Abstract
Background
Growing evidence suggests a mutual interaction between gut microbiome alterations and ALS pathogenesis. However, previous studies were susceptible to potential confounding factors and reverse causation bias, likely leading to inconsistent and biased results.
Objectives
To decipher the potentially mutual relationship between gut microbiota and ALS, we used a bidirectional two-sample MR approach to examine the associations between the gut microbiome and ALS.
Results
Using the inverse variance-weighted method, OTU10032 unclassified Enterobacteriaceae species-level OTU and unclassified Acidaminococcaceae were associated with a higher risk of ALS (per relative abundance: OR, 1.04; 95% CI, 1.01–1.07; P = 0.011 and OR, 1.02; 95% CI, 1.01–1.04; P = 0.009, respectively). Importantly, Gamma-Glu-Phe was showed potential deleterious effects on the risk of ALS (genetically predicted per a 1-standard deviation increase in the level of Gamma-Glu-Phe: OR, 1.96; 95% CI, 1.50–2.55; P = 0.012). Sensitivity analysis of the two candidate genera and metabolites using the MR-Egger and weighted-median methods produced similar estimates, and no horizontal pleiotropy or outliers were observed. Intriguingly, genetically predicted ALS was associated with an increase in the relative abundance of OTU4607_Sutterella (per 1-unit higher log odds: β, 2.23; 95% CI, 1.27–3.18; P = 0.020) and Lactobacillales_ORDER (per 1-unit higher log odds: β, 0.51; 95% CI, 0.09–0.94; P = 0.019).
Conclusions
Our findings provide novel evidence supporting the bidirectional relationship between the gut microbiota and ALS. These results may contribute to designing microbiome- and microbiome-dependent metabolite interventions in future ALS clinical trials.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12883-021-02522-z.
Keywords: Amyotrophic lateral sclerosis, Gut microbiota, Gamma-glutamyl amino acids, Bidirectional relationships, Two-sample Mendelian randomization
Background
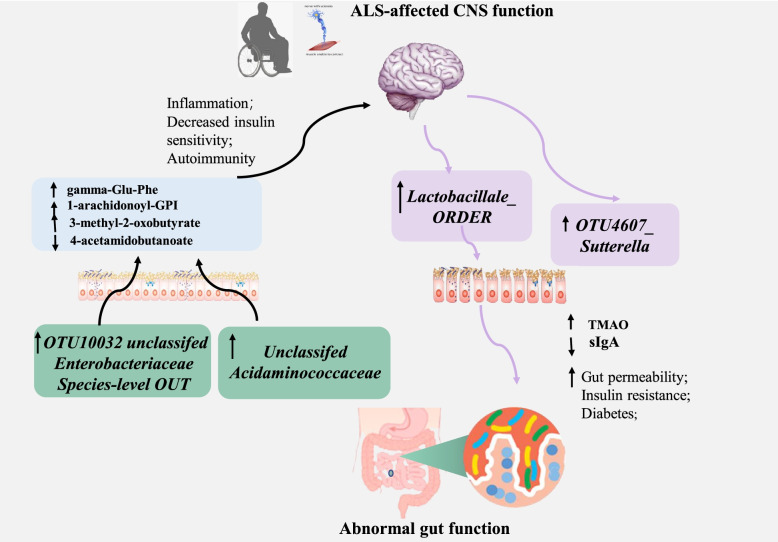
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative motor neuron disease accompanied by both systemic and central nervous system–specific inflammation as well as energy dysmetabolism [1–3]. Structural components of the bacteria and various metabolites (pro-inflammatory cytokines or anti-inflammatory) secreted by the gut microbiota can stimulate or inhibit a cascade of inflammatory pathways on both a local and systemic scale [4]. Additionally, by-products of metabolic processes in bacteria, including some short-chain fatty acids, can play a role in inhibiting inflammatory processes [5]. These local and systemic inflammatory, which in turn could lead to perturbed gut-microbiota (dysbiosis) and increased intestinal permeability (leaky-gut) [6]. These potential pathogenetic factors have recently been found to mutually interact with the gut microbiota [7, 8], suggesting that the gut microbiota could be involved in the development of the disease and be affected by the disease vice versa (Fig. 1).
Fig. 1.
Graphic Abstract
Observational studies have shown that the interface between the host and the gut microbiome may be altered in mouse models of ALS [9, 10], including impaired gut barrier function and a dysbiotic microbiome configuration that can be partially corrected by butyrate supplementation [10]. Studies of whether gut microbiome dysbiosis occurs between ALS patients and healthy controls have yielded conflicting results [11–13]. Notably, a recent study [14] of 11 distinct commensal bacteria based on their individual supplementation into antibiotic-treated Sod1-Tg mice found that Akkermansia muciniphila (AM) and AM-associated nicotinamide ameliorate symptoms of ALS. In humans, distinct microbiome and metabolite configurations have been observed in a small preliminary study that compared 37 patients with ALS with household controls [14].
Growing but conflicting evidence is attractive, raising the hypothesis of a mutual interaction between gut microbiome alterations and ALS pathogenesis. However, it has been difficult to determine whether these changes in the intestinal microbiota are causative of ALS disease, an exacerbating factor for disease, or a consequence of disease. The composition and diversity of the gut microbiome can be easily altered as a result of bacterial infections, antibiotic treatment, lifestyle changes, surgery, and long-term changes in diet [4]. Available evidence is in large part inadequate, as observational studies are susceptible to these potential confounding and reverse causation biases, which can lead to inconsistent and biased results [15–17]. To some extent, data from antibiotic-treated Sod1-Tg mice could demonstrate causal relationships but are scarce, and the number of commensal bacteria that have been investigated is limited [14].
The Mendelian randomization (MR) approach is a widely used genetic epidemiological method for assessing causal associations between risk factors and disease by exploiting genetic variants as instrumental variables (IVs) for exposure [18–20]. This approach is less likely to be affected by the confounding or reverse causation bias that exists in observational findings.
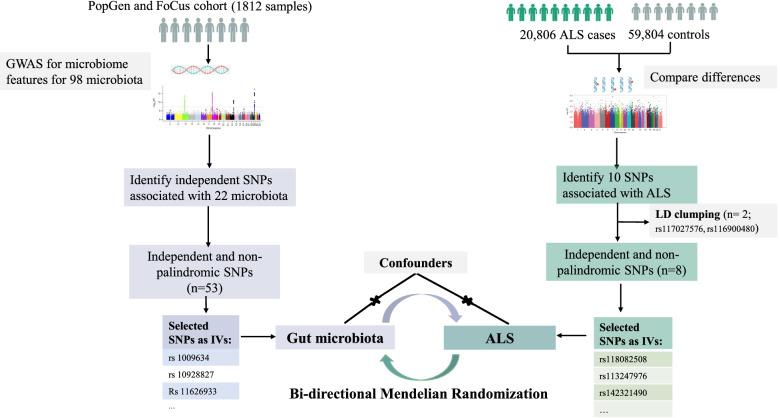
Therefore, to decipher the potentially mutual relationship between the gut microbiota and ALS, we used a bidirectional two-sample MR approach to examine the associations between the gut microbiome and ALS (Fig. 2). Notably, the gut microbiome is remote from the disease site of ALS, it is suggested that a potential systemic influx of microbiome-regulated metabolites may affect the susceptibility of motor neurons in ALS. We also estimated the effects of potential metabolites on ALS in MR design.
Fig. 2.
Schematic representation of the study
Methods
The detailed approach of selection of IVs for exposures, genome-wide association study (GWAS) summary statistics for ALS, and MR analysis were previously described [21]. The MR approach we used was based on the following three assumptions: 1) genetic variants (single nucleotide polymorphisms (SNPs)) used as IVs are associated with exposures; 2) genetic variants are not associated with confounders; and 3) genetic variants influence the risk of outcomes only through interested exposures, not through other pathways [22] (Fig. 2). The IVs (F statistic > 10) for all the exposures were sufficiently informative [23].
Genetically predicted gut microbiota genera
Genetic instruments of the abundance of 98 genera of gut microbiota at the level of genome-wide significance (P < 5 × 10− 8) were obtained from available GWAS data of stool samples in humans [24]. As a result, independently significant SNPs were identified for 22 genera of the gut microbiota, but no significant genetic variants were found for the remaining 76 genera of the gut microbiota.
If an SNP was not available for an outcome, a highly correlated proxy SNP (r2 > 0.9) (https://ldlink.nci.nih.gov/) was used instead, if available. We checked the phenotypes of selected SNPs using comprehensive genotype-to-phenotype cross-references (GWAS Catalog [25]) and repeated the analysis with potentially pleiotropic SNPs excluded. We calculated SNP-specific F statistics as a quotient of squared SNP-genus association and its variance [26].
Genetically predicted gut microbial metabolites
A transsynaptic, glutaminergic, excitotoxic mechanism (the so-called dying-forward hypothesis) has been proposed as a pathophysiological biomarker in ALS [27]. We therefore used 18 potential blood metabolites that might have causal effects on the development of ALS, including a group of gamma-glutamyl amino acids [28]. The candidate metabolites were identified among 486 untargeted serum metabolites from Shin’s study [29]. A total of 7824 adult individuals from 2 European cohorts were included in the GWAS analysis. Metabolomics data were acquired based on nontargeted mass spectrometry analysis of human fasting serum [29].
For each of the metabolites, we selected SNPs that showed an association at P < 1 × 10− 5 as candidate IVs of the specific metabolite. Then, a clumping procedure was conducted with European 1000G as a reference panel to identify the independent variants, with a linkage disequilibrium threshold of r2 < 0.01 in a 500-kb window.
Genetically predicted ALS
We drew on summary statistics from the largest and most recent GWAS of ALS [30] patients who were defined as having been diagnosed with probable or definite ALS according to the El Escorial criteria (Brooks, 1994) by a neurologist specializing in ALS. This GWAS of ALS involving 20,806 patients and 59,804 controls of European ancestry identified 10 independent genome-wide significant SNPs at the level of P < 5 × 10− 8 [30].
Statistical analysis
For each direction of the potential relationship, we combined MR estimates using an inverse variance-weighted method (IVW) meta-analysis, which essentially translates to a weighted regression of SNP outcome effects on SNP exposure effects where the intercept is constrained to zero. The IV assumptions can be biased if instrument SNPs show horizontal pleiotropy, influencing the outcome through causal pathways other than exposure [22]. Therefore, other established MR methods, including weighted, weighted median mode, and MR Egger regression, were also applied to confirm the IVW results (number of SNPs ≥3) because their estimates are known to be relatively robust to horizontal pleiotropy, although at the cost of reduced statistical power [31]. MR Egger regression allows the intercept to be freely estimated as an indicator of average pleiotropic bias. Effect estimates are reported in β values when the outcome is continuous (i.e., the abundance of each genus of gut microbiota) and are converted to ORs when the outcome is dichotomous (i.e., ALS status).
To assess the robustness of significant results, we conducted further tests for horizontal pleiotropy using meta-analytic methods to detect heterogeneous outcomes, including leave-1-SNP-out analyses and the MR Egger intercept test of deviation from the null [32].
The analyses were performed with R version 3.1.1 (R foundation) and Stata version 11.2 (Stata Corp, College Station, TX). All human research was approved by the relevant institutional review boards and conducted according to the Declaration of Helsinki. Ethical approval was obtained from relevant Research Ethics Committees and from the review boards of Peking University Third Hospital.
Results
Effects of genetically predicted gut microbiota on ALS
The resulting lists of instrument SNPs for each genus of gut microbiota are given in Table 1.
Table 1.
Characteristics of selected SNPs for core gut microbiota
| Core gut microbiota | SNP | Chr. | Locus start | Locus end | A1 | A2 | P | Beta | SE | β-div P | Nearest gene | Genes in locus | Variance explained |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs1009634 | 12 | 4,779,313 | 4,900,344 | G | A | 7.12E-09 | −1.3 | 0.23 | 0.93 | AKAP3 | NDUFA9, GALNT8, RP11-234B24.2 | 0.0183 |
| Bacilli class | rs10928827 | 2 | 129,426,740 | 129,473,850 | G | A | 1.02E-08 | −0.2 | 0.04 | 0.19 | HS6ST1 | – | 0.0180 |
| Lactobacillales order | rs10928827 | 2 | 129,426,740 | 129,473,850 | G | A | 4.19E-09 | −0.2 | 0.04 | 0.19 | HS6ST2 | – | 0.0189 |
| Unclassifed Erysipelotrichaceae | rs11626933 | 14 | 90,681,816 | 90,810,659 | G | A | 1.83E-08 | −0.2 | 0.04 | 0.55 | C14orf102 | C14orf102 | 0.0173 |
| Marinilabiliaceae family | rs11724031 | 4 | 77,441,448 | 77,467,405 | G | A | 2.44E−10 | −1 | 0.15 | 0.68 | SHROOM3 | SHROOM3 | 0.0219 |
| Unclassifed Marinilabiliaceae | rs11724031 | 4 | 77,441,448 | 77,467,405 | G | A | 2.44E-10 | -1 | 0.15 | 0.68 | SHROOM3 | SHROOM3 | 0.0219 |
| Erysipelotrichaceae family | rs11877825 | 18 | 10,566,345 | 10,595,758 | G | T | 2.82E-11 | −0.3 | 0.04 | 0.34 | NAPG | – | 0.0242 |
| Erysipelotrichia class | rs11877825 | 18 | 10,566,345 | 10,595,758 | G | T | 2.82E-11 | −0.3 | 0.04 | 0.34 | NAPG | – | 0.0242 |
| Erysipelotrichales order | rs11877825 | 18 | 10,566,345 | 10,595,758 | G | T | 2.82E-11 | −0.3 | 0.04 | 0.34 | NAPG | – | 0.0242 |
| Marinilabiliaceae family | rs11915634 | 3 | 1,452,602 | 1,517,331 | T | C | 2.99E-10 | −1.3 | 0.21 | 0.14 | CNTN6 | – | 0.0217 |
| Unclassifed Marinilabiliaceae | rs11915634 | 3 | 1,452,602 | 1,517,331 | T | C | 2.99E-10 | −1.3 | 0.21 | 0.14 | CNTN7 | – | 0.0217 |
| OTU10032 unclassifed Enterobacteriaceae | rs12149695 | 16 | 27,205,994 | 27,293,886 | A | T | 1.82E-09 | 0.61 | 0.10 | 0.23 | FLJ21408 | NSMCE1, FLJ21408, KDM8 | 0.0198 |
| OTU15355 Dialister Species-level OTU | rs12442649 | 15 | 37,968,393 | 38,035,538 | G | A | 3.72E-08 | −1.5 | 0.27 | 0.85 | TMCO5A | – | 0.0166 |
| EscherichiaShigella | rs13096731 | 3 | 58,014,818 | 58,089,851 | A | G | 2.55E-08 | −0.4 | 0.08 | 0.12 | FLNB | FLNB | 0.0170 |
| OTU10032 unclassifed Enterobacteriaceae | rs13276516 | 8 | 56,589,428 | 56,596,140 | A | G | 5.54E-09 | − 0.6 | 0.10 | 0.41 | TGS1 | – | 0.0186 |
| Lactobacillales order | rs1362404 | 16 | 51,955,443 | 52,017,380 | T | G | 1.56E-08 | 0.23 | 0.04 | 7.50E-05 | TOX3 | – | 0.0175 |
| Bacilli class | rs148330122 | 19 | 38,497,288 | 38,631,252 | C | T | 1.32E-09 | −0.5 | 0.08 | 0.18 | SIPA1L3 | SIPA1L3 | 0.0201 |
| OTU10032 unclassifed Enterobacteriaceae | rs17085775 | 9 | 71,165,704 | 71,167,878 | C | T | 2.06E-08 | −1 | 0.18 | 0.54 | C9orf71 | – | 0.0172 |
| Erysipelotrichaceae family | rs17421787 | 4 | 131,293,675 | 131,512,291 | C | G | 3.60E-08 | −0.3 | 0.05 | 0.16 | RP11-22 J15.1 | – | 0.0166 |
| Erysipelotrichales order | rs17421787 | 4 | 131,293,675 | 131,512,291 | C | G | 3.60E-08 | −0.3 | 0.05 | 0.16 | RP11-22 J15.2 | – | 0.0166 |
| Erysipelotrichia class | rs17421787 | 4 | 131,293,675 | 131,512,291 | C | G | 3.60E-08 | −0.3 | 0.05 | 0.16 | RP11-22 J15.3 | – | 0.0166 |
| Unclassifed Acidaminococcaceae | rs17661843 | 7 | 48,381,902 | 48,433,594 | T | C | 3.72E-14 | −1.4 | 0.18 | 0.26 | ABCA13 | ABCA13 | 0.0312 |
| Bacilli class | rs2071199 | 20 | 43,030,809 | 43,037,422 | T | C | 1.24E-08 | −0.3 | 0.06 | 0.58 | HNF4A–AS1 | HNF4A | 0.0178 |
| OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs2318350 | 8 | 139,889,972 | 139,942,500 | T | C | 3.65E-09 | −1.2 | 0.19 | 0.95 | COL22A1 | COL22A1 | 0.0190 |
| OTU10032 unclassifed Enterobacteriaceae | rs249733 | 5 | 141,877,862 | 141,911,748 | T | C | 4.74E-10 | −0.7 | 0.10 | 0.68 | SPRY4 | – | 0.0212 |
| Actinobacteria class | rs34613612 | 21 | 32,184,901 | 32,204,347 | C | G | 6.34E-10 | 0.25 | 0.04 | 9.87E-03 | KRTAP8–1 | KRTAP8–1 | 0.0209 |
| Actinobacteria phylum | rs34613612 | 21 | 32,184,901 | 32,204,347 | C | G | 6.34E-10 | 0.25 | 0.04 | 9.87E-03 | KRTAP8–1 | KRTAP8–1 | 0.0209 |
| Enterobacteriaceae family | rs35275482 | 15 | 60,027,987 | 60,128,040 | C | A | 3.72E-11 | −0.5 | 0.08 | 0.06 | BNIP2 | – | 0.0239 |
| Enterobacteriales order | rs35275482 | 15 | 60,027,987 | 60,128,040 | C | A | 3.72E-11 | −0.5 | 0.08 | 0.06 | BNIP3 | – | 0.0239 |
| OTU10032 unclassifed Enterobacteriaceae Species-level OTU | rs3925158 | 3 | 38,161,078 | 38,313,688 | C | G | 6.29E-09 | −1 | 0.17 | 0.78 | SLC22A13 | SLC22A13, MYD88, DLEC1, ACAA1, OXSR1 | 0.0185 |
| Gammaproteobacteria class | rs4621152 | 2 | 217,857,450 | 217,924,261 | C | T | 1.40E-08 | −0.3 | 0.05 | 0.79 | AC007557.1 | – | 0.0176 |
| Blautia genus | rs4669413 | 2 | 9,801,744 | 9,818,596 | T | C | 1.20E-08 | −0.2 | 0.03 | 0.75 | RP11–521D12.1 | – | 0.0178 |
| Bacilli class | rs479105 | 12 | 3,357,596 | 3,393,503 | T | C | 1.21E-08 | −0.2 | 0.04 | 0.48 | PRMT8 | – | 0.0178 |
| Unclassifed Acidaminococcaceae | rs56006724 | 2 | 228,486,044 | 228,523,585 | A | G | 6.35E-10 | −0.9 | 0.14 | 0.93 | C2orf83 | C2orf83 | 0.0209 |
| Lactobacillales order | rs59042687 | 3 | 95,359,287 | 95,823,523 | T | G | 6.22E-09 | −0.2 | 0.04 | 0.02 | LINC00879 | – | 0.0185 |
| OTU13305 Fecalibacterium Species-level OTU | rs597205 | 1 | 112,379,026 | 112,415,622 | T | C | 7.68E-09 | −0.6 | 0.11 | 0.85 | C1orf183 | C1orf183 | 0.0183 |
| Lactobacillales order | rs62295801 | 3 | 162,444,724 | 163,236,170 | G | T | 5.32E-10 | −0.3 | 0.04 | 0.21 | LINC01192 | LINC01192 | 0.0211 |
| Lactobacillales order | rs7083345 | 10 | 7,020,329 | 7,044,987 | T | C | 2.89E-09 | 0.24 | 0.04 | 0.02 | RP11-554I8.2 | – | 0.0199 |
| Bacilli class | rs7083345 | 10 | 7,020,329 | 7,044,987 | T | C | 3.38E-10 | 0.25 | 0.04 | 0.02 | RP11-554I8.2 | – | 0.0209 |
| Lactobacillales order | rs7113056 | 11 | 122,091,502 | 122,154,110 | C | T | 1.72E-13 | −0.5 | 0.07 | 0.07 | RP11-166D19.1 | – | 0.0296 |
| Unclassifed Acidaminococcaceae | rs75036654 | 1 | 37,717,219 | 37,780,821 | C | T | 4.94E-10 | −1.4 | 0.22 | 0.06 | LINC01137 | – | 0.0212 |
| Bacilli class | rs7646786 | 3 | 185,729,634 | 185,742,372 | T | C | 2.29E-08 | −0.2 | 0.04 | 0.5 | LOC344887 | – | 0.0171 |
| Unclassifed Porphyromonadaceae | rs7656342 | 4 | 9,721,358 | 9,895,176 | A | G | 2.80E-09 | 0.39 | 0.07 | 0.22 | DRD5 | SLC2A9, DRD5 | 0.0193 |
| Blautia genus | rs79387448 | 2 | 103,099,953 | 103,239,356 | C | T | 7.68E-11 | −0.3 | 0.05 | 0.66 | SLC9A2 | SLC9A2 | 0.0231 |
| Unclassifed Porphyromonadaceae | rs9291879 | 5 | 66,515,817 | 66,550,855 | C | T | 3.51E-09 | −0.6 | 0.10 | 0.08 | CD180 | – | 0.0191 |
| Gammaproteobacteria class | rs9300430 | 13 | 98,269,478 | 98,306,405 | C | T | 1.30E-09 | −0.6 | 0.10 | 0.12 | RAP2A | – | 0.0201 |
| Proteobacteria phylum | rs9323326 | 14 | 58,476,448 | 58,532,709 | A | G | 8.76E-10 | −0.2 | 0.03 | 0.02 | SLC35F4 | C14orf37 | 0.0206 |
| Unclassifed Enterobacteriaceae | rs938295 | 1 | 16,087,164 | 16,124,985 | C | T | 2.34E-08 | −0.5 | 0.09 | 0.76 | FBLIM1 | FBLIM1 | 0.0171 |
| Unclassifed Marinilabiliaceae | rs9831278 | 3 | 98,879,786 | 98,942,990 | C | T | 2.53E-08 | −1.2 | 0.21 | 0.49 | LINC00973 | – | 0.0170 |
| Marinilabiliaceae family | rs9831278 | 3 | 98,879,786 | 98,942,990 | C | T | 2.53E-08 | −1.2 | 0.21 | 0.49 | LINC00974 | – | 0.0170 |
| Unclassifed Acidaminococcaceae | rs986417 | 14 | 60,787,269 | 61,122,040 | C | T | 2.63E-09 | −1.4 | 0.23 | 0.47 | SIX6 | SIX6, C14orf39, SIX1 | 0.0194 |
| Marinilabiliaceae family | rs9996716 | 4 | 77,441,448 | 77,467,405 | G | A | 5.58E-09 | −0.7 | 0.12 | 0.2 | SHROOM3 | SHROOM3 | 0.0186 |
| Unclassifed Marinila-biliaceae | rs9996716 | 4 | 77,441,448 | 77,467,405 | G | A | 5.58E-09 | −0.7 | 0.12 | 0.2 | SHROOM3 | SHROOM3 | 0.0186 |
The 53 associations with bacterial abundance are grouped into 40 loci on the basis of LD. SNP single-nucleotide polymorphisms, Chr chromosome, A1 effect allele, A2 non-effect allele, P meta-analysis P value for A1, Beta meta-analysis coeffcient for A1, SE standard error, β-div P P value for association with β diversity.
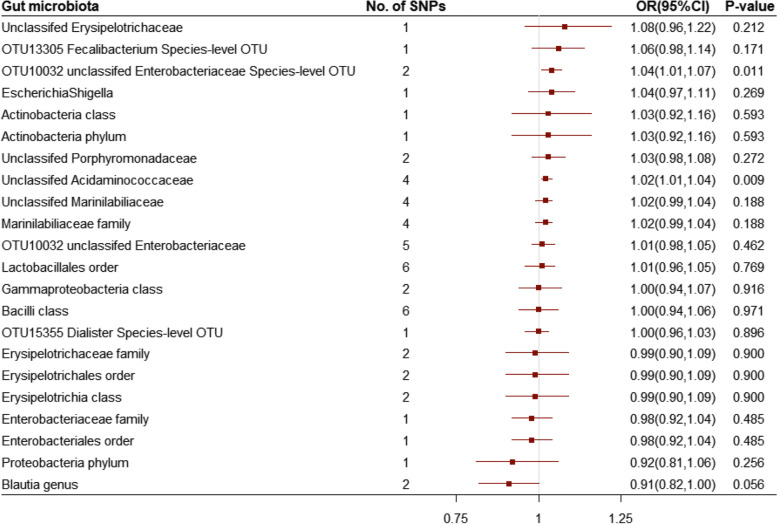
On the basis of 2 independent SNPs, OTU10032 unclassified Enterobacteriaceae was associated with a higher risk of ALS (per relative abundance: OR, 1.04; 95% CI, 1.01–1.07; P = 0.011) (Fig. 3, eFigure 1). Additionally, on the basis of 4 uncorrelated SNPs, unclassified Acidaminococcaceae was associated with a higher risk of ALS (per relative abundance: OR, 1.02; 95% CI, 1.01–1.04; P = 0.009) (Fig. 3, eFigure 2). The independent SNPs for two genera with r 2 = 0 are listed in eTable 1. Sensitivity analysis for the two candidate genera using the MR-Egger and weighted-median methods produced similar estimates, and no horizontal pleiotropy or outliers were observed (eTable 2–3).
Fig. 3.
Odds ratio for association of genetically predicted gut microbiota with amyotrophic lateral sclerosis. OR: odds ratio; CI: confidence internal. OR (95% CI) means risk of amyotrophic lateral sclerosis per 1-allele increase in single nucleotide polymorphisms related to greater abundance of gut microbiota
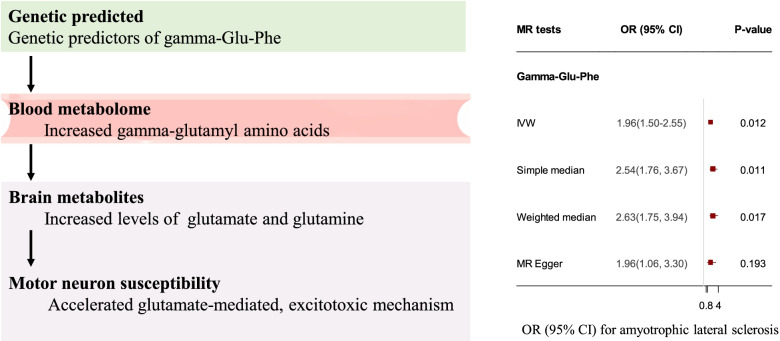
Importantly, gamma-glutamyl amino acids showed potential deleterious effects on the risk of ALS. Gamma-glutamylphenylalanine (Gamma-Glu-Phe), a peptide in the gamma-glutamyl pathway, showed a significantly increased risk of ALS (genetically predicted per 1-standard deviation (SD) increase in the level of Gamma-Glu-Phe: OR, 1.96; 95% CI, 1.50–2.55; P = 0.012) (Fig. 4). In addition, two metabolites, 1-arachidonoyl-GPI and 3-methyl-2-oxobutyrate, were also estimated to be associated with a higher risk of ALS, with a genetically predicted per 1-SD increase in levels: OR, 1.64; 95% CI, 1.37–1.96; P = 0.005 for 1-arachidonoyl-GPI and OR, 2.78; 95% CI, 1.98–3.90; P = 0.003 for 3-methyl-2-oxobutyrate. The results also showed that a genetically predicted increase in the levels of 4-acetamidobutanoate may lower the risk of ALS (per 1-SD increase in levels: OR, 0.49; 95% CI, 0.36–0.66; P = 0.020). Sensitivity analysis for the metabolites using the MR-Egger and weighted-median methods produced similar estimates, and no horizontal pleiotropy or outliers were observed (eTable 4).
Fig. 4.
Causal effect of microbiome-dependent metabolites on the risk of ALS. OR: odds ratio; CI: confidence internal
Effects of genetically predicted ALS on gut microbiota
On the basis of 2 independent SNPs, genetically predicted ALS was associated with an increase in the relative abundance of OTU4607_Sutterella (per 1-unit higher log odds: β, 2.23; 95% CI, 1.27–3.18; P = 0.020). The risk of ALS on each OTU4607_Sutterella-related SNP effect was estimated and is shown in eFigure 3. Similarly, on the basis of 2 independent SNPs, genetically predicted ALS was associated with an increase in the relative abundance of Lactobacillales_order (per 1-unit higher log odds: β, 0.51; 95% CI, 0.09–0.94; P = 0.019). Single Lactobacillales_ORDER-related SNP effect was estimated and is shown in eFigure 4. The estimated effects of ALS on the microbiota of each genus are listed in eTable 5. No horizontal pleiotropy or outliers were observed.
Discussion
This study assessed the causal effects of potential microbiome modulators of human ALS and added intriguing evidence implicating some genera of the gut microbiome in modifying susceptibility to ALS. These genera attenuate ALS risk through gamma-glutamyl-related metabolite levels, supporting that a trans-synaptic, glutaminergic, excitotoxic mechanism could provide a pathogenic basis for ALS. These results may contribute to designing microbiome- and microbiome-dependent metabolite interventions in future ALS clinical trials. We further provide genetic evidence that the pathophysiology of ALS is associated with an altered relative abundance of the microbiota, strengthening the bidirectional relationship between the gut microbiota and ALS.
The gut microbiome is a source of these potentially disease-modifying bioactive metabolites and has recently been suggested to contribute to the pathogenesis of neurological disorders [33, 34]. The family Enterobacteriaceae includes over 30 genera and 120 species of Enterobacteriaceae, but more than 95% of clinically significant strains fall into 10 genera and fewer than 25 species. All members of the Enterobacteriaceae family ferment glucose with acid production and nitrogen metabolism. Glutamine synthetases (GSs) are key enzymes of nitrogen metabolism, and their activity is modulated by nitrogen repression [35]. Acidaminococcaceae, an important glutamate-fermenting family of microbes, produces ammonia as the major end product through glutamate fermentation [36]. It is possible that alterations in the microbiomes of the two genera lead to changes in gamma-glutamyl-related metabolite levels. Circulating bioactive gamma-glutamyl-related metabolite levels produced by the gut microbiome permeate the blood–brain barrier, after which they can play important roles in the pathogenesis of brain-related diseases [37].
Our study showed that higher ALS susceptibility was associated with a higher relative abundance of OTU4607_Sutterella and Lactobacillales_ORDER. In previous studies, gut dysbiosis, particularly reduced levels of butyrate-producing bacteria and higher E. coli and Enterobacteria abundance, was also found in ALS mice and ALS patients [9, 38]. Furthermore, butyrate and short-chain fatty acids (SCFAs) produced by gut microbiota have been proposed as promising potential therapeutic agents affecting ALS progression [39, 40]. However, unravelling the interplay between the gut microbiome and ALS is imperative, and more direct evidence and results are needed to clarify how the gut microbiota improves or aggravates ALS.
There are several strengths in the present study, including the assessment of genera of gut microbiota and promising metabolites in relation to ALS, the use of data from the largest GWASs to date and bidirectional MR design. This design technique minimizes confounding by known and unknown factors and avoids reverse causation. In addition, consistent results from several sensitivity analyses, including the use of weighted mode, weighted median, and MR-Egger methods, indicate the robustness of our findings. Several limitations merit consideration. First, we used a limited number of gut microbiota and ALS SNPs as IVs; we cannot exclude that our findings might have been affected by weak instrument bias, although all genetic instruments were associated with exposure (F-statistic > 10). Second, another potential source of bias in MR analyses is population stratification. We reduced this bias because the dataset for gut microbiota, metabolites and ALS was restricted to individuals of European ancestry. Replication with functionally relevant genetic prediction of gut microbiota is warranted given the substantial difference in gut microbiota composition among different populations. Finally, 16S rRNA gene sequencing only permits resolution from the genus to the phylum level rather than at a more specific level, resulting in biased results if some specific species contributed to ALS.
Conclusion
Our findings provide novel evidence supporting the bidirectional relationship between the gut microbiota and ALS and highlight that a transsynaptic, glutaminergic, excitotoxic mechanism could provide a pathogenic basis for ALS. These results may contribute to designing microbiome- and microbiome-dependent metabolite interventions in future ALS clinical trials.
Supplementary Information
Additional file 1: eTable 1. Correlation Matrixes for Single Nucleotide Polymorphisms Predicting (a)OTU10032 unclassifed Enterobacteriaceae Species-level OUT and (b)Unclassifed Acidaminococcaceae From SNiPA Pairwise LD.
Additional file 2: eTable 2. Associations between gut microbiota and amyotrophic lateral sclerosis in sensitivity analyses.
Additional file 3: eTable 3. Associations between gut microbiota and amyotrophic lateral sclerosis in in a leave-one-out approach.
Additional file 4: eTable 4. Associations between gut microbiota and amyotrophic lateral sclerosis in sensitivity analyses.
Additional file 5: eTable 5. Effect estimates for association of genetically predicted amyotrophic lateral sclerosis with gut microbiota using inverse variance weighting method.
Additional file 6: eFigure 1. Association of genetically predicted OTU10032 unclassified Enterobacteriaceae species-level OTU with amyotrophic lateral sclerosis. Squares represent the odd ratios of amyotrophic lateral sclerosisper 1-allele increase in single nucleotide polymorphisms related to greater abundance of OTU10032 unclassified Enterobacteriaceae Species-level OTU; horizontal lines represent 95% confidence intervals (CIs); diamond represent the overall odds ratio with its 95% CI.
Additional file 7: eFigure 2. Association of genetically predicted unclassified Acidaminococcaceae with amyotrophic lateral sclerosis. Squares represent the odd ratios of amyotrophic lateral sclerosisper 1-allele increase in single nucleotide polymorphisms related to greater abundance of unclassified Acidaminococcaceae; horizontal lines represent 95% confidence intervals (CIs); diamond represent the overall odds ratio with its 95% CI.
Additional file 8: eFigure 3. Association of genetically predicted amyotrophic lateral sclerosis with OTU4607 Sutterella. Squares represent the effect estimates of the relative abundance ofOTU4607 Sutterellaper 1-unit higher log odds of amyotrophic lateral sclerosis; horizontal lines represent 95% confidence intervals (CIs); diamond represent the effect size with its 95% CI.
Additional file 9: eFigure 4. Association of genetically predicted amyotrophic lateral sclerosis with Lactobacillalesorder. Squares represent the effect estimates of the relative abundance ofLactobacillalesorderper 1-unit higher log odds of amyotrophic lateral sclerosis; horizontal lines represent 95% confidence intervals (CIs); diamond represent the effect size with its 95% CI.
Acknowledgments
We are grateful that the summary data for all the gut microbiome, blood metabolite and AVS consortium studies are publicly available, and we thank all the investigators and participants who contributed to those studies.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- MR
Mendelian randomization
- AM
Akkermansia muciniphila
- IVs
Instrumental variables
- IVW
Inverse variance-weighted method
- SNPs
Single nucleotide polymorphisms
- GWASs
Genome-wide association studies
- OR
Odds ratio
- GS
Glutamine synthetases
Authors’ contributions
D.F. and T.H. conceived and designed the study. L.Z., Z.Z. and G.Z. contributed to the acquisition and analysis of data. L.Z. wrote the manuscript. D.F. and T.H. reviewed and edited the manuscript. All authors read and approved the manuscript.
Authors’ information
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (81030019, 81873784) and Beijing Key Laboratory of Biomarker and Translational Research in Neurodegenerative disorders.
Availability of data and materials
Data-set used in the current study is publicly available and not anonymized in this study. The summary datasets analyzed during the current study are available in the http://als.umassmed.edu/#sumstats. The rest datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All human research was approved by the relevant institutional review boards and conducted according to the Declaration of Helsinki. Ethical approval was obtained from relevant Research Ethics Committees and from the review boards of Peking University Third Hospital (IRB 00006761). Informed consent was obtained from all subjects and/or their legal guardian(s).
Consent for publication
The work is original research article, is not under consideration by another journal, and has not been published previously. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Linjing Zhang, Zhenhuang Zhuang, Tao Huang and Dongsheng Fan contributed equally to this work.
Contributor Information
Linjing Zhang, Email: zhanlinjing@bjmu.edu.cn.
Zhenhuang Zhuang, Email: 1510306133@pku.edu.cn.
Gan Zhang, Email: nmzhanggan@yahoo.com.
Tao Huang, Email: huangtao@bjmu.edu.cn.
Dongsheng Fan, Email: dsfan2010@aliyun.com.
References
- 1.Turner MR, Hardiman O, Benatar M, Brooks BR, Chio A, de Carvalho M, Ince PG, Lin C, Miller RG, Mitsumoto H, et al. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. Lancet Neurol. 2011;10:253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- 3.Dupuis L, Pradat PF, Ludolph AC, Loeffler JP. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 4.Al Bander Z, Nitert MD, Mousa A, Naderpoor N. The gut microbiota and inflammation: an overview. Int J Environ Res Public Health. 2020;17. [DOI] [PMC free article] [PubMed]
- 5.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, Herrema H. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. 2020;11:571731. doi: 10.3389/fimmu.2020.571731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 8.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Yi J, Zhang YG, Zhou J, Sun J. Leaky intestine and impaired microbiome in an amyotrophic lateral sclerosis mouse model. Physiol Rep. 2015;3. [DOI] [PMC free article] [PubMed]
- 10.Zhang YG, Wu S, Yi J, Xia Y, Jin D, Zhou J, Sun J. Target intestinal microbiota to alleviate disease progression in amyotrophic lateral sclerosis. Clin Ther. 2017;39:322–336. doi: 10.1016/j.clinthera.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang X, Wang X, Yang S, Meng F, Wang X, Wei H, Chen T. Evaluation of the microbial diversity in amyotrophic lateral sclerosis using high-throughput sequencing. Front Microbiol. 2016;7:1479. doi: 10.3389/fmicb.2016.01479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzini L, Mogna L, De Marchi F, Amoruso A, Pane M, Aloisio I, Cionci NB, Gaggia F, Lucenti A, Bersano E, et al. Potential Role of Gut Microbiota in ALS Pathogenesis and Possible Novel Therapeutic Strategies. J Clin Gastroenterol. 2018;52(Suppl 1):S68–s70. doi: 10.1097/MCG.0000000000001042. [DOI] [PubMed] [Google Scholar]
- 13.Brenner D, Hiergeist A, Adis C, Mayer B, Gessner A, Ludolph AC, Weishaupt JH. The fecal microbiome of ALS patients. Neurobiol Aging. 2018;61:132–137. doi: 10.1016/j.neurobiolaging.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 15.Sattar N, Preiss D. Reverse causality in cardiovascular epidemiological research: more common than imagined? Circulation. 2017;135:2369–2372. doi: 10.1161/CIRCULATIONAHA.117.028307. [DOI] [PubMed] [Google Scholar]
- 16.Phillips AN, Smith GD. How independent are "independent" effects? Relative risk estimation when correlated exposures are measured imprecisely. J Clin Epidemiol. 1991;44:1223–1231. doi: 10.1016/0895-4356(91)90155-3. [DOI] [PubMed] [Google Scholar]
- 17.Fewell Z, Davey Smith G, Sterne JA. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol. 2007;166:646–655. doi: 10.1093/aje/kwm165. [DOI] [PubMed] [Google Scholar]
- 18.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 19.Ding M, Huang T, Bergholdt HK, Nordestgaard BG, Ellervik C, Qi L. Dairy consumption, systolic blood pressure, and risk of hypertension: Mendelian randomization study. Bmj. 2017;356:j1000. doi: 10.1136/bmj.j1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng T, Smith CE, Li C, Huang T. Childhood BMI and adult type 2 diabetes, coronary artery diseases, chronic kidney disease, and Cardiometabolic traits: a Mendelian randomization analysis. Diabetes Care. 2018;41:1089–1096. doi: 10.2337/dc17-2141. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Tang L, Huang T, Fan D. Life course adiposity and amyotrophic lateral sclerosis: a Mendelian randomization study. Ann Neurol. 2020;87:434–441. doi: 10.1002/ana.25671. [DOI] [PubMed] [Google Scholar]
- 22.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen FA, Rühlemann MC, Szymczak S, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkasir R, Li J, Li X, Jin M, Zhu B. Human gut microbiota: the links with dementia development. Protein Cell. 2017;8:90–102. doi: 10.1007/s13238-016-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geevasinga N, Menon P, Özdinler PH, Kiernan MC, Vucic S. Pathophysiological and diagnostic implications of cortical dysfunction in ALS. Nat Rev Neurol. 2016;12:651–661. doi: 10.1038/nrneurol.2016.140. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Lv X, Du H, Wu D, Wang M. Causal effects of serum metabolites on amyotrophic lateral sclerosis: a Mendelian randomization study. Prog Neuro-Psychopharmacol Biol Psychiatry. 2020;97:109771. doi: 10.1016/j.pnpbp.2019.109771. [DOI] [PubMed] [Google Scholar]
- 29.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron. 2018;97:1268–1283. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–r208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, Zink EM, Casey CP, Taylor BC, Lane CJ, et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell. 2019;177:1600–1618. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen P, Weinstock A, Wagner A, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gough EK, Stephens DA, Moodie EE, Prendergast AJ, Stoltzfus RJ, Humphrey JH, Manges AR. Linear growth faltering in infants is associated with Acidaminococcus sp and community-level changes in the gut microbiota. Microbiome. 2015;3:24. doi: 10.1186/s40168-015-0089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Mazzini L, Mogna L, De Marchi F, Amoruso A, Pane M, Aloisio I, Cionci NB, Gaggìa F, Lucenti A, Bersano E, et al: Potential Role of Gut Microbiota in ALS Pathogenesis and Possible Novel Therapeutic Strategies. J Clin Gastroenterol. 2018;52 Suppl 1:S68-s70, Proceedings from the 9th Probiotics, Prebiotics and New Foods, Nutraceuticals and Botanicals for Nutrition & Human and Microbiota Health Meeting, held in Rome, Italy from September 10 to 12, 2017. [DOI] [PubMed]
- 39.Erber AC, Cetin H, Berry D, Schernhammer ES. The role of gut microbiota, butyrate and proton pump inhibitors in amyotrophic lateral sclerosis: a systematic review. Int J Neurosci. 2020;130:727–735. doi: 10.1080/00207454.2019.1702549. [DOI] [PubMed] [Google Scholar]
- 40.Ghadge GD, Kay BK, Drigotas C, Roos RP. Single chain variable fragment antibodies directed against SOD1 ameliorate disease in mutant SOD1 transgenic mice. Neurobiol Dis. 2019;121:131–137. doi: 10.1016/j.nbd.2018.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: eTable 1. Correlation Matrixes for Single Nucleotide Polymorphisms Predicting (a)OTU10032 unclassifed Enterobacteriaceae Species-level OUT and (b)Unclassifed Acidaminococcaceae From SNiPA Pairwise LD.
Additional file 2: eTable 2. Associations between gut microbiota and amyotrophic lateral sclerosis in sensitivity analyses.
Additional file 3: eTable 3. Associations between gut microbiota and amyotrophic lateral sclerosis in in a leave-one-out approach.
Additional file 4: eTable 4. Associations between gut microbiota and amyotrophic lateral sclerosis in sensitivity analyses.
Additional file 5: eTable 5. Effect estimates for association of genetically predicted amyotrophic lateral sclerosis with gut microbiota using inverse variance weighting method.
Additional file 6: eFigure 1. Association of genetically predicted OTU10032 unclassified Enterobacteriaceae species-level OTU with amyotrophic lateral sclerosis. Squares represent the odd ratios of amyotrophic lateral sclerosisper 1-allele increase in single nucleotide polymorphisms related to greater abundance of OTU10032 unclassified Enterobacteriaceae Species-level OTU; horizontal lines represent 95% confidence intervals (CIs); diamond represent the overall odds ratio with its 95% CI.
Additional file 7: eFigure 2. Association of genetically predicted unclassified Acidaminococcaceae with amyotrophic lateral sclerosis. Squares represent the odd ratios of amyotrophic lateral sclerosisper 1-allele increase in single nucleotide polymorphisms related to greater abundance of unclassified Acidaminococcaceae; horizontal lines represent 95% confidence intervals (CIs); diamond represent the overall odds ratio with its 95% CI.
Additional file 8: eFigure 3. Association of genetically predicted amyotrophic lateral sclerosis with OTU4607 Sutterella. Squares represent the effect estimates of the relative abundance ofOTU4607 Sutterellaper 1-unit higher log odds of amyotrophic lateral sclerosis; horizontal lines represent 95% confidence intervals (CIs); diamond represent the effect size with its 95% CI.
Additional file 9: eFigure 4. Association of genetically predicted amyotrophic lateral sclerosis with Lactobacillalesorder. Squares represent the effect estimates of the relative abundance ofLactobacillalesorderper 1-unit higher log odds of amyotrophic lateral sclerosis; horizontal lines represent 95% confidence intervals (CIs); diamond represent the effect size with its 95% CI.
Data Availability Statement
Data-set used in the current study is publicly available and not anonymized in this study. The summary datasets analyzed during the current study are available in the http://als.umassmed.edu/#sumstats. The rest datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.