Abstract
The c-myc proto-oncogene encodes a transcription factor that participates in the regulation of cellular proliferation, differentiation, and apoptosis. Ectopic overexpression of c-Myc has been shown to sensitize cells to apoptosis. We report here that cells lacking c-Myc activity due to disruption of the c-myc gene by targeted homologous recombination are defective in DNA damage-initiated apoptosis in the G2 phase of the cell cycle. The downstream effector of c-Myc is cyclin A, whose ectopic expression in c-myc−/− cells rescues the apoptosis defect. The kinetics of the G2 response indicate that the induction of cyclin A and the concomitant activation of Cdk2 represent an early step during commitment to apoptosis. In contrast, expression of cyclins E and D1 does not rescue the apoptosis defect, and apoptotic processes in G1 phase are not affected in c-myc−/− cells. These observations link DNA damage-induced apoptosis with cell cycle progression and implicate c-Myc in the functioning of a subset of these pathways.
Mutations affecting the c-myc proto-oncogene are among the most common genetic lesions found in a variety of human and animal cancers (19, 39). The c-Myc protein is a transcription factor that functions as an obligate heterodimer with its partner Max and binds the core recognition sequence CACGTG (E-box) and a number of related sequences (28). Genes whose expression is regulated by c-Myc have been intensively sought (8), but our understanding of downstream signal transduction pathways remains fragmentary (7, 47). It is now well established that c-Myc can repress, as well as activate transcription, but the relative contribution of these mechanisms to the variety of physiological responses mediated by c-Myc is not yet clear (6, 13).
The role of c-Myc as a positive effector of the cell cycle has been extensively documented (40). Under appropriate circumstances, both repression and overexpression of c-Myc can lead to apoptosis. For example, in a variety of transformed cell types c-myc antisense oligonucleotides cause growth inhibition, which in some (but not all) cases is associated with the onset of apoptosis (51). On the other hand, there are many examples where c-Myc is required, to a greater or lesser degree, for the efficient induction of apoptosis by a variety of stimuli (42). Overexpression of c-Myc augments the apoptotic program and rapidly induces cell death when cells are deprived of survival factors (3, 12).
The tumor suppressor gene p53 has been implicated as a target of c-Myc regulation (44, 45). c-Myc-induced apoptosis requires p53 in some (20, 53) but not all (46, 52) cases. Likewise, Bcl-2 exerts a sparing effect on some (54, 55) but not all (52) c-Myc-induced apoptotic responses. To explain such discrepancies, it has been proposed that c-Myc acts to sensitize the cell to a variety of apoptotic stimuli, both p53 dependent and p53 independent, that can be counteracted by survival signals (11). Considerable evidence supports a dual function for c-Myc as a coordinate activator of both proliferation and apoptosis. According to this model, both functions would be intrinsic to c-Myc and may involve distinct apoptosis priming and triggering pathways, at least some of which may be mechanistically distinct from the promotion of proliferation (42). Indeed, recent work is beginning to uncover c-Myc targets or effectors, such as p19ARF1 (57) and Bin1 (42), which appear to function in apoptosis but do not affect proliferation.
The majority of studies on c-Myc have employed overexpression paradigms. In some cases antisense or dominant-defective approaches have been used, but their interpretation is complicated by the incomplete inhibition of c-Myc expression, as well as uncertainties pertaining to the mechanisms of dominant-defective action. We have isolated c-myc null cell lines (31) and have initiated an investigation of their proliferative phenotypes (32). In this study we use the c-myc−/− cell lines for the first time to investigate the function of c-Myc in apoptotic pathways initiated by DNA-damaging agents. This is a topic of considerable interest because of its applications to the chemotherapy of cancer but, in contrast to studies of survival signal withdrawal, it has received very little attention (4, 9, 38).
MATERIALS AND METHODS
Cell lines and culture conditions.
TGR-1 is a subclone of the immortalized rat embryo fibroblast cell line Rat-1 (43). The c-myc null cell lines have been described (31). The c-myc, temperative-sensitive p53 (36), cyclin A, cyclin E, and cyclin D1 cDNA transgenes were introduced in the retrovirus vector LXSH (37). Clonal cell lines were selected with hygromycin and screened by immunoblotting. TGR/p53-4 is a derivative of TGR-1 and expresses the temperature-sensitive p53 transgene. HO/myc3, HO/p53-5, HO/cycA-2, HO/cycA-4, HO/cycA-7, HO/cycE-2, and HO/cycD-2 are derivatives of HO15.19 and express c-myc, temperature-sensitive p53, cyclin A, cyclin E, and cyclin D1 transgenes, respectively. Cell lines HO/cycE-2 and HO/cycD-2 were previously described as cell lines HO15E2 and HO15D2, respectively (31). Cells were cultured in Dulbecco modified Eagle medium containing glutamine, pyruvate, high glucose, and 3.7 g of sodium bicarbonate per liter, supplemented with 10% calf serum and penicillin-streptomycin. Cells were maintained in a 5% CO2 atmosphere at 37°C. Drugs were added directly to the culture medium at the times indicated in the figures at the following concentrations: etoposide, 2 μM; cisplatin, 10 μg/ml; staurosporine, 5 μM; aminopurvalanol, 10 μM. The cyclin-dependent kinase (Cdk) inhibitor aminopurvalanol (5) was a kind gift of Peter G. Schultz (The Scripps Research Institute, La Jolla, Calif.).
Apoptotic assays.
The dose response of TGR-1 cells to etoposide and cisplatin treatment was determined by measuring the extent of apoptosis at the end of a 48-h treatment, with the following results: for etoposide, 0 μM, 0.5%; 0.5 μM, 38.4%; 0.75 μM, 58.6%; 1 μM, 76.8%; 1.5 μM, 86.2%; and 2 μM, 88.4%; and for cisplatin, 0 μg/ml, 2%; 0.5 μg/ml, 35.6%; 1 μg/ml, 52.6%; 2.5 μg/ml, 77.8%; 5 μg/ml, 85.2%; 7.5 μg/ml, 87.4%; and 10 μg/ml, 88.9%. A 72-h treatment elicited 100% apoptosis in the range of 1 to 2 μM etoposide and 2.5 to 10 μg of cisplatin per ml. We chose 2 μM etoposide and cisplatin at 10 μg/ml as doses representative of the early to mid plateau phase of each response. Cisplatin elicited a strong apoptotic response in c-myc−/− cells in the 1 to 10 μg/ml range. The percentage of cells undergoing apoptosis was calculated as the ratio of apoptotic and total cells. Apoptotic TGR-1 cells become detached from the substratum; thus, the extent of apoptosis can be determined by counting floating and adherent cells. To perform in situ TUNEL (terminal deoxynucleotidyl-transferase-mediated dUTP-biotin nick end labeling) assays, floating cells were recovered from the medium by centrifugation and pooled with the adherent cells collected with trypsin. Cells were spun down onto polylysine-coated slides, and 3′-end elongation was catalyzed using TdT enzyme and digoxigenin-11-dUTP, which was detected with rhodamine-conjugated anti-digoxigenin antibody. Hoechst 33258 was used at 0.5 μg/ml in phosphate-buffered saline. The Apo-Direct kit (PharMingen) was used according to manufacturer's instructions. At least two independent assays of apoptosis were used in all experiments. The results of all apoptosis assays were always in agreement. All experiments were repeated on at least two independent occasions with consistent results. Error bars indicate the standard deviations of a minimum of three datum points. The absence of error bars indicates that the errors were smaller than the datum point symbols.
Immunoblotting and kinase assays.
Samples were prepared by rapid lysis of whole cells in Laemmli sample buffer supplemented with protease inhibitors (10 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride) and subjected to immunoblotting analysis using standard methods (32). Equivalent loading of lanes was carefully determined by running pilot gels loaded with various dilutions of extracts and analyzing images of Coomassie-stained gels using the Gel-Doc (Bio-Rad) digital gel documentation system and the Molecular Analysis software package (27, 32). Adjustments in sample volumes were made based on this analysis, and the process was repeated until all the lanes were equivalently loaded. This method can reliably establish equivalent loading within a 10 to 20% error value for all lanes. For immunoblotting, polyclonal antibodies to p53 (Novacastra), cyclin A (Upstate Biotechnology), and poly-ADP-ribosyl polymerase (PARP; clone C-2-10, SA-250; Biomol Research Laboratories) were used at 1 μg/ml. c-Myc antibody was provided by Steve Hann (Vanderbilt). Signals were visualized with ECL chemiluminescence (Amersham). Cdk activity was assayed using Tween 20 lysis conditions (33), immunoprecipitation with the cyclin A (C-19) antibody (Santa Cruz), and histone H1 as substrate as described earlier (32).
RESULTS
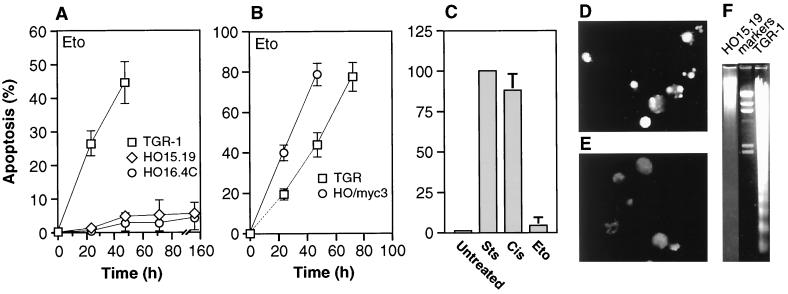
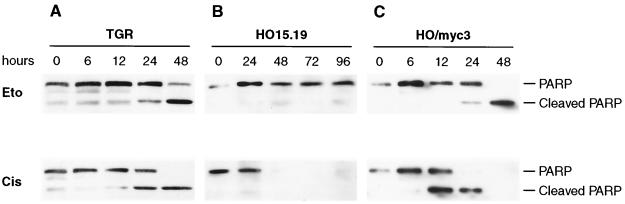
The role of c-Myc in DNA damage-triggered apoptotic processes was first investigated by treating c-myc+/+ (TGR-1) and two independently derived c-myc−/− cell lines (HO15.19 and HO16.4) with the topoisomerase II inhibitor etoposide, a drug known to elicit double-stranded DNA breaks (see Materials and Methods for the dose response to drug treatment). All cultures were in the exponential phase of growth and subconfluent at the time of drug addition. In contrast to the robust apoptosis observed in c-myc+/+ cultures, c-myc−/− cells were severely impaired in their apoptotic response, even after extended periods of incubation in the presence of drugs (Fig. 1A). Restoration of c-Myc activity by introducing a c-myc transgene on a retrovirus vector reversed the apoptosis defect (Fig. 1B). Apoptotic death in c-myc+/+ cells was confirmed by in situ TUNEL assay (Fig. 1D and E), nucleosomal laddering (Fig. 1F), nuclear condensation visualized by Hoechst dye staining (data not shown), and cleavage of endogenous poly-ADP-ribosyl polymerase (PARP) protein (Fig. 2A). The apoptotic defect observed in c-myc−/− cells was not general, because a variety of stimuli that do not involve DNA damage, such as the protein kinase inhibitor staurosporine (Fig. 1C) or the proteasome inhibitor MG132 (data not shown), were capable of eliciting a normal apoptotic response. Furthermore, not all DNA damage-induced responses were impaired, because cisplatin (Fig. 1C and 2B) and UV irradiation (data not shown) were capable of eliciting a strong apoptotic response.
FIG. 1.
(A) Apoptosis elicited in c-myc+/+ and c-myc−/− cells by treatment with etoposide (Eto). (B) Rescue of apoptosis by reconstitution of c-Myc activity. (C) Apoptosis elicited in c-myc−/− cells by a variety of agents. Sts, staurosporine; Cis, cisplatin; Eto, etoposide. Cells were treated for 48 h. (D and E) TUNEL assay of c-myc+/+ (D) and c-myc−/− (E) cells. TGR-1 and HO15.19 cells were harvested after 24 and 96 h of treatment with etoposide, respectively. (F) Nucleosomal laddering assay of c-myc+/+ and c-myc−/− cells. Cells were treated as in panels D and E, and 3 μg of DNA was loaded into each lane. Cell lines: TGR-1, c-myc+/+; HO15.19, HO16.4, c-myc−/−; HO/myc3, c-myc−/− expressing ectopic c-myc. HO/myc3 cells express wild-type murine c-Myc at a level two- to threefold above that found in TGR-1 cells.
FIG. 2.
PARP cleavage elicited by drug treatment of c-myc+/+ and c-myc−/− cells. Exponentially growing cultures of the indicated cell lines were treated with drugs at the zero time point, collected at the indicated times, and analyzed by immunoblotting. Eto, etoposide treatment; Cis, cisplatin treatment. See Fig. 1 for a description of the cell lines.
c-myc−/− cells proliferate approximately threefold more slowly than c-myc+/+ cells, with cell cycle durations of approximately 50 and 18 h, respectively (31). The possibility that the apoptotic impairment in c-myc−/− cells may be caused primarily by generalized metabolic defects was therefore considered. We believe this explanation is unlikely for the following reasons. First, the initial onset of apoptosis occurred within one cell cycle of drug treatment in both c-myc+/+ and c-myc−/− cells, between 12 to 15 h and 48 to 56 h after drug addition, respectively. Thus, c-myc−/− cells initiated the apoptotic response at a time commensurate with their reduced growth rate, but apoptosis failed to proceed to normal levels. Second, 80 to 100% of c-myc+/+ cells became apoptotic within three cell cycle durations, whereas the extent of apoptosis in c-myc−/− cultures did not exceed 10 to 15% at the commensurate times (i.e., the 156- and 192-h time points). Third, the apoptotic profile of c-myc−/− cells did not change whether drug was added once at the time zero point or fresh medium with drug was replenished at 48-h intervals (data not shown).
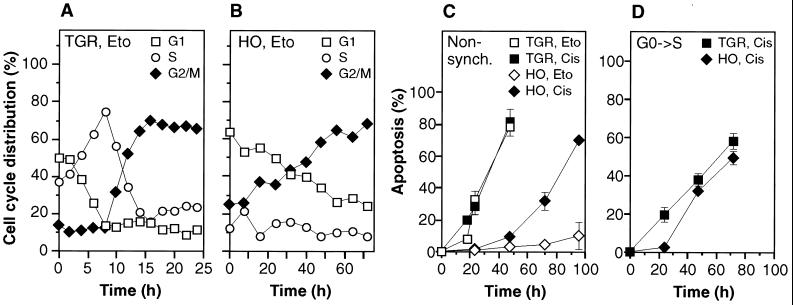
At the relatively low concentrations used in this study, etoposide elicits DNA damage primarily in the S phase, and the ensuing apoptosis occurs predominantly in the G2 phase (10). To investigate the relationship between cell cycle progression and apoptosis, drug-treated cultures were examined by flow cytometry (Fig. 3A and B). Both c-myc+/+ and c-myc−/− cultures showed a substantial (65 to 70%) accumulation of cells with G2/M DNA content, and this plateau was reached within 18 h (c-myc+/+) and 56 h (c-myc−/−) after addition of the drug. The total number of cells in both cultures ceased to increase after addition of the drug. These cell cycle profiles are consistent with the accumulation of DNA damage during passage through S phase and a subsequent arrest in G2, and the kinetics of the progression are in good agreement with the doubling times of c-myc+/+ and c-myc−/− cells, 18 and 50 h, respectively (31). In c-myc+/+ cultures the accumulation of cells with a G2/M DNA content coincided with the initial appearance of apoptosis 12 to 18 h after addition of the drug. Likewise, in c-myc−/− cells the first apoptotic cells were detected at 48 to 56 h after addition of the drug, at which time the G2/M content of the cultures had risen to 57 to 62%. However, although both the extent of G2/M accumulation and the initial onset of apoptosis were commensurate with cell cycle progression in c-myc+/+ and c-myc−/− cells, c-myc+/+ cultures became quickly consumed by a wave of apoptosis, whereas in c-myc−/− cultures apoptosis did not exceed 10 to 15% even at very late times (156 to 192 h). Neither c-myc+/+ or c-myc−/− cells could be rescued by removal of the drug (data not shown). c-myc−/− cultures could not be maintained beyond the 192-h time point because the remaining nonapoptotic cells gradually degenerated with signs of necrosis (data not shown).
FIG. 3.
(A and B) Cell cycle progression of c-myc+/+ (A) and c-myc−/− (B) cells following treatment with etoposide (Eto). Samples were collected concurrently with the experiment shown in Fig. 1A and analyzed by flow cytometry. The G2/M plateau remained constant until the termination of the experiments (48 h for TGR-1 cells, 96 h for HO15.19 cells). (C) Apoptosis elicited by cisplatin (Cis). (D) Apoptosis elicited in synchronized cells by cisplatin. Quiescent cultures were stimulated with serum at the zero time point and treated with cisplatin 2 h later.
To investigate the effect of drugs that damage DNA irrespective of cell cycle position, cultures were treated with cisplatin (Fig. 3C), a drug that causes interstrand cross-links (see Materials and Methods for the dose response to drug treatment). In c-myc+/+ cells cisplatin caused the same degree of apoptosis as had etoposide and, after an initial lag, cisplatin also elicited a strong apoptotic response in c-myc−/− cells. The progression of apoptosis was approximately twofold slower in c-myc−/− cultures but proceeded to the same extent as in c-myc+/+ cultures. The twofold delay in c-myc−/− cultures may be related to their reduced rate of proliferation and macromolecular synthesis (31). It is therefore apparent that c-myc−/− cells, given an appropriate stimulus, are capable of mounting a significant apoptotic response to DNA damage. To investigate selectively apoptosis in the G1 phase, cells were synchronized in G0 by serum deprivation and treated with cisplatin for 2 h after serum-induced release into the cell cycle (Fig. 3D). After a short initial lag both the kinetics and the extent of apoptosis were identical in c-myc+/+ and c-myc−/− cultures. Flow cytometry showed that both c-myc+/+ and c-myc−/− cells became arrested with a G1 DNA content (data not shown). UV light treatment gave the same results as cisplatin (data not shown). Treatment of G0 synchronized cells with etoposide did not elicit a G1 arrest; in contrast to cisplatin, the cells progressed through S phase and accumulated with a G2/M DNA content (data not shown). Again, the kinetics of G2/M accumulation were commensurate with cell cycle progression rates in c-myc+/+ and c-myc−/− cells, but the extent of G2/M accumulation was the same in both. Finally, the same apoptotic response as that seen in exponentially cycling cells was observed: c-myc+/+ cultures were quickly consumed by apoptosis, whereas in c-myc−/− cells apoptosis did not exceed 10 to 15% even at very late times (156 h). Thus, c-myc−/− cells show a dramatic defect in apoptosis in G2/M but appear capable of essentially normal apoptosis in G1.
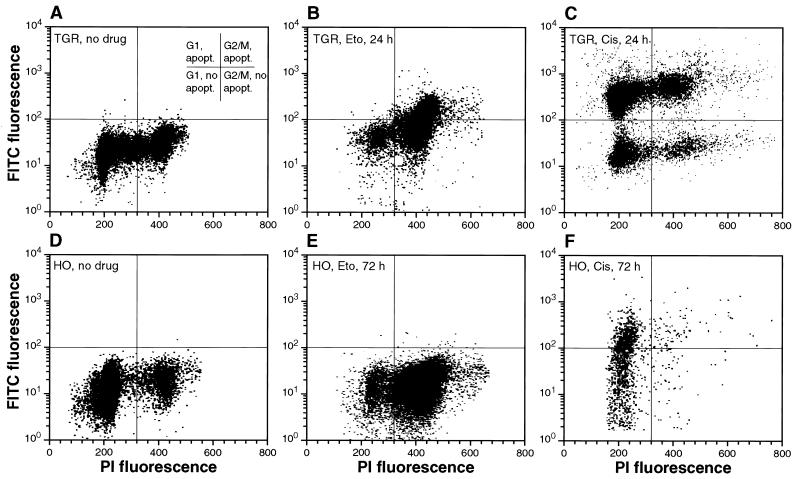
To further investigate the relationship between cell cycle position and apoptosis, TUNEL and cell cycle assays were combined by two-parameter flow cytometry. As previously seen, in c-myc+/+ cultures etoposide caused an accumulation of cells in G2/M (Fig. 4B). In addition, the two-parameter analysis showed that the majority of TUNEL-positive cells had a G2/M DNA content, indicating that apoptosis was occurring directly in the G2/M-arrested cells. In c-myc−/− cells etoposide also caused an accumulation in G2/M, but very few TUNEL-positive cells were seen (Fig. 4E). Cisplatin caused strong apoptosis both in G1 and G2/M in c-myc+/+ cells (Fig. 4C); this result confirms that cisplatin can cause DNA damage irrespective of cell cycle position and that the Rat-1 cells used in this study can undergo apoptosis in both G1 and G2/M phases. In contrast, c-myc−/− cultures became depleted of G2/M cells and accumulated in G1 (Fig. 3F), and TUNEL-positive cells were seen predominantly with a G1 DNA content. Thus, in c-myc−/− cells even a drug that causes DNA damage irrespective of cell cycle position elicits apoptosis predominantly in G1. It therefore appears that c-myc−/− cells are compromised in their apoptotic response in the G2/M phase of the cell cycle.
FIG. 4.
Concurrent analysis of apoptosis and cell cycle progression. Exponentially growing cultures of c-myc+/+ (A, B, and C) and c-myc−/− (D, E, and F) cells were treated with etoposide (Eto) or cisplatin (Cis) as indicated, harvested at the indicated time points, processed for TUNEL using the Apo-Direct kit (Pharmingen), counterstained with propidium iodide, and analyzed by two-parameter flow cytometry.
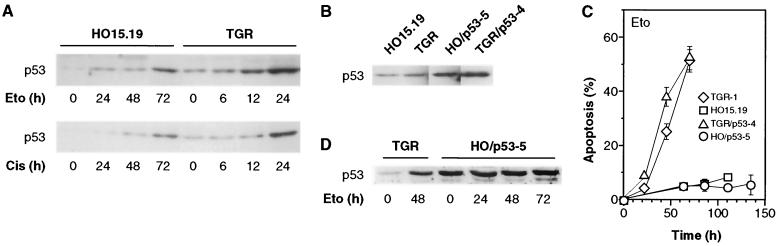
Treatment of c-myc+/+ or c-myc−/− cells with etoposide, cisplatin, or UV light (Fig. 5A and data not shown) in all cases caused a marked accumulation of p53 protein. This effect is known to occur primarily by the stabilization of the p53 protein, as well as by a relatively minor induction of the mRNA (15, 29). The Rat-1 cell line used in this study has been reported to express wild-type p53 protein (30). In c-myc+/+ cells the increase in p53 protein levels after drug treatment was greater than 10-fold, while induction of the mRNA was only 2- to 3-fold (data not shown). Although the increase in p53 protein expression was somewhat variable from experiment to experiment, c-myc−/− cells reproducibly displayed a modest (two- to threefold) defect in p53 induction as detected by immunoblotting (Fig. 5A) or immunoprecipitation (data not shown). It should be noted that because of the decreased growth rate of c-myc−/− cells, data spanning the critical first cell cycle after addition of drug are presented for both cell lines. For example, the 24-h time point for c-myc+/+ cells (ca. 1.3 cell cycles) is most appropriately compared to the 72-h time point for c-myc−/− cells (1.4 cell cycles). p53 protein expression in c-myc−/− cells did not increase beyond the 72-h time point and actually declined at later times (data not shown).
FIG. 5.
Relationship of p53 expression and apoptosis. (A) Expression of p53 after DNA damage. Exponentially growing cells were treated with drugs at the zero time point, collected at the indicated times, and analyzed by immunoblotting. (B) Expression of p53 in cells ectopically expressing a temperature-sensitive mutant of p53. Subconfluent cells at 37°C in the absence of drugs were analyzed by immunoblotting. (C) Influence of p53 overexpression on apoptosis elicited by etoposide. All cells were grown at 37°C prior to the experiment. Exponentially growing subconfluent cells were treated with etoposide at the zero time point and shifted to 32.5°C. Cell cycle profiles, monitored by flow cytometry, were similar to those shown in Fig. 2A and B. (D) p53 expression in the experiment shown in panel C. Samples were collected at the indicated times and analyzed by immunoblotting.
To address whether the small defect in p53 accumulation was responsible for the apoptosis defect, stable cell lines expressing a temperature-sensitive mutant of p53 were isolated. The temperature-sensitive p53 protein displays wild-type activity at the permissive temperature (32.5°C) and mutant activity at the nonpermissive temperature (37°C) (36). Since overexpression of wild-type p53 protein causes cell cycle arrest, this strategy allowed the propagation of p53-overexpressing cells at 37°C, as well as the analysis of functional p53 overexpression by shiftdown to 32.5°C. Overexpression of the introduced temperature-sensitive p53 protein was documented by immunoblotting (Fig. 5B). Furthermore, the introduced p53 protein was biologically active because at 32.5°C it accelerated apoptosis in response to all drugs in c-myc+/+ cells, and in response to cisplatin and UV light in c-myc−/− cells (data not shown). Exponentially growing cells at 37°C were treated with etoposide and shifted to 32.5°C to activate the p53 protein (Fig. 5C). Overexpression of p53 did not restore apoptosis, even at late times during incubation with etoposide. Apoptosis was also not restored when the shiftdown to 32.5°C was delayed (up to 60 h) relative to the addition of the drug (data not shown). Immunoblotting analysis showed that p53 protein was overexpressed during the course of the experiment (Fig. 5D). Thus, the small defect in p53 expression seen in c-myc−/− cells does not appear to be causally linked to the apoptotic defect.
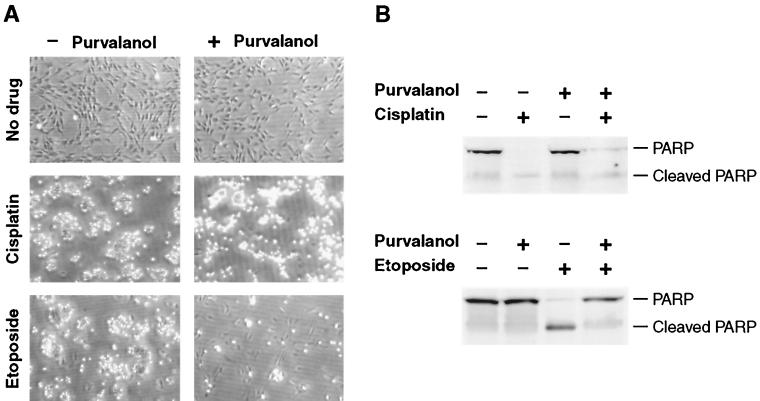
Position in the cell cycle may be important for susceptibility to apoptosis, and Cdks have been implicated in apoptotic processes in several experimental systems (2, 14, 18, 21, 34, 35). The fact that we have previously documented activation defects of several cyclin-Cdk complexes in c-myc−/− cells made this a compelling line of investigation. The recent development of a highly specific and potent class of Cdk inhibitors, purvalanol and its derivatives (5, 16), allowed us to rapidly evaluate pharmacologically the involvement of these pathways in DNA damage-induced apoptosis. Purvalanols, unlike earlier Cdk inhibitors such as roscovitine and olomoucine, display a high degree of both selectivity and potency for Cdk1 and Cdk2 (5, 16). Exponentially growing c-myc+/+ cells were treated with etoposide, cisplatin, or left without drug and at the same time either exposed to aminopurvalanol or left untreated. The extent of apoptosis was assessed after 48 h either by microscopic observation of the cultures (Fig. 6A) or by harvesting the cells and immunoblotting for PARP (Fig. 6B). As shown previously, in the absence of aminopurvalanol, both etoposide and cisplatin elicited a strong apoptotic response that was accompanied by PARP cleavage. Strikingly, aminopurvalanol was strongly protective against etoposide-induced apoptosis, while it had no effect on the response elicited by cisplatin.
FIG. 6.
Effect of the Cdk inhibitor aminopurvalanol on apoptosis induced by etoposide and cisplatin. Exponentially growing c-myc+/+ cells were treated with drugs as indicated and incubated for 48 h. At the end of this incubation the cultures were photographed (A), and the cells were recovered and processed for immunoblotting (B) with anti-PARP antibody.
We have previously shown that exponentially growing c-myc−/− cells display a modest (two- to threefold) defect in cyclin A expression, as well as in cyclin A-associated Cdk activity (32). In c-myc+/+ cells treatment with etoposide caused a strong accumulation of cyclin A protein as early as 6 h after drug addition, whereas in c-myc−/− cells the magnitude of the induction was reduced and the kinetics were delayed (Fig. 7A). Cyclin B expression peaked at 24 h in c-myc+/+ cells and appeared at a low level at the 72-h time point in c-myc−/− cells (data not shown). Thus, as would be expected, cyclin B expression followed kinetically that of cyclin A. It is of interest to note that cyclin A induction was rapid and preceded the accumulation of p53 (compare results with Fig. 5A). Cyclin A-associated kinase activity after etoposide treatment peaked at 12 h in c-myc+/+ cells (Fig. 7D) and also clearly preceded the onset of apoptosis at 18 to 24 h. The peak of cyclin A-associated kinase activity in c-myc−/− cells was significantly delayed as well as reduced in magnitude. Cyclin B-associated kinase activity peaked at 24 h in c-myc+/+ cells and was also significantly delayed and dampened in c-myc−/− cells (data not shown).
FIG. 7.
Relationship of cyclin A expression and apoptosis. (A) Expression of cyclin A after etoposide treatment. Exponentially growing cells were treated with drugs at the zero time point, collected at the indicated times, and analyzed by immunoblotting. (B) Expression of cyclin A in cells ectopically expressing cyclin A. Subconfluent cells in the absence of drugs were analyzed by immunoblotting. (C) Cyclin A expression in the experiment shown in panel E analyzed by immunoblotting. (D) Cyclin A-associated Cdk activity after etoposide treatment. Exponentially growing cells were treated with drugs at the zero time point, collected at the indicated times, immunoprecipitated with anti-cyclin A antibody, and assayed for histone H1 kinase activity. (E) Influence of cyclin A overexpression on apoptosis elicited by etoposide. Exponentially growing subconfluent cells were treated with etoposide at the zero time point. (F) Influence of cyclin D and cyclin E overexpression on apoptosis elicited by etoposide. The experiment was performed as indicated in panel E. (G) Cell cycle progression profiles in the experiment shown in panel E.
To address the functional relevance of reduced cyclin A expression, c-myc−/− cell lines stably overexpressing cyclin A were isolated. It should be noted that in the cell lines chosen for study the overexpression of cyclin A was modest (Fig. 7B), and no apoptosis was observed in the absence of DNA-damaging agents. However, the expression of ectopic cyclin A in c-myc−/− cells restored to a significant degree the ability of etoposide to elicit apoptosis (Fig. 7E). Control experiments showed that in cyclin A-overexpressing cell lines etoposide caused the expected G2/M arrest (Fig. 7G) and that cyclin A was overexpressed at the time points when apoptosis was occurring (Fig. 7C). Several features of the cyclin A rescue were notable. First, the rescue was dose dependent, such that the magnitude of the apoptotic response correlated positively with the expression levels of cyclin A. Second, although the rescue was significant (50% apoptosis in HO/cycA-2 cells versus 15% in HO15.19 cells), it was not complete and apoptosis occurred with slower kinetics than in c-myc+/+ cells (compare to Fig. 1A). In fact, the kinetics of the apoptotic response to etoposide in the cyclin A-rescued cells resembled the response elicited by cisplatin in parental c-myc−/− cells (compare to Fig. 3C).
Perhaps the most significant aspect of the cyclin A rescue of apoptosis is that cyclin A overexpression had no effect whatsoever on the slow-growth phenotype of c-myc−/− cells and did not affect progression through any phase of the cell cycle relative to the parental c-myc−/− cells. The cyclin A-expressing cell lines used here were part of a larger study to investigate the effects of cyclin overexpression on the growth phenotype of c-myc−/− cells (32). That study found that expression of neither cyclin D1, E, nor A (expressed singly) could rescue the proliferation defect of c-myc−/− cells. We subsequently tested cell lines ectopically expressing cyclin D1 or E for the rescue of etoposide-induced apoptosis and found that, in contrast to cyclin A, neither cyclin D1 nor E had any effect on either the extent or the kinetics of the apoptotic response (Fig. 7F). None of the cyclins tested (D1, E, or A) had an effect on the apoptotic response elicited by cisplatin (data not shown). These results further confirm that the apoptotic defect in c-myc−/− cells affects predominantly processes in G2 phase.
DISCUSSION
Although it has been known for some time that elevated c-Myc expression can predispose cells to apoptosis (3, 12) and that antisense oligonucleotide-mediated reduction of c-Myc expression can in some cases partially protect against apoptosis (9, 22, 25, 50), evidence that c-Myc is functionally required in any one DNA damage-induced apoptotic pathway has been lacking. We show here that c-Myc is necessary for the activation of DNA damage-induced apoptosis in G2 phase. c-myc+/+ cells treated with etoposide, a drug that causes DNA damage primarily in the S phase, accumulate in G2 and subsequently undergo apoptosis. c-myc−/− cells are strongly defective in this apoptotic response. In contrast, c-Myc is not required for apoptosis in G1, since cisplatin, a drug that causes DNA damage irrespective of cell cycle position, causes robust apoptosis in c-myc−/− cells. It is important to note that while cisplatin elicits apoptosis both in G1 and G2 in c-myc+/+ cells, in c-myc−/− cells apoptosis is restricted to the G1 compartment. This result argues that the apoptotic defect in c-myc−/− cells is due to a failure to either initiate or execute apoptosis rather than to a defect in the accumulation of DNA damage. UV light elicited the same response at cisplatin. Restoration of c-Myc expression in c-myc−/− cells completely rescued the apoptotic defect, making it unlikely that the cells have accumulated secondary mutations that affect apoptosis. Furthermore, independently gene targeted c-myc−/− clones displayed the same apoptotic phenotype.
The G2 checkpoint that monitors the integrity of the genome is operational in all cells (17), including tumor cells that are defective in the p53-dependent G1 checkpoint (24). Overexpression of wild-type p53 in c-myc−/− cells did not rescue the G2 apoptotic defect. Although this is a negative result, it was obtained in clonal cell lines stably overexpressing a temperature-sensitive p53 protein, which was biologically active because it accelerated G1 apoptosis in response to cisplatin. The failure of p53 to rescue is not consistent with c-Myc acting functionally upstream of p53 in the G2 apoptotic pathway. The result is consistent with a model (11) in which c-Myc acts downstream of p53 to sensitize the cell to apoptotic stimuli. It is formally also possible that the G2 DNA damage-triggered apoptotic pathway defective in c-myc−/− cells does not involve p53.
We have identified cyclin A as one critical effector of c-Myc-dependent G2 apoptosis: cyclin A levels were reduced in c-Myc knockout cells, and restoration of cyclin A expression significantly rescued apoptosis. In contrast, overexpression of cyclin A had no effect whatsoever on the slow growth of c-myc−/− cells (32). This result argues strongly that the apoptotic defect in c-myc−/− cells is not caused by the cell cycle defects observed in these cells. Other observations are also consistent with this interpretation. First, c-myc−/− cells show an approximately equivalent lengthening of both G1 and G2 phases (31), whereas the apoptotic defect is confined to G2. This argues that slow cell cycle progression per se is not the cause of the apoptotic defect. Furthermore, the duration of the S phase, the period when the majority of etoposide-elicited lesions are accumulated, is essentially identical in c-myc−/− and c-myc+/+ cells (31). Second, the kinetics of apoptosis in response to cisplatin in G1 are very similar in c-myc+/+ and c-myc−/− cells, indicating that the major pathways for the execution of apoptosis are likely to be intact in both cell lines. Third, it takes approximately one cell cycle interval (18 h) for c-myc+/+ cells to accumulate in G2 and approximately two additional cell cycle intervals (36 h) for apoptosis to be virtually completed (50 to 80%). In contrast, while c-myc−/− cells arrest in G2 within one cell cycle interval (50 h), incubation for up to 192 h after the addition of drug (three additional cell cycle intervals) results in minimal (<15%) apoptosis. Thus, the kinetics of the G2 arrest and ensuing apoptosis are also not consistent with the slow growth rate of c-myc−/− cells being the primary cause of the observed apoptotic defect.
The activation of Cdk kinases has been temporally associated with apoptosis (2, 34), and inhibition of Cdk activation has been shown to exert an apoptosis-sparing effect (35, 49). We show here that aminopurvalanol, a highly selective and potent inhibitor of Cdk activity (5), blocks the induction of apoptosis by etoposide but not cisplatin. Thus, aminopurvalanol exerts the same effect on these responses as the loss of c-Myc, which we have previously shown results in the downregulation of Cdk activity (32). However, since both aminopurvalanol and loss of c-Myc affect the activity of both G1 and G2 kinases, we performed rescue experiments by ectopically expressing individual cyclin genes in c-myc−/− cells. These studies clearly implicated cyclin A as the relevant downstream effector of c-Myc: cyclin A significantly rescued etoposide-induced apoptosis, while cyclin E and cyclin D1 were without effect. We were unable, in spite of repeated attempts, to isolate cell lines overexpressing cyclin B. It is important to note that none of the overexpressed cyclins rescued the proliferation defect of c-myc−/− cells (32), thus providing further evidence that the apoptosis defect observed in these cells is not due to their slow growth.
In most (18), but not all (14) cases, the Cdk activity implicated in apoptotic processes has been associated with cyclin A. The fact that overexpression of cyclin A alone can in some cases drive cells into apoptosis (21) is consistent with an important role in apoptotic processes. It should be noted that the c-myc−/− cell lines that we have engineered to express ectopic cyclin A grow normally and do not display any signs of apoptosis in the absence of drug treatment. Cyclin A thus joins p19ARF1 (57) as a putative c-Myc target gene that is specific for mediating proapoptotic functions. Although a positive effect of c-Myc overexpression on cyclin A expression was noted some time ago (23), it is unlikely that the cyclin A gene is a direct transcriptional target of c-Myc: the promoter does not contain c-Myc binding sites, and the major regulator responsible for cell cycle dependent expression has been identified as E2F (48). The cyclin A promoter has also been shown to be actively repressed by E2F-Rb complexes in G0 and early G1 (41). These observations provide a good explanation for the observed reduction of cyclin A expression in c-myc−/− cells, which display a significant defect in the expression of the E2F-1, -2, and -3 genes, as well as persistence of unphosphorylated Rb in late G1 (32).
The expression of cyclin A and associated Cdk activity in response to DNA damage displayed the characteristics of a DNA damage-inducible response that occurred independently of the changes in cell cycle distribution. Etoposide caused a rapid induction of cyclin A that somewhat preceded the progression into S and G2/M (compare Fig. 3A and 7A). More importantly, cisplatin (Fig. 7A) and UV light (data not shown) caused a robust induction of cyclin A in spite of the fact that the cell cycle distribution of the cultures did not change after treatment (compare Fig. 4A and C and Fig. 7A). Cyclin A induction has also been reported to accompany apoptosis in postmitotic cardiomyocytes (1), and transfection of a dominant-defective Cdk2 protected against apoptosis in this cell type. Etoposide-stimulated cyclin A-Cdk activity in c-myc+/+ cells decayed rapidly and was below basal levels at the time of maximum apoptosis. This contrasts with apoptosis induced by growth or survival factor deprivation, which typically occurs in G1, and where the induction of cyclin A/Cdk activity has been shown to be a late event (18, 26). The expression of c-Myc was not affected by DNA damage (data not shown), but its presence was clearly required for the normal induction of cyclin A. There are examples in other cell types where cytotoxic drugs can elicit an induction of c-Myc expression (56).
Although apoptosis in response to etoposide occurred at times of maximum G2 content of the cultures, the resolution of the flow cytometry data is not sufficient to rule out that some apoptosis was also taking place in the S phase. This possibility is also raised by the apparent involvement of cyclin A-Cdk2 complexes, which are known to be active during the S phase. Nevertheless, the kinetics of cell cycle progression and apoptosis in etoposide-treated cultures indicate that the majority of apoptosis was occurring only after most of the cells reached late S or G2.
In summary, we show here that the loss of c-Myc expression results in an impairment of DNA damage-initiated apoptosis in the G2 phase of the cell cycle. We propose a model wherein DNA damage-induced cyclin A-Cdk2 activity is required to initiate the apoptotic process. Apoptosis in G1 does not appear to require c-Myc and does not seem to be influenced by G1 cyclin-Cdk complexes in the cell type studied here. The kinetics of the G2 response indicate that the induction of cyclin A and the concomitant activation of Cdk2 represent an early step during commitment to apoptosis. Although c-Myc in not absolutely required for the induction of cyclin A, in its absence the magnitude of this response is significantly dampened. The involvement of c-Myc in these processes sheds new light on the mechanisms by which this important oncogene acts to sensitize cells to a wide variety of apoptotic stimuli.
ACKNOWLEDGMENTS
We thank P. Schultz and Y.-T. Chang for providing aminopurvalanol.
This work was supported by NIH grant R01-GM41690 to J.M.S. and NSF grant MCB-9630362 to J.H.W. A.J.O. was supported in part by a postdoctoral fellowship from the Ministerio Education y Cultura de Espana.
REFERENCES
- 1.Adachi S, Ito H, Tamamori-Adachi M, Ono Y, Nozato T, Abe S, Ikeda M, Marumo F, Hiroe M. Cyclin A/cdk2 activation is involved in hypoxia-induced apoptosis in cardiomyocytes. Circ Res. 2001;88:408–414. doi: 10.1161/01.res.88.4.408. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J A, Lewellyn A L, Maller J L. Ionizing radiation induces apoptosis and elevates cyclin A1-Cdk2 activity before but not after the midblastula transition in Xenopus. Mol Biol Cell. 1997;8:1195–1206. doi: 10.1091/mbc.8.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 4.Canelles M, Delgado M D, Hyland K M, Lerga A, Richard C, Dang C V, Leon J. Max and inhibitory c-Myc mutants induce erythroid differentiation and resistance to apoptosis in human myeloid leukemia calls. Oncogene. 1997;14:1315–1327. doi: 10.1038/sj.onc.1200948. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y T, Gray N S, Rosania G R, Sutherlin D P, Kwon S, Norman T C, Sarohia R, Leost M, Meijer L, Schultz P G. Synthesis and application of functionally diverse 2,6,9-trisubstituted purine libraries as CDK inhibitors. Chem Biol. 1999;6:361–375. doi: 10.1016/S1074-5521(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 6.Claassen G, Hann S R. Myc-mediated transformation: the repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- 7.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 8.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J, Naito M, Tsuruo T. c-Myc plays a role in cellular susceptibility to death receptor-mediated and chemotherapy-induced apoptosis in human monocytic leukemia U937 cells. Oncogene. 1997;15:639–647. doi: 10.1038/sj.onc.1201237. [DOI] [PubMed] [Google Scholar]
- 10.Dubrez L, Goldwasser F, Genne P, Pommier Y, Solary E. The role of cell cycle regulation and apoptosis triggering in determining the sensitivity of leukemic cells to topoisomerase I and II inhibitors. Leukemia. 1995;9:1013–1024. [PubMed] [Google Scholar]
- 11.Evan G I, Littlewood T. A matter of file and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 12.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 13.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 14.Fotedar R, Flatt J, Gupta S, Margolis R L, Fitzgerald P, Messier H, Fotedar A. Activation-induced T-cell death is cell cycle dependent and regulated by cyclin B. Mol Cell Biol. 1995;15:932–942. doi: 10.1128/mcb.15.2.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsche M, Haessler C, Brander G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 16.Gray N S, Wodicka L, Thunnissen A-M W H, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, Kim S-H, Lockhart D J, Schultz P G. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 18.Harvey K J, Blomquist J F, Ucker D S. Commitment and effector phases of the physiological cell death pathway elucidated with respect to Bcl-2, caspase, and cyclin-dependent kinase activities. Mol Cell Biol. 1998;18:2912–2922. doi: 10.1128/mcb.18.5.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 20.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 21.Hoang A T, Cohen K J, Barrett J F, Bergstrom D A, Dang C V. Participation of cyclin A in Myc-induced apoptosis. Proc Natl Acad Sci USA. 1994;91:6875–6879. doi: 10.1073/pnas.91.15.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janicke R U, Lin X Y, Lee F H, Porter A G. Cyclin D3 sensitizes tumor cells to tumor necrosis factor-induced, c-Myc-dependent apoptosis. Mol Cell Biol. 1996;16:5245–5253. doi: 10.1128/mcb.16.10.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, Wessbecher J, Draetta G, Eilers M. Differential modulation of cyclin gene expression by Myc. Proc Natl Acad Sci USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 25.Klefstrom J, Vastrik I, Saksela E, Valle J, Eilers M, Alitalo K. c-Myc induces cellular susceptibility to the cytotoxic action of TNF-alpha. EMBO J. 1994;13:5442–5450. doi: 10.1002/j.1460-2075.1994.tb06879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levkau B, Koyama H, Raines E W, Clurman B E, Herren B, Orth K, Roberts J M, Ross R. Cleavage of p21(CIP1/WAF1) and p27(KIP1) mediates apoptosis in endothelial cells through activation of Cdk2—role of a caspase cascade. Mol Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 27.Lu K K, Bazarov A V, Yoon L S, Sedivy J M. Isolation of temperature-sensitive mutations in the c-raf-1 catalytic domain and expression of conditionally active and dominant-defective forms of Raf-1 in cultured mammalian cells. Cell Growth Differ. 1998;9:367–380. [PubMed] [Google Scholar]
- 28.Luscher B, Larsson L G. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncogenes: function and regulation. Oncogene. 1999;18:2955–2966. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- 29.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 30.Marhin W W, Chen S, Facchini L M, Fornace A J, Jr, Penn L Z. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 31.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 32.Mateyak M K, Obaya A J, Sedivy J M. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J V. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meikrantz W, Gisselbrecht S, Tam S W, Schlegel R. Activation of cyclin A-dependent protein kinases during apoptosis. Proc Natl Acad Sci USA. 1994;91:3754–3758. doi: 10.1073/pnas.91.9.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meikrantz W, Schlegel R. Suppression of apoptosis by dominant negative mutants of cyclin-dependent protein kinases. J Biol Chem. 1996;271:10205–10209. doi: 10.1074/jbc.271.17.10205. [DOI] [PubMed] [Google Scholar]
- 36.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 37.Miller A D, Miller D G, Garcia J V, Lynch C M. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 38.Nesbit C E, Fan S, Zhang H, Prochownik E V. Distinct apoptotic responses imparted by c-Myc and Max. Blood. 1998;92:1003–1010. [PubMed] [Google Scholar]
- 39.Nesbit C E, Tersak J M, Prochownik E V. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 40.Obaya A J, Mateyak M K, Sedivy J M. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 41.Philips A, Huet X, Plet A, Le Cam L, Vie A, Blanchard J M. The retinoblastoma protein is essential for cyclin A repression in quiescent cells. Oncogene. 1998;16:1373–1381. doi: 10.1038/sj.onc.1201655. [DOI] [PubMed] [Google Scholar]
- 42.Prendergast G C. Mechanisms of apoptosis by c-Myc. Oncogene. 1999;18:2967–2987. doi: 10.1038/sj.onc.1202727. [DOI] [PubMed] [Google Scholar]
- 43.Prouty S M, Hanson K D, Boyle A L, Brown J R, Shichiri M, Follansbee M R, Kang W, Sedivy J M. A cell culture model system for genetic analyses of the cell cycle by targeted homologous recombination. Oncogene. 1993;8:899–907. [PubMed] [Google Scholar]
- 44.Reisman D, Elkind N B, Roy B, Beamon J, Rotter V. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ. 1993;4:57–65. [PubMed] [Google Scholar]
- 45.Roy B, Beamon J, Balint E, Reisman D. Transactivation of the human p53 tumor suppressor gene by c-Myc/Max contributes to elevated mutant p53 expression in some tumors. Mol Cell Biol. 1994;14:7805–7815. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamuro D, Eviner V, Elliott K J, Showe L, White E, Prendergast G C. c-myc induces apoptosis in epithelial cells by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:2411–2418. [PubMed] [Google Scholar]
- 47.Sakamuro D, Prendergast G C. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 48.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi L, Nishioka W K, Th'ng J, Bradbury E-M, Litchfield D W, Greenberg A H. Premature p34cdc2 activation required for apoptosis. Science. 1994;263:1143–1145. doi: 10.1126/science.8108732. [DOI] [PubMed] [Google Scholar]
- 50.Shi Y, Glynn J M, Guilbert L J, Cotter T G, Bissonnette R P, Green D R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- 51.Thompson E B. The many roles of c-Myc in apoptosis. Annu Rev Physiol. 1998;60:575–600. doi: 10.1146/annurev.physiol.60.1.575. [DOI] [PubMed] [Google Scholar]
- 52.Trudel M, Lanoix J, Barisoni L, Blouin M J, Desforges M, L'Italien C, D'Agati V. c-Myc-induced apoptosis in polycystic kidney disease is Bcl-2 and p53 independent. J Exp Med. 1997;186:1873–1884. doi: 10.1084/jem.186.11.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 54.Wagner A J, Small M B, Hay N. Myc-mediated apoptosis is blocked by ectopic expression of Bcl-2. Mol Cell Biol. 1993;13:2432–2440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Szekely L, Okan I, Klein G, Wiman K G. Wild-type p53-triggered apoptosis is inhibited by Bcl-2 in a v-myc-induced T-cell lymphoma line. Oncogene. 1993;8:3427–3431. [PubMed] [Google Scholar]
- 56.Zhan Y, Cleveland J L, Stevens J L. A role for c-myc in chemically induced renal cell death. Mol Cell Biol. 1997;17:6755–6764. doi: 10.1128/mcb.17.11.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]