Abstract
Gene vectors targeting CNS endothelial cells allow to manipulate the blood-brain barrier and to correct genetic defects in the CNS. Because vectors based on the adeno-associated virus (AAV) have a limited capacity, it is essential that the DNA sequence controlling gene expression is short. In addition, it must be specific for endothelial cells to avoid off-target effects. To develop improved regulatory sequences with selectivity for brain endothelial cells, we tested the transcriptional activity of truncated promoters of eleven (brain) endothelial-specific genes in combination with short regulatory elements, i.e., the woodchuck post-transcriptional regulatory element (W), the CMV enhancer element (C), and a fragment of the first intron of the Tie2 gene (S), by transfecting brain endothelial cells of three species. Four combinations of regulatory elements and short promoters (Cdh5, Ocln, Slc2a1, and Slco1c1) progressed through this in-vitro pipeline displaying suitable activity. When tested in mice, the regulatory sequences C-Ocln-W and C-Slc2a1-S-W enabled a stronger and more specific gene expression in brain endothelial cells than the frequently used CAG promoter. In summary, the new regulatory elements efficiently control gene expression in brain endothelial cells and may help to specifically target the blood-brain barrier with gene therapy vectors.
Keywords: Synthetic promoter, gene therapy, transcriptional targeting, 5′-flanking gene region, cell-specific promoter, lysosomal diseases
Introduction
Currently, many basic, preclinical and clinical research projects use adeno-associated viral (AAV) vectors for treating neurological diseases or for investigating cerebral physiology and pathophysiology.1,2 AAV vectors enable the long-term expression of genes in proliferating and post-mitotic cells. For a tailored intervention, AAV vectors have been modified to target specific cell types in the brain.3–9 Endothelial cells are of special interest because of their eminent role in common cerebrovascular, neuroinflammatory and neurodegenerative diseases.10,11 Their function is disrupted in some monogenetic disorders.12–14 Moreover, as a gateway to the CNS endothelial cells may enable cross-correction of lysosomal disorders.9,15,16 By expressing and secreting missing enzymes, vector-transduced brain endothelial cells rescue enzyme-deficient brain cells.
Despite an overall favorable safety profile, AAV-based gene therapy can lead to side effects that result from gene expression in off-target cells and tissues. 17 A warning observation is the association of AAV infections with hepatocellular carcinoma. The risk may further increase with strong and ubiquitously active promoters inserting close to oncogenes.18,19 Therefore, it is important to limit expression in off-target cells and tissues. Restricting gene expression to a specific cell population can be achieved by transductional targeting, using a cell type-specific capsid, 20 and by transcriptional targeting, using a cell type-specific promoter. Combining these strategies has further enhanced specificity for other cell populations.21,22 In the case of brain endothelial cells, transductional targeting has already been successful. Screening of a peptide display library led to the discovery of the AAV2-derived mutant capsid AAV-BR1 that has an unprecedented selectivity for transducing brain endothelial cells when injected intravenously in mice. This vector has considerable potential for gene therapy.8,9,20 However, despite its specificity, AAV-BR1 transduces some off-target cells in the liver and in other organs as well as in the brain. 20 How AAV vectors penetrate the blood-brain barrier is still a matter of debate. 23
For transcriptional targeting endothelial cells of all organs, several 5′-flanking promoter sequences with pan-endothelial specificity were used in genetically modified mice. 24 In the CNS, endothelial cells show a highly characteristic pattern of gene expression. 25 In accordance with the notion that transcriptional control may enable specificity for this cell population, regulatory genomic sequences direct gene expression selectively to CNS endothelial cells of genetically modified mice. 26 To use this strategy for AAV vectors, the length of regulatory sequences is one of the key aspects to consider, because the packaging capacity of AAV is limited to approximately 4,700 bp. Until now, only few pan-endothelial promoter sequences fitting into the confinement of an AAV vector have been identified.27–29 The regulatory sequences Ple34 and Ple261 afford specificity for brain endothelial cells although their lengths of 3,854 or 2,963 bp, respectively, leave little additional space for the target gene within the packaging capacity of AAV vectors. 30
Therefore, the aim of the present study was to develop short regulatory sequences that enable selective expression of target genes in brain endothelial cells. Based on the comparison of the transcriptome of endothelial cells in brain, liver and lung by Daneman and colleagues, 25 we investigated the promoters of 8 brain endothelial-specific genes and 3 pan-endothelial genes in vitro. To enhance gene expression, we tested additional regulatory elements, i.e. a sequence from exon 1 and intron 1 of the Tek receptor tyrosine kinase (Tie2) gene, the CMV enhancer, and the Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), leading to short synthetic regulatory sequence that enable expression in brain endothelial cells with improved specificity.
Material and methods
Animals
We used C57BL/6NCrl mice which were obtained from Charles River (Strain Code: 027). All mice were housed in individually ventilated cages (IVCs) with 12-h light/dark cycles with food and water ad libitum. Vectors were injected in male mice at the age of 30 days and organs were extracted for analysis 14-15 days later. Primary endothelial cells were prepared from brains of male and female mice older than 42 days. Primary cortical astrocytes were prepared from brains of male and female mice at day P0/1. Investigators were blinded for treatment of mice in all experiments. All animal experiments were performed in accordance with the EU Directive 2010/63/EU, the German Animal Welfare Act, and the ARRIVE guidelines. All efforts were made to minimize pain and discomfort of animals. Animal experiments were approved by the responsible local authorities and ethics review boards (Ministerium für Energiewende, Landwirtschaft, Umwelt, Natur und Digitalisierung, Kiel, Schleswig-Holstein, Germany).
Selection of promoter sequences with putative blood-brain barrier specificity
We based the selection of putatively brain endothelial-specific genes on a transcriptome analysis by Daneman et al. 25 These authors investigated the transcriptome of endothelial cells from brain, liver, and lung in comparison to other cell types of the brain. Out of the genes that were among the 50 most enriched in (1) brain endothelial cells compared to brain parenchyma (Table 1 in Daneman et al.), 25 (2) blood-brain barrier compared to liver endothelial cells (Table 2A in Daneman et al.), 25 or (3) blood-brain barrier compared to lung endothelial cells (Table 2B in Daneman et al.), 25 we selected those genes that were characterized by a ratio of vascular expression in brain to liver >150, a ratio of expression in brain endothelial to brain parenchymal cells >5, and a ratio of vascular expression in brain and lung >1. For the resulting 38 genes (Supplementary Table 1), we consulted the database DropViz displaying Single Cell expression profiles analyzed by Drop-seq-analysis of adult mouse brain. 31 Based on this database, we excluded genes that did not reveal a mainly endothelial expression pattern (>10/100,000 transcripts in endothelial cluster). Moreover, genes were screened for pan-endothelial expression and whether they belong to the most enriched pericyte transcripts (Table 1B in Daneman et al.). 25 Based on this analysis, we identified 26 genes that qualified as brain endothelial-specific (Supplementary Table 1). These genes were analyzed for binding sites of the transcription factors Erg, SoxF, Foxq1, Mecom, Foxf2, Sox18, Bcl6b, Sox7, Meox1, and Zic3 that are known to enhance gene expression in endothelial cells. 32 This analysis was performed on the sequence 1,000 bp upstream and 100 bp downstream of the transcription start site using the transcription factor database JASPAR CORE 2018 vertebrates and a p-value of 0.001. Finally, we focused on 24 genes containing the largest number of endothelial-specific transcription factor binding sites and picked the following 8 genes for further reporter gene assays in this study: Foxq1, Ocln, Itih5, Tbx3, Slc2a1, Slco1c1, Pglyrp1, and Sgpp2. Moreover, the promoter regions of the Tie2 (Tek) as well as the Cdh5 gene, which are known as pan-endothelial sequences were included in the analysis. In addition, we examined gene expression under the regulation of the promoter of Plvap, which is typically not expressed in endothelial cells of the blood-brain barrier. 33
Using the Eucaryotic Promoter Database, we retrieved 5′-flanking promoter sequences of genes, including about +100 bp downstream and −600 to −1,000 bp upstream of the transcription start site. 34 Within these boundaries, the selection of individual 5′-flanking sequences was guided by the attempt to optimize the number of endothelial-specific transcription factor binding sites. Deviating from this procedure, 2,099 bp of the mouse Tie2 promoter and −289/+24 bp of the mouse Cdh5 gene were used for reporter gene assays in vitro and in vivo.
Plasmids containing short regulatory elements and promoters
To analyze the effect of short regulatory elements and promoter sequences on gene expression, we generated 61 plasmids. The plasmids encoded either eGFP, firefly luciferase, Renilla luciferase or mCherry as reporters. Expression of the reporter genes was controlled by 49 different combinations of promoter sequences with or without the following short regulatory enhancer elements: a long (L) or short (S) fragment of the first exon and the first intron of the Tie2 gene, the CMV enhancer (C), and the Woodchuck posttranscriptional regulatory element (WPRE; W). Whereas the long and short Tie2 intron were amplified by PCR using mouse genomic DNA as a template, the CMV enhancer was amplified from the plasmid pUF-C-MLC1500-eGFP. 21 The promoters Tie2, Foxq1, Ocln, Itih5, Slc2a1, Slco1c1, Pglyrp1, Sgpp2, Tbx3, and Plvap were amplified from mouse genomic DNA. The Cdh5 promoter was isolated from the plasmid pDS-Cdh5-eGFP and corresponded to −289/+24 bp of the mouse gene. 35 The CAG promoter and the T81 promoter were used as control promoters. The T81 promoter contains the thymidine kinase promoter of herpes simplex virus 1 (-81 to +52 upstream of the transcription start, distal GC + CCAAT deleted). 36
p-6p1 (hSYN-eGFP-W) was used for the generation of the new plasmids. 37 Briefly, we replaced the hSYN promoter by the (brain) endothelial-specific promoters and regulatory elements as described in Supplementary Table 2. The coding sequence of eGFP or the firefly luciferase was placed downstream of the corresponding promoter sequence with or without WPRE. The sequence lengths of the promoters as well as the downstream and upstream sequences are presented in Supplementary Table 3. Information about the investigated short regulatory elements are provided in Supplementary Table 4. We confirmed all DNA constructs by sequencing (Lightrun).
Vector production, injection, and quantification
To investigate the effect of regulatory sequences on eGFP gene expression in vivo, we packaged the constructs in AAV-BR1 that has a higher tropism for neurovascular endothelial cells than AAV2 or other serotypes. 20 AAV-BR1 vectors were produced by transfecting HEK293T cells as previously described. 20 We used p179 as adenoviral helper plasmid, 38 pXX187-NRGTEWD encoding the AAV-BR1 capsid and the plasmids p-CAG-eGFP, p-S-C-Cdh5-eGFP-W, p-C-Ocln-eGFP-W, p-C-Slc2a1-S-eGFP-W, p-C-Slco1c1-eGFP-W, p-Tie2S-eGFP-W, p-Tie2-eGFP-W (containing inverted terminal repeats of AAV2) to be packaged. 20 HEK293T cells were transfected by the calcium-phosphate method. 39 Three days after transfection, cells were harvested and lysed in TNT extraction buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100). The vectors were purified via affinity column purification (Hi Trap™ AVB sepharose™, GE Healthcare). Genomic titers were determined by quantitative real-time PCR using eGFP- and WPRE-specific primers (eGFP-qPCR-F, 5′-TGA CCC TGA AGT TCA TCT GC-3′, eGFP-qPCR-R, 5′-AAG TCG TGC TGC TTC ATG TG-3′, WPRE-F, 5‘-ACT GTG TTT GCT GAC GCA AC-3‘and WPRE-R, 5‘-CAA-CAC CAC GGA ATT GTC AG-3‘), the SYBR Green I-based FastStart Essential DNA Green Master (Roche), and the Light Cycler Nano system (Roche). An initial denaturation of the probes (95 °C, 10 min) was followed by 45 cycles of amplification (95 °C/67 °C/72 °C; 30 s each; rampage 5 °C/s) and a final melting curve analysis (60 °C to 97 °C with 0.1 °C/s). AAV vectors were injected into the tail vein of C57BL/6NCrl mice at a dose of 1.8 × 10 11 genomic particles (gp)/mouse under anesthesia with 2.5% isoflurane. Fourteen days after AAV injection, mice were sacrificed, and organs were collected (see “Organ extraction and immunostaining”).
Isolation and transfection of endothelial cells
Primary brain endothelial cells of mice were isolated and cultured in 24-well plates as described previously. 40 On day 6 after isolation, cells were transfected using the Lipofectamine 3000 Reagent (Thermo Fisher Scientific). For lipofection, medium free of antibiotics, antimycotics, heparin, and puromycin was used.
Porcine brain endothelial cells were prepared from brains of slaughtered pigs as described previously. 41 After isolation, the cells were cultured in 24-well-plates in DMEM high glucose medium (Invitrogen) supplemented with bovine plasma-derived serum (20%), penicillin-streptomycin (100 µg/ml), heparin (125 µg/ml), and puromycin (8 µg/ml). The lipofection was performed 4 days after cell isolation at 80% confluence after removal of penicillin-streptomycin, heparin, and puromycin from the medium.
Human brain endothelial cells of the cell line hCMEC/D3 (Merck #SCC066) 42 were cultured in 24-well-plates coated with rat tail collagen type I (1:20 in 1x PBS) in EndoGRO-MV medium (Merck #SCME004). The lipofection was performed at 60% confluence. Ten hours before the lipofection, the EndoGRO-MV medium was exchanged with medium supplemented with bovine plasma-derived serum (2%), L-glutamine (10 mM), rh EGF (5 ng/mL) and EndoGro-LS supplement (0.2%).
For one well of a 24-well plate, 800 ng plasmid DNA containing eGFP or firefly luciferase as reporter genes were used. To control for transfection efficiency, we added 40 ng of the plasmid p-CAG-RLuc. Analysis of plasmid encoded gene expression via immunostaining and Dual Luciferase Assay was performed 4 to 5 days after lipofection.
Human umbilical endothelial cells (HUVECs) were isolated from umbilical cords (approved by Local Ethics Committee, 18-325) by washing with PBS, degrading the extracellular matrix with dispase (1 mg/ml), and suspending the endothelial cells in medium as described before. 43 The cells were cultured on 48-well plates that were coated with gelatin in Gibco Medium 199 supplemented with fetal calf serum (FCS, 10%), penicillin-streptomycin (100 U/ml, 100 µg/ml), large vessel endothelia supplement (1%, Thermo Fisher Scientific), and heparin (5,000 U/ml).
We transfected HUVECs with eGFP encoding plasmids (500 ng per well of a 48-well plate) and a control plasmid (500 ng per well) expressing mCherry under regulation of the CAG promoter by using the Lipofectamin 3000 reagent. eGFP and mCherry expression was measured as fluorescence intensity of living single cells 24 hours after lipofection by fluorescence microscopy in 6 independent experiments.
Isolation and transfection of primary cortical astrocytes
Primary cortical astrocytes were prepared from brains of C57BL/6NCrl mice at the age P0/1. Mice were sacrificed and brain cortices were isolated under a binocular. Meninges were removed and the cortex was homogenized by pressing the tissue through 20-G, 21-G, and 25-G needles with 4 ml DMEM (Thermo Fisher Scientific, #41960-029), supplemented with 10% FCS and antibiotics/antimycotics (100 µg/ml). Finally, medium (46 ml) was added. Cortical astrocytes were cultured in 24-well-plates. Medium was changed every 3 days. Twenty-four hours before the lipofection, at day 21 after cell isolation, the medium was exchanged with medium free of antibiotics and antimycotics. Analysis of plasmid encoded gene expression via immunostaining was performed 4 days after lipofection.
Measurement of reporter gene expression
To measure firefly and Renilla luciferase activity we used the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was determined with the microplate reader CLARIOstar for 48 sec (measurements every 0.5 sec). eGFP reporter gene expression was determined by immunostaining of transfected cells as described below.
Organ extraction and immunostaining
Brain, liver, lung, heart, kidney, and spleen were dissected for analysis 14 days after intravenous vector administration. Animals were intracardially perfused with Ringer solution under anesthesia. For immunostaining, organs were fixed in paraformaldehyde (4%) for 24 hours and subsequently stored in PBS.
For the immunohistochemical analysis organs were embedded in 3% low melting agarose (Merck, dissolved in PBS). Sections (50 µm) were prepared using a vibratome and were stored in PBS, supplemented with sodium azide (0.02%), at 4 °C. For immunostaining, tissues were incubated in blocking buffer (5% BSA in TBS-buffer supplemented with 0.3% Triton-X-100 (TBS-Tx)) and then overnight at 4 °C in the following primary antibody solutions: chicken anti-GFP (1:2,000; Abcam #ab13970), rat anti-CD31 (1:400; BD Pharmingen #557355), mouse anti-NeuN, clone A60 (1:1000; Merck #MAB377), rabbit anti-GFAP (1:2000; Dako #ZO334), rabbit anti-Olig2 (1:1000; Millipore #AB6910), and rabbit anti-Iba1 (Wako Chemicals #019-19741). All primary antibodies were diluted in blocking buffer. After washing with TBS-Tx twice, the following secondary antibodies were added for 2 hours at room temperature in the dark: goat anti-chicken IgG Alexa Fluor 488 (1:2,000, abcam #ab150169), donkey anti-rat Cy3 (1:400, Jackson ImmunoResearch #712-165-150), donkey anti-mouse Cy3 (1:400, Jackson ImmunoResearch #715-165-140), and donkey anti-rabbit Cy3 (1:400, Jackson ImmunoResearch #711-165-152). All secondary antibodies were prepared in blocking buffer. Nuclei were stained with DAPI (1 µg/ml). After removing the antibody and washing, the sections were mounted with Mowiol 4-88 containing 1,4-diazabicyclo-(2,2,2)octane.
Organ sections were analyzed using a confocal microscope (SP5, Leica; objective, HCX PLAPO CS 20x070 IMM UV corrected; aperture, 0.7; microscopic zoom, 1; scanning frequency, 400 Hz; average: 3 times and pinhole, 0.5 AU with a z-stack over ∼ 50 µm with a step size of ∼1 µm). Images were analyzed with ImageJ software. Images were not manipulated in any way except to make brightness and contrast adjustments. The final figures showing immunofluorescence images were assembled using the Illustrator (Adobe) software.
For immunostaining of isolated cells, we used the following primary antibodies: chicken anti-GFAP (1:500, Millipore #AB5541), mouse anti-NeuN, clone A 60 (1:2000, Millipore #MAB377), and rabbit anti-Iba1 (1:400, Wako Chemicals #019-19741) to determine cell composition of astrocyte culture. The following secondary antibodies were used: goat anti-chicken Cy3 (1:400, abcam #ab97147), goat anti-mouse Alexa 488 (1:400, Invitrogen #A-31619), and donkey anti-rabbit Alexa 488 (1:400, Invitrogen #A-21206). Fluorescence images were taken at room temperature with the fluorescent microscope DMI 6000 B (Leica), the objective HCXPL FLUOTAR 10.0× (aperture 0.3, immersion; dry), the Leica DFC360FX camera, and the acquisition software LAS AF.
Quantitative RT-PCR
RNA was isolated from liver samples using the Nucleo Spin RNA Kit (Macherey-Nagel) according to the manufacturer’s instructions. RNA was transcribed with Moloney Murine Leukemia Virus Reverse Transcriptase and random hexamer primers (Promega). Quantitative real-time PCR was performed using Ppia- and eGFP-specific primers (Ppia-qPCR-F, 5′-AGG TCC TGG CAT CTT GTC CAT-3′, Ppia-qPCR-R, 5′-GAA CCG TTT GTG TTT GGT CCA-3′; eGFP-qPCR-F, 5′-TGA CCC TGA AGT TCA TCT GC-3′ and eGFP-qPCR-R, 5′-AAG TCG TGC TGC TTC ATG TG-3′), the SYBR Green I-based FastStart Essential DNA Green Master (Roche), and the Light Cycler Nano system (Roche). An initial denaturation of the probes (95 °C, 10 min) was followed by 40 cycles of amplification (95 °C/67 °C/72 °C; 30 s each; rampage 5 °C/s) and a final melting curve analysis (60 °C to 97 °C with 0.1 °C/s). Quantified results were normalized to Ppia using the ΔΔCt method.
Statistical analysis
Statistical analysis was performed using the software GraphPad Prism 7 (GraphPad Software, San Diego, CA, USA). All data were analyzed for normality using Shapiro-Wilk-test for n ≤ 7 and D’Agostino and Pearson omnibus normality test for n ≥ 8. Moreover, we tested for outliers using Grubbs‘outlier test before applying parametric statistical tests. Outliers were omitted from further analysis. Datasets were analyzed either by one-way ANOVA or two-way ANOVA, followed by Tukey’s multiple comparison test. Correlations were calculated by determining the Pearson correlation coefficient. P-values < 0.05 were considered significant. P-values for all experiments with indicated statistical significance are reported in the figure legends. Data are represented as means ± SD.
Results
Short regulatory elements enhance gene expression in cultured brain endothelial cells
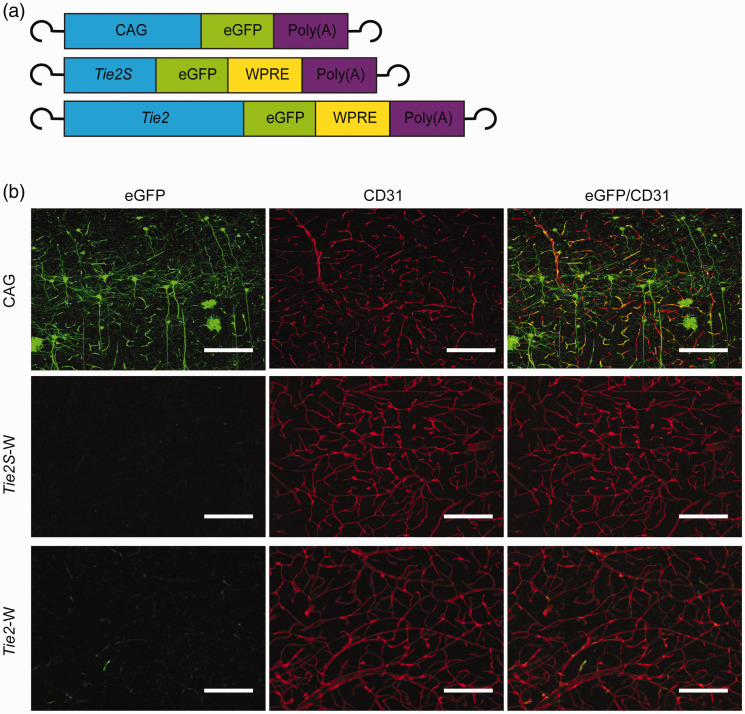
With the AAV-BR1 capsid, vectors are able to transduce brain endothelial cells in vivo with a certain selectivity over other cell populations. 20 To enhance endothelial specificity of gene expression, we used the pan-endothelial Tie2 promoter to control the expression of the reporter eGFP in AAV-BR1 vectors. When we compared a short 485-bp (Tie2S) and a long 2,099-bp fragment of the Tie2 promoter (Tie2) 44 in conjunction with the WPRE, only the long sequence resulted in eGFP expression in mouse brain vessels demonstrating that the additional 1,614 bp of the long promoter fragment are required to drive gene expression. However, the expression was lower than with the ubiquitously active CAG promoter (Figure 1).
Figure 1.
The short promoter sequence of the Tie2 promoter (Tie2S) resulted in a lower gene expression in vivo than the long promoter sequence (Tie2). (a) The vectors AAV-BR1-Tie2S-eGFP-W (Tie2S-W) and AAV-BR1-Tie2-eGFP-W (Tie2-W) express eGFP under the control of the short or the long sequence of the Tie2 promoter, respectively; the vector AAV-BR1-CAG-eGFP (CAG) expresses eGFP under control of the ubiquitous CAG promoter. (b) Fifteen days after the intravenous injection of vectors, brain sections were immunostained for eGFP (green) and CD31 (red). Representative images of the cortex of the three mice per sequence are presented. Scale bar, 150 µm.
To search for stronger regulatory sequences, we went back to cultured brain endothelial cells. We transfected primary mouse and pig brain endothelial cells and the human brain endothelial cell line hCMEC/D3 with eGFP or luciferase reporter plasmids containing various regulatory elements. Again, the pan-endothelial long Tie2 promoter and a previously described Cdh5 promoter 35 in conjunction with the WPRE (Tie2-W or Cdh5-W) had a weaker effect on eGFP expression than the CAG promoter in endothelial cells of the three species (Supplementary Figure 1). The short T81 promoter served as a negative control.
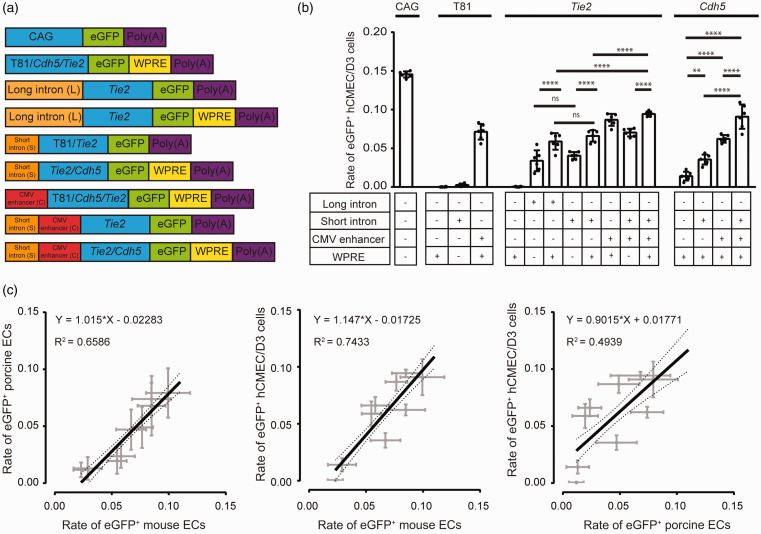
To achieve a stronger induction, we evaluated whether the following regulatory sequences would enhance gene expression: a 0.3-kbp (short Tie2 intron, S) or a 1.7-kbp (long Tie2 intron, L) fragment of exon 1/intron 1 of the Tie2 gene and the CMV enhancer sequence (C), corresponding to the enhancer and promoter region of the major immediate-early enhancer gene of human cytomegalovirus (-598/-68; Figure 2(a)). In addition, we investigated the effect of the WPRE (W).
Figure 2.
The long (L) and short Tie2 intron (S), the CMV enhancer (C), and the WPRE (W) enhance gene expression in endothelial cells. (a) Schematic representation of the investigated DNA constructs. (b) eGFP expression driven by various combinations of short regulatory elements. The relative number of eGFP-positive cells of the human brain endothelial hCMEC/D3 cell line are shown. After transfecting cells with the indicated plasmids, immunostainings were performed. Data are presented as mean ± SD analyzed by one-way ANOVA (plasmid, F(15/79)=162, p < 0.0001), **p < 0.0021, ****p < 0.0001 (Tukey’s multiple comparison test, n = 5-6 transfected wells per plasmid). (c) The short regulatory elements have a similar transcriptional effect in human, pig, and mouse brain endothelial cells, as shown by the close correlation between eGFP-positive cells in the three species. The results of Pearson correlation are given in the graphs.
When transfecting human brain endothelial hCMEC/D3 cells, the long and the short Tie2 intron had a similar effect on gene expression driven by the long Tie2 promoter (compare L-Tie2-eGFP with S-Tie2-eGFP and L-Tie2-eGFP-W with S-Tie2-eGFP-W in Figure 2(b)). In combination with the Tie2 promoter and one of the Tie2 introns, the WPRE further enhanced gene expression (compare S-Tie2-eGFP-W with S-Tie2-eGFP, L-Tie2-eGFP-W with L-Tie2-eGFP, and S-C-Tie2-eGFP-W with S-C-Tie2-eGFP in Figure 2(b)). Finally, we investigated the effect of the CMV enhancer on gene expression. In the context of the Tie2 promoter with or without short Tie2 intron or WPRE, the CMV enhancer increased gene expression, even in combination with the minimal T81 promoter (Figure 2(b)). Due to the comparable effect on the gene expression of the short and long Tie2 intron, we decided to use the short Tie2 intron for further experiments. To confirm the synergistic effects of the regulatory elements we additionally tested the combinations with the Cdh5 promoter. In presence of the WPRE, the short Tie2 intron, the CMV enhancer and their combination enhanced the gene expression driven by the Cdh5 promoter (Figure 2(b)). These data suggest that in the context of the pan-endothelial Tie2 and Cdh5 promoters the short Tie2 intron, CMV enhancer, and WPRE increase gene expression with the CMV enhancer having the strongest effect.
Testing the transcriptional activity of regulatory elements in primary mouse and pig brain endothelial cells led to similar results as in human hCMEC/D3 cells, as shown by the close correlation between eGFP-positive cells in the three species (Figure 2(c)). We repeated the experiments with firefly luciferase as reporter gene (Supplementary Figure 2(a)). The data confirmed that the combination of short Tie2 intron, CMV enhancer, and WPRE had the strongest effect on gene expression. Importantly, the transcriptional activity of regulatory elements closely correlated between experiments in which we used eGFP or firefly luciferase as reporter plasmids, supporting the validity of the assay (Supplementary Figure 2(b)).
In the previous experiments, the short Tie2 intron was positioned 5′ to the regulatory elements because we had detected a significantly higher expression of eGFP when inserting the short Tie2 intron on the 5′-side of the CMV enhancer instead of on the 3′-side of the pan-endothelial Cdh5 promoter (Supplementary Figure 3(a-c)). However, the ideal position of the Tie2 intron depends on the specific promoter. When we re-analyzed the position effect of the short Tie2 intron in the context of the brain endothelial-specific Foxq1 promoter, we obtained the opposite result (Supplementary Figure 3(c)). Due to this finding and since the usual order of elements is enhancer, promoter, intron, and coding sequence in 5′-to-3′ direction,17,45 we positioned the short Tie2 intron between promoter and coding sequence in subsequent experiments using brain endothelial-specific promoters.
Transcriptional activity of brain endothelial-specific promoters
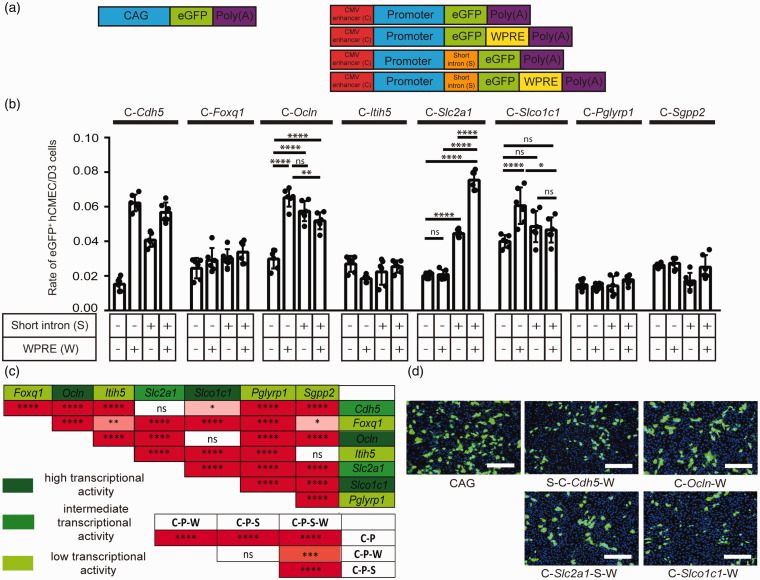
Having found that the CMV enhancer and, to a lesser extent, the short Tie2 intron and WPRE increased gene expression driven by pan-endothelial promoters, we asked whether the short Tie2 intron and WPRE would increase expression driven by promoters that are presumably brain endothelial-specific in comparison to the pan-endothelial Cdh5 promoter. For this purpose, we selected 8 genes that scored highly for brain endothelial-specific expression in a transcriptomic analysis of the blood-brain barrier: Foxq1, Ocln, Itih5, Slc2a1, Slco1c1, Pglyrp1, Sgpp2, and Tbx3 (for details of the selection, see Methods and Supplementary Table 1). 25 The postulated short promotor fragments of these genes were combined with the CMV enhancer, the short Tie2 intron, and the WPRE in reporter genes (Figure 3(a)).
Figure 3.
Transcriptional activity of short regulatory elements and promoters (P) in transfected human endothelial hCMEC/D3 cells. (a) Schematic representation of the investigated DNA constructs. For short regulatory elements, we used the CMV enhancer (C) in all constructs as well as the short Tie2 intron (S) and the WPRE (W) as indicated. (b and c) Rate of eGFP-positive hCMEC/D3 cells transfected with plasmids containing different regulatory sequences. Five days after transfection, cells were immunostained for eGFP. Data are presented as mean ± SD and were analyzed by two-way ANOVA (short regulatory element F(3/160)=171, p < 0.0001; promoter F(7/160)=151.9, p < 0.0001; interaction F(21/160)=34.34, p < 0.0001), ns not significant, *p < 0.0332, **p < 0.0021, ****p < 0.0001 (Tukey’s multiple comparison test, n = 6 transfected wells per plasmid). (d) Representative images of hCMEC/D3 cells transfected with plasmids expressing eGFP under the regulation of the synthetic promoter sequences CAG, S-C-Cdh5-W, C-Ocln-W, C-Slc2a1-S-W and C-Slco1c1-W. The images were stained for eGFP (green) and DNA (blue). Scale bar, 100 µm.
The reporter fusion genes were tested in hCMEC/D3 cells (Figure 3(b) to (d)). Two-way ANOVA showed that the promoters of Cdh5, Ocln and Slco1c1 had the strongest effect on gene expression (Figure 3(c)). An intermediate effect was observed for the Slc2a1 promoter, while the promoters of Foxq1, Itih5, Pglyrp1, and Sgpp2 had a small effect on gene expression (Figure 3(c)). The Tbx3 promoter was not different from the promoter of Plvap, a marker gene of vessels lacking the blood-brain barrier that served as negative control (Supplementary Figure 3(d) to (f)).
The statistical analysis also revealed that the short Tie2 intron, the WPRE and the combination of short Tie2 intron plus WPRE increased gene expression further in the presence of the CMV enhancer (Figure 3(c)). A posthoc analysis showed that the WPRE increased expression in the context of the C-Ocln and the C-Slco1c1 enhancer-promoter combination, the short Tie2 intron in the context of the C-Ocln and C-Slc2a1 enhancer-promoter, and the short Tie2 intron plus WPRE only in the context of the C-Slc2a1 enhancer-promoter (Figure 3(b)). Thus, C-Ocln-W, C-Slco1c1-W, and C-Slc2a1-S-W emerged as regulatory sequences with a high activity in hCMEC/D3 brain endothelial cells. Interestingly, the new regulatory sequences were much less active in peripheral endothelial cells. After transfecting HUVECs, the plasmids p-S-C-Cdh5-eGFP-W, p-C-Ocln-eGFP-W, p-C-Slc2a1-S-eGFP-W, and p-C-Slco1c1-eGFP-W displayed a lower transcriptional activity than CAG-eGFP (Supplementary Figure 4).
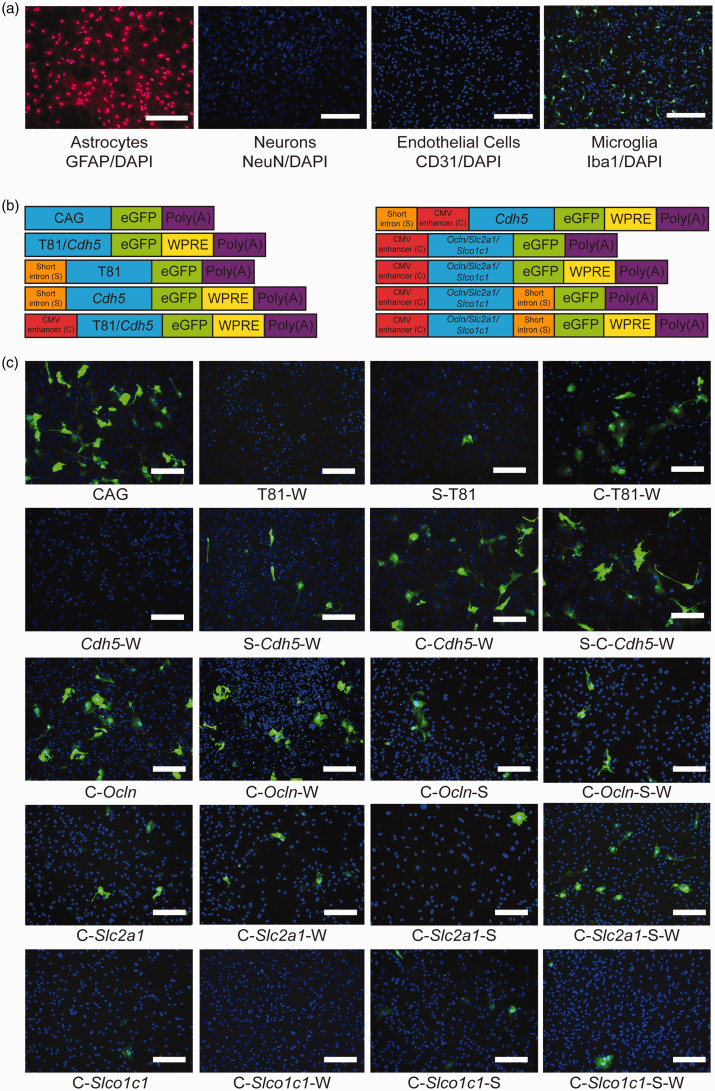
Assessing cell specificity in vitro
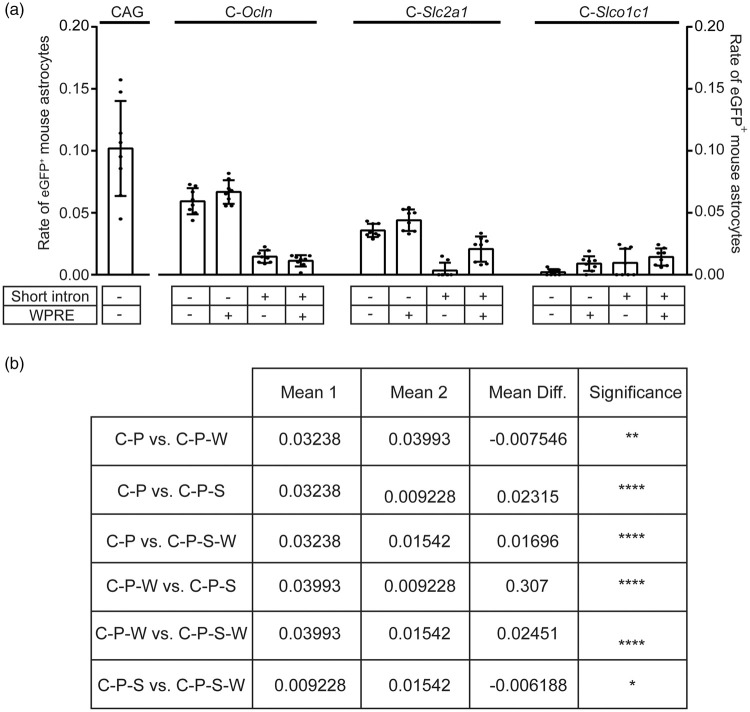
AAV-BR1 and other AAV vectors targeting the brain have the tendency to transduce also astrocytes (Figure 1). 20 To investigate cell specificity of the short regulatory sequences, reporter gene assays were performed in primary mouse astrocytes. About 96% of the cells were astrocytes, as confirmed by immunostainings for the astrocytic marker GFAP, the endothelial marker CD31, the neuronal marker NeuN, and the microglial marker Iba1 (Figure 4(a)). We focused on the Ocln, Slc2a1 and the Slco1c1 promoters with the highest transcriptional activity in endothelial cells (Figure 3) as well as on the pan-endothelial Cdh5 promoter in combinations with regulatory elements (Figure 4(b)). CAG and T81 promoters were used as positive and negative controls, respectively. While the WPRE enhanced eGFP expression in astrocytes, adding the short Tie2 intron to the regulatory sequence significantly reduced eGFP expression in primary astrocytes (Figures 4(b) and (c) and Figure 5). Thus, the short Tie2 intron promotes transcriptional activity specifically in endothelial cells. In addition to the WPRE and the short Tie2 intron, the promoter had a significant effect on astrocytic expression. In astrocytes, the transcriptional activity of promoters decreased in the order Ocln, Slc2a1, and Slco1c1 (Figure 5).
Figure 4.
Reporter gene expression in transfected primary astrocytes depends on short regulatory elements. (a) Representative microscopic images to determine the purity of the cortical astrocyte culture. Cortical astrocytes were isolated from brains of mice of age P0/1. The purity of the cortical astrocyte culture was determined 21 days after isolation of the cells by immunostaining for GFAP, NeuN, CD31 and Iba1 as well as DNA. The purity of the culture was determined to be 96%. Neurons and endothelial cells were not detected. However, approximately 4% of the cultured cells were stained positive for Iba1 as a marker for microglia. Scale bar, 100 µm. (b) Schematic representation of the investigated constructs indicating the position of the short Tie2 intron (S), the CMV enhancer (C), and the WPRE (W) relative to the promoter sequence. (c) Representative images of cortical astrocytes transfected with plasmids expressing eGFP under the regulation of the indicated regulatory sequences 21 days after isolation of the cells. Analysis of plasmid encoded gene expression via immunostaining was performed four days after lipofection. The images were stained for eGFP (green) and DNA (blue). Scale bar, 100 µm.
Figure 5.
Quantification of eGFP expression in primary astrocytes shown in Figure 4. (a) Determination of the rate of eGFP-positive astrocytes revealed that adding the WPRE to the synthetic promoter sequence significantly enhanced eGFP-expression in astrocytes. Moreover, adding the short Tie2 intron significantly reduced eGFP-expression in primary astrocytes. Data are presented as mean ± SD analyzed by two-way ANOVA (short regulatory elements, F(3/84)=83.29, p < 0.0001; promoter F(2/84)=116.8, p < 0.0001; interaction F(6/84)=39.06, p < 0.0001; n = 8 analyzed microscopic images per plasmid). *p < 0.0332, **p < 0.0021, ****p < 0.0001 (Tukey's multiple comparison test). (b) The table presents the results of the two-way ANOVA for the impact of the short regulatory elements in astrocytes.
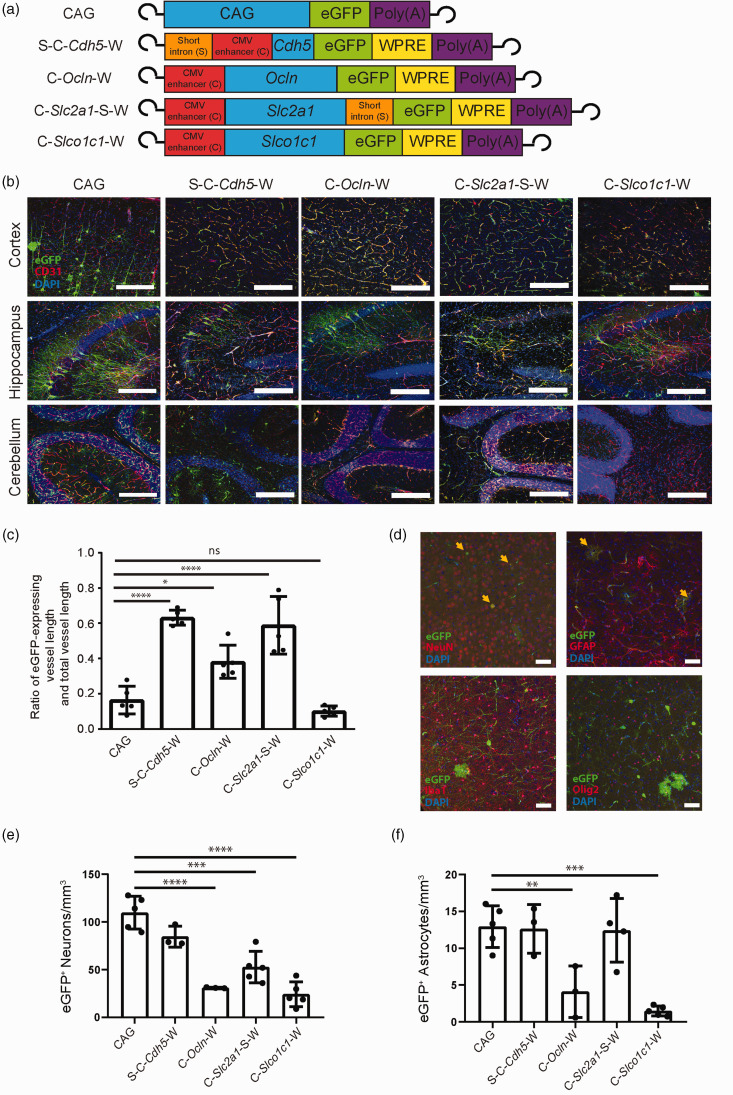
Assessing cell specificity of short regulatory sequences in vivo
Based on the transcriptional activity in endothelial cells in vitro, we selected four of the strongest synthetic regulatory sequences (Figure 3(d)) for investigation in vivo: S-C-Cdh5-eGFP-W, C-Ocln-eGFP-W, C-Slc2a1-S-eGFP-W, and C-Slco1c1-eGFP-W (Figure 6(a)). Because the short Tie2 intron did not reproducibly enhance gene expression in the context of the Ocln and Slco1c1 promoters (Figure 3(b) and (c)) but increases the overall length of the constructs, we selected synthetic regulatory sequences containing the Ocln and Slco1c1 promoter but without the short Tie2 intron. CAG-eGFP served as a control (Figure 6(a)).
Figure 6.
Synthetic regulatory sequences enhance blood-brain barrier endothelial cell specificity in vivo. (a) Schematic representation of the investigated AAV-BR1 constructs indicating the inverted terminal repeats at both ends as well as the position of the short Tie2 intron and the CMV enhancer relative to the promoter sequence (CAG, Cdh5, Ocln, Slc2a1, Slco1c1, blue) controlling the expression of the reporter gene eGFP. The CAG promoter served as control. (b) Representative immunostainings of the vector-expressed eGFP (green) and CD31 (red) in the cortex, hippocampus, and cerebellum, 14 to 15 days after injecting the indicated vectors. Nuclei were stained with DAPI (blue). Scale bar, 100 µm. (c) The ratio of the length of the vessels expressing eGFP to the total vessel length was determined. Each vector was administered to 2 to 5 animals (AAV-BR1-CAG-eGFP (n = 3), AAV-BR1-S-C-Cdh5-eGFP-W (n = 2), AAV-BR1-C-Ocln-eGFP-W (n = 2), AAV-BR1-C-Slc2a1-S-eGFP-W (n = 5), AAV-BR1-C-Slco1c1-eGFP-W (n = 3)). Data are presented as mean ± SD analyzed by one-way ANOVA (vector, F(4/20) = 29.86, p < 0.0001), *p < 0.0332, ****p < 0.0001 (Tukey’s multiple comparison test, n = 5 images per vector). (d to f) The vector-encoded expression of the reporter gene eGFP in non-targeted cell types of the brain was reduced by short regulatory sequences. Representative immunostainings (d) of the expressed eGFP (green) and NeuN, GFAP, Iba1, Olig2 (red) in the cortex 14 to 15 days after injecting the vector AAV-BR1-CAG-eGFP. Arrows indicate neurons or astrocytes expressing eGFP. Nuclei were stained with DAPI (blue). Scale bar, 20 µm. Quantification of eGFP-expressing neurons (e) and astrocytes (f) per mm3 in the cortex 14 to 15 days after injecting the vectors from (a). Data are presented as mean ± SD analyzed by one-way ANOVA. For neurons (e), F(4/15) = 29.22, p < 0.0001; for astrocytes (f), F(4/15) = 14.22, p < 0.0001. **p < 0.01, ***p < 0.001, ****p < 0.0001 (Tukey’s multiple comparison test, n = 3-5 animals per vector).
For investigation in vivo, AAV-BR1 vectors were produced for intravenous administration. Fourteen days after mice received a dose of 1.8 × 1011 gp/mouse (i.v.), organs were extracted and analyzed for eGFP expression by immunostaining. To quantify the transduction rate in brain endothelial cells, we determined the ratio of eGFP-expressing vessel length to total vessel length in the cortex. The ratio of transduced vessels was significantly increased after injecting vectors containing S-C-Cdh5-eGFP-W, C-Ocln-eGFP-W, or C-Slc2a1-S-eGFP-W in comparison to CAG-eGFP. The vector C-Slco1c1-eGFP-W afforded a higher endothelial selectivity than the CAG-eGFP control, but the transduced vessel length was similar (Figure 6(b) to (f)). In comparison to the CAG-eGFP control, C-Slco1c1-eGFP-W and C-Ocln-eGFP-W were associated with a lower eGFP expression in neurons or astrocytes (Figure 6(e) and (f)). C-Slc2a1-S-eGFP-W led to a reduced number of eGFP-positive neurons, while the S-C-Cdh5-eGFP-W vector did not differ from CAG-eGFP with respect to directing eGFP expression to neurons and astrocytes. With all short regulatory elements tested, no eGFP expression could be found in microglia or oligodendrocytes (Figure 6(d)).
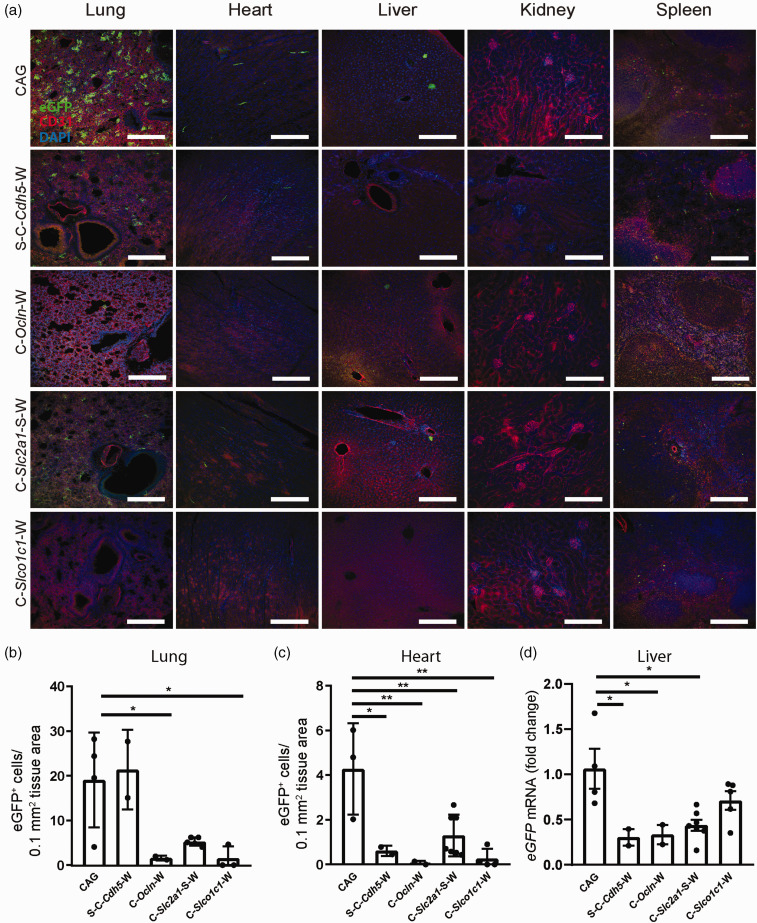
In order to evaluate the effect of the synthetic regulatory sequences on tissue tropism, other organs were examined for off-target gene expression. After injecting CAG-eGFP, considerable expression of eGFP was observed in the lung (Figure 7(a)). In contrast, when S-C-Ocln-eGFP-W or C-Slco1c1-eGFP-W drove eGFP expression from AAV-BR1 vectors, off-target expression in the lung was clearly reduced (Figure 7(a) and (b)). In cardiomyocytes, eGFP expression was detected when mice were treated with CAG-eGFP but significantly lower when S-C-Cdh5-eGFP-W, C-Ocln-eGFP-W, C-Slc2a1-S-eGFP-W or C-Slco1c1-eGFP-W were administered (Figure 7(a) and (c)). As we detected rare eGFP-positive cells in the liver after injecting CAG-eGFP, it seemed more reliable to compare expression at the mRNA level. eGFP mRNA levels in the liver were lower in animals that received S-C-Cdh5-eGFP-W, C-Ocln-eGFP-W, or C-Slc2a1-S-eGFP-W than in mice treated with CAG-eGFP (Figure 7(d)). In summary, the regulatory sequences C-Ocln-eGFP-W and C-Slc2a1-S-eGFP-W resulted in a strong expression of the reporter gene in brain endothelial cells and reduced vector-encoded gene expression in non-targeted cells and tissues in vivo, particularly in neurons, astrocytes, and cells of the lung, liver, and heart. When using the regulatory sequence S-C-Cdh5-eGFP-W, strong expression of the reporter gene was likewise obtained in brain endothelial cells, but with lower cell specificity.
Figure 7.
The vector-encoded expression of the reporter gene eGFP in non-targeted cell types of extracerebral tissues is reduced by short regulatory sequences. (a) Representative images from lung, heart, liver, kidney and spleen 14 to 15 days after vector injection. AAV-transduced cells expressing eGFP under the control of the regulatory promoter sequences (green) were reduced compared to the results obtained with the control vector CAG-eGFP. CD31 was stained as a marker of endothelial cells (red). Scale bar, 100 µm. (b and c) The number of eGFP-positive cells in the lung (b) or heart (c) 14 to 15 days after vector injection are shown. Data are presented as means ± SD and were analyzed by one-way ANOVA (lung, F(4/11) = 6.645, p = 0.0057; heart, F(4/13) = 7.919, p = 0.0018), *p < 0.05, **p < 0.01 (Tukey’s multiple comparison test, n = 2–5 mice per vector). (d) Relative eGFP mRNA in liver 14 to 15 days after vector injection. Data are presented as means ± SD and were analyzed by one-way ANOVA (F(4/15) = 5.353, p = 0.007), *p < 0.0354 (Tukey’s multiple comparison test, n = 2-5 mice per vector).
Discussion
In this study we have identified a series of short regulatory sequences that help target brain endothelial cells and have the potential to improve gene therapy of cerebrovascular diseases and lysosomal disorders. Three of the short regulatory sequences, S-C-Cdh5-[coding sequence]-W, C-Slc2a1-S-[coding sequence]-W, and C-Ocln-[coding sequence]-W had a stronger effect on brain endothelial gene expression in vivo than the universal promoter CAG. Additionally, C-Slc2a1-S-[coding sequence]-W and C-Ocln-[coding sequence]-W revealed a higher specificity for this cell population of the brain and peripheral organs.
Building on a previous transcriptomic study, 25 we tested the efficacy of 49 combinations of regulatory elements with 2 different reporters in vitro. The various combinations were based on the promoters of 3 genes expressed in all endothelial cells (Tie2, Cdh5) or in fenestrated endothelium (Plvap), and of 8 genes with a brain endothelial-specific expression (Foxq1, Ocln, Itih5, Slc2a1, Slco1c1, Pglyrp1, Sgpp2, Tbx3). Transcriptional activity varied from promoter to promoter but, as expected, cell specificity was higher with most of the endothelial promoters than with the ubiquitously active CAG promoter. Both activity and specificity of gene expression were enhanced, at least in the context of some promoters, by the Tie2 intron. This element had been identified in transgenic mice and has been utilized in lentiviral but not in AAV vectors.44,46,47 The likely reason is that the original report by Schläger et al. mainly focused on a 1,667-bp sequence that would exceed the capacity of AAV vectors. 44 However, these authors also noted that a 0.3-kbp fragment of the intron is sufficient to afford endothelial expression in transgenic mice. Our data confirm the finding that a short 0.3-kbp fragment of the Tie2 intron has a similar activity as the full length and suggest the short Tie2 intron as a suitable element for AAV targeting of endothelial cells. Introns may enhance mRNA stability, promote mRNA transport, 17 or function as intronic enhancers. The latter seems to apply to the Tie2 intron that contains binding sites for members of the ETS family and other transcription factors and enhances gene expression when positioned upstream of the core Cdh5 promoter (Supplementary Figure 3).44,48 To enhance expression further, we tested the WPRE. As noted by others, 49 the effect of the WPRE depended on the promoter context. Finally, transcriptionally activity was strongly increased by the CMV enhancer that also promoted AAV-mediated gene expression in the heart. 21
The regulatory sequences in S-C-Cdh5-[coding sequence]-W add up to an overall length of about 1.9 kbp and in C-Slc2a1-S-[coding sequence]-W to about 2.8 kbp, including the WPRE (Supplementary Table 4). Thus, they are only slightly longer than the frequently used unspecific CAG promoter with 1.6 kbp, but considerably shorter than Ple34 with 4.4 kbp or Ple261 with 3.6 kbp, including WPRE. 30 The relative shortness of the regulatory sequences increases the length of coding DNA that can be expressed in brain endothelial cells. Under the control of S-C-Cdh5-[coding sequence]-W, the target proteins that are directed into brain endothelial cells can be up to about 930 amino acids long. This includes a large segment of expressed genes. Depending on the target gene, a low expression level may be better. With the four short regulatory sequences that provide a high brain endothelial specificity, it will be possible to adjust gene expression to the required level. C-Slco1c1-[coding sequence]-W results in low expression, C-Ocln-[coding sequence]-W in an intermediate, and S-C-Cdh5-[coding sequence]-W or C-Slc2a1-S-[coding sequence]-W in a high expression.
Interestingly, optimal combinations of regulatory elements did not quite achieve the transcriptional activity of the CAG promoter in vitro but were better in vivo. This observation is probably explained by the downregulation of brain endothelial-specific genes when primary cells are cultured in vitro. 50 In the latter study, genes of which we used the promoter markedly decreased in vitro, illustrating the need to perform in vivo experiments for a realistic picture.
The main advantage of transcriptional over transductional targeting is that the cell specificity of promoters is often independent of species.51–53 Our data confirm that the activity of short regulatory sequences was similar in endothelial cells of three species in vitro. Therefore, we expect that the sequences will also confer brain endothelial specificity in humans. Cell type-specific regulatory sequences have been developed for many other cell types, including hepatocytes, muscle cells, or astrocytes, and have improved gene therapy.17,54 The regulatory sequences developed here may provide the means to target brain endothelial cells as key players in cerebrovascular diseases or as a gateway to the brain.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211039617 for Short regulatory DNA sequences to target brain endothelial cells for gene therapy by Hanna Graßhoff, Helge Müller-Fielitz, Godwin K Dogbevia, Jakob Körbelin, Jacqueline Bannach, Carl MG Vahldieck, Kristina Kusche-Vihrog, Olaf Jöhren, Oliver J Müller, Ruben Nogueiras, Vincent Prevot and Markus Schwaninger in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
We are grateful to Dr. Rolf Sprengel, University of Heidelberg, Germany, for providing the plasmid p179, to Dr. Mazahir Hasan, Achucarro Basque Center for Neuroscience, Spain, for providing the plasmid pAAV-CAG-iCre-2A-eGFP, to Dr. Sebastian Kügler, University Medical Center Göttingen, Germany, for providing the plasmid p-6p1 (hSYN-eGFP-W), and to Dr. Urban Deutsch, University of Bern, Switzerland, for advice on the short Tie2 intron. We also thank Frauke Spiecker, Wiebke Brandt, Ines Stölting, and Christine Eichholz, University of Lübeck, for expert technical support.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the European Research Council (ERC) Synergy Grant-2019-WATCH-810331 to VP, RN, and MS and by grants of the Deutsche Forschungsgemeinschaft (DFG, SCHW 416/5-3, 9-1).
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: HG, MS, and GKD designed the experiments; HG, GKD, HMF, JK and JB performed the experiments; HG, GKD, HMF, OJ, OJM, RN, VP and MS analyzed the data; and HG and MS wrote the manuscript. All authors revised the manuscript for important intellectual content.
ORCID iDs: Helge Müller-Fielitz https://orcid.org/0000-0003-2815-4426
Markus Schwaninger https://orcid.org/0000-0002-4510-9718
Supplemental material: Supplemental material for this article is available online.
References
- 1.Kirschen GW, Kery R, Liu H, et al. Genetic dissection of the neuro-glio-vascular machinery in the adult brain. Mol Brain 2018; 11: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haery L, Deverman BE, Matho KS, et al. Adeno-associated virus technologies and methods for targeted neuronal manipulation. Front Neuroanat 2019; 13: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarty P, Rosario A, Cruz P, et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One 2013; 8: e67680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vagner T, Dvorzhak A, Wojtowicz AM, et al. Systemic application of AAV vectors targeting GFAP-expressing astrocytes in Z-Q175-KI huntington's disease mice. Mol Cell Neurosci 2016; 77: 76–86. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z, Parry M, Hou XY, et al. Gene therapy conversion of striatal astrocytes into GABAergic neurons in mouse models of huntington's disease. Nat Commun 2020; 11: 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coucha M, Barrett AC, Elgebaly M, et al. Inhibition of Ephrin-B2 in brain pericytes decreases cerebral pathological neovascularization in diabetic rats. PLoS One 2019; 14: e0210523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosario AM, Cruz PE, Ceballos-Diaz C, et al. Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol Ther Methods Clin Dev 2016; 3: 16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dogbevia GK, Töllner K, Körbelin J, et al. Gene therapy decreases seizures in a model of incontinentia pigmenti. Ann Neurol 2017; 82: 93–104. [DOI] [PubMed] [Google Scholar]
- 9.Dogbevia G, Grasshoff H, Othman A, et al. Brain endothelial specific gene therapy improves experimental Sandhoff disease. J Cereb Blood Flow Metab 2020; 40: 1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweeney MD, Zhao Z, Montagne A, et al. Blood-brain barrier: from physiology to disease and back. Physiol Rev 2019; 99: 21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med 2013; 19: 1584–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ridder DA, Wenzel J, Müller K, et al. Brain endothelial TAK1 and NEMO safeguard the neurovascular unit. J Exp Med 2015; 212: 1529–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M, Gao G, Rueda CB, et al. Brain microvasculature defects and Glut1 deficiency syndrome averted by early repletion of the glucose transporter-1 protein. Nat Commun 2017; 8: 14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vatine GD, Al-Ahmad A, Barriga BK, et al. Modeling psychomotor retardation using iPSCs from MCT8-Deficient patients indicates a prominent role for the Blood-Brain barrier. Cell Stem Cell 2017; 20: 831–843. e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YH, Chang M, Davidson BL. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med 2009; 15: 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YH, Claflin K, Geoghegan JC, et al. Sialic acid deposition impairs the utility of AAV9, but not peptide-modified AAVs for brain gene therapy in a mouse model of lysosomal storage disease. Mol Ther 2012; 20: 1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domenger C, Grimm D. Next-generation AAV vectors-do not judge a virus (only) by its cover. Hum Mol Genet 2019; 28: R3–R14. [DOI] [PubMed] [Google Scholar]
- 18.Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015; 47: 1187–1193. [DOI] [PubMed] [Google Scholar]
- 19.La Bella T, Imbeaud S, Peneau C, et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut 2020; 69: 737–747. [DOI] [PubMed] [Google Scholar]
- 20.Körbelin J, Dogbevia G, Michelfelder S, et al. A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol Med 2016; 8: 609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller OJ, Leuchs B, Pleger ST, et al. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res 2006; 70: 70–78. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds PN, Nicklin SA, Kaliberova L, et al. Combined transductional and transcriptional targeting improves the specificity of transgene expression in vivo. Nat Biotechnol 2001; 19: 838–842. [DOI] [PubMed] [Google Scholar]
- 23.Liu D, Zhu M, Zhang Y, et al. Crossing the blood-brain barrier with AAV vectors. Metab Brain Dis 2021; 36: 45–52. [DOI] [PubMed] [Google Scholar]
- 24.Assmann JC, Körbelin J, Schwaninger M. Genetic manipulation of brain endothelial cells in vivo. Biochim Biophys Acta 2016; 1862: 381–394. [DOI] [PubMed] [Google Scholar]
- 25.Daneman R, Zhou L, Agalliu D, et al. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One 2010; 5: e13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridder DA, Lang MF, Salinin S, et al. TAK1 in brain endothelial cells mediates fever and lethargy. J Exp Med 2011; 208: 2615–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu C, Chen Z, Kim KT, et al. Metformin alleviates hyperglycemia-induced endothelial impairment by downregulating autophagy via the hedgehog pathway. Autophagy 2019; 15: 843–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White SJ, Papadakis ED, Rogers CA, et al. In vitro and in vivo analysis of expression cassettes designed for vascular gene transfer. Gene Ther 2008; 15: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimuro J, Muramatsu S, Hakamada Y, et al. Recombinant adeno-associated virus vector-transduced vascular endothelial cells express the thrombomodulin transgene under the regulation of enhanced plasminogen activator inhibitor-1 promoter. Gene Ther 2001; 8: 1690–1697. [DOI] [PubMed] [Google Scholar]
- 30.de Leeuw CN, Korecki AJ, Berry GE, et al. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol Brain 2016; 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders A, Macosko EZ, Wysoker A, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 2018; 174: 1015–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Chen K, Sloan SA, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosma EK, van Noorden CJF, Schlingemann RO, et al. The role of plasmalemma vesicle-associated protein in pathological breakdown of blood-brain and blood-retinal barriers: potential novel therapeutic target for cerebral edema and diabetic macular edema. Fluids Barriers CNS 2018; 15: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eukaryotic Promoter Database. https://epd.epfl.ch//index.php (2020, accessed 2 August 2021).
- 35.Gory S, Vernet M, Laurent M, et al. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood 1999; 93: 184–192. [PubMed] [Google Scholar]
- 36.Nordeen SK. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques 1988; 6: 454–458. [PubMed] [Google Scholar]
- 37.Kügler S, Lingor P, Schöll U, et al. Differential transgene expression in brain cells in vivo and in vitro from AAV-2 vectors with small transcriptional control units. Virology 2003; 311: 89–95. [DOI] [PubMed] [Google Scholar]
- 38.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 1998; 72: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther 2003; 7: 839–850. [DOI] [PubMed] [Google Scholar]
- 40.Assmann JC, Müller K, Wenzel J, et al. Isolation and cultivation of primary brain endothelial cells from adult mice. Bio Protoc 2017; 7: e2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patabendige A, Skinner RA, Morgan L, et al. A detailed method for preparation of a functional and flexible blood-brain barrier model using porcine brain endothelial cells. Brain Res 2013; 1521: 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weksler BB, Subileau EA, Perrière N, et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. Faseb J 2005; 19: 1872–1874. [DOI] [PubMed] [Google Scholar]
- 43.Drüppel V, Kusche-Vihrog K, Grossmann C, et al. Long-term application of the aldosterone antagonist spironolactone prevents stiff endothelial cell syndrome. Faseb j 2013; 27: 3652–3659. [DOI] [PubMed] [Google Scholar]
- 44.Schlaeger TM, Bartunkova S, Lawitts JA, et al. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci U S A 1997; 94: 3058–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Z, Sun J, Zhang T, et al. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol Ther 2008; 16: 280–289. [DOI] [PubMed] [Google Scholar]
- 46.De Palma M, Venneri MA, Naldini L. In vivo targeting of tumor endothelial cells by systemic delivery of lentiviral vectors. Hum Gene Ther 2003; 14: 1193–1206. [DOI] [PubMed] [Google Scholar]
- 47.Pariente N, Mao SH, Morizono K, et al. Efficient targeted transduction of primary human endothelial cells with dual-targeted lentiviral vectors. J Gene Med 2008; 10: 242–248. [DOI] [PubMed] [Google Scholar]
- 48.Dube A, Akbarali Y, Sato TN, et al. Role of the Ets transcription factors in the regulation of the vascular-specific Tie2 gene. Circ Res 1999; 84: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 49.Klein R, Ruttkowski B, Knapp E, et al. WPRE-mediated enhancement of gene expression is promoter and cell line specific. Gene 2006; 372: 153–161. [DOI] [PubMed] [Google Scholar]
- 50.Sabbagh MF, Nathans J. A genome-wide view of the de-differentiation of central nervous system endothelial cells in culture. Elife 2020; 9: e51276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dimidschstein J, Chen Q, Tremblay R, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 2016; 19: 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jüttner J, Szabo A, Gross-Scherf B, et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat Neurosci 2019; 22: 1345–1356. [DOI] [PubMed] [Google Scholar]
- 53.Meireles-Filho ACA, Stark A. Comparative genomics of gene regulation—conservation and divergence of cis-regulatory information. Curr Opin Genet Dev 2009; 19: 565–570. [DOI] [PubMed] [Google Scholar]
- 54.Kuzmin DA, Shutova MV, Johnston NR, et al. The clinical landscape for AAV gene therapies. Nat Rev Drug Discov 2021; 20: 173–174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211039617 for Short regulatory DNA sequences to target brain endothelial cells for gene therapy by Hanna Graßhoff, Helge Müller-Fielitz, Godwin K Dogbevia, Jakob Körbelin, Jacqueline Bannach, Carl MG Vahldieck, Kristina Kusche-Vihrog, Olaf Jöhren, Oliver J Müller, Ruben Nogueiras, Vincent Prevot and Markus Schwaninger in Journal of Cerebral Blood Flow & Metabolism