Abstract
Ischemic stroke (IS) is a detrimental neurological disease with limited treatments options. It has been challenging to define the roles of brain cell subsets in IS onset and progression due to cellular heterogeneity in the CNS. Here, we employed single-cell RNA sequencing (scRNA-seq) to comprehensively map the cell populations in the mouse model of MCAO (middle cerebral artery occlusion). We identified 17 principal brain clusters with cell-type specific gene expression patterns as well as specific cell subpopulations and their functions in various pathways. The CNS inflammation triggered upregulation of key cell type-specific genes unpublished before. Notably, microglia displayed a cell differentiation diversity after stroke among its five distinct subtypes. Importantly, we found the potential trajectory branches of the monocytes/macrophage’s subsets. Finally, we also identified distinct subclusters among brain vasculature cells, ependymal cells and other glia cells. Overall, scRNA-seq revealed the precise transcriptional changes during neuroinflammation at the single-cell level, opening up a new field for exploration of the disease mechanisms and drug discovery in stroke based on the cell-subtype specific molecules.
Keywords: Single cell RNA-seq, ischemic stroke, mouse model, cellular heterogeneity, neuroinflammation
Introduction
As the second leading cause of preventable deaths worldwide, 1 ischemic stroke (IS) can lead to long-term neurological and psychiatric disorders. The complexity of the brain structure and the diversity of cell types in the organ prevent us from understanding the molecular basis underlying stroke process. Given that regulation of gene expression plays a decisive role in the complex biological phenomenon incorporated by cells, emerging high-throughput sequencing technologies could contribute to interrogating the molecular underpinnings for pathological processes in IS, including necrosis and apoptosis of nerve cells, ischemic injury, treatment response, and inflammatory cell infiltration. Previous transcriptomic studies on IS 2 were generally focused on bulk tissue mixtures of cell aggregation, masking fateful changes of gene expression spectrum in the most susceptible cell subsets. Here, we performed single-cell transcriptomic analyses with the 10x genomics, 3 using thousands of individual brain cells from the animal with MCAO-commonly used as the model for ischemic stroke. As the stroke occurs suddenly, develops rapidly, and displays narrow treatment time window, early intervention in the acute phase would be particularly important for the treatment. Meanwhile, the above clinical characteristics of IS can explain why we collected the ischemic hemispheres at 24 hours after ischemia reperfusion.
To the best of our knowledge, this study is the first one to employ single-cell RNA sequencing to explore the different transcriptomic patterns among the brain cells as well as distinct gene expression properties of individual cells from the adult brain with ischemic insult. Our findings provided novel insights into the cellular and molecular heterogeneity in the brain at the acute phase of MCAO and unveiled known and novel markers for each of the main brain cell types. According to the analysis of different gene expression patterns, we identified multiple subpopulations within each cell type. Moreover, comparing with the sham samples, ischemic injury up- and downregulated numerous genes in each cellular subpopulation. Furthermore, we sought to determine the potential vulnerable cell types and new candidate biomarkers serving as molecular signatures of ischemic insult, contributing to a better understanding of the signaling pathways associated with therapeutic intervention. More importantly, the transcriptional regulatory network analysis uncovered distinct ischemia-associated regulatory modules in various microglia subtypes. Overall, this study provides a valuable large-scale resource for further exploring the molecular and cellular basis of pathogenesis in the mouse model of IS.
Materials and methods
All animal experimental procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Animal Experiments Ethics Committee of Tianjin Medical University General Hospital, which were in compliance with the ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. 4 The tissue dissociation, scRNA-seq, immunostaining, and computational methods are described in supplementary materials.
Results
Identification of major brain cell types and specific genes
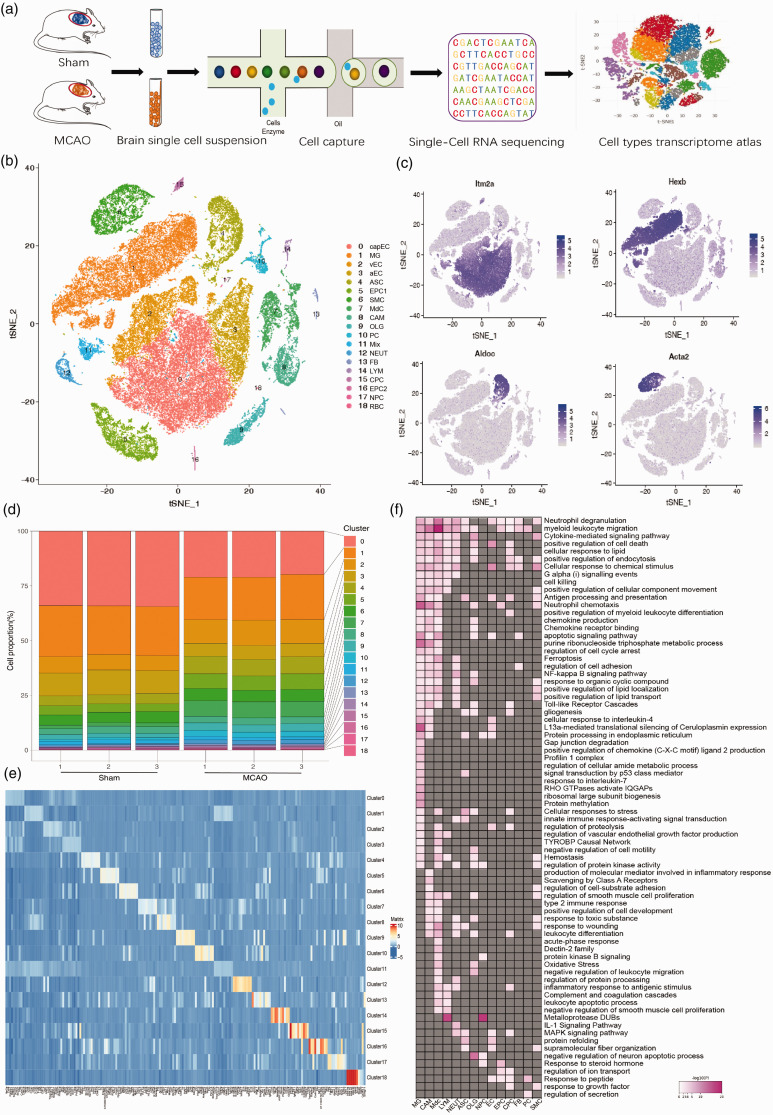
To examine cell heterogeneity and ischemic insult associated cellular alterations in gene expression profile following MCAO, we analyzed mRNA transcripts of single cells extracted from the brain using the 10x Genomics technology (Figure 1(a)). In the experiments, we collected the same left cerebral hemispheres from three sham mice without neurological disorders as well as three mice with MCAO 24 h after the modeling. To avoid any bias associated with the use of cell surface markers that may be differentially expressed in special populations, we isolated all single cells from the brain of MCAO and sham animals, respectively (Figure S1(a)). Data were subjected to a quality control (Figure S1(b) and (c)), and a total of 58,528 cells with a mean of 1,295 genes per cell and a mean of 2,610 unique molecular identifiers (UMIs) per cell were recovered. As depicted in Figure S2, all single cells were projected on t-distributed stochastic neighbor embedding (t-SNE) plot 5 after batch-effect correction 6 using Seurat 3, 7 and each individual cell from either MCAO or sham animals were distinctly assigned into different clusters. Detection of known cell type markers led us to identify 17 transcriptionally distinct clusters (Figure 1(b) and (e)), including vascular smooth muscle cells (SMC); perivascular fibroblast-like cells (FB); central nervous system (CNS)-associated macrophages (CAM); monocyte-derived cells (MdC); venous endothelial cells (vEC); capillary endothelial cells (capEC); arterial endothelial cells (aEC); pericytes (PC); choroid plexus capillary endothelial cells (CPC); ependymocytes (EPC); microglia (MG); neutrophils (NEUT); astrocytes (ASC); oligodendrocytes (OLG); neural progenitor cells (NPC);lymphocytes(LYM); red blood cell(RBC).
Figure 1.
Single-cell RNA sequencing of mouse brain recapitulates transcriptome atlas in ischemic stroke. (a) Graphical diagram of single cell isolation and RNA-seq experimental setup. (b) t-SNE plot visualizing clustering of single cells colored by cell types. (c) t-SNE plots visualizing the expression distribution of a set of selected marker genes, including Itm2a, Hexb, Aldoc, and Acta2. (d) Bar plot of the proportion of cells in each sample for each cell cluster identified. (e) Heatmap representing the top 10 most highly expressed genes within each cluster relative to the rest of clusters. (f) The pathway and biological process analyses between MCAO and sham group for each cell type cluster. Pink color indicates upregulation, Gray indicates no significant change. vascular smooth muscle cells (SMC); perivascular fibroblast-like cells (FB); CNS boreder-associated macrophages (CAM); monocyte-derived cells (MdC); venous endothelial cells (vEC); capillary endothelial cells (capEC); arterial endothelial cells (aEC); pericytes (PC); choroid plexus epithelial cells (CPC); ependymocytes (EPC), microglia (MG); neutrophils (NEUT); astrocytes (ASC); oligodendrocytes (OLG); neural progenitor cells (NPC); lymphocytes (LYM); red blood cell (RBC).
As shown in Figures 1(c) and S3(a), the cell clustering in this study can be validated by the majority of the previously published marker genes, for example, Hexb 8 for microglia; Aldoc 9 for astrocyte; Plp1 10 for oligodendrocytes; Ttr 11 for EPC; Itm2a for ECs; Acta2 for SMC. Furthermore, we showed selected marker genes identified for each cell cluster in violin plots (Figure S3(b)). To determine brain cell types susceptible to ischemic insult, we carried out a comprehensive comparison of the varying composition proportions among each cell type identified from both sham and MCAO groups. As shown in Figure 1(d), the infiltrating number of MdCs markedly increased in the MCAO group compared with sham counterpart (the cell ratio was shifted from 2% to 16%). The significant increase in recruiting MdCs to the ischemic hemisphere 12 was closely correlated with the key characteristics of MdCs during acute ischemic insult, including innate immunity, cell adhesion, and phagocytosis. Moreover, the comparative analysis showed that while the proportions of neutrophils, and pericytes increased considerably following stroke, a slight decrease in the percentages of vascular endothelial cells, perivascular fibroblast-like cells was evident. 13 Finally, the pathway and process enrichment analysis based on marker genes of each cell cluster revealed cell-type specific or shared biological and molecular function (Figure S4).
Overall assessment of DEGs and pathways associated with ischemic stroke
To investigate the DEGs and pathways or biological processes involved in stroke pathogenesis within each cell type identified above, we compared the significant gene changes between MCAO and sham groups, and summarized the top representative pathways and processes enriched from each cell-subset specific DEGs (Supplemental Table S2). Most strikingly, a total of 275 DEGs between MCAO and Sham samples with p-value <0.05 were identified in microglia, ranking microglia at the top of the list. This finding suggested that as innate immune cells of the brain, microglia may dominate prominent neuro-inflammation after ischemic stroke. Moreover, varied pathways or biological processes enrichment analysis of DEGs (up and downregulation genes) between MCAO and sham group were assigned to the following distinct cell types (Figures 1(f) and S5): neutrophil chemotaxis and apoptosis signaling pathway for microglia; signal transduction for astrocytes; regulation of neuron apoptotic process for oligodendrocytes; transport of anion and small molecules for endothelial cells; Response to steroid hormone for ependymal cells. Taken together, this study provided a context for a comprehensive understanding of cell-type specific DEGs associated with IS, as well as related pathways or biological processes in the brain inflammation following stroke.
In acute ischemic stroke, cell death was induced within minutes under hypoxia, and ischemic cascade reaction including glial activation, infiltration of peripheral immune cells, and release of inflammatory cytokines and chemokines occurred swiftly throughout the entire brain. 14 In this study (Supplemental Table S2), we detected an up-regulation in Ccl7 and Ccl12 in microglia, Ccl4 and Cdkn1a in astrocyte, and Ccl4 in ependymal cells, providing a large amount of evidence for the molecular and cellular foundations of inflammatory responses post MCAO. Secretion of these chemokines and cytokines from glia cells was initiated by the release of damage-associated molecular patterns (DAMPs) from the damaged tissues. 15 Subsequently, numerous infiltrating peripheral myeloid cells and lymphocytes characterized by increased expression of Ccrl2, Cxcl3, Ccl7, Ccl22 and many other inflammatory factors further aggravated brain injury by enhancing cell excitotoxicity. In sum, the present study provided a characterization of the full-scale global brain inflammation after stroke involving almost all specific cell types.
Cell-type specific gene expression changes following ischemic stroke
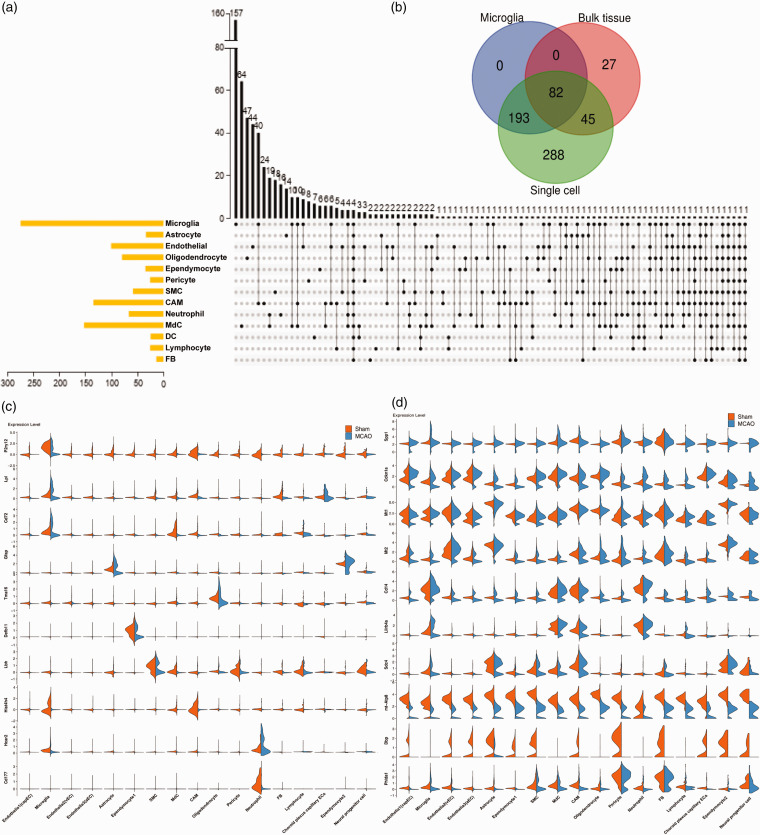
As shown in Figure 2(a), a large number of the DEGs identified after stroke changed in a cell type-dependent manner. Strikingly, the overlap DEGs analysis between the single-cell and bulk tissue revealed that approximately 80% of DEGs identified using single cell type were undetected at the whole brain level (Figure 2(b)). Moreover, shared DEGs between bulk tissue and the single cell displayed a broad spectrum of microglial DEGs (>50%), providing more evidence that microglia emerged as a central player in the inflammation of ischemic stroke. Meanwhile, microglia harbored 157 unique DEGs among all cell types and were located at the top of the list, followed by MdCs, oligodendrocyte, EC and CAMs. Besides, microglia and CAMs shared the most common DEGs post MCAO among all cell types. It has been reported that CAMs and microglia orchestrated the neuroinflammation in either cellular morphology or molecular function in CNS physiology and the pathogenesis of CNS diseases. While many cell types with a small population such as pericytes and FB were shown to interact with brain inflammatory factors, cell-type specific DEGs among these cell clusters may be undetected in whole tissue analysis. Hence, single-cell RNA sequencing has a great advantage over traditional microarrays and bulk RNA-seq, especially in recovering the rare cell types within a cell population that might play a key part in the regulation of diseases.
Figure 2.
The differential expression analysis of each cluster defined between Sham and MCAO groups. (a) UpSet plots showing unique or overlapping DEGs derived from the comparison between MCAO and Sham samples within each cluster. The black dots represent the DEGs that are shared by more than two cell types. The black bar graph above each plot represents the number of DEGs for each category and the yellow bars represent the total number of DEGs between MCAO and sham groups of each cell type. (b) The overlapping DEGs at the bulk tissue (calculated by averaging the gene expression between all single cells from MCAO group and all single cells from Sham group in our scRNA seq data), microglia and single cell levels. (c, d) Violin plots showing the normalized expression of cell-type specific (c) and shared-brain (d) DEGs between Sham and MCAO groups. The orange plots represent samples from Sham groups; blue plots represent samples from MCAO groups.
To investigate the potential molecular markers for diagnosis and treatment of the stroke, we sought to identify the unique and shared DEGs within each cell type between the sham and MCAO animals (Figure 2(c)). In the experiments, we observed that one P2 purinergic receptor gene P2ry12, known as a marker gene for microglia, was specifically down-regulated in microglia post-MCAO. This observation was consistent with the previous reports that the expression of P2ry12 was decreased upon microglia activation with an amoeboid round shape following inflammatory stimulation. 16 Moreover, we found that Lpl and CD72 presumably involved in scavenging, recycling of lipid debris, and regulation of proliferation of microglia 17 were specifically upregulated in microglia after MCAO, while the expression of GFAP, a crucial marker for reactive astrogliosis was increased primarily in astrocytes. 18 In addition, we identified the following typical DEGs across multiple cell types post-MCAO(Figure 2(d)): Spp1- osteopontin (OPN) associated with neuroprotective effects after ischemic stroke 19 ; Lilrb4a, one of the most enriched genes in microglia associated with neurodegeneration diseases 20 ; CXCL2, a ligand of CXCR2 in neutrophil infiltration after ischemic stroke; Saa3, serum amyloid A3 in response to injured tissues 21 ; MT-ATP8 encoding key enzyme of oxidative phosphorylation in the electron transport chain. 22
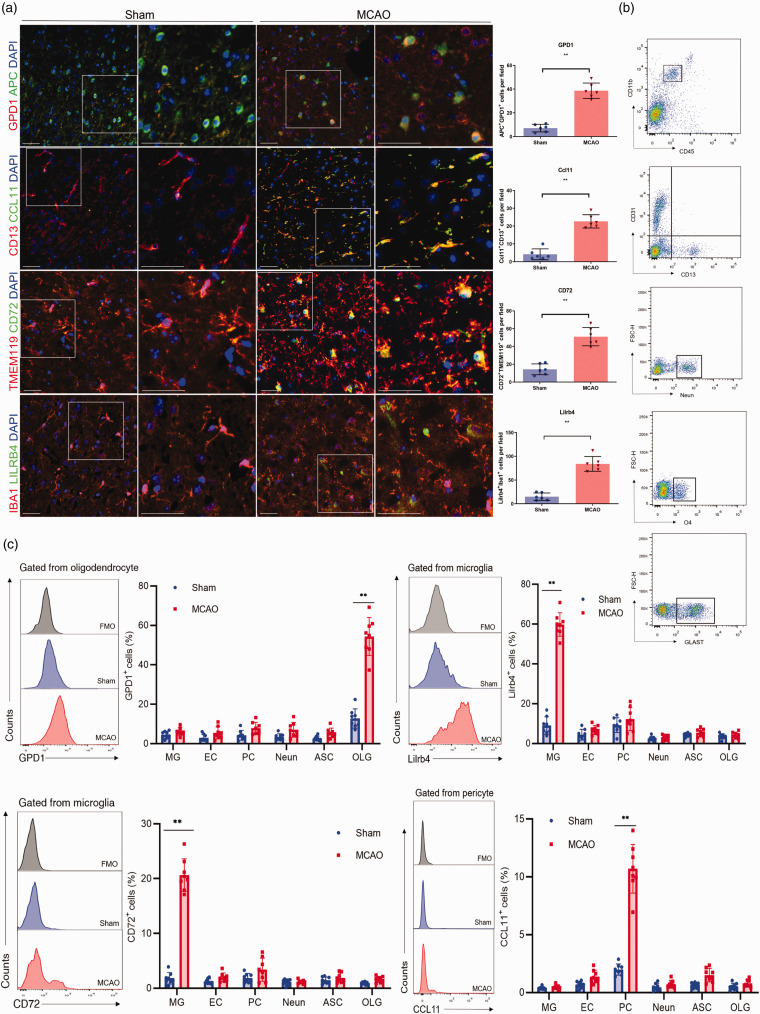
Moreover, to further identify whether our altered gene expression can be detected changes at the protein level, we also conducted immunofluorescence staining (Figure 3(a)) and flow cytometry analysis (Figure 3(b)). As a result, we indeed discovered the specific ischemic injury-related upregulation of GPD1 in oligodendrocyte, CCL11 in pericyte, CD72 and LILRB4 in microglia (Figure 3(c)). In sum, the cell-type specific DEGs perturbed by ischemic injury could be used as unique biomarkers for tracing the trajectory of the progression of the stroke, while the pan-brain DEGs across multiple cell types may provide the most vulnerable targets for the therapeutic interventions.
Figure 3.
Validation of cell-type-specific and shared ischemic injury-related DEGs. (a) Representative immunofluorescence staining showing the ischemic injury-related upregulation of GPD1 in oligodendrocyte (APC), CCL11 in pericyte (CD13), CD72 and LILRB4 in microglia (TMEM119, IBA1) at 24 h after MCAO. Scale bars,50 um. n = 6 per group. **P < 0.01 by unpaired two-tailed t test. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution. (b, c) Flow cytometry plots (b) and quantification (c) showed that ischemic injury-related oligodendrocyte highly expressed GPD1, pericyte highly expressed CCL11, microglia highly expressed CD72 and LILRB4 obtained from mice brain tissues at 24 h after MCAO. n = 8 mice per group. **P < 0.01 by unpaired two-tailed t test. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution.
Unique gene expression signatures of microglia subclusters
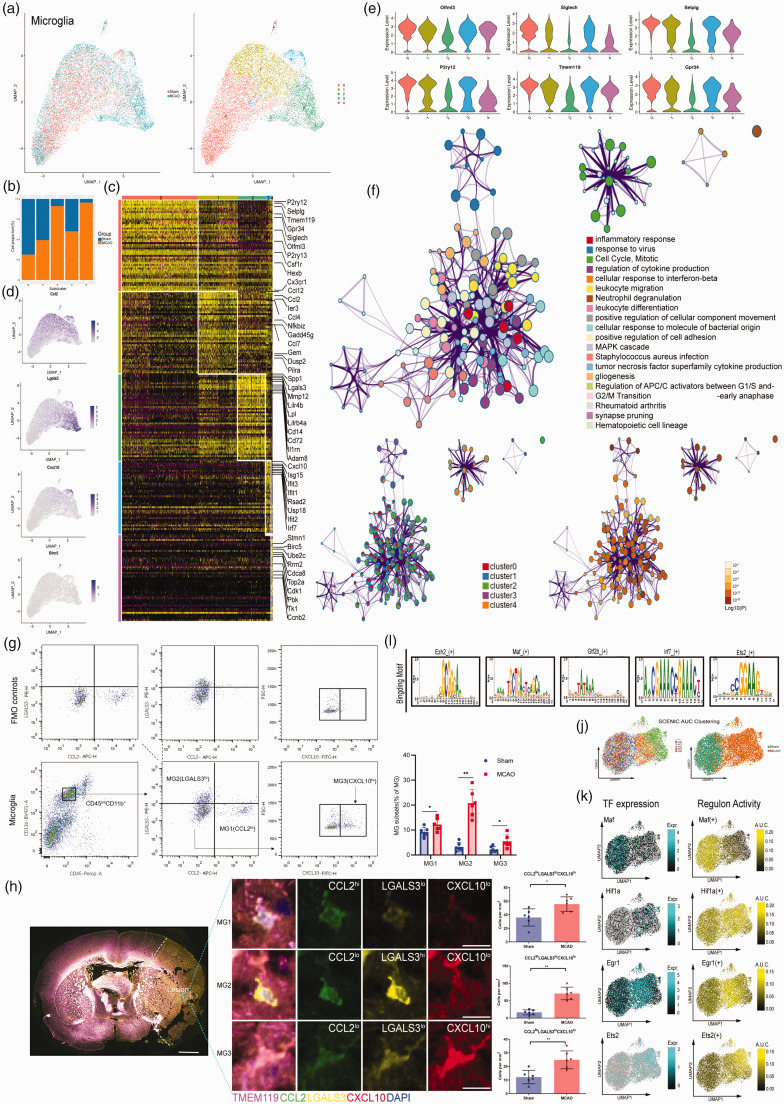
As the resident immune cells in the brain, microglia (MG) are responsible for carrying out remarkable and multiple functions during ischemic stroke. 23 Recently, numerous studies with scRNA-seq or CyTOF revealed microglia heterogeneity in development, homeostasis, and diseases. 24 In our study, we identified five unique cell clusters (Figure 4(a)). MG0 contained cells primarily from the sham group, and three clusters were mainly made up of cells from the MCAO group (MG 2, 3, 4). MG 1, however, was composed of equal proportion of cells from both the sham and MCAO group (Figure 4(b)). All MG subclusters expressed core microglial markers including Gpr34, Olfml3, P2ry12, TMEM119, Selplg and Siglech, which were relatively low expression in the most inflammatory ischemic injury-associated microglial subclusters (MG 2,3,4) (Figure 4(e)). This phenotype has also been observed in other injuries and diseases. 25 The MG1 cluster expressed not only the monocyte chemotactic protein (MCP) family genes such as CCL12, CCL2, and CCL7, 26 but also IER3 associated with hypoxia. 27 The MCP family genes have been shown to play a key role in pro-inflammatory reactions through chemotaxis of monocyte-derived macrophages and other peripheral immune cells to the ischemic brain. Although it contained nearly half amount of the cells from the sham group, the gene expression level of which was relatively lower than the MCAO group (Figure 4(d)). Notably, as the most inflammatory ischemic-associated subcluster, the MG2 cluster shared a list of significant genes with the neurodegeneration-associated microglial subset, 28 which includes Lgals3, Lilrb4, Lpl, Spp1 and Fth1. We also detected a high expression of matrix metalloproteinases 12 (MMP12) unique in MG2 cluster. MMP12 has been reported to cause great damage to the blood–brain barrier (BBB) after ischemic stroke. 29 Likewise, ADAM8 was found to be exclusively up-regulated in MG2. As a member of A Disintegrin and Metalloproteinase (ADAM) family, ADAM8 may modulate shedding of TNF-R1 in microglia-mediated neuroprotective effects. 30 Morever,the MG4 cluster was also involved in the most complex regulatory network of inflammatory response (Figure 4(f)). The ischemic lesions associated MG3 cluster highly expressed genes involved in IFN (interferon) signaling pathway such as Isg15, Ifit3, Irf7 and Cxcl10.And the gene ontology (GO) network analysis for MG3 was also enriched for response to virus and cellular response to interferon-beta(Figure 4(f)). MG4 cluster expressed genes related to proliferation markers, such as Birc5, Ube2c, Top2a and Stmn1, suggesting that they are a proliferating pool of microglia, which was consistent with the gene enrichment result of the cell cycle, mitotic and G2/M transition as shown in Figure 4(f).The further verification for the most inflammatory reaction subsets could be confirmed in flow cytometry (Figure 3(g)) and immunohistochemistry (Figure 4(h)) with MG1 as CCL2hiLGALS3loCXCL10lo, MG2 as CCL2loLGALS3hiCXCL10lo, and MG3 as CCL2loLGALS3loCXCL10hi.
Figure 4.
Single-cell transcriptome analysis of the microglia. (a) UMAP showing the disease origin of each individual cell (left panel) and the distribution of each subtype of microglia (right panel). (b) The bar graph showing the composition of cells in subclusters by disease state. (c) A heatmap showing the top 50 marker genes of the subclusters. (d) UMAP plots showing the expression level of marker genes for each microglia subcluster(m1-m4). (e) Violin plot depicting the expression levels of known core signature genes for each subtype of microglia. (f) Representative enriched ontology network of the five microglia subtypes with P-value. (g) FACS and staining strategy for microglia, MG1(CCL2+LGALS3−CXCL10−), MG2 (CCL2−LGALS3+CXCL10−), and MG3 (CCL2−LGALS3−CXCL10+). Fluorescence minus one (FMO) control (top) were used to control for spillover-related contribution to background in each channel. n = 6 mice per group. *P < 0.05, **P < 0.01 by unpaired two-tailed t test. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution. (h) (Left) Representative immunofluorescence pictures of coronal overview of the spatial evolution of the microglia subsets at 24 h after MCAO. Scale bars, 1 mm (overview) and 10 µm (inset). (Right)Representative immunofluorescence pictures depicting MG1(CCL2hiLGALS3loCXCL10lo), MG2 (CCL2loLGALS3hiCXCL10lo), and MG3 (CCL2loLGALS3loCXCL10hi) subsets within the core of ischemic lesions of the brain. Scale bars,10 µm. n = 6 per group, *P < 0.05, **P < 0.01 by unpaired two-tailed t test. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution. (i) The representative motif logo for the regulon in five microglia subclusters. (j) UMAP plots based on the SCENIC regulon activity-barcodes, colored by the subtypes of microglia identified above (left panel) and disease of origin (right panel). (k) The expression level of the indicated TFs and activity of the corresponding regulon in individual microglia as determined by SCENIC analysis and projected onto the UMAP plot representing the dataset.
To explore microglia regulatory network inference, we also applied SCENIC 31 analysis, which revealed distinct transcription factor (TFs) for these five subsets (Figure 4(i) and (j)). For example, we identified Dbp, Klf2 and Maf as candidate TFs shared by the MG0, 1, 2, and 3 from the sham group (Figure S6). In cluster MG0-4 mainly from the MCAO group, we observed preferential activity of regulons corresponding to e.g. hypoxia sensor Hif1a (Figure 4(k)), as well as Spi1, a transcription factor with a critical role in enhancing monocyte‑specific promoter activity and affecting NF‑κ B associated TNF-α signaling pathway. 32 Some TFs associated with immediate early gene family, were significantly upregulated as early as 3 h after ischemia reperfusion and rapidly induced at the onset of hypoxic conditions, 33 such as Fos, Egr1 and cebpb, which were identified in MG0 and 1 cluster from MCAO group. Noteworthy, we also revealed candidate regulons for the major ischemic injury associated MG2 cluster such as Creb3, Sp3 and Ets2 (Figure S6). As a transcriptional repressor, Sp3 was reported to be involved in the regulation of the NCX1 expression in the brain ischemia. 34 It is well reported that Ets2 was crucial for TNFα-induced expression of MCP1, IL6, and vascular cell adhesion molecule 1 and induction of an intraplaque proinflammatory phenotype in endothelial cells. 35 Regulons related to type I IFN response had enriched activity in MG3 cluster (Stat1, Stat2 and Irf7). Furthermore, MG4 cluster cells were enriched for Ezh2, Mybl2 and Nfyb activities (Figure S6). Ezh2 was required during cell mitosis and responsible for regulating development and differentiation. 36
Uncovering CNS border-associated macrophages subsets during ischemic stroke
The neuroinflammation in the CNS was not only attributed to the parenchymal microglia, but also associated with the non–parenchymal macrophages at the CNS borders, including meningeal, perivascular and choroid plexus macrophages (collectively known as CAMs). 37 We identified six distinct subsets of CAMs (Figure S7(a)),which all expressed the core signature genes(Lyv1,Cd163,Mrc1,Cbr2) (Figure S7(c)). According to the cell proportion transition upon ischemic injury among the six subsets (Figure S7(b)),we could easily infer the CAM4 mainly from the MCAO group with higher levels of MHCII-related antigen presentation molecules(such as H2-Aa, H2-Ab1 and Cd74) (Figure S7(d)).Besides, we also found a distinctive cluster primarily origin from the MCAO group-CAM5 characterized with oxidative phosphorylation and respiratory electron transport chain expressing Cox7b,Cox8a and Uqcr11. We also recovered brain resident macrophages populations-CAM1 enriched in the process of endocytosis, and predicted that the expression of Mgl2 and Timd4 38 might help distinguish these cells from recruited monocyte-derived macrophages. In addition, other clusters including CAM0 with higher levels chemotaxis genes (Ccl2, Cxcl1 and Cxcl2), CAM2 showing a similar expression pattern (Spp1, Lpl, Lilrb4,Lgals3) with the CPepi-BAMs (choroid plexus epithelial border-associated macrophages) 39 and CAM3 expressing Type I IFN response genes(Ifit3,Isg15,Rsad2 and Irf7).
Heterogeneity of HSC-derived circulating myeloid cells during ischemic injury
To further investigate the potential interaction between hematopoietic stem cell (HSC) derived myeloid subsets, including the major infiltrated APCs (antigen presenting cells including monocytes/macrophages (Mon/Mφ), dendritic cells(DCs) and lymphocytes (T, B and NK cells), we extracted them and examined in a new clustering analysis (Figure S8(a)). Analyzing the cell numbers and proportion (Figure S8(b)), we found that the majority of infiltrating cells were Mon/Mφ, followed by DCs. Mon/Mφ were highly actived during IS and exhibited a markedly different phenotypic signature as we identified including Mo/Mφ_Spp1(Fn1, Spp1, Ctsl), Mo_Cxcl2(Thbs1, Cxcl2), Ly6chiMo(Plac8,Ly6c2), Ly6cloMo(Ace,Eno3,Ear2), Res-like Mφ(C1qa,C1qb,C1qc),MHCII Mφ(Cd74,H2-Ab1,H2-Aa) and IFNIC Mo(Cxcl10,Ifit1,Isg15) (Figure S8(c)).
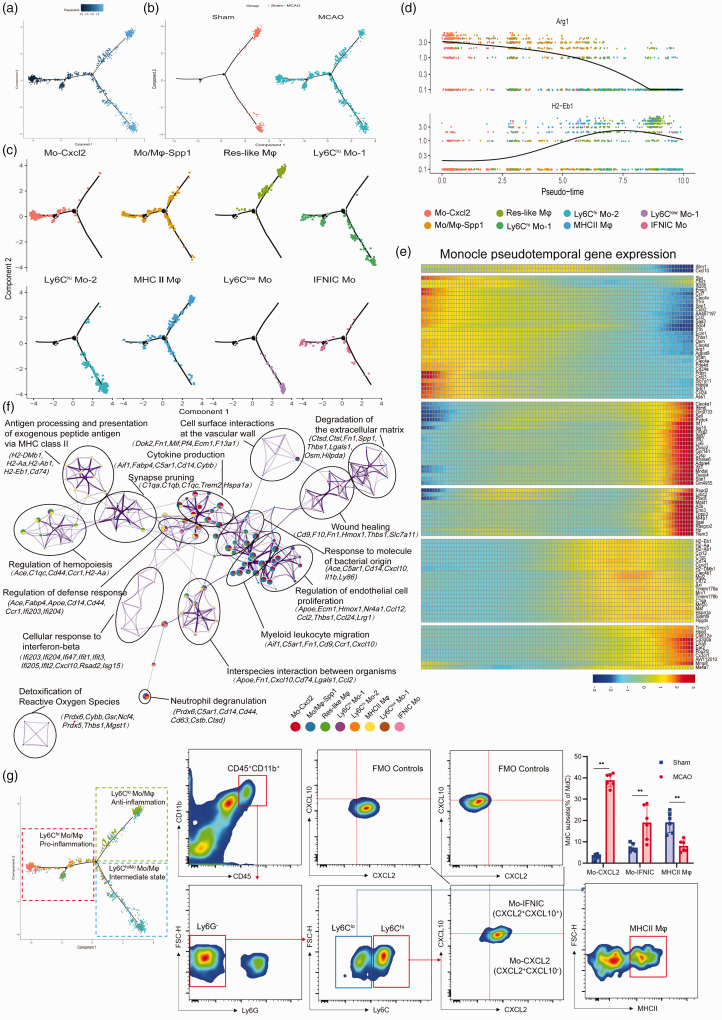
To identify ischemic insult associated Mon/Mφ on the homeostatic state and distinguish differentiated cell states, we performed BEAM algorithm in Monocle 2. 40 In the experiments, each dot represents a single-cell expression profile ordered in “pseudotime”, while the line connecting the dots outlines a fate trajectory of relatedness in their expression patterns (Figure 5(a)). We found the Mon/Mφ from the sham group occupied two separate trajectory branches at the original point, whereas the MCAO subclusters straddled all three branches (Figure 5(b)). As depicted in Figure 5(c), the pseudotime analysis of the cells ordered mainly based on Seurat-defined Mon/Mφ subclusters (as mentioned in Figure S8) revealed a distinct differentiation trajectory diverging along three major bifurcation. Mon/Mφ in the Ly6clo Mo and Ly6chi Mo_2 clusters occupied a separate trajectory branch, while the Res-like Mφ occupied a second branch. Interestingly, Mo_Cxcl2, IFNIC Mo and Mo/Mφ occupied the residual branches. Then, the continuous fluctuation of differentially expressed genes were displayed in accordance with the pseudotime order to track changes via different Mon/Mφ fates. The expression level of Arg1, Clec4d, and Osm expression were low in the Ly6clo Mo and Ly6chi Mo_2 clusters, higher in the Mo/Mφ_Spp1 cluster and even greater in the Mo_Cxcl2 cluster (Figure 5(d) and (e)), indicating the dynamic changes of these genes followed the Mon/Mφ states. In contrast, major histocompatibility complex (MHC) molecules (such as H2-Eb1, H2-Aa and Cd74) seemed to lose the expression along from the MHCII Mφ cluster to the Mo_Cxcl2 cluster. The IFNIC Mo cluster highly expressing genes associated with type I interferons (IFNs)-mediated signaling showed an intermediate transition state. Exploring the changes of gene expression profile according to Mon/Mφ fates suggested the potential on/off switches across different Mon/Mφ clusters and the dynamics of the various cellular processes. Given that ischemic injury-associated Mon/Mφ exhibit distinct phenotypes, we then constructed gene ontology (GO) networks based on each unique subcluster marker genes. We found that the eight subclusters were not separated from each other. Instead, they shared common gene sets and related enriched ontology terms across subcluster-specific genes to a certain extent (Figure 5(f)). Mo_Cxcl2 and Mo/Mφ_Spp1 clusters-enriched genes were associated with the process of myeloid leukocyte migration and cytokine production, while universally upregulated MHCII Mφ cluster genes were linked to antigen presentation and presentation. Genes that were restricted to Ly6chi Mo_2 cluster revealed cluster-specific gene ontologies, including detoxification of reactive oxygen species. Furthermore, flow cytometry also identified the dynamic change of the Mon/Mφ subclusters between the sham and the MCAO group (Figure 5(g)).
Figure 5.
Single-cell trajectory analysis of monocyte/macrophage clusters. (a) Pseudotime analysis of monocytes/macrophages using using Monocle, display based on (b) tissue origin and (c) Seurat-based clusters. (d) Dot plots showing differential expression genes according to pseudotime order, overlapped with Seurat’s original cluster colors superimposed. (e) Heatmap of differentially expressed genes, ordered based on their similar dynamic trend following pseudotime process. (f) Gene ontology network analysis of monocytes/macrophages clusters relied on DEGs between them ((adjusted P < 0.01, log 2 (FC) > 1)). Each node corresponds a gene set. Each color represents a monocytes/macrophages cluster. Representative genes of each gene set are showed. (g) FACS and staining strategy for monocyte-derived cells, Mo-CXCL2+ (CXCL2+), MHCII Mφ (MHCII+), and IFNIC Mo (CXCL10+CXCL2+). Fluorescence minus one (FMO) control (top) were used to control for spillover-related contribution to background in each channel. n = 6 mice per group. **P < 0.01 by unpaired two-tailed t test. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution.
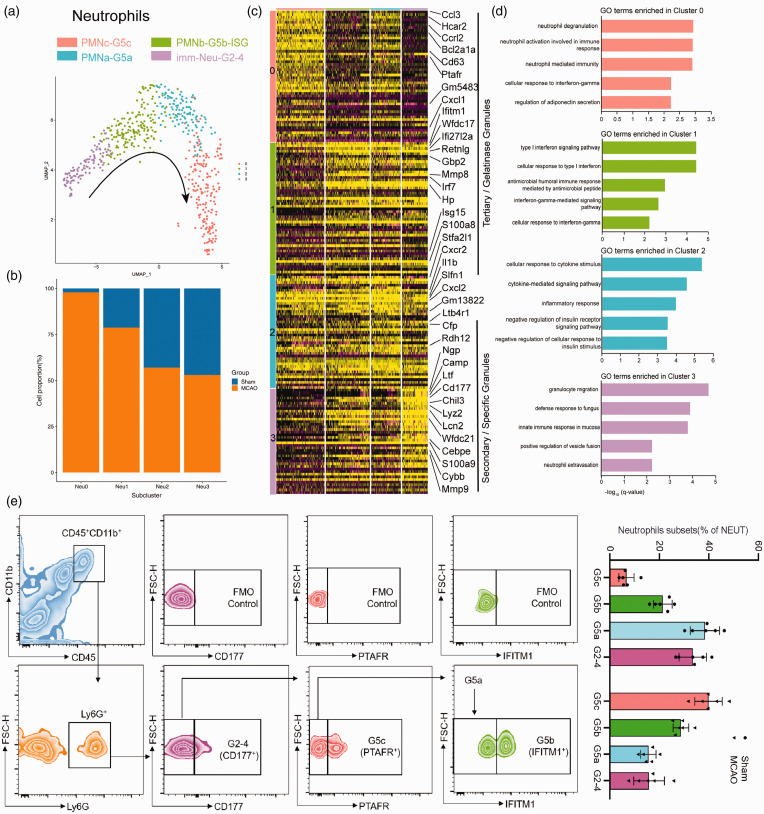
We detected four distinct neutrophil clusters, NEUT0(PMNc-G5c), NEUT1(PMNb-G5b-ISG), NEUT2(PMNa-G5a) and NEUT3(immNeu-G2-4) (Figure 6(a)). As the most mature neutrophils with typical polymorphonuclear morphology (PMN) infiltrating into the brain mainly from the MCAO group (Figure 6(b)), NEUT0 significantly expressed much higher mobilizing signals (Cxcl1) for promoting the release of the mature neutrophils from the BM(bone marrow) into the circulation, 41 as well as the upregulation of Cd63,Ptafr and Hcar2 corresponding to the neutrophil mediated immunity(Figure 6(c) and (d)). On the other hand, NEUT1 featured higher expression of type I interferon (IFN) stimulated genes (ISGs), such as Ifitm1, Gbp2, Isg15 and Irf7. NEUT2 cells highly expressed Stfa2l1, Cxcr2 and Ltb4r1, as well as genes related to cellular response to cytokine stimulus (Trem1, Fpr1 and Ccr1). Noticeably, a list of secondary granules associated genes previous labelled preNeu and imNeu 42 were identified mostly within NEUT3, such as Ltf, Camp, Cybb, Cd177,along with the strong upregulation of Cebpe expression, which was the crucial transcription factors (TFs) for early neutrophil differentiation. 43 Moreover, ischemic injury altered the cell composition ratio of the various neutrophil subclusters. The dynamic conversion among each subcluster was also identified by flow cytometry in our study (Figure 6(e)).
Figure 6.
Differential gene expression patterns in neutrophils clusters. (a) UMAP visualization of subclusters of the neutrophils. (b) The bar graph showing the fraction of the neutrophils subclusters originating from disease state. (c) Heatmap showing single-cell gene expression of the neutrophils subcluster-specific genes. (d) GO biological-process terms enriched in various neutrophils subclusters. (e) Flow cytometry analysis of proportion of the neutrophils subsets transitions obtained from mice brain tissues at 24 h after MCAO. n = 6 mice per group. Data are presented as mean ± SD. P > 0.05 by Shapiro-Wilk test for normal distribution.
We also proceed the lymphocytes sub-clustering analysis and revealed six subsets (Figure S9(a)), including NK1 cells (Ifng, Klrb1c), NK2 cells (Car2, S100a1), B1 cells (Ly6d, Cd79a), B2 cells (Ramp1, Lmo4), T1 cells(CD3d, Ms4a6b, Cd160) and proliferative T cells(Stmn1,Birc5,Ube2c) (Figure S9(d)). Remarkably, we found increased expression of chemokine genes (Ccl3, Ccl4, Ccl5) and cytokine gene (Ifng) in NK cells, cytotoxic genes (Cd63, Gzmk,Gzma,Gzmb) in NK cells and proliferative T cells within the MCAO brain (Figure S9(e)).
Gene expression analysis of types diversity in the brain vasculature
We found six vascular endothelial cells (ECs) subclusters (Figure S10(a)) according to the established markers of ECs in previous studies.44,45 Ischemia-induced inflammation and oxidative stress increased endothelial cell death, which directly resulted in the decrease of the cell percentage in the MCAO group, while, the cells from BBB associated clusters increased (Figure S10(b))with a list of core BBB dysfunction module associated genes, 46 such as Adamts4, Upp1,Timp1 and Pdlim1 (Figure S10(d)). Interestingly, we identified two ECs clusters including capillary (capEC-IFNhi) and arterial (aEC- IFNlo) expressed IFN-I signaling genes (such as Ifit3, Isg15, Usp18), whereas the cell proportion decreased in the MCAO group, which displayed consistent result with a reference of bulk RNA-seq dataset of brain ECs. 47 In an attempt to explain the ECs heterogeneity after stroke, we used the GSVA analysis to reveal the different biological processes among the six subclusters (Figure S10(e)). Indeed, the BBB enriched subcluster-EC3 highly involved in the KRAS gene charging for the regulation of RAS/MAPK pathway, which was closely related to the apoptosis of cerebral microvascular endothelial cells in ischemic stroke. 48 In addition, the venous EC5 involved in the hypoxia and oxidative phosphorylation. In a word, the identity of each ECs subpopulation was still existed after acute ischemic injury, however, ischemia up- and downregulated numerous genes encoding inflammatory cytokines (such as Spp1, Cxcl2, Ccl4, Cd14) in each ECs subpopulation (Figure S10(f)).
We also identified distinct vascular mural cells subsets (three PCs and six SMCs subsets) occurring in ischemia condition (Figure S10(g) and (l)). SMCs highly expressed gene sets involved in the specific markers to arteriole(aaSMC) cluster0(Acta2,Tagln), arterial SMC (aSMC) cluster1,2and 3(Cnn1,Ptgis,Eln), venous (vSMC-IFN)-cluster5(Car4,Art3) (Figure S10(j)), in line with previous reports. 44 Besides, we also found an activated SMC cluster4 specific enriched in the neutrophil mediated immunity and regulated exocytosis (Figure S10(k)), in agreement with their known role in atherosclerosis by neutrophil-driven SMC lysis and death. 49 Moreover,the type I interferon signaling pathway related SMC subcluster5(vSMC-IFN) also showed a decreased cell proportion in the MCAO group (Figure S10(h)), in line with the expression trend of ECs. In addition to SMCs, we also identified three PCs subsets (Figure S10(l)). PC0 subset was mainly enriched in the process of the potassium, sodium, calcium ion transport (Figure S10(o)) and were important for maintaining BBB normal functions.PC1 subset highly expressed gene sets involved in immune functions after ischemic stroke, including Oncostatin-M-produced BBB breakdown, HIF-1 and cytokine-mediated signaling pathway. PC3 subset with specific high expression of Acta2 in regulating cerebral blood flow was enriched in the pathway of muscle contraction and syndecans 1 pathway related to the multi-potential differentiation capacity of pericytes.
Perivascular fibroblast-like cells (FBs) are a newly defined subsets with unknown function that occupy the perivascular space in the brain. 44 We found three subclusters of FBs (Figure S10(p)). Among the FBs, FB0 specifically expressed some genes related to membrane transporters (e.g., Slc22a6,Slc6a13, Slc7a11, and Slc1a3) and collagen fibril organization genes(Col1a1, Col1a2, Col12a1). FB1 was enriched in the process of extracellular matrix organization (Vcam1,Lum,Dcn) and response to interferon-beta (Ifitm1,Ifitm2,Gsn).FB2 highly expressed genes encoding pumps (e.g., Fxyd5 and Atp1b1), carboxylic acid transport (e.g., Slc38a2,Slc16a11, Slc6a6), and regulation of cellular pH (e.g., Slc26a2,Slc4a10) (Figure S10(s)). The differential expression genes (DEGs) for each FBs subcluster was examined between MCAO and sham groups (Figure S10(u)), which revealed subsets-specific expression pattern.
Heterogeneity of other brain cells upon ischemic injury
In the experiments, we projected the oligodendrocytes (OLGs), astrocytes (ASCs), neural progenitor cells (NPCs) into the tSNE space, respectively. The graph-based clustering algorithm were employed to identify the subclusters within each cell type (Figure S11(a), (e) and (i)). Moreover, we found that each cell type from MCAO and sham groups was mostly segregated into distinct subclusters, indicating that firm and sound ischemic injury-associated gene expression patterns changes across almost each cell type. Meanwhile, each subcluster of a given cell type could be distinguished by specific marker genes (Figure S11(c), (g) and (k)). Besides, the enrichment analysis of DEGs for each cell type after ischemic stroke led to an identification of specific functional categories (Figure S11(d), (h) and (l)).
Besides, we also investigated the ependymal cells (EPCs) and choroid plexus capillary ECs (CPCs) after IS (Figure S12(a), (e) and (i)). EPCs were divided into two subtypes according to the previous study 11 and gene expression pattern: EPC1- secretory ependymal cells highly expressed Prlr and Enpp2 genes that are related to the cellular secretion and EPC2-ciliated ependymal cells are featured with the expression of Ccdc153, Hydin and Dnali1 genes that are associated with cilium movement. The six EPC1 subsets and three EPC2 subsets nearly all showed the gradual changes in gene expression distributed into three patterns respectively (Figure S12(c) and (h)). Notably, the subcluster0 of EPC1 mainly from the MCAO group highly expressed immune response genes (e.g., Hspa1a, Ctss, Ccl3 and Ctsb). On the other hand, the CPCs were divided into four subsets, one of which representing subcluster0 also expressed type I interferon genes (e.g., Irf7, Isg15, Ifit3 and Rsad2) and the subcluster1 mainly from the MCAO group selectively expressed chemotaxis and immune response genes (e.g., Cd14, Ccl2, Ccl4 and Cts3)
Ischemic stroke diversifies cell–cell communication in the brain
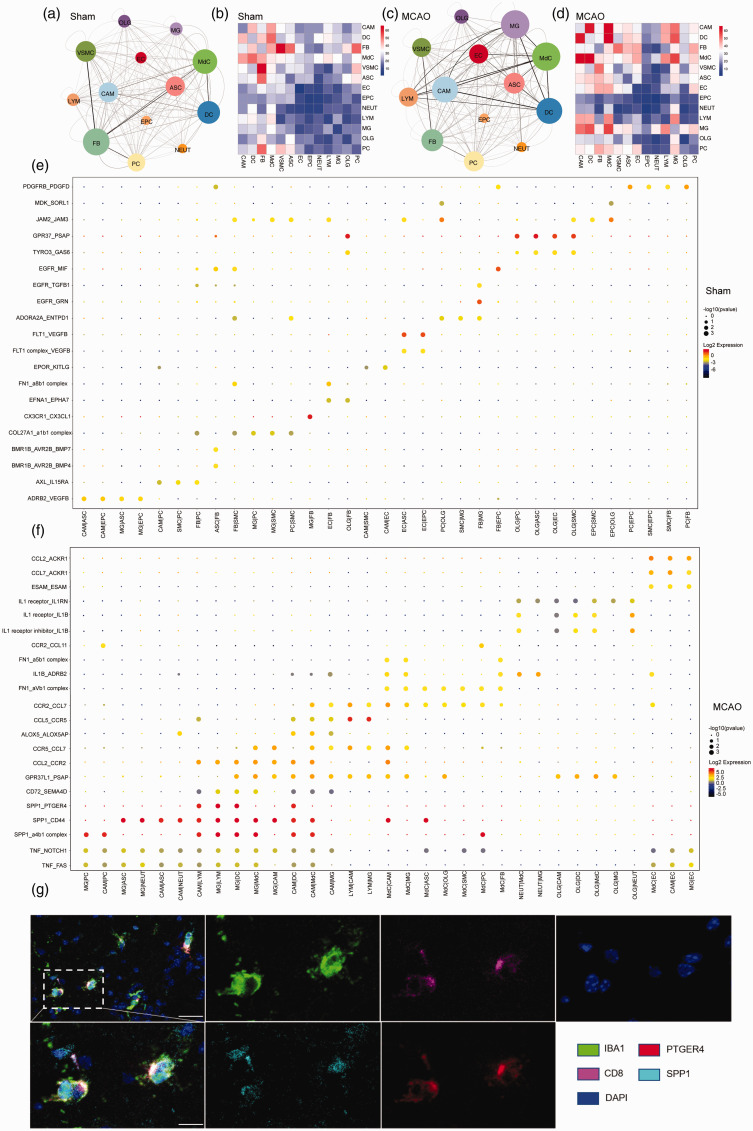
Ischemic brain injury is characterized by the neuroinflammatory responses of both the innate and the adaptive immune system, which relies on sophisticated interactions between brain resident and myeloid immune cells, both through adhesion of molecules on the surface of adjacent cells or extracellular matrix, and chemical signaling molecules. Here, based on the ligand-receptor interactions, we infer the potential cell -cell communication in the brain and identify their variation during ischemic stroke using the CellPhoneDB database. While some interactions are unvaried, some were particular to the ischemic injury or sham groups. In sham groups, the cell -cell interaction landscape of the brain was dominated by FB communicating with each other (Figure 7(a)and (b)). In the MCAO groups, the number of inferred communications between FB and other cells was reduced. However, the cell -cell interactions dominated by microglia and CAM increased significantly after stroke, especially the predicted interactions between microglia and other immune cells, astrocytes, pericytes and oligodendrocytes (Figure 7(c) and (d)).
Figure 7.
Comparative cell-cell interactions in sham and MCAO mice. (a, c) Interaction networks analysis of brain cells. nodes represent cell types; edges represent interactions between cells; Node’s size correlates with the total number of interactions of each cell type. The thickness and transparency of edges correlates to the number of interactions among the cell types. (b, d) Heatmap showing the number of all possible interactions between the clusters analyzed. (e, f) Dot plot depicting selected ligand–receptor interactions using CellPhoneDB. Size indicates p values, and color indicates the scaled mean expression level of the receptor-ligand pairs between 2 clusters. (g) SPP1+ microglia and Ptger4+ CD8+T cells are colocalized in the brain of mice at 24 h after MCAO under confocal observation. n = 6 per group. [Scale bars, 40 µm (overview) and 20 µm (Insets).]
The results of the inferred cell -cell communication among brain cells in sham controls suggested plenty of growth factor signaling pathways, such as the epidermal growth factor receptor (EGFR), transforming growth factor (TGF), platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) pathways (Figure 7(e)). Cell–cell interactions unique to stroke included chemokines, cytokines, and TNF families, such as CCL2, CCL5, IL1A, IL1RN, and TNF, all of which are known to play significant roles in enhancing peripheral immune cell infiltration into ischemic brain tissue (Figure 7(f)). Notably, we also identified SPP1(osteopontin, OPN) mainly expressed by macrophages and activated microglia, with receptor such as CD44 and integrin receptors(α4β1), which were expressed broadly across brain resident and infiltrated immune cells, suggesting potential neuroprotective effects via diverse signaling pathways as previous before. Apart from discovering these widely known interactions, we identified novel microglia–lymphocytes interactions in stroke: SPP1-PTGER4, which were involved in macrophage-exhausted CD8+ T cells interactions in lung adenocarcinoma influencing T cell function to maintain the balance between immune activation and exhaustion before. 50 We also verified that the SPP1+ microglia were within close proximity to PTGER4+ CD8+T cells in the periinfarct area of brain sections at 24 h after MCAO (Figure 7(g)). All in all, the intercellular interactions indicate a close relationship between peripheral immune cell dynamics and molecular features of brain resident cells, which might influence the prognostic and therapeutic responses in ischemic stroke.
Discussion
In this study, single-cell RNA sequencing provided a full-scale data resource for exploring the ischemic stroke at the single-cell transcriptomic level, unprecedentedly uncovering the brain cell phenotypic heterogeneity and identifying diverse functional changes at the acute stages in ischemic stroke (24 hr post MCAO).
The cerebral stroke usually occurred under hypoxic-ischemic condition due to the occlusion of major brain arteries. A rapid inflammatory response in the stroke was characterized by the activation of brain resident cells and the recruitment of peripheral immune cells. The proportion of the infiltrated cells fluctuated at different time points. 51 Here, we showed a temporal composition of influx of inflammatory cells with varied increase in the number of monocytes, neutrophils, and lymphocytes within 24 hours after reperfusion. Strikingly, every infiltrated immune cell harbored a detailed transcriptional information, and these cells could be classified into distinct subtypes. In comparison with the previous studies in which only sorted cells with limited number of markers were explored in flow cytometry, high-dimensional unbiased single-cell sequencing in this study defined a relatively accurate cell heterogeneity and cell subtype specific molecular regulators. The precise identity of these cell subsets might enable us to investigate the potential key subcluster of the immune cell repertoire that could be the target of therapeutic intervention in stroke.
In our results, we depicted the single cell transcriptional maps of the brain cells during normal condition versus ischemic injury, uncovering cell-type specific DEGs, pathways and the special molecular and cellular subtypes. Although most genes showed distinct cell-type specific expression patterns, many of the top DEGs were found to be involved in related biological processes across cell types. Specifically, comprehensive prediction of the cell–cell interactions between the sham and MCAO group suggests the dominance of microglia interacting with resident brain cell and infiltrating immune cells. Hence, cell-type specific regulatory genes, especially those involved in subcluster associated diseases, should be considered for biomarkers in stroke, and ischemic insult caused alteration of DEGs across multiple cell types may have a potential implication for the intervention. In agreement, Guo et al. 52 recently profiled the single-cell transcriptional analysis of the mouse brain cortex penumbra area 24 h after MCAO, which identified 13 major cell types, each showing similar gene expression patterns to our study. However, the composition ratio of the cell types was different between the two studies (Guo et al. versus our study), probably owing to several reasons, including in the subclusters classification of cell types, the different brain tissues sampled (cortex penumbra versus cerebral hemisphere), varying enzymatic methods for tissue dissociation and whether samples were mixed or not for library construction (yes versus no) and so on.
As residential immune cells, microglia have been studied for understanding of their heterogeneous responses to many neurological diseases. 53 So far, numerous basic studies and clinical trials have attempted to regulate microglia activation through either suppressing microglia (M1 phenotype) or promoting microglia polarization to M2 phenotype to secrete anti-inflammatory cytokines, exerting a protective role in ischemic stroke. 54 Unfortunately, none of the above projects have proceeded to the clinical practice. Recently, accumulating evidence points that M1/M2 dichotomy for microglia/macrophages polarization was oversimplified, 55 thus further application of single-cell techniques to investigate the microglia multipolarization is imperative. 56 Moreover, a recent study also revealed that four clusters of TFs were involved in regulating microglial phenotypes in a rat model of ischemic stroke, 55 which suggested that novel phenotypes of activated microglia extending beyond the M1/M2-like patterns. Here, we did not observe a fully overlap between the microglia subsets and classical M1 or M2 subsets after stroke at the single cell level. Instead, we identified five distinct microglial subtypes in the sham and MCAO groups. In agreement, Guo et al. 52 recently also identified 14 microglia subclusters in cortex penumbra, while, none of which fully expressed M1 or M2-type marker genes. In addition, the core marker genes of microglia including Gpr34, P2ry12, and Cx3cr1 were also downregulated after MCAO in both Guo et al and our study. Besides, we also found the diversity of CAMs, including non -parenchymal meningeal, perivascular, and choroid plexus macrophages, and which showed different expression patterns with microglia. Furthermore, the trajectory analysis of the Mon/Mφ revealed the potential differential trajectory post ischemic injury. All these findings prompted us to infer that regional disparity of immune cells located in “core” of the infarct or the “penumbra” and the different durations after reperfusion exhibited diverse pathophysiological features, suggesting spatial and temporal interplays. With the emerging of a next-generation molecular imaging technology, the Visium Spatial Gene Expression solution could be helpful in globally classifying immune cells based on the total mRNA content or characterizing novel gene expression profiles of cells by focusing on ischemic region of interest.
In addition to main myeloid cells, we also explored the brain vasculature subsets during ischemic injury. Although the proportion of each cell subsets nearly unchanged, the ischemia also changed numerous immune responses associated genes among each subcluster between the sham and MCAO group. The DEGs related functions will be explored in our next work. Moreover, we uncovered the transcriptional basis of the gradual phenotypic change (zonation) along the ependymal axis in each subset. And we will examine the gene regulatory network activities each subcluster after ischemic injury.
The present study had the following limitations. First, the results could be influenced by the timing for collecting samples and the severity of the animal model for stroke, due to the fact that the inflammatory reaction during ischemic stroke displays a dynamic pattern of progression. Second, this study represented the transcriptome landscape of the stroke at the early stage (24 hr). The transcriptome landscapes at advanced stages of the disease (3 days or 7 days after the onset) would be more valuable in understanding the development of stroke. Besides, stroke largely occurs in the elderly, while our data obtained from young adults. In fact, emerging transcriptomic analysis57,58 showed that inflammatory responses, immune cell chemotaxis, angiogenesis and tissue remodeling in aged microglia was fully suppressed post stroke. Thus, further studies at single-cell resolution in age brain after stroke might clarified potential mechanisms underlying age-related changes. Lastly, there is still a drawback to identify the molecular mechanisms of human disease by applying the animal model because of the challenge in gaining critical human brain tissues with stroke. Thus, studies on the surgery samples or donated brain tissues from the dead patients with stroke need to be done to elucidate the underlying mechanism.
In conclusion, our research mapped the first global single-cell RNA sequencing picture of brain cells post ischemic stroke, broadening our vision of cell heterogeneity and cell-type unique gene expression pattern of the stroke. Identification of the cell-type specific genes could subsequently promote the exploitation and utilization of molecular imaging studies on biomarkers for dynamic disease surveillance among stroke patients. Particularly, this study offers new insights into the diversity in molecular and cellular subtypes underlying cerebral ischemia-reperfusion injury in mouse model of MCAO and lays a foundation for the in-depth research on human brain after ischemic stroke.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-2-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-3-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism
Footnotes
Data availability: The raw and analyzed data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE174574 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174574). More detailed information for this paper can be found in the supplementary materials. Additional data information is available upon reasonable request from the authors.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Major Program of the National Natural Science Foundation of China (82090043 to J. Hao), the National Outstanding Youth Science Fund Project of National Natural Science Foundation of China (81825008 to J. Hao), National Natural Science Foundation of China (81801197 to W. Jiang), and Natural Science Foundation of Tianjin (17JCZDJC35500 to J. Hao, 19JCQNJC10500 to W. Jiang).
Acknowledgements: We thank Ye Liu for help with fluorescence-activated cell sorting (FACS) and Dongshan Wan for technical support.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: K.Z. participated in the experimental design, analyzed sequencing datasets, and wrote the paper. L.L, X.Z. and L.C. conducted animal and performed scRNA-seq experiments. Y.R. and Q.Z. conducted the bioinformatics analyses. W.J. supervised this project. J.H. designed experiments and revised the manuscript.
Supplemental material: Supplemental material for this article is available online.
References
- 1.Johnson W, Onuma O, Owolabi M, et al. Stroke: a global response is needed. Bull World Health Organ 2016; 94: 634–634A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dergunova LV, Filippenkov IB, Stavchansky VV, et al. Genome-wide transcriptome analysis using RNA-Seq reveals a large number of differentially expressed genes in a transient MCAO rat model. BMC Genomics 2018; 19: 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017; 8: 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Percie Du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. J Cereb Blood Flow Metab 2020; 40: 1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun 2019; 10: 5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 2019; 15: e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019; 177: 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sousa C, Golebiewska A, Poovathingal SK, et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep 2018; 19: e46171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisel A, Muñoz-Manchado AB, Codeluppi S, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015; 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
- 10.Marques S, Zeisel A, Codeluppi S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016; 352: 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gokce O, Stanley GM, Treutlein B, et al. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-Seq. Cell Rep 2016; 16: 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kronenberg G, Uhlemann R, Richter N, et al. Distinguishing features of microglia- and monocyte-derived macrophages after stroke. Acta Neuropathol 2018; 135: 551–568. [DOI] [PubMed] [Google Scholar]
- 13.Ritzel RM, Patel AR, Grenier JM, et al. Differences between microglia and monocytes after ischemic stroke. J Neuroinflammation 2015; 12: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi K, Tian D-C, Li Z-G, et al. Global brain inflammation in stroke. Lancet Neurol 2019; 18: 1058–1066. [DOI] [PubMed] [Google Scholar]
- 15.Li P, Stetler RA, Leak RK, et al. Oxidative stress and DNA damage after cerebral ischemia: potential therapeutic targets to repair the genome and improve stroke recovery. Neuropharmacology 2018; 134: 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarruk JG, Greenhalgh AD, David S. Microglia and macrophages differ in their inflammatory profile after permanent brain ischemia. Exp Neurol 2018; 301: 120–132. [DOI] [PubMed] [Google Scholar]
- 17.Sawano T, Watanabe F, Ishiguchi M, et al. Effect of Sema4D on microglial function in MCAO mice. Glia 2015; 63: 2249–2259. [DOI] [PubMed] [Google Scholar]
- 18.Liddelow SA, Barres BA. Reactive astrocytes: production, function, and therapeutic potential. Immunity 2017; 46: 957–967. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Q, Luo X, Zhang J, et al. Osteopontin as a potential therapeutic target for ischemic stroke. Curr Drug Deliv 2017; 14: 766–772. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd AF, Miron VE. The pro-remyelination properties of microglia in the central nervous system. Nat Rev Neurol 2019; 15: 447–458. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Zhu H, Taheri S, et al. Serum amyloid A-mediated inflammasome activation of microglial cells in cerebral ischemia. J Neurosci 2019; 39: 9465–9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjan A, Hauert T, Hussain M, et al. Abstract WP161: sovateltide promotes regeneration and functional recovery of ischemic brain by improving mitochondrial function. Stroke 2020; 51: AWP161-AWP161. [Google Scholar]
- 23.Hu X, Leak RK, Shi Y, et al. Microglial and macrophage polarization – new prospects for brain repair. Nat Rev Neurol 2015; 11: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geirsdottir L, David E, Keren-Shaul H, et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 2019; 179: 1609–1622.e16. [DOI] [PubMed] [Google Scholar]
- 25.Jordao MJC, Sankowski R, Brendecke SM, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 2019; 363: eaat7554. [DOI] [PubMed] [Google Scholar]
- 26.Singhal G, Baune BT. Microglia: an interface between the loss of neuroplasticity and depression. Front Cell Neurosci 2017; 11: 270–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Pan X, Christiansen LI, et al. Necrotizing enterocolitis is associated with acute brain responses in preterm pigs. J Neuroinflammation 2018; 15: 180–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat Rev Neurosci 2018; 19: 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chelluboina B, Klopfenstein JD, Pinson DM, et al. Matrix metalloproteinase-12 induces blood-brain barrier damage after focal cerebral ischemia. Stroke 2015; 46: 3523–3531. [DOI] [PubMed] [Google Scholar]
- 30.Bartsch JW, Wildeboer D, Koller G, et al. Tumor necrosis factor-alpha (TNF-alpha) regulates shedding of TNF-alpha receptor 1 by the metalloprotease-disintegrin ADAM8: evidence for a protease-regulated feedback loop in neuroprotection. J Neurosci 2010; 30: 12210–12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aibar S, Gonzalez-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nature methods 2017; 14: 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harari OA, Liao JK. NF-κB and innate immunity in ischemic stroke. Ann N Y Acad Sci 2010; 1207: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y-Y, Wang K, Liu Y-E, et al. Identification of key transcription factors associated with cerebral ischemia‑reperfusion injury based on gene‑set enrichment analysis. Int J Mol Med 2019; 43: 2429–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Formisano L, Guida N, Valsecchi V, et al. Sp3/REST/HDAC1/HDAC2 complex represses and Sp1/HIF-1/p300 complex activates ncx1 gene transcription, in brain ischemia and in ischemic brain preconditioning, by epigenetic mechanism. J Neurosci 2015; 35: 7332–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng C, Tempel D, Den Dekker WK, et al. Ets2 determines the inflammatory state of endothelial cells in advanced atherosclerotic lesions. Circ Res 2011; 109: 382–395. [DOI] [PubMed] [Google Scholar]
- 36.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 2010; 35: 323–332. [DOI] [PubMed] [Google Scholar]
- 37.Kierdorf K, Masuda T, Jordao MJC, et al. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat Rev Neurosci 2019; 20: 547–562. [DOI] [PubMed] [Google Scholar]
- 38.Rizzo G, Vafadarnejad E, Arampatzi P, et al. Single-cell transcriptomic profiling maps monocyte/macrophage transitions after myocardial infarction in mice. 2020. DOI: 10.1101/2020.04.14.040451.
- 39.Van Hove H, Martens L, Scheyltjens I, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 2019; 22: 1021–1035. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017; 14: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devi S, Wang Y, Chew WK, et al. Neutrophil mobilization via plerixafor-mediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J Exp Med 2013; 210: 2321–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evrard M, Kwok IWH, Chong SZ, et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity 2018; 48: 364–379. e368. [DOI] [PubMed] [Google Scholar]
- 43.Hock H, Hamblen MJ, Rooke HM, et al. Intrinsic requirement for zinc finger transcription factor gfi-1 in neutrophil differentiation. Immunity 2003; 18: 109–120. [DOI] [PubMed] [Google Scholar]
- 44.He L, Mäe MA, Andrae J, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018; 554: 475–480. [DOI] [PubMed] [Google Scholar]
- 45.Kalucka J, de Rooij L, Goveia J, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 2020; 180: 764–779. e720. [DOI] [PubMed] [Google Scholar]
- 46.Munji RN, Soung AL, Weiner GA, et al. Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module. Nat Neurosci 2019; 22: 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Androvic P, Kirdajova D, Tureckova J, et al. Decoding the transcriptional response to ischemic stroke in young and aged mouse brain. Cell Reports 2020; 31: 107777. [DOI] [PubMed] [Google Scholar]
- 48.Yong YX, Yang H, Lian J, et al. Up-regulated microRNA-199b-3p represses the apoptosis of cerebral microvascular endothelial cells in ischemic stroke through down-regulation of MAPK/ERK/EGR1 axis. Cell Cycle 2019; 18: 1868–1881. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Silvestre-Roig C, Braster Q, Wichapong K, et al. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature 2019; 569: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020; 11: 2285–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gelderblom M, Leypoldt F, Steinbach K, et al. Temporal and spatial dynamics of cerebral immune cell accumulation in stroke. Stroke 2009; 40: 1849–1857. [DOI] [PubMed] [Google Scholar]
- 52.Guo K, Luo J, Feng D, et al. Single-cell RNA sequencing with combined use of bulk RNA sequencing to reveal cell heterogeneity and molecular changes at acute stage of ischemic stroke in mouse cortex penumbra area. Front Cell Dev Biol 2021; 9: 624711–624703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyu J, Jiang X, Leak RK, et al. Microglial responses to brain injury and disease: functional diversity and new opportunities. Transl Stroke Res 2021; 12: 474–495. [DOI] [PubMed] [Google Scholar]
- 54.He Y, Ma X, Li D, et al. Thiamet G mediates neuroprotection in experimental stroke by modulating microglia/macrophage polarization and inhibiting NF-kappaB p65 signaling. J Cereb Blood Flow Metab 2017; 37: 2938–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng W, Mandeville E, Terasaki Y, et al. Transcriptomic characterization of microglia activation in a rat model of ischemic stroke. J Cereb Blood Flow Metab 2020; 40: S34–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu X. Microglia/macrophage polarization: fantasy or evidence of functional diversity? J Cereb Blood Flow Metab 2020; 40: S134–S136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang L, Mu H, Xu F, et al. Transcriptomic and functional studies reveal undermined chemotactic and angiostimulatory properties of aged microglia during stroke recovery. J Cereb Blood Flow Metab 2020; 40: S81–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L, Rocha M, Zhang W, et al. Genome-wide transcriptomic analysis of microglia reveals impaired responses in aged mice after cerebral ischemia. J Cereb Blood Flow Metab 2020; 40: S49–S66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-2-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-3-jcb-10.1177_0271678X211026770 for Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke by Kai Zheng, Lingmin Lin, Wei Jiang, Lin Chen, Xiyue Zhang, Qian Zhang, Yi Ren and Junwei Hao in Journal of Cerebral Blood Flow & Metabolism