Abstract
Background
Observational studies and previous Mendelian randomization (MR) studies have shown that genetically low 25-hydroxyvitamin D (25OHD) levels are associated with a high susceptibility to multiple sclerosis (MS). The present MR study aims to update the causal estimates for the effects of 25OHD levels on MS risk.
Methods
To date, the largest genome-wide association study (GWAS) for serum 25OHD (n = 401,460) and MS (14,498 MS cases and 24,091 controls) was used to assess the effect of serum 25OHD levels on MS. All participants were of European ancestry. The MR-egger_intercept test and Cochran’s Q statistic were used to determine the pleiotropy and the heterogeneity, respectively. MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode, and weighted mode methods were used to evaluate the causal association of serum 25OHD levels with MS. Finally, the effect of a single 25OHD SNP (single nucleotide polymorphism) on MS was used to test the SNP bias.
Results
One hundred and fifteen newly identified serum 25OHD genetic variants were extracted from a large-scale serum 25OHD GWAS dataset. The 20 most effective and independent 25OHD genetic instrumental variables were extracted from the MS GWAS summary statistics. Pleiotropy analysis suggested no significant pleiotropic variant among the 20 selected 25OHD genetic instrument variants in MS GWAS datasets. As serum levels of 25OHD based on genetic changes increased, the risk of MS decreased using MR-egger (Beta = − 0.940, p = 0.001; OR = 0.391), weighted median (Beta = − 0.835, p = 0.000; OR = 0.434), IVW (Beta = − 0.781, p = 0.000; OR = 0.458), simple mode (Beta = − 1.484, p = 0.016; OR = 0.227), and weighted mode (Beta = − 0.913, p = 0.000; OR = 0.401). Our results were robust, with no obvious bias based on investigating the single 25OHD SNP on MS.
Conclusions
Our analysis suggested a causal association between genetically increased serum 25OHD levels and reduced MS in the European population.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-021-03205-6.
Keywords: Multiple sclerosis, 25OHD, Genome-wide association study, Mendelian randomization, Single nucleotide polymorphism
Background
Multiple sclerosis (MS) is an inflammatory autoimmune disease in the central nervous system [1–4]. The prevalence of MS is rising globally [1]. In the past decades, better understanding of the disease mechanisms of MS has led to several disease-modifying therapy development [5]. However, current therapeutic treatments for MS still remain disappointing [5]. It is necessary to explore the pathogenic mechanisms driving MS [5]. It is known that MS is mediated by the interaction between genetic and environmental factors and well-powered genome-wide association studies (GWAS) have been explored to investigate the genetic risk factors in MS [6].
The active form of vitamin D, 1,25-Dihydroxyvitamin D3 (1,25(OH)2D3), suppresses autoimmune diseases such as MS by reducing the production of proinflammatory cytokines such as IFN (interferon)-γ, IL (interleukin)-2, and IL-17, as well as enhancing the secretion of anti-inflammatory cytokines such as IL-4 and IL-10 [7–11]. A recent retrospective case–control study showed that the effect of vitamin D3 on MS susceptibility was not mediated by regulatory T cells (Tregs) [12]. Increased evidences suggest that decreased vitamin D levels is associated with an abnormal immune response in MS [13, 14]. Low vitamin D levels have been shown to correlate with disease activity in various autoimmune disorders and neurological diseases including MS [13–19]. Numerous epidemiological studies have strongly suggested that vitamin D insufficiency contributes to MS risk [20]. One SNP, rs10766197 CYP2R1 which regulates serum 25-OH-vitamin D3 (25OHD) levels, is reported to relate with MS risk [21, 22]. Thus, hypovitaminosis D was considered as a possible risk factor for MS, with multiple decades of research on the association of vitamin D status and MS [23].
Mendelian randomization (MR) studies have been widely used to assess the causal link between an exposure and an outcome by an analytical method using genetic variants as instrumental variables [13, 24–26]. Four previous MR studies used three or four serum 25OHD genetic instrumental variables (IVs) and suggested a link between predicted serum 25OHD and the risk of MS [27–30]. MR studies have many limitations, including low statistical power. Thus, to improve statistical power, 20 serum 25OHD genetic IVs from the largest GWAS for serum 25OHD (n = 401,460) were used to update the effect of serum 25OHD levels on MS risk.
Methods
Ethics approval and consent to participate
Our study was approved by the Ethics Committee of Beijing Institute of Brain Disorders in Capital Medical University. This article contains human participants collected by several studies to report the large-scale GWAS for the serum 25OHD [31] and MS [32]. All participants gave informed consent in all the corresponding original studies, as described in the Methods.
Study design
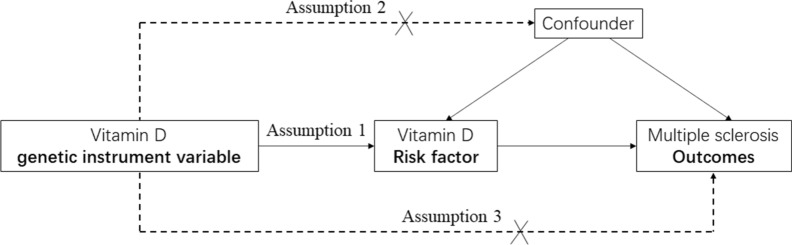
Three principal assumptions were prerequisites in the MR study [24, 25] and are described in Fig. 1. Assumption 1 is that genetic instrument variants are reliably associated with serum 25OHD levels. Assumption 2 is that genetic instrument variants should undoubtedly not be associated with any confounders. Assumption 3 is that genetic instrument variants firmly influence the risk of MS through the 25OHD and not through other pathways.
Fig. 1.
Design flow chart for the MR study. MR assumptions: assumption 1, 2 and 3. Solid line represents direct causal effects that 25OHD genetic instrument variants are reliably associated with 25OHD levels and influence the risk of MS through the 25OHD in assumption 1. The dotted line represents that 25OHD genetic instrument variants are not associated with any measured or unmeasured confounders and do not influence the risk of MS through other pathways in assumption 2 and 3, respectively
Genetic instrument variants for serum 25OHD
Sixty-nine independent 25OHD genetic variant loci were identified using imputed genotypes from 401,460 white British UK Biobank participants with available serum 25OHD levels [31]. The TwoSampleMR R package was used to perform a two-sample MR study. One hundred and fifteen independent 25OHD genetic variants with each trait at p < 5 × 10−8 are demonstrated in Additional file 1: Table S1 and were selected as “suggestive” instruments in our MR studies, as described in recent studies [24, 25]. To obtain a more accurate effect size estimate with these variants, the following rules was used to remove some variants. (1) Variants with a linkage disequilibrium (LD) (r2 > 0.05) were removed by using LDlink (https://ldlink.nci.nih.gov/?tab=ldmatrix, CEU). (2) Palindromic variants resulting in potential strand ambiguity were removed. (3) To ensure unconfounded instruments only affected MS via the relevant exposure, variants associated with possible exposure-outcome confounders (e.g. age, smoking, and socioeconomic position) were also removed. Finally, 20 effective and independent 25OHD-associated single nucleotide polymorphisms (SNPs) were selected as the potential IVs and are shown in Table 2.
Table 2.
Association of 20 vitamin D genetic variants in MS GWAS dataset
| SNP | Serum 25OHD GWAS | MS GWAS | ||||
|---|---|---|---|---|---|---|
| EA | NEA | EAF | Beta | SE | p val | |
| rs8063565 | C | G | 0.733 | − 0.002 | 0.019 | 0.928 |
| rs11542462 | A | G | 0.130 | 0.026 | 0.025 | 0.292 |
| rs964184 | C | G | 0.134 | − 0.014 | 0.024 | 0.570 |
| rs1260326 | C | T | 0.601 | 0.005 | 0.017 | 0.749 |
| rs1800588 | T | C | 0.219 | − 0.005 | 0.020 | 0.786 |
| rs10908469 | C | A | 0.278 | − 0.021 | 0.018 | 0.262 |
| rs1532085 | G | A | 0.607 | − 0.006 | 0.017 | 0.717 |
| rs2131925 | G | T | 0.318 | 0.005 | 0.020 | 0.805 |
| rs7528419 | A | G | 0.784 | 0.025 | 0.020 | 0.225 |
| rs804281 | G | A | 0.576 | 0.028 | 0.017 | 0.098 |
| rs532436 | A | G | 0.210 | 0.042 | 0.020 | 0.034 |
| rs10822145 | T | C | 0.472 | − 0.014 | 0.016 | 0.398 |
| rs1660818 | A | G | 0.317 | 0.000 | 0.018 | 0.999 |
| rs2276360 | C | G | 0.785 | − 0.105 | 0.018 | 0.000 |
| rs12317268 | G | A | 0.162 | 0.037 | 0.022 | 0.084 |
| rs11182428 | C | T | 0.514 | 0.017 | 0.017 | 0.302 |
| rs61003750 | C | G | 0.300 | 0.029 | 0.018 | 0.122 |
| rs142158911 | A | G | 0.120 | − 0.071 | 0.026 | 0.006 |
| rs62115743 | T | C | 0.077 | − 0.056 | 0.031 | 0.071 |
| rs1841850 | C | A | 0.114 | − 0.051 | 0.025 | 0.040 |
SNP single-nucleotide polymorphism, EA effect allele, NEA non-effect allele, EAF effect allele frequency; Beta is the regression coefficient based on the vitamin D raising effect allele, SE standard error
MS GWAS dataset
The MS GWAS dataset is from the International Multiple Sclerosis Genetics Consortium (IMSGC) and consists of 14,498 MS subject and 24,091 healthy control individuals of European ancestry [32]. The TwoSampleMR R package was used to perform a two-sample MR study. Twenty effective and independent 25OHD genetic variants were extracted from MS GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ieu-a-1025/) by using functions “extract_outcome_data” and “harmonise_data”. Table 1 provides the sample profiles for the MS GWAS datasets.
Table 1.
The sample information in the MS GWAS datasets
| Population | Sex | MS cases | Control | Number of SNPs |
|---|---|---|---|---|
| European | Males and females | 14,498 | 24,091 | 156,632 |
Pleiotropy test
The MR-egger_intercept test was used to determine the pleiotropy test [33]. The TwoSampleMR R package was used to perform the MR-egger_intercept test. The MR-egger_intercept test was performed using function “mr_pleiotropy_test”. P < 0.05 is the level of statistical significance.
Heterogeneity test
Cochran’s Q statistic was used to determine the heterogeneity test [34, 35]. The TwoSampleMR R package was used to perform Cochran’s Q statistic. The heterogeneity test was performed by using function “mr_heterogeneity”. P < 0.05 is the level of statistical significance.
MR analysis
The TwoSampleMR R package was used to perform the MR analysis. The MR analysis was performed by using function “mr”. We selected five MR analysis methods including MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode, weighted mode [33, 36, 37]. The effect size (beta) and 95% confidence interval (CI) correspond to 1 standard deviation (SD) in 25OHD levels. P < 0.05 is the level of statistical significance.
Single SNP effect analysis
The TwoSampleMR R package was used to analyze the effect of serum single 25OHD SNP on MS. This analysis was performed by using functions “mr_singlesnp” and “mr_leaveoneout”. Forest plot were prepared by using functions “mr_forest_plot” and “mr_leaveoneout_plot”.
Results
Serum 25OHD genetic instrument variants in MG GWAS dataset
To explore the effect of serum 25OHD genetic instrument variants on MS, we used the MS GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ieu-a-1025/), described in Table 1, to extract 20 effective and independent 25OHD genetic variants from the MS GWAS dataset by using functions “extract_outcome_data” and “harmonise_data”. Table 2 showed the detailed information about 20 25OHD genetic instrument variants in the MS GWAS dataset.
Pleiotropy analysis
MR-egger_intercept test was performed to determine the pleiotropy test by using function “mr_pleiotropy_test”. Table 3 shows detailed information about the pleiotropy analysis of 20 25OHD genetic instrument variants in MG GWAS dataset. The statistical P = 0.409 suggests that there is no significant pleiotropic variant among the selected 20 25OHD genetic instrument variants in the MS GWAS datasets. Thus, we established the assumption that genetic instrument variants should not be associated with any confounders. Therefore, all these 20 25OHD genetic variants could be used as the effective instrumental variables in this MR analysis.
Table 3.
Pleiotropy test of 20 serum 25OHD genetic variants in MS GWAS dataset
| GWAS dataset | MR-Egger intercept | ||
|---|---|---|---|
| Egger_intercept | SE | p-val | |
| MS | 0.007 | 0.008 | 0.409 |
SE standard error. p < 0.05 is set as the significant threshold. p-val = 0.409 represents no significant pleiotropy
Heterogeneity test
Cochran’s Q statistic was performed to test the heterogeneity by using function “mr_heterogeneity”. Table 4 shows detailed information about the heterogeneity analysis of 20 25OHD genetic instrument variants in the MS GWAS dataset. Both MR-egger and inverse variance weighted methods demonstrated the statistical P = 0.04. P < 0.05 suggests significant heterogeneity among the 20 25OHD genetic instrument variants in MG GWAS datasets. Therefore, we mainly use the random effect model to estimate the MR effect size.
Table 4.
Heterogeneity test of 20 serum 25OHD genetic variants in MS GWAS dataset
| Method | MR-Egger intercept | ||
|---|---|---|---|
| Q | Q_df | Q_pval | |
| MR Egger | 29.446 | 18 | 0.043 |
| Inverse variance weighted | 30.614 | 19 | 0.044 |
p < 0.05 is set as the significant threshold. Q_pval = 0.04 represents significant heterogeneity
MR analysis
We found that as 25OHD levels increased due to genetic differences, the risk of MS decreased. We used MR-egger (Beta = − 0.940, p = 0.001; OR = 0.391), weighted median (Beta = − 0.835, p = 0.000; OR = 0.434), IVW (Beta = − 0.781, p = 0.000; OR = 0.458), simple mode (Beta = − 1.484, p = 0.016; OR = 0.227), and weighted mode (Beta = − 0.913, p = 0.000; OR = 0.401) (Table 5). The individual MR estimates demonstrate that as the effect of a single SNP on 25OHD increased, the suppressive effect of a single SNP on MS increased, as determined using five MR analysis methods including MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode, and weighted mode (Fig. 2). Collectively, our data suggest causal association of increased 25OHD levels from greater genetic variation with decreased risk of MS.
Table 5.
The causal association of serum 25OHD levels with MS
| Method | nsnp | Beta | SE | p val | OR | OR_lci95 | OR_uci95 |
|---|---|---|---|---|---|---|---|
| MR Egger | 20 | − 0.940 | 0.251 | 0.001 | 0.391 | 0.239 | 0.639 |
| Weighted median | 20 | − 0.835 | 0.162 | 0.000 | 0.434 | 0.316 | 0.596 |
| IVW | 20 | − 0.781 | 0.165 | 0.000 | 0.458 | 0.331 | 0.633 |
| Simple mode | 20 | − 1.484 | 0.560 | 0.016 | 0.227 | 0.076 | 0.679 |
| Weighted mode | 20 | − 0.913 | 0.165 | 0.000 | 0.401 | 0.290 | 0.554 |
IVW Inverse variance weighted. IVW was based on multiplicative random effects. nsnp the number of single-nucleotide polymorphism. Beta: the regression coefficient based on 25OHD raising effect allele. SE: standard error. p < 0.05 represents the causal association of serum 25OHD levels with MS. OR Odds ratio. OR_lci95 Lower limit of 95% confidence interval for OR. OR_uci95 Upper limit of 95% confidence interval for OR
Fig. 2.
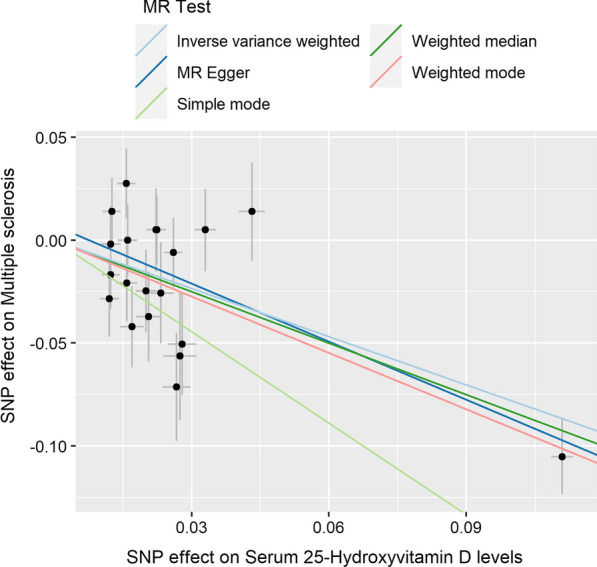
Individual estimates about the causal effect of serum 25OHD levels on MS. The x-axis shows the SNP (single nucleotide polymorphism) effect and SE (standard error) on each of the 20 serum 25OHD SNPs from 25OHD GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90000615/). The y-axis shows the SNP effect and SE on multiple sclerosis (MS) from MS GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ieu-a-1025/). The regression lines for the MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode, weighted mode method are shown
Single SNP effect analysis
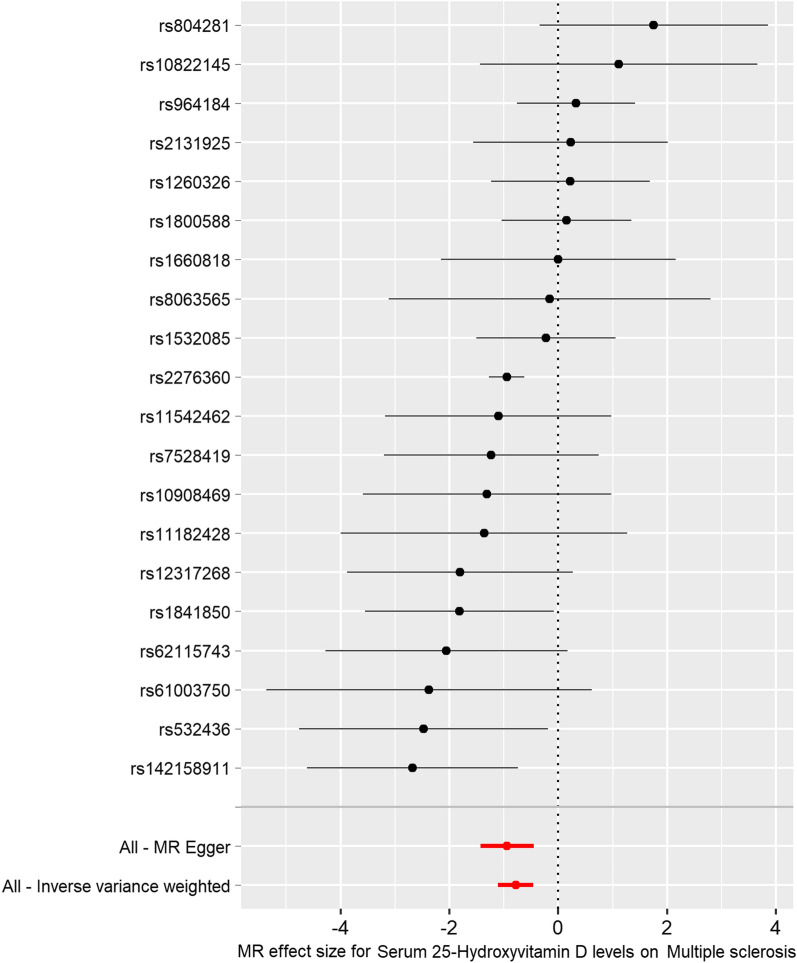
A single effect of serum 25OHD SNP was performed by using functions “mr_singlesnp” and “mr_leaveoneout”. Forest plots were prepared by using functions “mr_forest_plot” and “mr_leaveoneout_plot”. A forest plot of the 20 serum 25OHD SNPs associated with risk of MS is demonstrated in Fig. 3. MR leave-one-out sensitivity analysis for the effect of the 20 serum 25OHD SNPs on MS is shown in Fig. 4. These findings indicate that our results are robust and have no obvious bias based on the effect of the single 25OHD SNP on MS.
Fig. 3.
Forest plot of the 20 serum 25OHD SNPs associated with risk of MS. The x-axis shows the MR effect size for the 25OHD GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90000615/) on multiple sclerosis (MS) from the MS GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ieu-a-1025/). The y-axis shows the analysis for each of the SNPs and for the SNPs in total using the MR-egger and inverse variance weighted (multiplicative random effects) methods
Fig. 4.
MR leave-one-out sensitivity analysis for the effect of the 20 serum 25OHD SNPs on MS. The x-axis shows the MR leave-one-out sensitivity analysis for serum 25OHD GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST90000615/) on multiple sclerosis (MS) from the MS GWAS dataset (https://gwas.mrcieu.ac.uk/datasets/ieu-a-1025/). The y-axis shows the analysis for leave-one-out of SNPs and the effect of the total SNPs on MS
Discussion
Epidemiological evidence suggests an association between 25OHD deficiency and risk and disease progression of MS [13, 20, 38–44]. This relationship is further proven by four previous MR studies using three or four serum 25OHD genetic IVs [27–30]. In the first MR study in 2015, Mokry et al. [28] identified an odds ratio [OR] of 2 by using four serum 25OHD genetic IVs obtained from a serum 25OHD GWAS (N = 33,996) [45], as well as a MS GWAS involving 14,498 MS cases and 24,091 healthy controls from International Multiple Sclerosis Genetic Consortium (IMSGC) [32]. In the second MR study in 2016, Rhead et al. [29] identified an OR = 0.79 using three serum 25OHD genetic IVs. These samples were obtained from a serum 25OHD GWAS (N = 4501) [46] and MS GWAS from members of Kaiser Permanente Medical Care Plan, Northern California Region (KPNC) (1,056 MS cases and 9,015 controls) and the Epidemiological Investigation of Multiple Sclerosis and Genes and Environment in Multiple Sclerosis MS case–control studies (6335 cases and 5762 controls). In the third MR study in 2020, Jacobs et al. [27] identified an OR = 0.57 using six serum 25OHD genetic IVs obtained from a serum 25OHD GWAS (N = 79,366) [47] and IMSGC discovery phase GWAS (14,802 MS, 26,703 controls). In the fourth MR study in 2021, Harroud et al. [30] identified an OR = 0.72 using four serum 25OHD genetic IVs obtained from a serum 25OHD GWAS (N = 33,996) and IMSGC discovery phase GWAS (14,802 MS, 26,703 controls). In the present study, we identified an OR = 0.22 ~ 0.45 using 20 serum 25OHD genetic IVs from to date, the largest serum 25OHD GWAS (n = 401,460) [31] and MS GWAS (14,498 MS cases and 24,091 controls) [32]. Thus, our MR analysis further strengthens evidence for a causal link between 25OHD levels and MS risk.
Among 115 newly identified serum 25OHD genetic variants, 20 effectively independent 25OHD genetic instrumental variables were extracted from MS GWAS summary statistics. We did not identify any significant pleiotropic variant among the selected 20 25OHD genetic instrument variants in the MS GWAS datasets by pleiotropy analysis. Of note, five MR analysis methods demonstrated that 25OHD genetic instrument variants firmly influence the risk of MS through 25OHD but not through other pathways. We found that increased 25OHD levels due to genetic changes were significantly associated with reduced MS risk. For every 1 SD, genetically increased 25OHD levels could reduce MS risk 0.940, 0.835, 0.781, 1.484, and 0.913 using MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode and weighted mode, respectively. These results suggest a causal association between genetically increased 25OHD levels and MS risk.
Both MR-egger and inverse variance weighted methods demonstrated a significant heterogeneity among the 20 25OHD genetic instrument variants in the MS GWAS datasets. Thus, we mainly used the random effect model to estimate the MR effect size. Inverse variance weighted (multiplicative random effects) showed a reduced trend of MS with 1 SD genetically increased 25OHD levels (beta = − 0.781, 95% CI: [− 1.105, − 0.457], p = 2.34E−06). MR-egger, weighted median, simple mode and weighted mode also proved a causal link between 25OHD levels and MS risk. In addition, a single SNP effect analysis and leave-one-out sensitivity analysis for the effect of the 20 serum 25OHD SNPs on MS suggests that our results are robust with no obvious bias based on the effect of the single 25OHD SNP on MS.
25OHD deficiency is becoming an increasing problem worldwide. A recent finding led to a proposed link between 25OHD deficiency and autoimmune diseases [48]. It is reported that 25OHD deficiency is associated with MS risk [39]. Apart from the maintenance of healthy calcium metabolism, Vitamin D and its active product (1,25-dihydroxyvitamin D) have additional roles in immune and central nervous system cell homeostasis [38].
To date, there is evidence to suggest there are some benefits of vitamin D for patient-important outcomes among people with MS [40]. However, vitamin D did not affect recurrence of relapse and worsening of disability in patients with MS [40]. However, in clinical trials, while doses of up to 40,000 IU/day were tested and appeared safe as add-on therapy for short-term periods, chronic and high-dose therapy can lead to life-threatening complications related to vitamin D toxicity, including renal failure, cardiac arrythmia and status epilepticus [43].
There were several strengths to this MR study. First, this MR design was based on three principal assumptions and we used different methods to prove three principal assumptions. Second, we used the large-scale serum 25OHD GWAS dataset (n = 401,460) and MS GWAS summary statistics (14,498 MS cases and 24,091 controls) [31, 32]. With a large-scale GWAS, it is easy to find a causal association between 25OHD levels and MS risk. Third, the two GWAS from European ancestry reduce the influence of population stratification. Fourth, using independent statistical methods, we proved 20 independent genetic variants to be the effective instrumental variables. Fifth, we used five MR analysis methods including MR-egger, weighted median, inverse variance weighted (multiplicative random effects), simple mode, and weighted mode. Critically, all five MR methods proved the causal association between 25OHD levels and MS risk. Lastly, our results were robust, with no obvious bias based on the different methods.
Of course, our MR study had some limitations. First, our MR analysis is only based on European ancestry. Therefore, our conclusions need to be investigated in other ancestries. Second, the GWAS dataset from IMSGC is based on clinically diagnosed MS [32]. Different diagnostic criteria may produce different results. Third, among 115 newly identified serum 25OHD genetic variants [31], only 20 25OHD genetic instrumental variables were effective and independent. Fourth, although we proved that increased serum 25OHD levels could reduce MS risk, the mechanisms by which 25OHD reduced MS risk is still unclear. Fifth, the genetically increased serum 25OHD level does not necessarily reflect the active form of vitamin D, 1,25-Dihydroxyvitamin D3 (1,25(OH)2D3), in the serum [49]. Sixth, while our MR studies may provide the strongest evidence for a causal link between vitamin D levels and the risk of MS, our study does not address the role of vitamin D on disease activity in patients with established MS. Therefore, our study must expand to explore the causal association of vitamin D levels with the activity of MS disease using other databases.
Although MR study offers several advantages, it also has many limitations [50]. These limitations include pleiotropy, low statistical power, canalization, population stratification (which is confounding by ancestry), etc. [50]. To detect and correct for pleiotropy, probably the most challenging to address, a robust method MR-egger intercept was used in our MR study. To improve the statistical power, we used the, to date, largest large-scale GWAS for serum 25OHD [31] and MS [32]. To eliminate population stratification, both 25OHD GWAS and MS GWAS were chosen from European descent. Although we try to minimize the shortcomings, maybe there are still some limitations. Thus, our results need be reproduced in randomized controlled trials (RCTs).
Conclusions
In summary, our MR analysis strengthens evidence for the causal association between genetically increased serum 25OHD levels and reduced MS in European people.
Supplementary Information
Additional file 1: Table S1. Main characteristics of 115 independent vitamin D genetic variants.
Acknowledgements
We thank the International Multiple Sclerosis Genetics Consortium (IMSGC) and ieu open gwas project for providing summary results data for these analyses.
Abbreviations
- 25OHD
25-Hydroxyvitamin D
- MS
Multiple sclerosis
- GWAS
Genome-wide association study
- MR
Mendelian randomization
- SNP
Single nucleotide polymorphism
- IMSGC
The International Multiple Sclerosis Genetics Consortium
- IVW
Inverse variance weighted
Authors’ contributions
RW conceived and initiated the project. RW analyzed the data and wrote the manuscript. RW contributed to the interpretation of the results, critical revision of the manuscript, and approved the final version of the manuscript. RW read and approved the final manuscript. The author read and approved the final manuscript.
Funding
This study was supported by grants from National Natural Science Foundation of China (82071758 and 31770956). The funding bodies have no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
The summary statistics for the genetic associations of 25OHD (GWAS ID: ebi-a-GCST90000615) and MS GWAS datasets (GWAS ID: ieu-a-1025) can be found on the ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.
Declarations
Ethics approval and consent to participate
Our study was approved by the Ethics Committee of Beijing Institute of Brain Disorders in Capital Medical University. This article contains human participants collected by several studies to report the large-scale GWAS for the serum 25OHD [31] and MS [32]. All participants gave informed consent in all the corresponding original studies, as described in the Methods.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yamout BI, Alroughani R. Multiple sclerosis. Semin Neurol. 2018;38(2):212–225. doi: 10.1055/s-0038-1649502. [DOI] [PubMed] [Google Scholar]
- 2.Ma N, He YD, Xiao H, Han GC, Chen GJ, Wang Y, et al. BAFF maintains T-cell survival by inducing OPN expression in B cells. Mol Immunol. 2014;57(2):129–137. doi: 10.1016/j.molimm.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Ma N, Xing C, Xiao H, He YD, Han GC, Chen GJ, et al. BAFF suppresses IL-15 expression in B cells. J Immunol. 2014;192(9):4192–4201. doi: 10.4049/jimmunol.1302132. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Wei Y, Liu X, Xing C, Han G, Chen G, et al. IL-15-secreting gammadeltaT cells induce memory T cells in experimental allergic encephalomyelitis (EAE) mice. Mol Immunol. 2015;66(2):402–408. doi: 10.1016/j.molimm.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 5.Correale J, Gaitan MI, Ysrraelit MC, Fiol MP. Progressive multiple sclerosis: from pathogenic mechanisms to treatment. Brain. 2017;140(3):527–546. doi: 10.1093/brain/aww258. [DOI] [PubMed] [Google Scholar]
- 6.Howard J, Trevick S, Younger DS. Epidemiology of multiple sclerosis. Neurol Clin. 2016;34(4):919–939. doi: 10.1016/j.ncl.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Cho EB, Shin JH, Kwon S, Kim J, Seok JM, Kim BJ, et al. Effects of vitamin D and dexamethasone on lymphocyte proportions and their associations with serum concentrations of 25-hydroxyvitamin D3 in vitro in patients with multiple sclerosis or neuromyelitis optica spectrum disorder. Front Immunol. 2021;12:677041. doi: 10.3389/fimmu.2021.677041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kongsbak M, Levring TB, Geisler C, von Essen MR. The vitamin d receptor and T cell function. Front Immunol. 2013;4:148. doi: 10.3389/fimmu.2013.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Royal W, 3rd, Mia Y, Li H, Naunton K. Peripheral blood regulatory T cell measurements correlate with serum vitamin D levels in patients with multiple sclerosis. J Neuroimmunol. 2009;213(1–2):135–141. doi: 10.1016/j.jneuroim.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Bivona G, Agnello L, Ciaccio M. Vitamin D and immunomodulation: is it time to change the reference values? Ann Clin Lab Sci. 2017;47(4):508–510. [PubMed] [Google Scholar]
- 11.Bivona G, Agnello L, Lo Sasso B, Scazzone C, Butera D, Gambino CM, et al. Vitamin D in malaria: more hypotheses than clues. Heliyon. 2019;5(2):e01183. doi: 10.1016/j.heliyon.2019.e01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scazzone C, Agnello L, Lo Sasso B, Salemi G, Gambino CM, Ragonese P, et al. FOXP3 and GATA3 polymorphisms, vitamin D3 and multiple sclerosis. Brain Sci. 2021;11(4):415. doi: 10.3390/brainsci11040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sintzel MB, Rametta M, Reder AT. Vitamin D and multiple sclerosis: a comprehensive review. Neurol Ther. 2018;7(1):59–85. doi: 10.1007/s40120-017-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Cai Y, Liu M, Zhu D, Guan Y. The potential immunoregulatory roles of vitamin D in neuromyelitis optica spectrum disorder. Mult Scler Relat Disord. 2020;43:102156. doi: 10.1016/j.msard.2020.102156. [DOI] [PubMed] [Google Scholar]
- 15.Scazzone C, Agnello L, Sasso BL, Ragonese P, Bivona G, Realmuto S, et al. Klotho and vitamin D in multiple sclerosis: an Italian study. Arch Med Sci. 2020;16(4):842–847. doi: 10.5114/aoms.2019.86969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bivona G, Gambino CM, Iacolino G, Ciaccio M. Vitamin D and the nervous system. Neurol Res. 2019;41(9):827–835. doi: 10.1080/01616412.2019.1622872. [DOI] [PubMed] [Google Scholar]
- 17.Bivona G, Agnello L, Bellia C, Iacolino G, Scazzone C, Lo Sasso B, et al. Non-skeletal activities of vitamin D: from physiology to brain pathology. Medicina (Kaunas) 2019;55(7):341. doi: 10.3390/medicina55070341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bivona G, Lo Sasso B, Iacolino G, Gambino CM, Scazzone C, Agnello L, et al. Standardized measurement of circulating vitamin D [25(OH)D] and its putative role as a serum biomarker in Alzheimer's disease and Parkinson's disease. Clin Chim Acta. 2019;497:82–87. doi: 10.1016/j.cca.2019.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Bivona G, Lo Sasso B, Gambino CM, Giglio RV, Scazzone C, Agnello L, et al. The role of vitamin D as a biomarker in Alzheimer's disease. Brain Sci. 2021;11(3):334. doi: 10.3390/brainsci11030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierrot-Deseilligny C, Souberbielle JC. Vitamin D and multiple sclerosis: an update. Mult Scler Relat Disord. 2017;14:35–45. doi: 10.1016/j.msard.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Scazzone C, Agnello L, Ragonese P, Lo Sasso B, Bellia C, Bivona G, et al. Association of CYP2R1 rs10766197 with MS risk and disease progression. J Neurosci Res. 2018;96(2):297–304. doi: 10.1002/jnr.24133. [DOI] [PubMed] [Google Scholar]
- 22.Agnello L, Scazzone C, Lo Sasso B, Bellia C, Bivona G, Realmuto S, et al. VDBP, CYP27B1, and 25-hydroxyvitamin D gene polymorphism analyses in a group of sicilian multiple sclerosis patients. Biochem Genet. 2017;55(2):183–192. doi: 10.1007/s10528-016-9783-4. [DOI] [PubMed] [Google Scholar]
- 23.Scazzone C, Agnello L, Bivona G, Lo Sasso B, Ciaccio M. Vitamin D and genetic susceptibility to multiple sclerosis. Biochem Genet. 2021;59(1):1–30. doi: 10.1007/s10528-020-10010-1. [DOI] [PubMed] [Google Scholar]
- 24.Went M, Cornish AJ, Law PJ, Kinnersley B, van Duin M, Weinhold N, et al. Search for multiple myeloma risk factors using Mendelian randomization. Blood Adv. 2020;4(10):2172–2179. doi: 10.1182/bloodadvances.2020001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Zhang Y, Zhang H, Wang L, Wang T, Han Z, et al. Effect of plasma vitamin C levels on Parkinson's disease and age at onset: a Mendelian randomization study. J Transl Med. 2021;19(1):221. doi: 10.1186/s12967-021-02892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs BM, Noyce AJ, Giovannoni G, Dobson R. BMI and low vitamin D are causal factors for multiple sclerosis: a Mendelian Randomization study. Neurol Neuroimmunol Neuroinflamm. 2020;7(2):e662. doi: 10.1212/NXI.0000000000000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokry LE, Ross S, Ahmad OS, Forgetta V, Smith GD, Goltzman D, et al. Vitamin D and risk of multiple sclerosis: a mendelian randomization study. PLoS Med. 2015;12(8):e1001866. doi: 10.1371/journal.pmed.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhead B, Baarnhielm M, Gianfrancesco M, Mok A, Shao X, Quach H, et al. Mendelian randomization shows a causal effect of low vitamin D on multiple sclerosis risk. Neurol Genet. 2016;2(5):e97. doi: 10.1212/NXG.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harroud A, Manousaki D, Butler-Laporte G, Mitchell RE, Davey Smith G, Richards JB, et al. The relative contributions of obesity, vitamin D, leptin, and adiponectin to multiple sclerosis risk: a Mendelian randomization mediation analysis. Mult Scler. 2021;27(13):1994–2000. doi: 10.1177/1352458521995484. [DOI] [PubMed] [Google Scholar]
- 31.Manousaki D, Mitchell R, Dudding T, Haworth S, Harroud A, Forgetta V, et al. Genome-wide association study for vitamin D levels reveals 69 independent loci. Am J Hum Genet. 2020;106(3):327–337. doi: 10.1016/j.ajhg.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Multiple Sclerosis Genetics C, Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–60. [DOI] [PMC free article] [PubMed]
- 33.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926–2940. doi: 10.1002/sim.6522. [DOI] [PubMed] [Google Scholar]
- 35.Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y, et al. PICALM gene rs3851179 polymorphism contributes to Alzheimer's disease in an Asian population. Neuromolecular Med. 2013;15(2):384–388. doi: 10.1007/s12017-013-8225-2. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Zhao Y, Jin S, Hu Y, Wang T, Tian R, et al. Circulating vitamin E levels and Alzheimer's disease: a Mendelian randomization study. Neurobiol Aging. 2018;72(189):e1–e9. doi: 10.1016/j.neurobiolaging.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smolders J, Torkildsen O, Camu W, Holmoy T. An update on vitamin D and disease activity in multiple sclerosis. CNS Drugs. 2019;33(12):1187–1199. doi: 10.1007/s40263-019-00674-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin L, Clarke L, Khalilidehkordi E, Butzkueven H, Taylor B, Broadley SA. Vitamin D for the treatment of multiple sclerosis: a meta-analysis. J Neurol. 2018;265(12):2893–2905. doi: 10.1007/s00415-018-9074-6. [DOI] [PubMed] [Google Scholar]
- 40.Jagannath VA, Filippini G, Di Pietrantonj C, Asokan GV, Robak EW, Whamond L, et al. Vitamin D for the management of multiple sclerosis. Cochrane Database Syst Rev. 2018;9:CD008422. doi: 10.1002/14651858.CD008422.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jagannath VA, Fedorowicz Z, Asokan GV, Robak EW, Whamond L. Vitamin D for the management of multiple sclerosis. Cochrane Database Syst Rev. 2010;12:CD008422. doi: 10.1002/14651858.CD008422.pub2. [DOI] [PubMed] [Google Scholar]
- 42.Govind N. Vitamin D for the management of multiple sclerosis. Br J Community Nurs. 2020;25(6):304–305. doi: 10.12968/bjcn.2020.25.6.304. [DOI] [PubMed] [Google Scholar]
- 43.Feige J, Moser T, Bieler L, Schwenker K, Hauer L, Sellner J. Vitamin D supplementation in multiple sclerosis: a critical analysis of potentials and threats. Nutrients. 2020;12(3):783. doi: 10.3390/nu12030783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berezowska M, Coe S, Dawes H. Effectiveness of vitamin D supplementation in the management of multiple sclerosis: a systematic review. Int J Mol Sci. 2019;20(6):1301. doi: 10.3390/ijms20061301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, et al. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet. 2010;19(13):2739–2745. doi: 10.1093/hmg/ddq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X, O'Reilly PF, Aschard H, Hsu YH, Richards JB, Dupuis J, et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun. 2018;9(1):260. doi: 10.1038/s41467-017-02662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bizzaro G, Antico A, Fortunato A, Bizzaro N. Vitamin D and autoimmune diseases: is vitamin D receptor (VDR) polymorphism the culprit? Isr Med Assoc J. 2017;19(7):438–443. [PubMed] [Google Scholar]
- 49.Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Curr Opin Pharmacol. 2010;10(4):482–496. doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Harroud A, Richards JB. Mendelian randomization in multiple sclerosis: a causal role for vitamin D and obesity? Mult Scler. 2018;24(1):80–85. doi: 10.1177/1352458517737373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Main characteristics of 115 independent vitamin D genetic variants.
Data Availability Statement
The summary statistics for the genetic associations of 25OHD (GWAS ID: ebi-a-GCST90000615) and MS GWAS datasets (GWAS ID: ieu-a-1025) can be found on the ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.