Abstract
Cutaneous melanoma is an aggressive malignant cancer associated with poor prognosis. Identification of reliable biomarkers for predicting prognosis of melanoma contributes to improved clinical outcome and disease management. Long non-coding RNAs (lncRNAs) serve a crucial regulatory role of oncogenesis and tumor suppression in melanoma. Using data from The Cancer Genome Atlas database, novel lncRNA 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1 (HSD11B1-AS1) was identified, which was significantly downregulated in malignant melanoma and its downregulation was significantly associated with poor clinicopathological characteristics, including advanced T and pathological stage, Clark level, Breslow depth and ulceration and worse prognosis. Multivariate analysis showed that HSD11B1-AS1, as well as N stage and Breslow depth, were independent prognostic factors in cutaneous melanoma, and nomograms suggested a good predictive value of 1-, 3- and 5-year overall survival, progression-free interval and disease-specific survival. In vitro experiments verified the decreased HSD11B1-AS1 expression in melanoma cell lines compared with human epidermal melanocytes. Moreover, cell experiments in vitro, including Cell Counting Kit-8, colony formation, wound healing and Transwell assay, suggested that overexpression of HSD11B1-AS1 significantly inhibited melanoma cell proliferation, migration and invasion. Functional enrichment showed significantly enriched pathways in IFN-γ and -α response, TNF-α signaling via NF-κB and IL-2/STAT-5 and IL-6/JAK/STAT-3 signaling. In addition, immune infiltration analysis demonstrated that HSD11B1-AS1 may function by accelerating immune response regulation and the immune cell infiltration of various immunocytes, especially T, T helper 1, activated dendritic and B cells. The present study revealed HSD11B1-AS1 as a potential therapeutic target and promising biomarker for diagnosis and prognosis of cutaneous melanoma.
Keywords: 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1, cutaneous melanoma, bioinformatics analysis, prognosis, quantitative PCR, overexpression, immune infiltration
Introduction
Melanoma, a deadly malignant cancer originating from melanocytes, accounts for 5% of all skin cancers but 75% of skin cancer-associated mortality (1,2). The prognosis of patients with high tumor burden, brain metastasis or elevated lactate dehydrogenase is poor with 3-year survival rate <10% (3,4). Advances have been made in drug therapy for advanced melanoma, especially in checkpoint inhibitor immunotherapy including anti-programmed cell death protein 1 and anti-cytotoxic T-lymphocyte antigen 4 (5). However, most patients with metastatic melanoma achieve only short-term benefits from immune therapy or show no response at all, potentially due to the low level of immune activation in the tumor microenvironment (6,7). Therefore, it is necessary to gain a broader view of cancer immunity and identify reliable biomarkers and therapeutic targets associated with clinical prognosis and immune activation.
Long non-coding RNAs (lncRNAs) are a group of ncRNAs >200 nucleotides in length that have no protein-coding capacity due to the lack of open reading frames (8). Numerous studies have illustrated that lncRNAs function as key regulatory molecules of tumor-causing pathways and tumor suppressors in cutaneous melanoma (9–13). For example, Leucci et al (14) demonstrated that a melanoma-specific lncRNA, survival-associated mitochondrial melanoma-specific oncogenic ncRNA, is a biomarker of malignancy and a highly selective and broad-spectrum anti-melanoma therapeutic target. Moreover, overexpression of BRAF-associated non-protein coding RNA is associated with worse prognosis and lower survival rate in patients with melanoma (11). Most lncRNAs are considered to function as tumor-promoting factors due to their roles in facilitating melanoma cell growth and metastasis; to the best of our knowledge, there are few reports of tumor-inhibiting lncRNAs (15).
To date, melanoma has been regarded as a highly immunogenic tumor because of the key role of the immune system in its development and progression. For example, it demonstrates immunogenicity attributed to the recognition of antigens expressed by melanocytes (16). Recently, emerging evidence has suggested that lncRNAs regulate cancer immunity by participating in diverse processes such as tumor antigen release/presentation, immune cell activation and infiltration and recognition and attack of cancer cells (17,18). Accordingly, lncRNAs are considered to be prognostic biomarkers and potential novel therapeutic targets to improve response to immunotherapy (19).
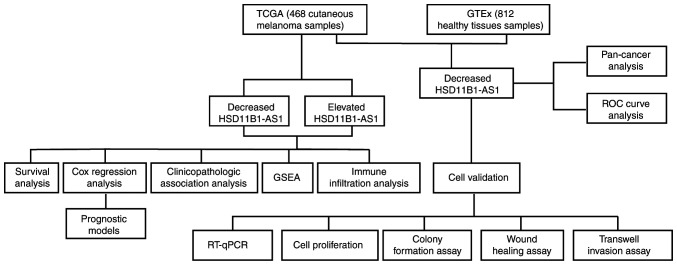
The present study used publicly available microarray data to identify a novel tumor-suppressing lncRNA, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1 (HSD11B1-AS1), which was validated via reverse transcription-quantitative PCR (RT-qPCR) to detect HSD11B1-AS1 expression in melanoma cell lines and melanocytes. Bioinformatic analysis was performed to determine differential expression, diagnostic and prognostic efficacy of HSD11B1-AS1 in cutaneous melanoma, which was validated in melanoma cell lines A375 and SK-MEL-1. The potential molecular mechanism and underlying signaling pathways were investigated using Gene Set Enrichment Analysis (GSEA) and immune infiltration analysis. The study design is presented in Fig. 1.
Figure 1.
Flow chart demonstrating how long non-coding RNA HSD11B1-AS1 acts as a protective prognostic biomarker in cutaneous melanoma. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; GTEx, Genotype Tissue Expression; RT-qPCR, reverse transcription-quantitative PCR; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis; ROC, receiver operating characteristic.
Materials and methods
RNA sequencing (seq) data and clinical characteristics
RNA-Seq data and clinical information were obtained from University of California Santa Cruz Xena databases (xenabrowser.net/datapages/). Cases without clinical features were excluded. In clinical information, the Clark Level, which has five levels from Level 1 to Level 5, is a staging system that describes the depth of melanoma as it grows in the skin, and the Breslow Depth is a more standardized method to measure how far melanoma has invaded the body, which requires an optical micrometer fitted to the ocular position of a standard microscope (20). A total of 812 healthy tissues and 468 cutaneous melanoma samples were included. Because the data was obtained from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx), it was not necessary to obtain informed consent or ethics approval. Expression of HSD11B1-AS1 was compared in unpaired healthy tissue and melanoma. Then, receiver operating characteristic (ROC) curve analysis was performed to determine the diagnostic value of HSD11B1-AS1 in discriminating melanoma from healthy tissue. Moreover, a pan-cancer analysis including 33 types of human cancer was performed to compare differential expression of HSD11B1-AS1 between cancer and healthy tissue using TCGA and GTEx gene expression data. Fig. 1 shows the experimental design.
Cell culture and transfection
Cutaneous melanoma cell lines and normal human epidermal melanocytes (HEMs) were cultured in RPMI-1640 medium containing 10% fetal bovine serum (HyClone; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin in a sterile and humidified incubator at 37°C with 5% CO2. The constructed HSD11B1-AS1 overexpression (pcDNA3.1-HSD11B1-AS1) and pcDNA3.1 empty vectors were obtained from Shanghai GenePharma Co., Ltd. To construct the pcDNA3.1-HSD11B1-AS1, primers (containing part of sequence of the pcDNA3.1 vector) were used as follows: Forward, 5′-ACCGAGCTCGGATCCCACACTCTGCTCACCACCCT-3′ and reverse, 5′-ATATCTGCAGAATTCGCAGCTTAACATAACAAAAG-3′. The melanoma cell lines A375 and SK-MEL-1 were transiently transfected using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol at concentration of 1 µg/ml at 37°C for 24 h. Transfection efficiency was confirmed by reverse transcription-quantitative (RT-q)PCR at 48 h post-transcription.
RT-qPCR
Total RNA was extracted from the A375, AK-MEL-1 and HEM cell lines using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) after which the RNA concentration was measured using NanoDrop 2000 (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed (10 µl reaction system) according to the instructions of PrimeScript™ RT reagent kit and SYBR Premix Ex Taq II (both Takara Biotechnology Co., Ltd.) was used to assess gene expression. The thermocycling conditions for RT-qPCR were as follows: Preheating for 10 min at 95°C; followed by 40 cycles at 95°C for 15 sec and 60°C for 60 sec. The following primers were used: HSD11B1-AS1 forward, 5′-GACAGCCTGAAGTGCTGGAC-3′ and reverse, 5′-GATAGGGCATGGCAGACCTC-3′ and GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′. The relative expression level of HSD11B1-AS1 was calculated using the 2−ΔΔCq formula (21).
Cell Counting Kit (CCK)-8 assay
Cell proliferation was assessed by CCK-8 assay (BBI Life Sciences Corporation) according to the manufacturer's protocol. At 0, 24, 48, 72 and 96 h after transfection, A375 and SK-MEL-1 cells (2×104/ml) were seeded in 96-well plates overnight and 10 µl CCK-8 reagent was added to each well before cells were incubated for 1 h. The absorbance value at 450 nm was detected using a microplate reader (EL800; BioTek Instruments, Inc.).
Colony formation assay
At 6 h after transfection, A375 and SK-MEL-1 melanoma cell lines were seeded into a 6 cm plastic culture dish (1,000 cells/plate). Following 14 days incubation at 37°C with 5% CO2, the colonies were fixed with 4% paraformaldehyde at room temperature for 10 min and stained with 0.1% crystal violet at room temperature for 5 min. Then, the number of colonies (>50 cells) was counted manually under an Olympus IX71 inverted microscope (magnification, ×40; Olympus Corporation).
Wound healing assay
A375 and SK-MEL-1 cell migration was assessed using scratch wound healing assay. In brief, when the monolayer reached ~70% confluence, a 1-ml sterile pipetting tip was used to vertically scratch a 6-well plate with a cell monolayer adhered to the wall. Then, the plates were washed three times with PBS to remove the detached serum-starved cells. Images were captured under an Olympus IX71 inverted microscope (magnification, ×100; Olympus Corporation) at 0 and 48 h, and the wound closure ratio was calculated.
Transwell invasion assay
Transwell assay was performed using Transwell chambers (Costar; Corning, Inc.; pore size, 8 µm) coated with Matrigel (Sigma-Aldrich; Merck KGaA) at 37°C for 4 h. For the assay, 100 µl A375 or SK-MEL-1 cell suspensions (5×105/ml) in serum-free medium was seeded into the upper chamber and 600 µl DMEM containing 10% fetal bovine serum (HyClone; Thermo Fisher Scientific, Inc.) was added to the bottom chamber at 37°C. After 24 h cell culture at 37°C with 5% CO2, the migrated cells were fixed with 4% paraformaldehyde at room temperature for 30 min and stained with 1% crystal violet solution at room temperature for 30 min. Cells in 10 random fields of view were counted and images were captured under an Olympus IX71 inverted microscope (magnification, ×100; Olympus Corporation).
Gene Set Enrichment Analysis (GSEA)
GSEA was performed using a gene expression matrix of melanoma extracted from RNA-Seq TCGA datasets to identify the distinct hallmarks of HSD11B1-AS1 (22). GSEA was performed using R package ClusterProfiler (version 3.14.3) (23), and each analysis included 1,000 permutations of the gene set. The HSD11B1-AS1 expression level was used as a phenotype label. H.all.v7.0.symbols.gmt (Hallmarks) (https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp#C2) was used as a gene set.
Immune infiltration analysis by single-sample gene set enrichment analysis
Immune infiltration analysis was performed by single-sample GSEA method using the R package Gene Set Variation Analysis (GSVA) (24) to examine the association between HSD11B1-AS1 expression and infiltration of immune cells reported by Bindea et al (25). Pearson's correlation coefficient was used to assess the association between HSD11B1-AS1 expression and the relative enrichment score of 24 types of immune cell. Wilcoxon rank sum test was performed to analyze the differential immune infiltration between high and low HSD11B1-AS1 expression groups.
Statistical analysis
All data were presented as the mean ± standard deviation and all tests were performed in triplicate. The comparison of the mean values between two factors was performed using an independent sample t-test when variances were homogeneous. When variances were not homogeneous, rank sum statistical analysis was performed. The comparison of the mean values of multiple factors was performed using one-way ANOVA followed by a Bonferroni post hoc test. All bioinformatics analysis was performed in R (v.3.6.2). Kaplan-Meier method using the R package Survminer and univariate and multivariate Cox regression analyses were used to assess the prognostic value of HSD11B1-AS1. All prognostic data, including overall survival, progression-free interval and disease-specific survival, were collected from an article published in Cell (26). Median age, years (inter-quartile range) was analyzed using Wilcoxon rank sum test, and Melanoma Clark levels were analyzed using a Fisher's exact test. Pearson's correlation coefficient was used to assess the association between HSD11B1-AS1 expression and the relative enrichment score of immune cells. Nomogram models were constructed using the R package rms based on independent prognostic factors identified in the multivariate Cox regression analysis. The accuracy of the nomograms was determined using the concordance index (C-index) and calibration curve. GraphPad Prism (version 7.0; GraphPad Software, Inc.) was used to assess the role of HSD11B1-AS1 in cell proliferation, invasion and migration. P<0.05 was considered to indicate a statistically significant difference. All hypothesis testing was two tailed.
Results
HSD11B1-AS1 is downregulated in melanoma and other types of malignancy
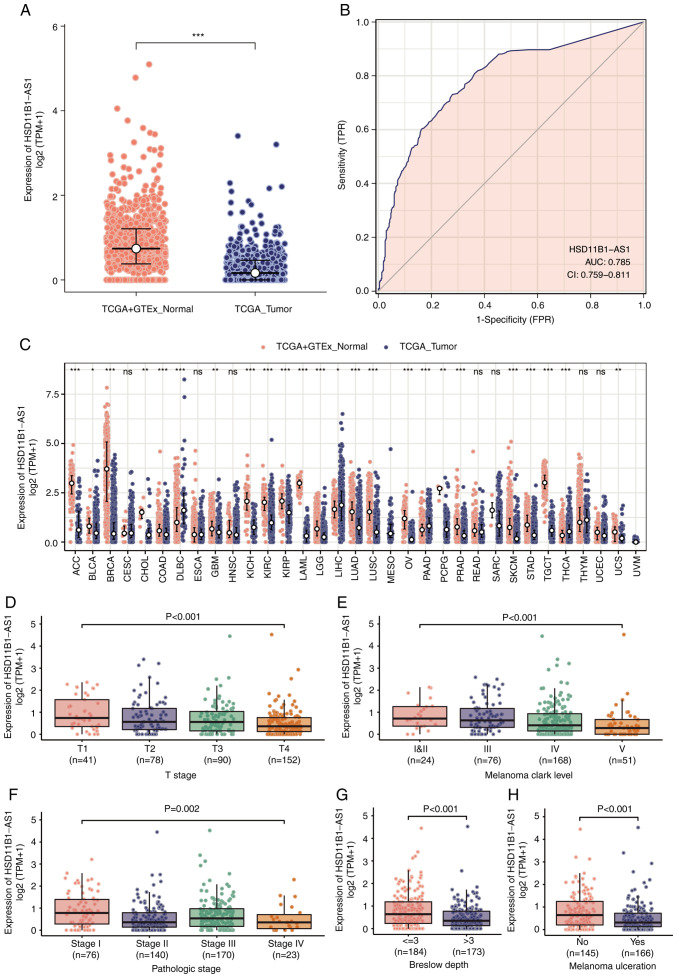
Significantly lower expression levels of HSD11B1-AS1 were observed in cutaneous melanoma than in healthy tissue (Fig. 2A). Furthermore, ROC curve analysis revealed that HSD11B1-AS1 had the potential to discriminate melanoma from healthy tissue, with an area under the curve of 0.785 (95% CI, 0.759-0.811; Fig. 2B). A pan-cancer analysis demonstrated that HSD11B1-AS1 expression was significantly lower in 19 other types of cancer, including breast, brain, and lung cancer and other types of tumor compared with melanoma (Fig. 2C).
Figure 2.
Differential expression of HSD11B1-AS1 and its association with patient clinicopathological characteristics. (A) Significantly decreased HSD11B1-AS1 levels were demonstrated in cutaneous melanoma compared with healthy tissues. (B) Receiver operating characteristic curve of HSD11B1-AS1 predicting a good diagnostic performance in discriminating melanoma from healthy tissues. (C) HSD11B1-AS1 expression levels were assessed in various types of human cancer. Decreased expression of HSD11B1-AS1 was significantly associated with advanced (D) T stage, (E) Clark levels, (F) pathological stage, (G) Breslow depth and (H) melanoma ulceration. *P<0.05, **P<0.01 and ***P<0.001. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; AUC, area under the curve; CI, confidence interval; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis; TPM, transcripts per million; TPR, true positive rate; FPR, false positive rate.
Decreased HSD11B1-AS1 expression is associated with poor clinicopathological variables
Association analysis of clinicopathological characteristics in low- and high-HSD11B1-AS1 expression groups demonstrated that HSD11B1-AS1 expression was significantly associated with age, Breslow depth, melanoma ulceration, T and pathological stage and Clark level. No significant differences between groups were observed for other clinicopathological characteristics (Table I).
Table I.
Association between HSD11B1-AS1 expression and clinicopathological characteristics in patients with cutaneous melanoma.
| HSD11B1-AS1 expression | |||
|---|---|---|---|
|
|
|||
| Characteristic | Low (n=234) | High (n=234) | P-value |
| Sex | 0.849 | ||
| Female | 88.00 (37.60) (%) | 91.00 (38.90) (%) | |
| Male | 146.00 (62.40) (%) | 143.00 (61.10) (%) | |
| Median age, years (IQR) | 60.00 (48.25-72.00) | 58.00 (47.00-68.00) | 0.036a |
| Breslow depth | 0.005a | ||
| ≤3 mm | 77.00 (43.80) (%) | 107.00 (59.10) (%) | |
| >3 mm | 99.00 (56.20) (%) | 74.00 (40.90) (%) | |
| Melanoma ulceration | 0.003a | ||
| No | 62.00 (38.30) (%) | 83.00 (55.70) (%) | |
| Yes | 100.00 (61.70) (%) | 66.00 (44.30) (%) | |
| T stage | 0.015a | ||
| T1 | 13.00 (7.30) (%) | 28.00 (15.30) (%) | |
| T2 | 37.00 (20.80) (%) | 41.00 (22.40) (%) | |
| T3 | 40.00 (22.50) (%) | 50.00 (27.30) (%) | |
| T4 | 88.00 (49.40) (%) | 64.00 (35.00) (%) | |
| N stage | 0.717 | ||
| N0 | 120.00 (59.10) (%) | 114.00 (54.80) (%) | |
| N1 | 36.00 (17.70) (%) | 38.00 (18.30) (%) | |
| N2 | 24.00 (11.80) (%) | 25.00 (12.00) (%) | |
| N3 | 23.00 (11.30) (%) | 31.00 (14.90) (%) | |
| M stage | 0.305 | ||
| M0 | 206.00 (93.20) (%) | 210.00 (95.90) (%) | |
| M1 | 15.00 (6.80) (%) | 9.00 (4.10) (%) | |
| Pathological stage | 0.003a | ||
| I | 27.00 (13.50) (%) | 49.00 (23.40) (%) | |
| II | 83.00 (41.50) (%) | 57.00 (27.30) (%) | |
| III | 76.00 (38.00) (%) | 94.00 (45.00) (%) | |
| IV | 14.00 (7.00) (%) | 9.00 (4.30) (%) | |
| Melanoma Clark level | 0.002a | ||
| I | 3.00 (1.90) (%) | 3.00 (1.90) (%) | |
| II | 4.00 (2.50) (%) | 14.00 (8.70) (%) | |
| III | 29.00 (18.40) (%) | 47.00 (29.20) (%) | |
| IV | 88.00 (55.70) (%) | 80.00 (49.70) (%) | |
| V | 34.00 (21.50) (%) | 17.00 (10.60) (%) | |
| Radiation therapy | 0.213 | ||
| No | 194.00 (85.10) (%) | 187.00 (80.30) (%) | |
| Yes | 34.00 (14.90) (%) | 46.00 (19.70) (%) | |
P<0.05. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; IQR, interquartile range.
Decreased HSD11B1-AS1 expression was significantly associated with poorer pathological staging, including T stage, Clark level, presence of melanoma ulceration and Breslow depth >3 mm (Fig. 2D-F). Moreover, univariate logistic regression indicated that lower HSD11B1-AS1 expression was significantly associated with negative features, including T stage [odds ratio (OR)=0.44 for T2, T3 and T4 vs. T1), pathological stage (OR=0.51 for stages IV, III and II vs. stage I), Clark level (OR=0.45 for IV and V vs. I, II and III), Breslow depth (OR=0.54 for >3 vs. ≤3) and melanoma ulceration (OR=0.49 for Yes vs. No; Table II). These results were consistent with those shown in Fig. 2D-F, indicating that tumors with lower HSD11B1-AS1 expression were more prone to progress to more advanced and invasive stages.
Table II.
HSD11B1-AS1 expression is associated with clinicopathological features (logistic regression).
| Characteristic | Total number of cases | OR | P-value |
|---|---|---|---|
| Sex, female vs. male | 468 | 1.06 (0.73-1.53) | 0.775 |
| Age, ≤60 vs. >60 years | 460 | 1.29 (0.90-1.87) | 0.170 |
| T stage, T2, 3 and 4 vs. 1 | 361 | 0.44 (0.21-0.86) | 0.019a |
| N stage, N1, 2 and 3 vs. 0 | 411 | 1.19 (0.81-1.76) | 0.378 |
| M stage, M1 vs. 0 | 440 | 0.59 (0.24-1.35) | 0.221 |
| Pathological stage, IV, III and II vs. I | 409 | 0.51 (0.30-0.85) | 0.011a |
| Melanoma Clark level, IV and V vs. I, II and III | 319 | 0.45 (0.27-0.72) | 0.001a |
| Breslow depth, >3 vs. ≤3 mm | 357 | 0.54 (0.35-0.82) | 0.004a |
| Melanoma ulceration, yes vs. no | 311 | 0.49 (0.31-0.77) | 0.002a |
| Radiation therapy, no vs. yes | 461 | 0.71 (0.44-1.16) | 0.172 |
P<0.05. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; OR, odds ratio.
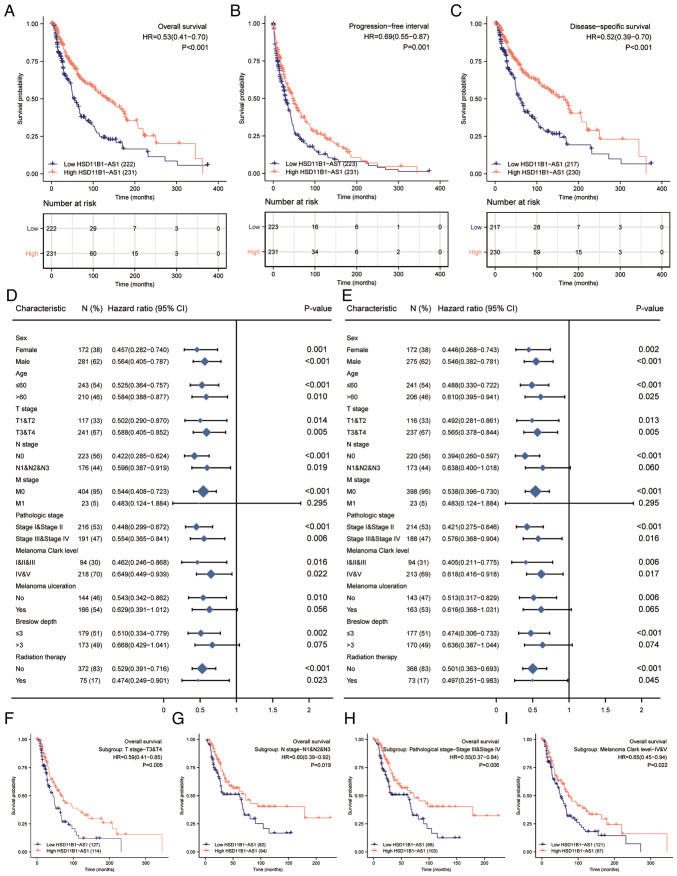
Increased HSD11B1-AS1 expression predicts favorable prognosis
Kaplan-Meier survival analysis indicated that increased HSD11B1-AS1 expression was associated with better overall survival [OS; hazard ratio (HR)=0.53; Fig. 3A], progression-free interval (PFI; HR=0.69; Fig. 3B) and disease-specific survival (DSS; HR=0.52; Fig. 3C). Increased HSD11B1-AS1 expression affected patient prognosis (OS and DSS) in different clinicopathological subsets, including sex, age, T and pathological stage and Clark level (Fig. 3D and E). Specifically, overexpression of HSD11B1-AS1 was significantly associated with favorable OS in advanced clinical stage subsets, including T3/4 [HR=0.59 (0.41-0.85)], N1, 2 and 3 [HR=0.60 (0.39-0.92)] and pathological stages III and IV [HR=0.55 (0.37-0.84)] and Clark level IV and V [HR=0.65 (0.45-0.94); Fig. 3F-I]. These findings indicated that HSD11B1-AS1 affected the prognosis of patients with melanoma with advanced clinicopathological features.
Figure 3.
Association of HSD11B1-AS1 expression and the clinical prognosis of patients with cutaneous melanoma. Elevated HSD11B1-AS1 expression was correlated with (A) an improved OS, (B) progression free interval and (C) DSS in patients with cutaneous melanoma. Overexpression of HSD11B1-AS1 was significantly associated with improved (D) OS and (E) DSS in various clinicopathological subsets. High HSD11B1-AS1 expression was associated with better OS in (F) T stage 3 and 4, (G) N1, N2 and N3 stages, (H) pathological stage III and IV, and (I) Clark levels IV and V. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; OS, overall survival; DSS, disease specific survival.
Development of prognostic models based on HSD11B1-AS1 expression and other clinicopathological characteristics
The univariate Cox regression analysis demonstrated that elevated HSD11B1-AS1 expression was significantly associated with favorable OS; other clinicopathological factors, such as age, T, N and pathological stage, Clark level, Breslow depth and melanoma ulceration were also significantly associated with OS (Table III). Furthermore, multivariate Cox regression analysis revealed that, similar to N stage and Breslow depth, HSD11B1-AS1 expression was independently associated with OS. Moreover, both the univariate and multivariate analyses showed that HSD11B1-AS1 expression was associated with PFI (Table IV) and DSS (Table V) as an independent prognostic factor. The contributions of other clinicopathological features to PFI and DSS are shown in Tables IV and V, respectively.
Table III.
Association between overall survival and clinicopathological features using univariate and multivariate Cox regression.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Characteristic | Total | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex, male vs. female | 453 | 1.164 (0.872-1.554) | 0.301 | - | - |
| Age, >60 vs. ≤60 years | 453 | 1.678 (1.266-2.225) | <0.001a | 1.166 (0.781-1.741) | 0.452 |
| T stage, T3 and 4 vs. 1 and 2 | 358 | 2.040 (1.468-2.836) | <0.001a | 0.909 (0.517-1.600) | 0.742 |
| N stage, N1, 2 and 3 vs. 0 | 399 | 1.711 (1.271-2.304) | <0.001a | 3.764 (1.140-12.425) | 0.030a |
| M stage, M1 vs. 0 | 427 | 1.734 (0.915-3.287) | 0.092 | - | - |
| Pathological stage, III and IV vs. I and II | 407 | 1.579 (1.177-2.118) | 0.002a | 0.598 (0.182-1.968) | 0.398 |
| Melanoma Clark level, IV and V vs. I, II and III | 312 | 2.117 (1.472-3.045) | <0.001a | 1.291 (0.797-2.090) | 0.299 |
| Melanoma ulceration, yes vs. no | 310 | 2.087 (1.494-2.916) | <0.001a | 1.315 (0.865-1.999) | 0.201 |
| Breslow depth, ≤3 vs. >3 mm | 352 | 0.386 (0.281-0.528) | <0.001a | 0.537 (0.314-0.916) | 0.023a |
| Radiation therapy, yes vs. no | 447 | 0.953 (0.674-1.348) | 0.785 | - | - |
| HSD11B1-AS1 expression, high vs. low | 453 | 0.534 (0.407-0.700) | <0.001a | 0.618 (0.423-0.903) | 0.013a |
P<0.05. HR, hazard ratio; HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1.
Table IV.
Association between with progression-free interval and clinicopathological features using univariate and multivariate Cox regression.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Characteristic | Total | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex, male vs. female | 454 | 1.027 (0.813-1.298) | 0.821 | - | - |
| Age, >60 vs. ≤60 years | 454 | 1.600 (1.258-2.035) | <0.001a | 1.398 (0.988-1.978) | 0.059 |
| T stage, T3 and 4 vs. 1 and 2 | 359 | 1.655 (1.259-2.175) | <0.001a | 0.903 (0.569-1.433) | 0.665 |
| N stage, N1, 2 and 3 vs. 0 | 400 | 1.853 (1.451-2.365) | <0.001a | 2.611 (0.991-6.880) | 0.052 |
| M stage, M1 vs. 0 | 428 | 1.942 (1.188-3.175) | 0.008a | 1.176 (0.481-2.877) | 0.722 |
| Pathological stage, III and IV vs. I and II | 408 | 1.717 (1.349-2.187) | <0.001a | 0.868 (0.322-2.343) | 0.781 |
| Melanoma Clark level, IV and V vs. I, II and III | 312 | 1.762 (1.302-2.386) | <0.001a | 0.956 (0.628-1.455) | 0.832 |
| Melanoma ulceration, yes vs. no | 310 | 1.635 (1.233-2.169) | <0.001a | 1.182 (0.831-1.681) | 0.351 |
| Breslow depth, ≤3 vs. >3 mm | 352 | 0.498 (0.378-0.655) | <0.001a | 0.566 (0.359-0.892) | 0.014a |
| Radiation therapy, yes vs. no | 448 | 1.201 (0.906-1.593) | 0.203 | - | - |
| HSD11B1-AS1 expression, high vs. low | 454 | 0.691 (0.552-0.866) | 0.001a | 0.704 (0.507-0.978) | 0.036a |
P<0.05. HR, hazard ratio; HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1.
Table V.
Association between disease-specific survival and clinicopathological features using univariate and multivariate Cox regression.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Characteristic | Total | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex, male vs. female | 447 | 1.151 (0.847-1.564) | 0.368 | ||
| Age, >60 vs. ≤60 years | 447 | 1.728 (1.278-2.337) | <0.001a | 1.096 (0.715-1.681) | 0.674 |
| T stage, T3 and 4 vs. 1 and 2 | 353 | 1.842 (1.308-2.594) | <0.001a | 0.913 (0.513-1.625) | 0.757 |
| N stage, N1, 2 and 3 vs. 0 | 393 | 1.620 (1.179-2.227) | 0.003a | 5.961 (1.369-25.957) | 0.017a |
| M stage, M1 vs. 0 | 421 | 2.013 (1.059-3.828) | 0.033a | 2.244 (0.755-6.665) | 0.146 |
| Pathological stage, III and IV vs. I and II | 402 | 1.495 (1.093-2.045) | 0.012a | 0.345 (0.078-1.536) | 0.163 |
| Melanoma Clark level, IV and V vs. I, II and III | 307 | 2.075 (1.419-3.035) | <0.001a | 1.382 (0.829-2.304) | 0.215 |
| Melanoma ulceration, yes vs. no | 306 | 1.949 (1.369-2.775) | <0.001a | 1.349 (0.871-2.089) | 0.179 |
| Breslow depth, ≤3 vs. >3 mm | 347 | 0.452 (0.323-0.633) | <0.001a | 0.571 (0.331-0.985) | 0.044a |
| Radiation therapy, yes vs. no | 441 | 0.966 (0.667-1.400) | 0.856 | ||
| HSD11B1-AS1 expression, high vs. low | 447 | 0.520 (0.389-0.695) | <0.001a | 0.623 (0.418-0.928) | 0.020a |
P<0.05. HR, hazard ratio; HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1.
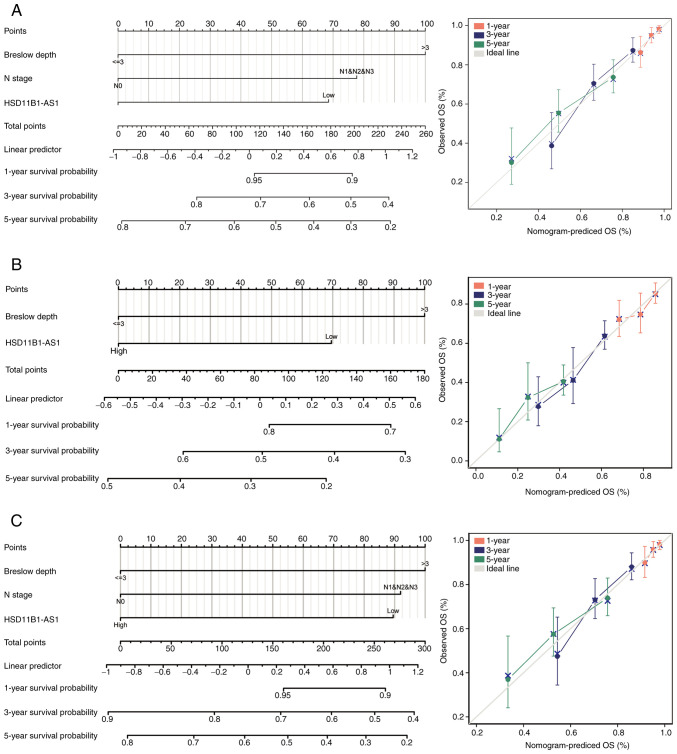
Nomograms for 1-, 3-, and 5-year OS, PFI and DSS were constructed based on the independent variables obtained from the multivariable analysis (Fig. 4A-C). The accuracy of the nomograms was validated using the C-index and calibration plots. The C-index values were 0.692 (95% CI, 0.671-0.713) for OS, 0.614 (95% CI, 0.595-0.634) for PFI and 0.675 (95% CI, 0.652-0.699) for DSS, indicating that the constructed nomograms had good predictive value. Calibration plots for the probability of 1-, 3- and 5-year OS, PFI, and DSS indicated notable agreement between the nomogram-predicted and observed survival.
Figure 4.
Prognostic nomogram and calibration plot to predict the 1-, 3- and 5-year OS, PFI and DSS probabilities in patients with cutaneous melanoma. Nomograms for predicting the probability of 1-, 3-, and 5-year (A) OS, (B) PFI and (C) DSS. Calibration curves of the prognostic nomograms for predicting OS, PFI and DSS at 1, 3, and 5 years demonstrated no distinct departure from the ideal lines. OS, overall survival; PFI, progression free interval; DSS, disease specific survival.
Expression levels of HSD11B1-AS1 in melanoma cell lines and determination of transfection efficiency
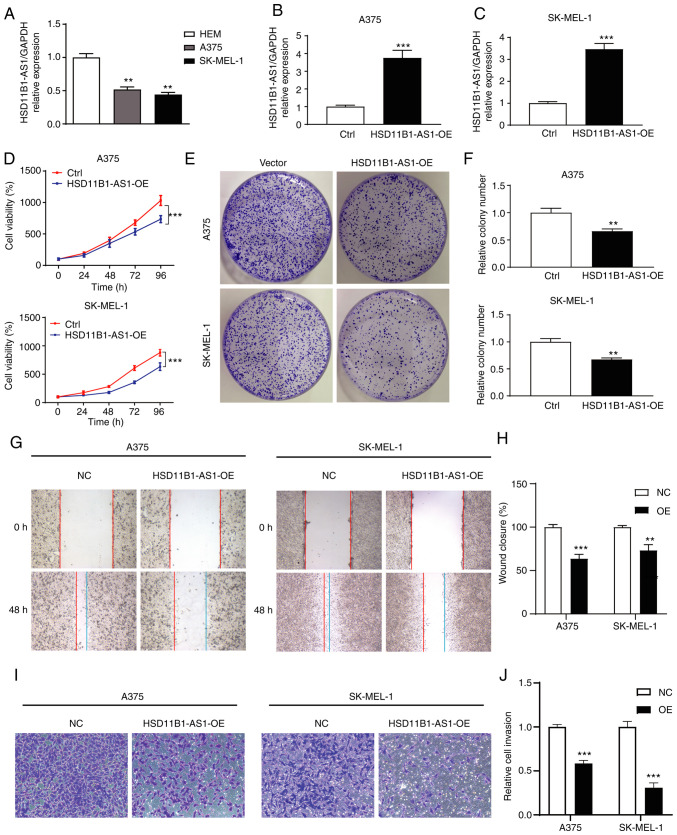
In vitro assay showed that the expression levels of HSD11B1-AS1 in melanoma cell lines were significantly lower than those in normal HEMs (Fig. 5A). Gain-of-function assays were performed by overexpressing HSD11B1-AS1 in A375 and SK-MEL-1 cell lines and transfection efficiency was determined using RT-qPCR. Transfection of pcDNA3.1-HSD11B1-AS1 significantly increased HSD11B1-AS1 expression levels compared with transfection of pcDNA3.1 empty vector (Fig. 5B and C).
Figure 5.
HSD11B1-AS1 inhibits the proliferation, migration and invasion of cutaneous melanoma cells in vitro. (A) Differential expression levels of HSD11B1-AS1 in normal human epidermal melanocytes and A375 and SK-MEL-1 cell lines. The transfection efficiency of HSD11B1-AS1 in (B) A375 and (C) SK-MEL-1 cell lines was verified by reverse transcription-quantitative PCR. (D) Overexpression of HSD11B1-AS1 significantly suppressed the viability of A375 and SK-MEL-1 cells. (E and F) Overexpression of HSD11B1-AS1 significantly decreased the number of colonies. (G and H) Effect of HSD11B1-AS1 on cell migration, as measured by a wound healing assay. (I and J) The effect of HSD11B1-AS1 on cell invasion was measured by performing a Transwell assay. **P<0.01 and ***P<0.001. HSD11B1-AS1, 11β-hydroxysteroid dehydrogenase type 1-antisense RNA 1; Ctrl, control; NC, negative control. OE, overexpression; HEM, human epidermal melanocyte.
Effect of HSD11B1-AS1 on proliferation of melanoma cells
CCK-8 assay showed that HSD11B1-AS1 overexpression significantly decreased proliferation of A375 and SK-MEL-1 cells (Fig. 5D). Colony formation assay showed that the number of A375 and SK-MEL-1 cell clones in the HSD11B1-AS1 overexpression group was significantly lower than in the control group (Fig. 5E and F). These results suggested that HSD11B1-AS1 inhibited the proliferation of melanoma cells.
Upregulation of HSD11B1-AS1 suppresses the metastatic ability of melanoma cells
Wound healing and Transwell migration assays were performed to assess the effect of HSD11B1-AS1 on the metastatic ability of A375 and SK-MEL-1 cells. Wound healing assay revealed that cells in the HSD11B1-AS1 overexpression group migrated a significantly shorter distance than cells in the control group (Fig. 5G and H). In the Transwell assay, the number of invading cells in the HSD11B1-AS1 overexpression group was significantly lower compared with cells transfected with empty vector (Fig. 5I and J).
HSD11B1-AS1-associated signaling pathways were associated with immunity and inflammation, as identified by GSEA
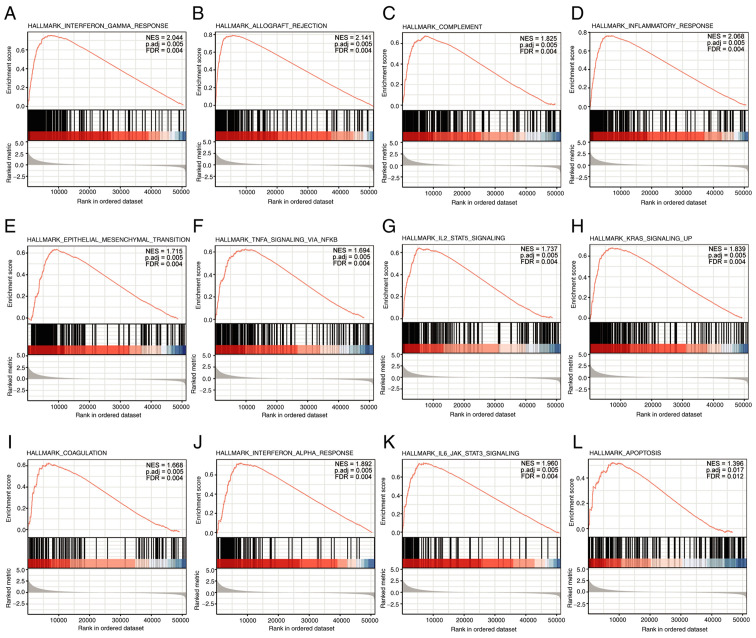
GSEA analysis revealed that 12 pathways were significantly enriched: ‘Interferon gamma response’, ‘IL-6/JAK/STAT-3 signaling’, ‘allograft rejection’, ‘inflammatory response’, ‘KRAS signaling up’, ‘complement’, ‘epithelial mesenchymal transition’, ‘TNF-α signaling via NF-κB, IL-2/STAT-5 signaling’, ‘coagulation’, ‘IFN-α response’, and ‘apoptosis’ (Fig. 6).
Figure 6.
Enrichment plots from Gene Set Enrichment Analysis. (A) Interferon gamma response, (B) allograft rejection, (C) complement, (D) inflammatory response, (E) epithelial mesenchymal transition, (F) TNF-α signaling via NF-κB, (G) IL-2/STAT-5 signaling, (H) KRAS signaling up, (I) coagulation, (J) IFN-α response, (K) IL-6/JAK/STAT-3 signaling, and (L) apoptosis were significantly enriched. The top 12 are presented and ordered by ascending adjusted P-value. Adj., adjusted. NES, normalized enrichment score; FDR, false discovery rate.
Association between HSD11B1-AS1 expression and immune infiltration
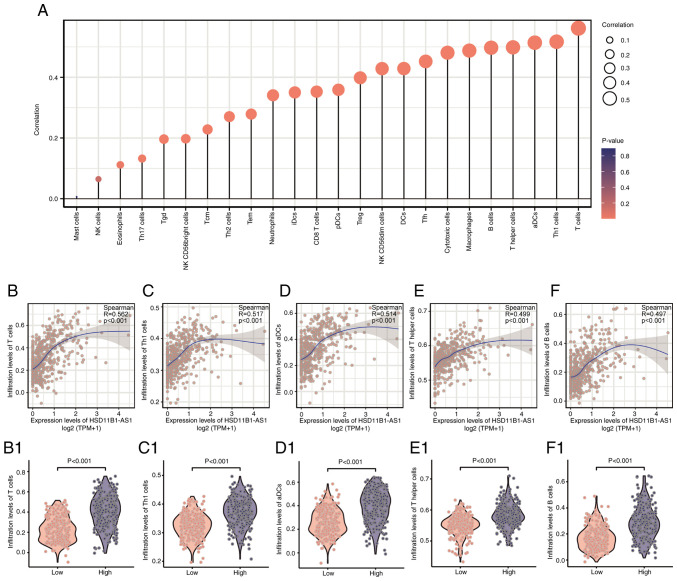
HSD11B1-AS1 expression levels exhibited a significant positive correlation with infiltrating immune cells, including T, T helper (h)1, activated dendritic cells (aDCs), Th, B, cytotoxic, T follicular h (Tfh), DCs, and natural killer (NK) CD56dim cells, as well as macrophages (R>0.40; Fig. 7A). The abundance of NK and mast cells was not significantly correlated with HSD11B1-AS1 expression levels. The association between HSD11B1-AS1 expression and the five most abundant infiltrating immune cells including T (R=0.562; Fig. 7B), Th1 (R=0.517; Fig. 7C), aDCs (R=0.514; Fig. 7D), Th (R=0.499; Fig. 7E) and B cells (R=0.497; Fig. 7F) was determined (Fig. 7B-F). T cells were most strongly associated with HSD11B1-AS1 expression (R=0.562; Fig. 7B); likewise, Wilcoxon rank sum test revealed a significantly higher enrichment score for T cells in the HSD11B1-AS1 overexpression group (Fig. 7B1). These findings illustrated that HSD11B1-AS1 was associated with immune cell infiltration in cutaneous melanoma.
Figure 7.
Association analysis of HSD11B1-AS1 expression levels and immune infiltration in the tumor environment. (A) Correlations between HSD11B1-AS1 expression levels and the relative infiltration levels of 24 types of immune cells. The colored dots indicate P-values, and the size of the dots indicates the absolute value of the Spearman correlation coefficient. Correlations between the relative enrichment score of (B) T cells, (C) Th1 cells, (D) aDCs, (E) T helper cells and (F) B cells and the expression of HSD11B1-AS1. Differential infiltration level of (B1) T cells, (C1) Th1 cells, (D1) aDCs, (E1) T helper cells and (F1) B cells between low and high HSD11B1-AS1 expression groups. Th, T-helper; aDCs, activated dendritic cells.
Discussion
Increasing evidence has suggested that lncRNAs serve as tumor suppressors or oncogenes and may be targets for developing selective anti-cancer therapeutic strategies owing to their cell type- and disease-specific expression profiles and their key role in tumor proliferation, migration and invasion (27,28). Identification of key biomarkers or therapeutic targets associated with cutaneous melanoma progression may contribute to better outcomes for patients with melanoma. Accordingly, using public data, the present study identified a novel immune-associated lncRNA, HSD11B1-AS1, which served as a tumor suppressor in cutaneous melanoma. HSD11B1-AS1, also known as lnc-LAMB3-1, was previously reported to be associated with better DFS in breast cancer (29), but its expression level, biological function and potential roles in prognosis of cutaneous melanoma are unknown. To the best of our knowledge, the present study is the first description of HSD11B1-AS1 in cutaneous melanoma.
The present study demonstrated that HSD11B1-AS1 is a valuable biomarker for diagnosis and prognosis and may serve as a therapeutic target in cutaneous melanoma. TCGA data demonstrated significantly decreased HSD11B1-AS1 expression levels in both cutaneous melanoma and other types of malignancy; this was validated in cutaneous melanoma cell lines and HEMs using RT-qPCR. The present study showed downregulated HSD11B1-AS1 was associated with poor clinicopathological characteristics, shorter survival time and worse prognosis in patients with melanoma. Moreover, elevated HSD11B1-AS1 expression was shown to affect the survival rate of patients with melanoma, including those with advanced clinicopathological variables. In vitro cell function experiments verified the role of HSD11B1-AS1 in cutaneous melanoma progression and metastasis. Overexpression of HSD11B1-AS1 was found to significantly inhibit cell proliferation, migration and invasion in melanoma cell lines A375 and SK-MEL-1. Therefore, HSD11B1-AS1 may be a promising biomarker and therapeutic target worthy of further investigation and clinical validation.
To assess the accuracy of HSD11B1-AS1 as a prognostic biomarker, nomogram and calibration plots were established based on multivariate Cox regression analysis. In the present study, a nomogram was established by combination of HSD11B1-AS1 with other independent clinical risk factors (N stage and Breslow depth). The C-index of nomogram was 0.692 (95% CI, 0.671-0.713) for OS, 0.614 (0.595-0.634) for PFI, and 0.675 (0.652-0.699) for DSS, indicating that the constructed nomograms had a good predictive value. Moreover, calibration plots for the probability of 1-, 3- and 5-year OS, PFI and DSS indicated notable agreement between the predicted and actual values. These results further demonstrated that HSD11B1-AS1 may serve as a valuable biomarker for the prognosis of patients with cutaneous melanoma.
To the best of our knowledge, the present study is the first to report that HSD11B1-AS1 mediated antitumor ability by regulating signaling pathways associated with immunity and inflammation, such as IFN-γ and -α response and IL-2/STAT-5 and IL-6/JAK/STAT-3 signaling. IFNs are pleiotropic cytokines critical for cancer immunosurveillance, quality of antitumor response and response to immunotherapy (30,31). Numerous studies have demonstrated that IFN signaling is downregulated in melanoma and decreased IFN-γ signaling limits immune cell recruitment and recognition of tumor cells by suppressing the production of IFN-γ-dependent chemokines and decreasing antigen presentation (32,33). IL-2 and IL-6 are also pleiotropic cytokines that serve key roles in inflammatory and immune response. IL-2 promotes proliferation of T cells and immunoglobulin synthesis by B cells; IL-6 exerts its effects via activation and differentiation of T and B lymphocytes (34). lncRNAs regulate innate and adaptive immunity in cancer by mediating the functional state of immunological cells, pathways and genes (35). For example, lncRNA colorectal neoplasia differentially expressed exacerbates IgA nephropathy progression by restraining the ubiquitination and degradation of NLRP3, and facilitating NLRP3 inflammasome activation in macrophages (36). Therefore, immune infiltration analysis was performed to assess tumor immune surveillance in cutaneous melanoma.
Immune infiltration analysis revealed that HSD11B1-AS1 overexpression exhibited a strong positive association with infiltration of various types of immune cell, most notably T (R=0.562), Th1 (R=0.517), aDCs (R=0.514), Th (R=0.499) and B cells (R=0.497). Studies have reported that immune cell infiltration, particularly high peritumoral levels of activated T cells, B lymphocytes, and mature DCs, is associated with favorable prognosis in melanoma (37,38). T cells are important in antitumor immune response and are the most abundant infiltrating cells in melanoma; increased infiltration of CD8+ T and Th1 cells is associated with favorable clinical outcomes in various types of cancer (37,39). B cells account for 15–20% of infiltrating lymphocytes, but growing evidence indicates that B cells are correlated with activated T cells and may promote tumor immunity by facilitating T cell infiltration into tumors, thus improving survival in melanoma (40,41). In addition to adaptive immune cells, DCs, as innate immune cells, serve an important role in bridging the innate and adaptive immune responses and exert their functions via antigen presentation to T cells. Similar to B cell infiltration, increased DC infiltration is associated with activated T cells and better prognosis (42). These results indicate a potential mechanism by which elevated HSD11B1-AS1 expression increases survival, which is consistent with results of the present study. Together, these findings suggest that overexpression of HSD11B1-AS1 may improve the prognosis of patients with melanoma by promoting immune response and cell infiltration.
Although the findings of the present study provide understanding about the functions of HSD11B1-AS1 in cutaneous melanoma, certain limitations should be noted. First of all, the present study focused on bioinformatics analysis; further studies should validate the molecular mechanism of HSD11B1-AS1 both in vitro and in vivo. Second, HSD11B1-AS1 was shown to have the potential to be a diagnostic and prognostic biomarker for cutaneous melanoma using expression profiles and clinical information obtained from TCGA, but large-sample, multicenter, randomized controlled trials need to be performed to determine whether HSD11B1-AS1 is a suitable molecular marker in practice.
To the best of our knowledge, the present study was the first to demonstrate that HSD11B1-AS1, which was significantly decreased in cutaneous melanoma compared with normal cells, may be a valuable diagnostic and prognostic biomarker and therapeutic target in cutaneous melanoma. Overexpression of HSD11B1-AS1 significantly suppressed melanoma cell proliferation, migration and invasion in melanoma cell lines A375 and SK-MEL-1. Elevated HSD11B1-AS1 in melanoma was correlated with favorable clinicopathological features and better prognosis, which may be caused by increased immune response and infiltration of immunocytes, particularly T and B cells and aDCs.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KL and LZ conceived and designed the present study. JZ and XL performed the literature review and analyzed the data. KL, LZ and XL drafted the initial manuscript and confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.McCarthy M. US melanoma prevalence has doubled over past 30 years. BMJ. 2015;350:h3074. doi: 10.1136/bmj.h3074. [DOI] [PubMed] [Google Scholar]
- 2.Rebecca VW, Somasundaram R, Herlyn M. Pre-clinical modeling of cutaneous melanoma. Nat Commun. 2020;11:2858. doi: 10.1038/s41467-020-15546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long GV, Grob JJ, Nathan P, Ribas A, Robert C, Schadendorf D, Lane SR, Mak C, Legenne P, Flaherty KT, Davies MA. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: A pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17:1743–1754. doi: 10.1016/S1470-2045(16)30578-2. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 5.Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Bastholt L, Grob JJ, Malvehy J, Newton-Bishop J, Stratigos AJ, et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline-Update 2016. Eur J Cancer. 2016;63:201–217. doi: 10.1016/j.ejca.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Luther C, Swami U, Zhang J, Milhem M, Zakharia Y. Advanced stage melanoma therapies: Detailing the present and exploring the future. Crit Rev Oncol Hematol. 2019;133:99–111. doi: 10.1016/j.critrevonc.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, Zhang H, Liu Y, Han D, Zhang N, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer. 2020;19:85. doi: 10.1186/s12943-020-01206-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Yu X, Zheng H, Tse G, Zhang L, Wu WKK. CASC2: An emerging tumour-suppressing long noncoding RNA in human cancers and melanoma. Cell Prolif. 2018;51:e12506. doi: 10.1111/cpr.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Yan Z, Hu F, Wei W, Yang C, Sun Z. Long non-coding RNA GAS5 accelerates oxidative stress in melanoma cells by rescuing EZH2-mediated CDKN1C downregulation. Cancer Cell Int. 2020;20:116. doi: 10.1186/s12935-020-01167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Zheng H, Tse G, Chan MT, Wu WK. Long non-coding RNAs in melanoma. Cell Prolif. 2018;51:e12457. doi: 10.1111/cpr.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richtig G, Ehall B, Richtig E, Aigelsreiter A, Gutschner T, Pichler M. Function and clinical implications of long Non-Coding RNAs in melanoma. Int J Mol Sci. 2017;18:715. doi: 10.3390/ijms18040715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riefolo M, Porcellini E, Dika E, Broseghini E, Ferracin M. Interplay between small and long non-coding RNAs in cutaneous melanoma: A complex jigsaw puzzle with missing pieces. Mol Oncol. 2019;13:74–98. doi: 10.1002/1878-0261.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 15.Wu K, Wang Q, Liu YL, Xiang Z, Wang QQ, Yin L, Liu SL. LncRNA POU3F3 contributes to dacarbazine resistance of human melanoma through the MiR-650/MGMT Axis. Front Oncol. 2021;11:643613. doi: 10.3389/fonc.2021.643613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attrill GH, Ferguson PM, Palendira U, Long GV, Wilmott JS, Scolyer RA. The tumour immune landscape and its implications in cutaneous melanoma. Pigment Cell Melanoma Res. 2021;34:529–549. doi: 10.1111/pcmr.12926. [DOI] [PubMed] [Google Scholar]
- 17.Wu M, Fu P, Qu L, Liu J, Lin A. Long noncoding RNAs, new critical regulators in cancer immunity. Front Oncol. 2020;10:550987. doi: 10.3389/fonc.2020.550987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Zhang L, Mi Y, Tang Y, Ren F, Liu B, Zhang Y, Zheng P. A ceRNA network and a potential regulatory axis in gastric cancer with different degrees of immune cell infiltration. Cancer Sci. 2020;111:4041–4050. doi: 10.1111/cas.14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Kryczek I, Nam J, Li X, Li S, Li J, Wei S, Grove S, Vatan L, Zhou J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23:526–537. doi: 10.1038/s41556-021-00672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appelbaum EN, Gross ND, Diab A, Bishop AJ, Nader ME, Gidley PW. Melanoma of the external auditory canal: A review of seven cases at a tertiary care referral center. Laryngoscope. 2021;131:165–172. doi: 10.1002/lary.28548. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leucci E, Coe EA, Marine JC, Vance KW. The emerging role of long non-coding RNAs in cutaneous melanoma. Pigment Cell Melanoma Res. 2016;29:619–626. doi: 10.1111/pcmr.12537. [DOI] [PubMed] [Google Scholar]
- 28.Ma W, Qiao J, Zhou J, Gu L, Deng D. Characterization of novel LncRNA P14AS as a protector of ANRIL through AUF1 binding in human cells. Mol Cancer. 2020;19:42. doi: 10.1186/s12943-020-01150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vishnubalaji R, Shaath H, Elkord E, Alajez NM. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFbeta and focal adhesion kinase (FAK) signaling. Cell Death Discov. 2019;5:109. doi: 10.1038/s41420-019-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 31.von Locquenghien M, Rozalen C, Celia-Terrassa T. Interferons in cancer immunoediting: Sculpting metastasis and immunotherapy response. J Clin Invest. 2021;131:e143296. doi: 10.1172/JCI143296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alavi S, Stewart AJ, Kefford RF, Lim SY, Shklovskaya E, Rizos H. Interferon signaling is frequently downregulated in melanoma. Front Immunol. 2018;9:1414. doi: 10.3389/fimmu.2018.01414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 35.Yu Y, Zhang W, Li A, Chen Y, Ou Q, He Z, Zhang Y, Liu R, Yao H, Song E. Association of long noncoding RNA biomarkers with clinical immune subtype and prediction of immunotherapy response in patients with cancer. JAMA Netw Open. 2020;3:e202149. doi: 10.1001/jamanetworkopen.2020.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen M, Pan X, Gao Y, Ye H, Zhang J, Chen Y, Pan M, Huang W, Xu X, Zhan Y, Jin L. LncRNA CRNDE exacerbates IgA nephropathy progression by promoting NLRP3 inflammasome activation in macrophages. Immunol Invest. 2021;7:1–13. doi: 10.1080/08820139.2021.1989461. [DOI] [PubMed] [Google Scholar]
- 37.Ladányi A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigment Cell Melanoma Res. 2015;28:490–500. doi: 10.1111/pcmr.12371. [DOI] [PubMed] [Google Scholar]
- 38.Ladányi A, Kiss J, Somlai B, Gilde K, Fejos Z, Mohos A, Gaudi I, Tímár J. Density of DC-LAMP(+) mature dendritic cells in combination with activated T lymphocytes infiltrating primary cutaneous melanoma is a strong independent prognostic factor. Cancer Immunol Immunother. 2007;56:1459–1469. doi: 10.1007/s00262-007-0286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi T, Hamaguchi Y, Hasegawa M, Fujimoto M, Takehara K, Matsushita T. B cells promote tumor immunity against B16F10 melanoma. Am J Pathol. 2014;184:3120–3129. doi: 10.1016/j.ajpath.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Garg K, Maurer M, Griss J, Brüggen MC, Wolf IH, Wagner C, Willi N, Mertz KD, Wagner SN. Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Hum Pathol. 2016;54:157–164. doi: 10.1016/j.humpath.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 42.Monti M, Consoli F, Vescovi R, Bugatti M, Vermi W. Human plasmacytoid dendritic cells and cutaneous melanoma. Cells. 2020;9:417. doi: 10.3390/cells9020417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.