicine, Faculty of Medicine, Chulalongkorn University and King Chulalongkorn Memorial Hospital, Thai Red Cross Society, Bangkok, Thailand.
References
- 1. Asrani SK, Devarbhavi H, Eaton J, et al. Burden of liver diseases in the world. J Hepatol 2019; 70: 151–171. [DOI] [PubMed] [Google Scholar]
- 2. Goldberg D, Ditah IC, Saeian K, et al. Changes in the prevalence of hepatitis C virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology 2017; 152: 1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018; 67: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Younossi Z, Tacke F, Arrese M, et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 2019; 69: 2672–2682. [DOI] [PubMed] [Google Scholar]
- 5. Eslam M, Newsome PN, Sarin SK, et al. A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J Hepatol 2020; 73: 202–209. [DOI] [PubMed] [Google Scholar]
- 6. Eslam M, Sanyal AJ, George J, et al. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 2020; 158: 1999–2014. [DOI] [PubMed] [Google Scholar]
- 7. Piccinino F, Sagnelli E, Pasquale G, et al. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68,276 biopsies. J Hepatol 1986; 2: 165–173. [DOI] [PubMed] [Google Scholar]
- 8. Kotronen A, Peltonen M, Hakkarainen A, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology 2009; 137: 865–872. [DOI] [PubMed] [Google Scholar]
- 9. Saadeh S, Younossi ZM, Remer EM, et al. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology 2002; 123: 745–750. [DOI] [PubMed] [Google Scholar]
- 10. Middleton MS, Van Natta ML, Heba ER, et al. Diagnostic accuracy of magnetic resonance imaging hepatic proton density fat fraction in pediatric nonalcoholic fatty liver disease. Hepatology 2018; 67: 858–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Besutti G, Valenti L, Ligabue G, et al. Accuracy of imaging methods for steatohepatitis diagnosis in non-alcoholic fatty liver disease patients: a systematic review. Liver Int 2019; 39: 1521–1534. [DOI] [PubMed] [Google Scholar]
- 12. Xiao G, Zhu S, Xiao X, et al. Comparison of laboratory tests, ultrasound, or magnetic resonance elastography to detect fibrosis in patients with nonalcoholic fatty liver disease: a meta-analysis. Hepatology 2017; 66: 1486–1501. [DOI] [PubMed] [Google Scholar]
- 13. Le Berre C, Sandborn WJ, Aridhi S, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology 2020; 158: 76–94.e72. [DOI] [PubMed] [Google Scholar]
- 14. Spann A, Yasodhara A, Kang J, et al. Applying machine learning in liver disease and transplantation: a comprehensive review. Hepatology 2020; 71: 1093–1105. [DOI] [PubMed] [Google Scholar]
- 15. Popa SL, Ismaiel A, Cristina P, et al. Non-alcoholic fatty liver disease: implementing complete automated diagnosis and staging. A systematic review. Diagnostics 2021; 11: 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Decharatanachart P, Chaiteerakij R, Tiyarattanachai T, et al. Application of artificial intelligence in chronic liver diseases: a systematic review and meta-analysis. BMC Gastroenterol 2021; 21: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021; 372: n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- 19. Review Manager (RevMan) [Computer program]. Version 5.3 ed. Copenhagen: The Nordic Cochrane Centre, the Cochrane Collaboration, 2014. [Google Scholar]
- 20. R: A language environment for statistical computing. Vienna: R Core Team, 2019. [Google Scholar]
- 21. Gallego-Duran R, Cerro-Salido P, Gomez-Gonzalez E, et al. Imaging biomarkers for steatohepatitis and fibrosis detection in non-alcoholic fatty liver disease. Sci Rep 2016; 6: 31421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuppili V, Biswas M, Sreekumar A, et al. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J Med Syst 2017; 41: 152. [DOI] [PubMed] [Google Scholar]
- 23. Byra M, Styczynski G, Szmigielski C, et al. Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg 2018; 13: 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biswas M, Kuppili V, Edla DR, et al. Symtosis: a liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed 2018; 155: 165–177. [DOI] [PubMed] [Google Scholar]
- 25. Shi X, Ye W, Liu F, et al. Ultrasonic liver steatosis quantification by a learning-based acoustic model from a novel shear wave sequence. Biomed Eng Online 2019; 18: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han A, Byra M, Heba E, et al. Noninvasive diagnosis of nonalcoholic fatty liver disease and quantification of liver fat with radiofrequency ultrasound data using one-dimensional convolutional neural networks. Radiology 2020; 295: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zamanian H, Mostaar A, Azadeh P, et al. Implementation of combinational deep learning algorithm for non-alcoholic fatty liver classification in ultrasound images. J Biomed Phys Eng 2021; 11: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma H, Xu CF, Shen Z, et al. Application of machine learning techniques for clinical predictive modeling: a cross-sectional study on nonalcoholic fatty liver disease in China. Biomed Res Int 2018; 2018: 4304376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Islam MM, Wu CC, Poly TN, et al. Applications of machine learning in fatty live disease prediction. Stud Health Technol Inform 2018; 247: 166–170. [PubMed] [Google Scholar]
- 30. Wu CC, Yeh WC, Hsu WD, et al. Prediction of fatty liver disease using machine learning algorithms. Comput Methods Programs Biomed 2019; 170: 23–29. [DOI] [PubMed] [Google Scholar]
- 31. Atabaki-Pasdar N, Ohlsson M, Viñuela A, et al. Predicting and elucidating the etiology of fatty liver disease: a machine learning modeling and validation study in the IMI DIRECT cohorts. PLoS Med 2020; 17: e1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen YS, Chen D, Shen C, et al. A novel model for predicting fatty liver disease by means of an artificial neural network. Gastroenterol Rep (Oxf) 2021; 9: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu YX, Liu X, Cen C, et al. Comparison and development of advanced machine learning tools to predict nonalcoholic fatty liver disease: an extended study. Hepatobiliary Pancreat Dis Int 2021; 20: 409–415. [DOI] [PubMed] [Google Scholar]
- 34. Naganawa S, Enooku K, Tateishi R, et al. Imaging prediction of nonalcoholic steatohepatitis using computed tomography texture analysis. Eur Radiol 2018; 28: 3050–3058. [DOI] [PubMed] [Google Scholar]
- 35. Uehara D, Hayashi Y, Seki Y, et al. Non-invasive prediction of non-alcoholic steatohepatitis in Japanese patients with morbid obesity by artificial intelligence using rule extraction technology. World J Hepatol 2018; 10: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Carretero R, Vigil-Medina L, Barquero-Perez O, et al. Relevant features in nonalcoholic steatohepatitis determined using machine learning for feature selection. Metab Syndr Relat Disord 2019; 17: 444–451. [DOI] [PubMed] [Google Scholar]
- 37. Docherty M, Regnier SA, Capkun G, et al. Development of a novel machine learning model to predict presence of nonalcoholic steatohepatitis. J Am Med Inform Assoc 2021; 28: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pournik O, Dorri S, Zabolinezhad H, et al. A diagnostic model for cirrhosis in patients with non-alcoholic fatty liver disease: an artificial neural network approach. Med J Islam Repub Iran 2014; 28: 116. [PMC free article] [PubMed] [Google Scholar]
- 39. Shahabi M, Hassanpour H, Mashayekhi H. Rule extraction for fatty liver detection using neural networks. Neur Comput Appl 2019; 31: 979–989. [Google Scholar]
- 40. Okanoue T, Shima T, Mitsumoto Y, et al. Artificial intelligence/neural network system for the screening of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatol Res 2021; 51: 554–569. [DOI] [PubMed] [Google Scholar]
- 41. Okanoue T, Shima T, Mitsumoto Y, et al. Novel artificial intelligent/neural network system for staging of nonalcoholic steatohepatitis. Hepatol Res 2021; 51: 1044–1057. [DOI] [PubMed] [Google Scholar]
- 42. Vanderbeck S, Bockhorst J, Komorowski R, et al. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum Pathol 2014; 45: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu F, Goh GB, Tiniakos D, et al. qFIBS: an automated technique for quantitative evaluation of fibrosis, inflammation, ballooning, and steatosis in patients with nonalcoholic steatohepatitis. Hepatology 2020; 71: 1953–1966. [DOI] [PubMed] [Google Scholar]
- 44. Sun L, Marsh JN, Matlock MK, et al. Deep learning quantification of percent steatosis in donor liver biopsy frozen sections. Ebiomedicine 2020; 60: 103029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teramoto T, Shinohara T, Takiyama A. Computer-aided classification of hepatocellular ballooning in liver biopsies from patients with NASH using persistent homology. Comput Methods Programs Biomed 2020; 195: 105614. [DOI] [PubMed] [Google Scholar]
- 46. Matteoni CA, Younossi ZM, Gramlich T, et al. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology 1999; 116: 1413–1419. [DOI] [PubMed] [Google Scholar]
- 47. Karlas T, Petroff D, Sasso M, et al. Individual patient data meta-analysis of controlled attenuation parameter (CAP) technology for assessing steatosis. J Hepatol 2017; 66: 1022–1030. [DOI] [PubMed] [Google Scholar]
- 48. Lee SS, Park SH, Kim HJ, et al. Non-invasive assessment of hepatic steatosis: prospective comparison of the accuracy of imaging examinations. J Hepatol 2010; 52: 579–585. [DOI] [PubMed] [Google Scholar]
- 49. Intraobserver interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. The French METAVIR Cooperative Study Group. Hepatology 1994; 20: 15–20. [PubMed] [Google Scholar]
- 50. Theodossi A, Skene AM, Portmann B, et al. Observer variation in assessment of liver biopsies including analysis by kappa statistics. Gastroenterology 1980; 79: 232–241. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tag-10.1177_17562848211062807 for Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: a systematic review and meta-analysis by Pakanat Decharatanachart, Roongruedee Chaiteerakij, Thodsawit Tiyarattanachai and Sombat Treeprasertsuk in Therapeutic Advances in Gastroenterology
Supplemental material, sj-docx-2-tag-10.1177_17562848211062807 for Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: a systematic review and meta-analysis by Pakanat Decharatanachart, Roongruedee Chaiteerakij, Thodsawit Tiyarattanachai and Sombat Treeprasertsuk in Therapeutic Advances in Gastroenterology