Abstract
The coronavirus disease 2019 (COVID-19) is still causing a wide range of infections and deaths due to the high variability of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Therefore, it is necessary to establish a reliable and convenient pseudovirus-based neutralization assay to develop drug targeted variants of SARS-CoV-2. Based on the HIV-1 backbone, we generated a high titer luciferase (Luc)-expressing pseudovirus packaging system. Three dominant S mutant substitution pseudovirus were also established and identified compared to wide type in hACE2-overexpressing HEK-293T cells (293T-ACE2 cells). Compared to serine protease inhibitor camostat mesylate, the cysteine protease inhibitor E-64d could significantly block all SARS-CoV-2 mutant S pseudovirus infection in 293T-ACE2 cells. Furthermore, the neutralization ability of two antibodies targeted receptor-binding domain (RBD) of SARS-CoV-2 spike protein (S) was evaluated, which showed different inhibition dose–effect curves among four types of S pseudovirus. Overall, we developed a pseudovirus-based neutralization assay for SARS-CoV-2, which would be readily adapted to SARS-CoV-2 variants for evaluating antibodies.
Keywords: COVID-19, SARS-CoV-2 variants, Pseudovirus, Neutralizing antibody, RBD
1. Introduction
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) is caused by SARS-CoV-2, which resulted in hundreds of millions of infections and millions of deaths [1]. The SARS-CoV-2, like other severe coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), can cause severe respiratory syndromes in humans, like fever, cough, and shortness of breath [2], [3]. Therefore, the quality of human life seriously declined, and the economic and social situation was severely disrupted by the pandemic worldwide.
As a membrane-enveloped virus, the spike (S) glycoprotein is expressed on the membrane of SARS-CoV-2. It binds to the human angiotensin-converting enzyme 2 (hACE2) receptor to mediate membrane fusion and virus entry into host cells [4], [5], [6]. The S protein is a homotrimer, which each monomer consists of a receptor-binding domain (RBD) subunit S1 and a membrane-fusion subunit S2 [7], [8]. The full-length S protein needs to be activated by cellular protease-mediated cleavage to S1 and S2, which the cysteine proteases cathepsin B and L (CatB/L) or trans-membrane protease serine 2 (TMPRSS2) is responsible [9], [10], [11]. Thus, the antibodies or inhibitors targeting S protein or cellular proteases could efficiently block viral entry [9]. However, the efficacy evaluation of antibodies or inhibitors with SARS-CoV-2 live virus has to be conducted under biosafety level 3 (BSL-3) conditions, limiting the development of SARS-CoV-2 drugs and therapeutics.
This study constructed the SARS-CoV-2 S pseudotyped virus based on an HIV-1 lentiviral packaging system incorporating luciferase reporter; thus, the S-mediated viral entry can be conveniently measured via luciferase activity. Protease inhibitors and human RBD-specific mAbs could inhibit the SARS-CoV-2 S pseudotyped virus infection. We established reliable and safe measurements of the SARS-CoV-2 S pseudotyped virus infection system for entry inhibition and neutralization assays, which could be conducted under BSL-2 conditions.
2. Materials and methods
Anti-Flag M2 antibody, polyethylenimine (PEI), lipofectamine 3000, and Polyethylene Glycol (PEG) 8000 were purchased from Sigma-Aldrich (St Louis, MO, USA). Anti actin and ACE2 antibodies were purchased from Proteintech (Wuhan, China). HIV-1 Gag-p24 antibody was purchased from Sino Biological (Beijing, China). Polybrene was purchased from Yeasen (Shanghai, China). E-64d and camostat mesylate were purchased from MedChem Express (NJ, USA). The anti-RBD monoclonal antibodies against the SARS-CoV-2 S protein were kindly provided by Zhangjiang Bio (Shanghai, China).
2.1. Cell lines
HEK-293T and HuH7 cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Hyclone, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), at 37 °C in 5% CO2. In addition, HEK-293T cells transfected with human ACE2 (293T-ACE2) were cultured under the same conditions with the addition of puromycin (0.5 μg/mL) to the medium.
2.2. Plasmid constructs
The S gene from the SARS-CoV-2 (previously 2019-nCoV) strain Wuhan-Hu-1 (GenBank: MN908947) with a C-terminal 19 amino acid deletion was codon-optimized, synthesized, and cloned into the NotI and XbaI sites of the pcDNA3.1–3 × Flag-C vector (pc-S for short) by Sangon Biotech Inc. (Shanghai, China). The translated amino acid sequence was identical to QHD43416. The primers 5′-GGTCCTCTATCAAGGCGTCAACTGTACG-3′ and 5′-CGTACAGTTGACGCCTTGATAGAGGACC-3′ were used to generate the plasmid pcDNA3.1-SARS-CoV-2-S-D614G (pc-S-D614G) encoding a mutant S protein-containing mutation D614G. The primers 5′-GGTTTCCAACCTACATATGGAGTAGGGTATC-3′ and 5′-GATACCCTACTCCATATGTAGGTTGGAAACC-3′ were used to create the plasmid pcDNA3.1-SARS-CoV-2-S-N501Y (pc-S-N501Y) encoding a mutant S protein-containing mutation N501Y. The S gene of Delta variant (T19R, G142D, E156del, F157del, R158G, L452R, T478K, D614G, P681R, D950N) was based on the codon-optimized sequence of pc-S construct, synthesized by Sangon Biotech Inc. (Shanghai, China) and sub-cloned into the pcDNA3.1 vector. The constructed recombinant SARS-CoV-2 plasmids containing the wide-type (pc-S) and mutant S variants were confirmed by DNA sequencing. The pLVX-Luc construct was derived from pLVX-Puro by inserting a luciferase reporter gene into the XhoI and BamHI sites. The pLVX-ACE2 construct was derived from pLVX-Puro by inserting the human ACE2 gene (pcDNA3.1-ACE2-3 × Flag) into the XhoI and XbaI sites. The plasmids of pcDNA3.1-ACE2-3 × Flag, pLVX-Puro, pMD2.G, and psPAX2 were obtained from Youbio (Hunan, China). The plasmid pNL4-3 was obtained from Hedgehogbio (Shanghai, China).
2.3. Generation human ACE2 over-expressing cells
To produce the human ACE2 over-expressing lentivirus, HEK-293T cells were seeded at a density of 4 × 105 cells/mL in 5 mL in a T25 flask. When cells reached 80% confluency the following day, they were transfected with the following mixture of plasmids: 4 μg of pLVX-ACE2, 4 μg of psPAX2, 2 μg of pMD2.G using lipofectamine 3000 reagents. After 8 h, the medium was changed, and the supernatants were collected at 48 h after the medium change, filtered with 0.45 μm filters, concentrated with PEG 8000, and stored at −80 °C.
For a generation of 293T-ACE2 cells, 5 × 105 HEK-293T cells were seeded into a six-well plate one day before viral transduction. Cells were transduced with the concentrated human ACE2 over-expressing lentivirus in the presence of 5 μg/mL polybrene overnight. The virus-containing supernatant was wholly removed the next day, and cells were cultured in their regular growth media for 48 h. After puromycin selection, HEK-293T cells expressing wild-type ACE2 protein were confirmed with Western blot.
2.4. Western blot
Cells were collected and lysed in RIPA buffer with a protease inhibitor cocktail. Cell lysates were clarified by centrifugation for 15 min, 12,000 rpm at 4 °C, and heated at 97 °C for 5 min in SDS loading buffer. Protein samples were carried out in 8%–16% Tris-Glycine Gels (Life Technologies, Carlsbad, CA, USA), electrophoresed, and transferred onto PVDF membranes. The chemiluminescence detection was performed according to the manufacturer's ECL Western detection kit (Amersham Pharmacia Biotech Europe, Freiburg, Germany).
2.5. Production and purification of pseudotype particles
HEK-293T cells (6 × 106) were plated in a T75 culture plate and transfected the next day when they were about 60% confluent with a combination of the following plasmids: 10 µg of pLVX-Luc, 10 µg of psPAX2, and 5 µg of pc-S, pc-S-D614G, pc-S-N501Y, pc-S-Delta or pMD2.G as control using PEI transfection reagent following manufacturer's protocols. The supernatant containing SARS-CoV-2 pseudoviruses was harvested 48–72 h after transfection. The supernatants were centrifuged for 15 min at 1,500 × g and then filtered through a 0.45 μm syringe filter. For pseudovirus purification and concentration, the supernatant was mixed at a 1:4 (v/v) ratio with 25% PEG 8000 solution and incubated at 4 °C overnight. The next day, lentiviral particles were concentrated by centrifugation at 3,000 × g for 30 min. Supernatants were removed and pellets resuspended in serum-free DMEM, and stored at −80 °C.
2.6. Quantification of pseudotyped virus particles
The titers of the pseudoviruses were calculated by determining the concentrations of viral RNA genomes using quantitative RT-PCR with primers targeting Luc gene LTR (5′-AGCCGCCTAGCATTTCATCA-3′ and 5′-AAAGTCCCCAGCGGAAAGTC-3′). Before quantification, viral RNAs were extracted from 5 μL of concentrated pseudoviruses using the TIANamp Virus RNA Kit (QIANGEN, Cat# DP315-R) and served as a template for reverse transcription using the FastKing RT Kit (QIANGEN, Cat# KR116). Then, virus quantification by real-time PCR was performed using the UltraSYBR Mixture (CWBIO, Cat# CW2601), following the supplier's instructions. The known quantity of pLVX-Luc was used to generate standard curves, with the viral copy number calculated accordingly. Finally, the titers of the pseudoviruses were adjusted to the same titer (copies/mL) for the pseudovirus-based inhibition and neutralization experiments.
2.7. Pseudovirus-based inhibition and neutralization assays
For the inhibition assay, the 293T-ACE2 cells (3 × 104 cells/100 μL) were pretreated with 50 μL, about 3-fold serially diluted (1, 3, 10, 30, 100 ∼ ) the protease inhibitors E-64d or camostat mesylate 1 h before infection. Then, 50 μL pseudoviruses (a signal about 1000-fold above the background luciferase activity) were added into the cells. For the neutralization assay, 50 μL pseudoviruses were incubated with 50 μL, about 3-fold serially diluted, starting from 30 μg/mL antibody for one h at 37 °C. The 293T-ACE2 cells (3 × 104 cells/100 μL) were seeded into the pseudoviruses in 96-well plates. 293T-ACE2 cells were in the presence of 5 μg/mL polybrene for both assays. After 24 h of infection, the medium was changed with fresh culture to each well. The relative light units (RLU) were measured 72 h after infection, and the percent neutralization was calculated using GraphPad Prism 7 software (GraphPad Software, San Diego, CA, USA). The relative luciferase activity (%) could be calculated as follows: [(mean RLU from each sample (virus + antibody/inhibitor) − mean RLU from cell-only control)/(mean maximum RLU from virus control − mean RLU from cell-only control) × 100. Results of neutralization assays were plotted by normalization to samples where no antibody was used, and the half-maximal inhibitory concentration (IC50) was calculated using 4-parameter non-linear regression.
2.8. Statistical analysis
All data are expressed as mean ± SE GraphPad Prism 7 software was used to perform all statistical analyses and prepare graphs. Statistical significance was determined using Student's t-tests to compare the two groups for unpaired observations or a two-way ANOVA for multiple comparisons with the Bonferroni correction. A P value of less than 0.05 was considered statistically significant.
3. Results
3.1. Identification of SARS-CoV-2 S protein expression
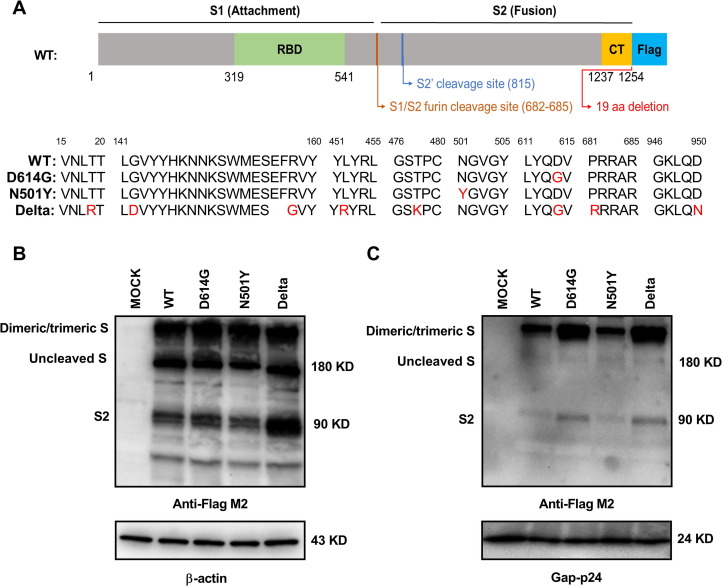
A codon-optimized cDNA encoding the S protein and 3 × FLAG tag was synthesized, with C-terminal 19 amino acid deletion to decrease endoplasmic reticulum (ER) location for facilitating incorporation of S protein into pseudovirons [12], [13]. We also generated S-D614G, S-N501Y, and S-Delta mutation plasmids based on pc-S (WT) plasmid, which exhibit the prevalent substitution mutations around 6,000 SARS-CoV-2 S variants globally [14], [15], [16]. The construction of SARS-CoV-2 S-WT protein expression plasmid and detailed information of different S mutant protein sequences are shown in Fig. 1 A. HEK-293T cells were transfected with SARS-CoV-2 S plasmids, and expression of SARS-CoV-2 S protein was determined by western blot with anti-FLAG M2 antibody. As shown in Fig. 1B, three major bands are detected. The band above 180 kD may indicate dimeric or trimeric S proteins. The 180 kD and 90 kD bands correspond to uncleaved S protein and cleaved S2 protein. However, the major size band of SARS-CoV-2 S proteins incorporation into pseudovirons showed above 180 kD (Fig. 1C), which indicated assembling dimeric or trimeric functional S proteins into the pseudovirons. And the S D614G and Delta of pseudovirons showed greater S expression than WT and N501Y, which is consistent with the D614G mutation increasing virion spike density.
Fig. 1.
Detection of SARS-CoV-2 spike (S) protein expression. A) Schematic illustration of the SARS-CoV-2 wt spike (S). Detail information of different S mutant protein sequences are shown below. The mutant amino acids were marked in red or removed. The RBD (receptor binding domain) is in subunit S1; the CT (cytoplasmic tail) are in subunit S2. The endoplasmic reticulum retrieval signals in the CT domain of S-FL were destroyed in S protein. B) Detection of SARS-CoV-2 S expression in HKE-293T cells by western blot using the anti-Flag monoclonal antibody. Actin served as a loading control. Cells were transfected with pc-S-WT, pc-S-D614G, pc-S-N501Y, pc-S-Delta plasmids or with an empty vector. C) Detection of SARS-CoV-2 S expression in pseudovirions by western blot using the anti-Flag monoclonal antibody. Gag p24 served as a loading control. Data in B) and C) are shown as one representative experiment of three independent experiments.
3.2. Comparison of the infectivity of SARS-CoV-2 S mutant pseudoviruses
Due to the highly infectious and pathogenic, the SARS-CoV-2 must be handled in a biosafety level 3 (BSL-3) laboratory. The lentiviral pseudovirus can be applied to replace the live virus for screening neutralizing antibodies [17]. We generated the SARS-CoV-2 pseudovirus based on HIV lentiviral system, which inserted the luciferase reporter gene into target cells. The effect of anti-SARS-CoV-2 entry inhibitors and neutralizing antibodies on S-mediated entry can be quantified by comparing the Luc signal intensity.
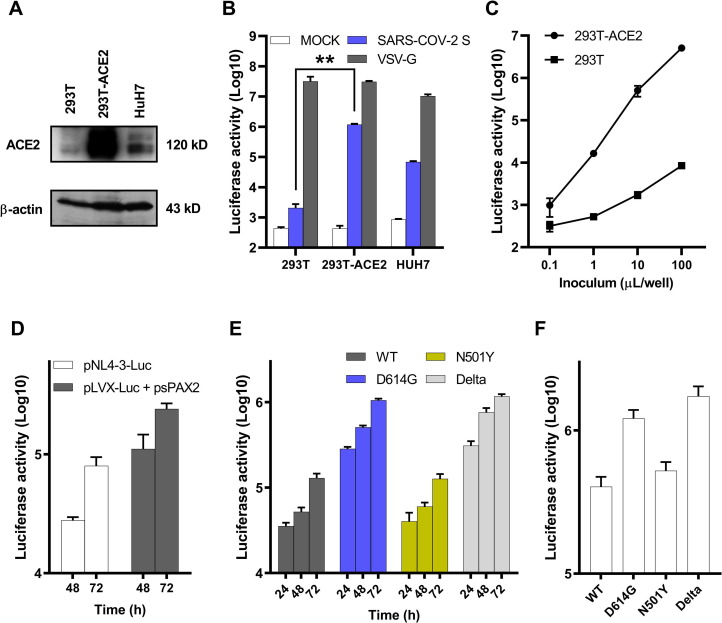
pLVX-Luc and psPAX2 were co-transfected with pc-S or pMD2.G respectively to package the SARS-CoV-2 S WT pseudovirus and VSV-G pseudovirus in HEK-293T cells. VSV-G pseudovirus was used as a control. Virus titers were determined by calculating the concentrations of viral RNA genomes using quantitative RT-PCR and measuring RLU based on a Luc assay [18]. The HEK-293T, established HEK-293T cells stably expressing human ACE2 (293T-ACE2) and HuH7 were used to test the correlation between ACE2 expression and pseudovirus susceptibility. Higher ACE2 expression was identified in 293T-ACE2 cells than HuH7 cells by western blot (Fig. 2 A). As shown in Fig. 2B, all the cells could be effectively transduced by VSV-G pseudovirus. However, 293T-ACE2 cells infected by SARS-CoV-2 pseudotyped viruses showed an approximately 400-fold increase in Luc activity compared to HEK-293T cells, a 20-fold increase in Luc activity compared to HuH7 cells, respectively, suggesting that the entry of SARS-CoV-2 S pseudovirus is highly dependent on ACE2 expression, and 293T-ACE2 cells are most efficiently transduced by SARS-CoV-2 pseudovirus. SARS-CoV-2 pseudotyped viruses infected ACE2-expressing HEK-293T cells, yielding a solid luminescence signal of up to 107 RLU, while unmanipulated parental cells were infected poorly (HEK-293T) (Fig. 2C). Next, we compared the titers of infectious particles of the established system with the classical pNL4-3 system. Both the two plasmids (pNL4-3-Luc) and three plasmids (pLVX-Luc + psPAX2) systems derived SARS-CoV-2 pseudotyped viruses yielded a strong luminescence signal after 48 h or 72 h infection in 293T-ACE2 cells (Fig. 2D). However, the titers of the three plasmids system were about 3–5 times of the pNL4-3 system.
Fig. 2.
Detection of SARS-CoV-2 S pseudotyped virus infectivity. A) Detection of ACE2 expression in 293T, 293T-ACE2 and HuH7 cells by western blot using the anti-ACE2 monoclonal antibody. Actin served as a loading control. B) Infectivity measurements of lentiviruses pseudotyped without (MOCK) or with vesicular stomatitis virus G (VSV-G) or SARS-CoV-2 S WT protein on the 293T, 293T-ACE2 and HuH7 cells after 72 h post inoculation. C) Infectivity measurements of lentiviruses pseudotyped with SARS-CoV-2 S WT protein on the 293T and 293T-ACE2 cells with the indicated volumes of concentrated pseudotyped viruses after 72 h post inoculation. D) Infectivity measurements of the two plasmids or three plasmids systems derived SARS-CoV-2 S WT pseudotyped viruses on the 293T-ACE2 cells after 48 h and 72 h post inoculation. E) Infectivity measurements of lentiviruses pseudotyped with SARS-CoV-2 S protein variants on the 293T-ACE2 cells after 24 h, 48 h and 72 h post inoculation. F) Infectivity measurements of lentiviruses pseudotyped with SARS-CoV-2 S protein variants with the same titer (copies/mL) on the 293T-ACE2 cells after 72 h post inoculation. Above the infectivity (luciferase activity) was quantified by measuring RLU following infection. Data in A) is shown as one representative experiment of three independent experiments. Data in B) C) D) E) and F) are shown as mean ± SE of three independent experiments.
To compare the viral entry efficiency meditated by S variants, we detected the Luc activity at different times post-infection with other S mutants. pLVX-Luc and psPAX2 were also co-transfected with pc-S, pc-S-D614G, pc-S-N501Y, pc-S-Delta, respectively, to package the SARS-CoV-2 S variants pseudovirus in HEK-293T cells. The highest transduction efficiency was observed in S-D614G and S-Delta pseudotyped virus (almost 1 × 106 RLU) at 72 h post-infection, approximately 8-fold higher than S-WT and S-N501Y pseudovirus (Fig. 2E). Furthermore, after normalizing to the same titer (copies/mL) of the S variants pseudovirus, S-D614G and S-Delta pseudotyped virus still yielded a stronger luminescence signal after 72 h infection, which was approximately 3-fold higher than S-WT and S-N501Y pseudovirus (Fig. 2F). As both S-D614G and S-Delta share the same D614G mutation, the higher luminescence signal suggested that the plasma membrane localization of S-D614G may facilitate lentivirus packaging and infection.
3.3. Pseudovirus-based inhibition and neutralization assays
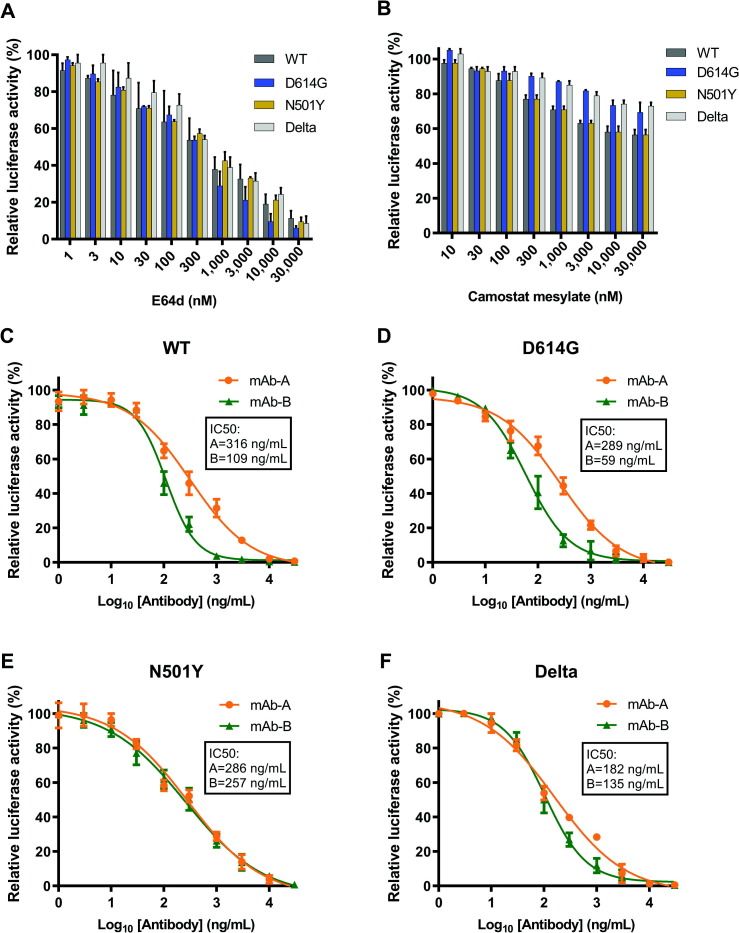
Since the protease-mediated proteolytic activation of S protein is required for coronavirus entry [19], we check the effect of the CatB/L protease inhibitor E-64d and TMPRSS2 inhibitor camostat mesylate on WT, D614G, N501Y, and Delta S protein pseudotyped lentiviral particles infectivity. As shown in Fig. 3 A, E-64d significantly inhibited these four pseudoviruses entry, and the IC50 was 0.34 μM for S-WT pseudovirus, 0.34 μM for S-D614G pseudovirus, 0.48 μM for S-N501Y pseudovirus, and 0.3 μM for S-Delta pseudovirus. However, the pseudoviruses were relatively insensitive to a TMPRSS2 inhibitor camostat mesylate in 293T-ACE2 cells (Fig. 3B). These results suggested that S-meditated viral entry into 293T-ACE2 cells is mainly endosomal cysteine proteases CatB/L dependent, as the 293T-ACE2 cells lack TMPRSS2 expression. Therefore, S-WT, S-D614G, S-N501Y, and S-Delta pseudovirus show similar sensitivity to the CatB/L inhibitor E-64d in 293T-ACE2 cells. Together, these data demonstrate that the pseudovirus-based inhibition assay could be an effective and convenient method to screen SARS-CoV-2 entry inhibitors.
Fig. 3.
Detection of entry inhibitors and antibodies against SARS-CoV-2 pseudovirus infection. A) E-64d blocked SARS-CoV-2 S variants pseudovirus entry. 293T-ACE2 cells were pre-incubated with E-64d (about 3-fold serially diluted) and subsequently inoculated with pseudovirions. RLU were detected after 72 h post-pseudovirus inoculation. B) Camostat mesylate slightly blocked SARS-CoV-2 S variants pseudovirus entry. 293T-ACE2 cells were pre-incubated with camostat mesylate (about 3-fold serially diluted) and subsequently inoculated with pseudovirions. RLU were detected after 72 h post-pseudovirus inoculation. Antibodies targeted RBD neutralized the SARS-CoV-2 S variants pseudovirus. The S WT C), D614G D), N501Y E) and Delta F) pseudovirus were pre-incubated with mAb-A or mAb-B (about 3-fold serially diluted), and subsequently inoculated with 293T-ACE2 cells. RLU were detected after 72 h post-pseudovirus inoculation. Data in A) B) C) D) E) and F) are shown as mean ± SE of three independent experiments.
Next, we tested the effect of two S RBD targeted antibodies on pseudotyped lentiviral particles infectivity. Both antibodies inhibited four types of pseudovirus in a concentration-dependent manner. Interestingly, each antibody showed similar IC50 to S-WT and S-D614G (S-WT: 316 ng/mL (mAb-A), 109 ng/mL (mAb-B); S-D614G: 289 ng/mL (mAb-A) and 59 ng/mL (mAb-B)), respectively. The IC50 of antibody to S-Delta are 182 ng/mL (mAb-A) and 135 ng/mL (mAb-B), respectively. However, the antibody B showed great loss of inhibition potential on S-N501Y (S- N501Y: 286 ng/mL (mAb-A), 257 ng/mL (mAb-B)) (Fig. 3C, D, E, F). These data indicated that decreased infectivity of the pseudotyped SARS-CoV-2 in the presence of these antibodies is associated with neutralizing activity of increased concentration on SARS-CoV-2 entry. Our data indicated that a safe and convenient assay had been established to test the entry inhibitors and neutralizing activity of antibodies against SARS-CoV-2.
4. Discussion
Under the highly infectious and operational risks of live SARS-CoV-2, the neutralization test needs to be done in a BSL-3 laboratory, limiting the availability of drugs against SARS-CoV-2. Our established S pseudovirus based on HIV-1 lentiviral packaging system could mimic the live virus by sharing the same S envelope protein and is much safer with a single round of replication and without other viral components, which can be performed in a single round of replication BSL-2 laboratory [20]. A live virus neutralization assay usually takes four days [21]. With a luciferase gene as the reporter, our neutralization assay takes only 48–72 h, costing less time. Furthermore, our essay could be performed in a 96-well plate with higher throughput than the conventional plaque-reduction neutralization test (PRNT), which must be performed in a 6-well plate. Therefore, our pseudovirus neutralization assay is safer, more time-saving, and has higher throughput than the PRNT. Although many researchers had successfully established an HIV-based pseudovirus system for SARS-CoV-2, they usually used specific plasmids which are not convenient to acquire in some areas [22], [23], [24], [25], [26]. The plasmids used in our system are constructed based on standard commercial plasmids that are easy to obtain, which benefit the pseudovirus research in these areas. Significantly, the low titer of pseudovirus usually limits their more comprehensive applications. According to our comparison, the titer of our pseudovirus is usually 3–5 times higher than the pseudovirus packaged by the classical pNL4-3 system, which contributes to the improvement of the concentration and purification efficiency. Our pseudovirus packaging system is much easier to establish and more efficient than the classical packaging system.
SARS-CoV-2 S plays a crucial role in viral infection and pathogenesis. Antibodies target S can block SARS-CoV-2 infectivity in vitro and in vivo. During budding, lentiviral particles (LVP) can incorporate cell membrane proteins in the viral envelope [27]. We created SARS-CoV-2 pseudovirus by replacing membrane proteins with S protein in LVP. However, the cytoplasmic tails of S protein contain an endoplasmic reticulum retrieval signal (ERRS), which is thought to accumulate S protein at the SARS-CoV-2 budding site to facilitate S protein incorporation into virions [13]. Considering the entire length of S protein is not in favour of lentivirus pseudotyping, we deleted the last 19 amino acids, which may increase the levels of S on the cell surface, resulting in a higher titer of pseudovirus. The D614G S mutant had become the dominant circulating strain globally by replacing the wide-type S strain, which replicates faster and is more transmissible [14]. Researchers demonstrated that the D614G mutation enhanced the replication of the mutated virus in the lung cells with higher viral load in the respiratory secretions [28], [29], [30]. Furthermore, structural analysis elucidated that the D614G mutation resulted in a more open ACE2 binding site in the RBD region, indicating that it enhanced the ability to attach to the ACE2, ultimately induced higher infectivity without influencing the neutralization potency of antibodies targeting the S protein RBD [31]. This function is consistent with the neutralization abilities of our two antibodies targeted RBD was not influenced by comparing WT with D614G S pseudovirus. After the emergence of the D614G substitution, the N501Y S mutant substitution occurred convergently in the United Kingdom (UK), South Africa, and Brazil, which is a significant determinant responsible for being more transmissible by improving the affinity of the viral spike protein for cellular receptors [32]. The antibodies targeting different regions of RBD showed a distinct change of neutralization potential against N501Y compared to WT [33]. The mAb B showed a loss of neutralization ability for N501Y compared to WT, indicating that mAb B may recognize the N501 amino acid. Currently, as the predominant strain of the virus, the Delta variant contained several mutations in S protein compared to the WT strain, including T19R, G142D, Δ156–157, R158G, L452R, T478K, D614G, P681R, and D950N. Lizhou Zhang showed D614G mutation indeed increases pseudovirus (PV) infectivity without influencing neutralization sensitivity [34], which is consistent with infectivity measurements results of D614G and Delta. Recent reports have indicated that Delta variant is resistant to certain antibodies, especially targeting RBD of SARS-CoV-2 S [35], [36], [37]. Our antibodies are engineered by fusing the antibodies which recognized the common RBD region of SARS-CoV, MERS-CoV and SARS-CoV-2 S to the ACE2 binding region, which can both competitive bind to receptor with coronavirus and directly bind to coronavirus to effenciently block the virus infection. Our data showed slightly increased sensitivity of SARS-CoV-2 variant Delta to mAb-A neutralization, which may contribute to broader applications.
In conclusion, we generate a replication-incompetent LVP carrying the SARS-CoV-2 S WT or mutant protein to study SARS-CoV-2 in a safer, more convenient, and higher throughput way. Furthermore, this pseudovirus neutralization assay could be applied to test the inhibitors that target infection progress by SARS-CoV-2 and neutralize antibodies, which could benefit the availability of inhibitors and antibodies against SARS-CoV-2.
Acknowledgments
Acknowledgements
This work was supported by grants 2020-SKT-14 and 2021-YKT-01 from the Shanghai Institute for Food and Drug Control, LX-2021-06 from the Shanghai Drug Administration, and 19DZ2294600 from the Shanghai Science and Technology Committee.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Author contributions
Sheng Wang: Conceptualization, Funding Acquisition, Methodology, Investigation, Writing – Original Draft. Lizhen Liu: Formal Analysis. Can Wang: Investigation, Funding Acquisition. Ziqiang Wang: Methodology. Xuhua Duan: Resources. Gang Chen: Supervision. Hu Zhou: Writing – Review & Editing. Hong Shao: Writing – Review & Editing, Project Administration.
References
- 1.Singh S.P., Pritam M., Pandey B., Yadav T.P. Microstructure, pathophysiology, and potential therapeutics of COVID-19: A comprehensive review. J. Med. Virol. 2021;93(1):275–299. doi: 10.1002/jmv.26254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohanty S.K., Satapathy A., Naidu M.M., Mukhopadhyay S., Sharma S., Barton L.M., Stroberg E., Duval E.J., Pradhan D., Tzankov A., et al. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19) - anatomic pathology perspective on current knowledge. Diagn. Pathol. 2020;15(1):103. doi: 10.1186/s13000-020-01017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chathappady House N.N., Palissery S., Sebastian H. Corona Viruses: A Review on SARS, MERS and COVID-19. Microbiol. Insights. 2021;14 doi: 10.1177/11786361211002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Luo Y., Yan B., Wang Y.Y., Xiao G.F., Shi Z.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laporte M., Raeymaekers V., Van Berwaer R., Vandeput J., Marchand-Casas I., Thibaut H.-J., Van Looveren D., Martens K., Hoffmann M., Maes P., Pöhlmann S., Naesens L., Stevaert A., Fouchier R.A.M. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021;17(4):e1009500. doi: 10.1371/journal.ppat.1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Sanchez-David R.Y., Giacca M., Davidson A.D., Matthews D.A., Barclay W.S., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 12.Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J. Virol. 2020;94(21):e01062–e1120. doi: 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou X., Liu Y., Lei X., Li P., Mi D., Chen J., Hu K., Jin Q., Wang J., Qian Z., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Smith N., Tucker R.M., Wang D., Wyles M.D., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennerstrand J., Palanisamy N. Global prevalence of adaptive and prolonged infections’ mutations in the receptor-binding domain of the SARS-CoV-2 spike protein. Viruses. 2021;13(10) doi: 10.3390/v13101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughan A. Delta to dominate world. New Sci. 2021;250(3341):9. doi: 10.1016/S0262-4079(21)01121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., Liu Q., Huang W., Li X., Wang Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018;28(1):e1963. doi: 10.1002/rmv.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mournetas V., Pereira S.M., Fernig D.G., Murray P. A descriptive guide for absolute quantification of produced shRNA pseudotyped lentiviral particles by real-time PCR. J. Biol. Methods. 2016;3(4):e55. doi: 10.14440/jbm.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson N.M., Alvarado A.F., Moffatt T.N., Edavettal J.M., Swaminathan T.A., Braun S.E. HIV-based lentiviral vectors: origin and sequence differences. Mol. Ther. Methods Clin. Dev. 2021;21:451–465. doi: 10.1016/j.omtm.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe K.T., Li Z., Samson R., Samavarchi-Tehrani P., Valcourt E.J., Wood H., Budylowski P., Dupuis A.P., Girardin R.C., Rathod B., et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5(19) doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang R., Huang B., A R., Li W., Wang W., Deng Y., Tan W. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosafety and Health. 2020;2(4):226–231. doi: 10.1016/j.bsheal.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neerukonda S.N., Vassell R., Herrup R., Liu S., Wang T., Takeda K., Yang Y.e., Lin T.-L., Wang W., Weiss C.D., Mantis N.J. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS ONE. 2021;16(3) doi: 10.1371/journal.pone.0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7(4):551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Robbiani D.F., Nussenzweig M.C., Rice C.M., Hatziioannou T., Bieniasz P.D., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217(11) doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5):513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen D.H., Hildreth J.E. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74(7):3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Kelly J.N., Wentworth D.E., Thiel V., Beer M., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- 30.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Widen S.G., Menachery V.D., Shi P.Y., Weaver S.C. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021 doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.J., Jang U.S., Soh S.M., Lee J.Y., Lee H.R. The Impact on Infectivity and Neutralization Efficiency of SARS-CoV-2 Lineage B.1.351 Pseudovirus. Viruses. 2021;13(4):633. doi: 10.3390/v13040633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Puech J., Mouquet H., Bruel T., Simon-Loriere E., Rey F.A., Schwartz O., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Arora P., Sidarovich A., Moldenhauer A.-S., Winkler M.S., et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3) doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]