Abstract
Development of a multicellular organism requires that mitosis and morphogenesis be coordinated. These processes must also be synchronized during the growth of unicellular organisms. In the yeast Saccharomyces cerevisiae, mitosis is dependent on the prior growth of a daughter cell in the form of a bud. Overexpression of wild-type Polo-like kinase Cdc5 or a catalytically inactive form resulted in the formation of multinucleate cells in budding yeast. Immunofluorescence analysis of these multinulceate cells showed that mitosis and bud formation were no longer linked. Others have shown that Swe1 is required for coupling mitosis to bud formation during a perturbed cell cycle. When the normal pathway of bud formation is perturbed, Swe1 functions to delay mitosis through negative regulation of Clb/Cdk. In cells lacking Swe1, multinucleate cells are formed in response to delays in bud formation. Affinity purification, two-hybrid analysis, and mutant characterization results suggested that Cdc5 and Swe1 interact. From these results, we conclude that multinucleate formation in response to Cdc5 overexpression is linked to titration of Swe1 function. These results also suggest that Cdc5 may be a negative regulator of Swe1.
The onset of mitosis in eukaryotic cells requires the activation of mitosis-promoting factor (MPF) (12). MPF is comprised of a cyclin-dependent kinase (Cdk) and B-type cyclin regulatory subunit (Clb/Cdk), and it is maintained in an inactive state during interphase through phosphorylation of Tyr15 on Cdk by the Wee1 kinase. MPF is activated in response to activation of the Tyr15 phosphatase Cdc25 and inactivation of the Tyr15 kinase Wee1 at the transition between G2 and mitosis (G2/M).
In addition to Clb/Cdk, Polo kinases are also key promoters of mitosis and have been implicated in coordinating the activation of Clb/Cdk. In vitro studies in Xenopus indicate that Cdc25 is activated when it binds to and is phosphorylated by the Polo kinase, Plx1 (23, 37). More recently, phosphorylation by the Polo kinase, Plk1, of a critical serine residue in the nuclear export signal sequence of cyclin B1 promotes accumulation of Clb/Cdk activity in the nucleus of vertebrate prophase cells (46). In Schizosaccharomyces pombe (3, 34), Drosophila (29), and vertebrate (15, 16) cells, Polo kinases have been localized to microtubule-organizing centers at the G2/M transition. Mutant studies in these systems show that Polo kinases play key roles in spindle pole duplication and formation of the bipolar spindle (15).
In Saccharomyces cerevisiae daughter cells originate as buds at the beginning of S phase (27). Bud formation is regulated by Cdc28, the yeast Cdk, in combination with the G1 (Cln) and mitotic (Clb) cyclins. The Cln-Cdc28 complex activates initiation of bud formation late in G1, while the Clb-Cdc28 complex inhibits rebudding later in the cell cycle (2, 25, 26, 40). Perturbations of the actin cytoskeleton prevent the normal pathway of bud formation and result in delayed mitosis through negative regulation of Clb/Cdk by the Wee1 family kinase member Swe1 (33). In response to bud formation, Swe1 is negatively regulated by the Nim1-like kinase Hsl1 (5, 24, 30, 31, 32). Swe1 is targeted to the bud neck after bud formation, and the neck localization requires Hsl1 and its interacting factor, Hs17 (30, 42; S. H. Woo and C. F. J. Hardy, unpublished data). In this manner, morphogenesis is linked to cell proliferation in budding yeast.
Previous studies have shown that Swe1 overexpression prevents spindle pole body (SPB) separation but not duplication (28). This suggests that Swe1 plays a role at SPBs. Endogenous Cdc5 is also present at SPBs before they separate (Woo and Hardy, unpublished), consistent with it playing a role similar to other Polo kinases in regulating spindle pole separation (see above). In this study, we have found that overproduction of either wild-type Cdc5 or a catalytically inactive form uncouples mitosis from bud formation. This results in the formation of multinucleate cells. Cdc5 interacts with the N-terminal region of Swe1, and overproduction of the catalytically inactive form of Cdc5 suppresses Swe1-dependent phenotypes associated with unregulated Swe1. In response to Cdc5 overproduction, Swe1 is modified and localized to SPBs. Taken together, our results suggest that multinucleate formation in response to Cdc5 overexpression is linked to titration of Swe1 function. The results in this report also suggest that Cdc5 may be a negative regulator of Swe1, possibly playing a role in regulating Swe1 function at SPBs prior to SPB separation.
MATERIALS AND METHODS
Strains.
Yeast strains and sources are listed in Table 1. Plasmid DNA was transformed into yeast by the lithium acetate method as described (19). The open reading frame of the endogenous SWE1 was disrupted with LEU2 by transforming strains with the integrating plasmid pSWE1-10g restricted with XbaI (a gift from R. Booher) (6). The open reading frame of the endogenous SWE1 was placed under control of the GAL1 promoter by transforming strains with the integrating plasmid pSWE1-41 restricted with PstI and BamHI (a gift from R. Booher) (6). Yeast strains were grown in YPD at 30°C unless noted otherwise in the text.
TABLE 1.
Strains used
| Strain | Genotype (source) |
|---|---|
| W303-1A | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 (R. Rothstein) |
| W303-1B | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 (R. Rothstein) |
| AA1120 | W303-1A cdh1/hct1::HIS3 PDS1-HA-LEU2::pds1 (A. Amon) |
| IAY18 | MATaspc42Δ1::LEU2 TRP1::SPC42-GFP (J. Kilmartin) |
| JC1195 | MATα leu2 ura3 his3-Δ200 (J. Cooper) |
| JC256 | W303-1A ura3::(GAL-CDC5-HA-URA3)3 (J. Charles) |
| JC278 | W303-1A ura3::(GAL-cdc5-N209A-HA-URA3)3(J. Charles) |
| K3371 | W303-A clb2::LEU2 trp1::GAL-CLB2-TRP1 (Kim Naysmith) |
| K6451 | W303-1A cdc5-3(msd2-1::URA3) omns PDS1::LEU2 (K. Nasmyth) |
| MAY1 | W303-1A hsl1Δ1::URA3 (M. Grunstein) |
| MAY2 | W303-1A hsl1Δ1::URA3 swe1::LEU2 (M. Grunstein) |
| MAY3 | W303-1A hsl7Δ1::URA3 (M. Grunstein) |
| MAY4 | W303-1A hsl7Δ1::URA3 swe1::LEU2 (M. Grunstein) |
| NEY100X | MATα hsl1::LEU2 ura3 leu2 |
| CH179 | MATα ura3::GFP-TUB1-URA3 his3 leu2 |
| CH199 | W303-1A bar1::URA3 CDC5-ProA-HIS3-URA3 |
| CH452 | MATα SWE1-GFP-HIS3 leu2 ura3 his3-Δ200 (GFP-tagged SWE1 in JC1195) |
| CH459 | MATabar1 ura3::(GAL-cdc5-N209A-HA-URA3)3 SWE1-ProA-URA3-HIS3 (ProA tag of SWE1 in JC278) |
| CH473 | MATaura3::(GAL-CDC5-HA-URA3)3 SWE1-ProA-URA3-HIS3 (ProA tag of SWE1 in JC256) |
| CH474 | W303-1A swe1::GAL-SWE1-HA-LEU2 (GAL-SWE1-HA integrated at the SWE1 locus in W303-1A using pSWE1-41, from D. Lew) |
| CH499 | W303-1A ura3::(GAL-cdc5-N209A-HA-URA3)3 swe1::(GAL-SWE1-HA-LEU2) (GAL-SWE1-HA integrated at the SWE1 locus in YJC278 using pSWE1-41, from R. Booher) |
| CH507 | MATα ura3::(GAL-CDC5-HA-URA3)3 hsl1::LEU2 (segregant of JC256 × NEY100X) |
| CH508 | MATaura3::(GAL-cdc5-N209A-HA-URA3)3 hsl1::LEU2 ADE2 his3 (segregant of NEY 100X × JC278) |
| CH572 | MATaura3::(GAL-cdc5-N209A-HA-URA3)3 SWE1-GFP-HIS3 his3 leu2 ura3 (segregant of JC278 × CH452) |
| CH573 | MATaura3::(GAL-CDC5-HA-URA3)3 SWE1-GFP-HIS3 his3 leu2 ura3 (segregant of JC256 × CH452) |
| CH618 | MATaura3::(GAL-cdc5-N209A-HA-URA3)3 hsl7Δ1::URA3 his3 leu2 ura3 trp1-1 (segregant of CH696 × MAY3) |
| CH667 | W303-1B cdc5-3(msd2-1::URA3) swe1::LEU2 (SWE1 disrupted with pSWE1-10g, from R. Booher) |
| CH696 | W303-1B ura3::(GAL-cdc5-N209A-HA-URA3)3 (segregant of JC278 × W303-1B) |
| CH745 | W303-1B cdc5-3(msd2-1::URA3) (segregant of KN6451 × W303-1α) |
| CH798 | W303-1A swe1::LEU2 (segregant of MAY4 × W303-1B) |
| CH810 | W303-1B cdh1(hct1)::HIS3 (segregant of AM1120 × W303-1B) |
| CH895 | W303-1A cdh1(hct1)::HIS3 swe1::LEU2 (segregant of CH798 × CH810) |
| CH896 | W303-1B cdh1(hct1)::HIS3 swe1::LEU2 (segregant of CH798 × CH810) |
| CH943 | MATα swe1::LEU2 leu2 ura3 his3-Δ200 (segregant of JC1195 × CH798) |
| CH964 | W303-1A cdh1(hct1)::HIS3 swe1::LEU2 pRS424 (CH895 transformed with pRS424) |
| CH965 | W303-1A cdh1/hct1::HIS3 swe1::LEU2 pRS424-SIC1 (CH895 transformed with pRS424-SIC1) |
| CH968 | W303-1A cdh1/hct1::HIS3 swe1:LEU2 ura3::GFP-TUB1-URA3 (GFP-tagged TUB1 using pAFS92, from A. Straight, in CH895) |
| CH1039 | MATa/α ura3::GFP-TUB1-URA3/ura3::(GAL-cdc5-N209A-HA-URA3)3 (JC278 × CH179) |
| CH1173 | W303-1A cdh1/hct1::HIS3 swe1::LEU2 clb2::LEU2 (segregant of CH896 × K3371) |
| CH1175 | MATaswe1::LEU2 GAL-CLB2::TRP1 (segregant of CH943 × K3371) |
| CH1201 | MATaspc42Δ1::LEU2 TRP1::SPC42-GFP ura3::(GAL-cdc5-N209A-HA-URA3)3(segregant of CH696 × IAY18) |
| CH1221 | W303-1A ura3::(GAL-cdc5-N209A-HA-URA3)3 cdh1/hct1::HIS3 (segregant of JC278 × CH810) |
| CH1224 | W303-1A ura3::3(GAL-CDC5-HA-URA3)3 cdh1/hct1::HIS3 (segregant of JC256 × CH810) |
Two-hybrid analysis.
Two-hybrid analysis was performed as described by James et al. (20) by using strain PJ69-4a, with the following variation. Following transformation of the reporter strain with the yeast library plasmids, the cells were washed and then resuspended into liquid synthetic complete medium (SC) lacking the amino acids histidine, uracil, and leucine but containing adenine; 200 μl of this solution was spread on 100-mm agar plates containing SC lacking histidine, uracil, leucine, and adenine. The extra initial adenine was essential to obtain transformants on the selective plates.
Immunofluorescence.
Indirect immunofluorescence was carried out as described (49). Yeast cells were grown to early log phase and prepared for immunofluorescence microscopy. For localization of hemagglutinin (HA)-tagged Cdc5, cell were fixed for 5 min. The cells were first incubated with affinity-purified anti-HA antibody (BAbCo) and then with rhodamine-conjugated donkey anti-mouse secondary antibody. For localization of Tub1, the cells were fixed for 30 min. The cells were first incubated with mouse anti-Tub1 and then with rhodamine-conjugated donkey anti-mouse secondary antibody. Green fluorescent protein gene (GFP)-TUB1 fusions were obtained by transforming strain C895 (cdh1/hct1 swe1) with pAFS92, which integrates, at the ura3 locus, a GFP fusion to the α-tubulin gene, TUB1, under control of the MET3 promoter. Tub1-GFP was induced by growing cells for 1 h in SC lacking methionine. Digital images were taken with a 100× objective on an Olympus microscope.
Generation of fusion proteins in yeast.
PCR was carried out to generate Swe1-protein A (ProA) and Swe1-GFP fusion sequences. The oligonucleotides are available on request. The ProA gene and adjacent HIS3 and URA3 markers were amplified by PCR using pProA-HIS3-URA3 (a gift from Mike Rout and John Aitchison) (1). GFP and the adjacent HIS3 marker were amplified by PCR using pYGFP3 (a gift from Brendan Cormack) (10). The codons in the mutant GFP (pYGFP3) were optimized for expression in Candida. The resulting PCR constructs contained SWE1 sequences fused in frame with sequence encoding GFP or ProA at their C-terminal ends. Strains were transformed with the resulting PCR products to generate a yeast strain expressing the desired fusion protein under its endogenous promoter. A strain expressing Spc42-GFP was previously obtained from J. Kilmartin (11). Strains lacking HSL1 produce elongated buds in a SWE1-dependent manner. We determined that buds in hsl1 SWE1-GFP and hsl1 SWE1-ProA strains are elongated and therefore the Swe1-ProA and Swe1-GFP constructs are functional.
Purification of GST and GST-Swe1 from Escherichia coli.
E. coli BL21 cells were transformed with either pGEX-5X or pGEX-5X-Swe1 and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 28°C. Bacteria were pelleted and then suspended in NETN buffer (0.5% NP-40, 20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) supplemented with protease inhibitors (Boehringer Mannheim) and were lysed with sonication. The lysate was clarified by centrifugation, and the clarified lysate was incubated with glutathione-agarose beads (Pharmacia) for 1 h at 4°C. Beads were pelleted and then washed extensively with NETN buffer with protease inhibitors, and bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by immunoblotting with anti-glutathione S-transferase (GST) antibodies.
Affinity chromatography.
Cells containing Cdc5 fused to ProA derived from strain C199 (CDC5-ProA) were pelleted, washed once in water, and lysed or frozen in liquid nitrogen. Pellets were resuspended in 0.3 ml of lysis buffer (L buffer) containing 5% glycerol, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 10 mM MgCl2, 0.3 M N2H8SO4, 1 mM dithiotheitol, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride 5 mg of leupeptin per ml, 2 mM pepstatin A, 50 mM NaF, 10 mM sodium pyrophosphate, and 0.5 mM NaVO4. Cells were lysed by adding 0.5 ml of acid-washed glass beads and vortexing in pulses until 90% lysis was achieved; 0.35 ml of immunoprecipitation buffer (1 M LiCl, 2% Triton X-100, 10% glycerol, 0.5 mM NaVO4, protease inhibitors as described for L buffer) was added, and the mixture was vortexed 1 min. The lysate was spun for 10 min at 3,000 rpm, and the supernatant was aliquoted and frozen in liquid nitrogen. Protein concentrations were determined with the Bio-Rad protein assay. Four hundred milligrams of lysate was incubated with 0.1 ml of GST and GST-Swe1 beads prepared as described above. After 2 h of incubation, beads were washed extensively with L buffer, and the bound proteins were resolved by SDS-PAGE and analyzed by immunoblotting.
Other methods.
YEP medium contained 1% yeast extract and 2% Bacto Peptone. Carbon sources (glucose, raffinose, and galactose) were all used at a 2% final concentration. α-Factor and hydroxyurea were obtained from Sigma and were used at final concentrations of 0.2 μM and 200 mM, respectively. Nocodazole was obtained from Aldrich and was added to medium from a 20-mg/ml stock solution in dimethyl sulfoxide. It was used at a final concentration of 20 μg/ml in 1% dimethyl sulfoxide as described by Jacobs et al. (19). The DNA content of cells was measured on a Becton Dickinson (San Jose, Calif.) FACSsan as described by Epstein and Cross (13). Movies were collected as described elsewhere (48). Briefly, mid-logarithmic-phase cells were placed on a slide with a thin agarose pad. A Z series of eight focal planes was collected over 8 s and projected onto a single two-dimensional image. Z series were collected every 5 min for 0.5 to 3 h. NIH image 1.62 (written by Wayne Rasband) was used for image acquisition. Cells were also visualized by differential interference contrast microscopy (DIC).
RESULTS
Cells overproducing Cdc5 or cdc5N209A are multinucleate.
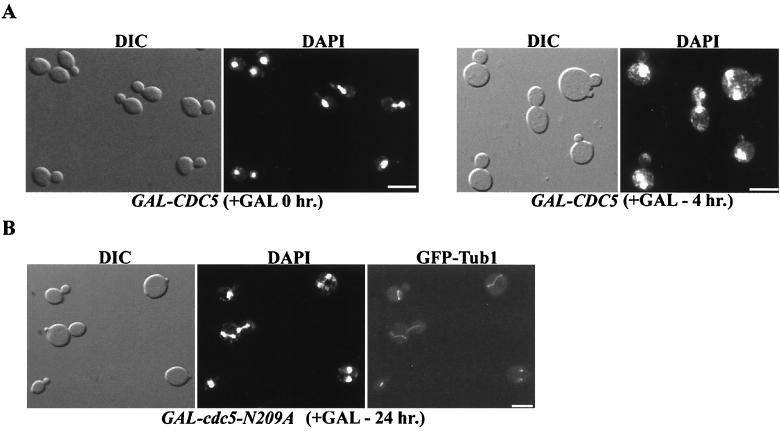
HeLa cells overproducing wild-type or catalytically inactive Plk1 are multinucleate (37). In an effort to further understand these results, we examined whether budding yeast became multinucleate in response to overproduction of Cdc5. To conduct the experiment, a strain expressing wild-type CDC5 from the inducible GAL1 promotor, GAL1-CDC5, was analyzed. The 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei in GAL1-CDC5 cells were counted after inducing CDC5 expression for 4 h through addition of galactose. Strikingly, 2.5% of the cells were multinucleate (Fig. 1A; Table 2). In addition, 0.9% of cells overexpressing a mutant version of CDC5 that lacks kinase activity (cdc5-N209A) were multinucleate (Fig. 1B; Table 2). In contrast, the multinucleate phenotype occurs very infrequently in wild-type cells (Table 2). To visualize the spindles, a GFP-tagged version of Tub1, the major α-tubulin, was expressed. Strikingly, elongated spindles were observed in unbudded and small budded cells coincident with Cdc5 (data not shown) or cdc5-N209A (Fig. 1B) overproduction. The advanced stage of the cell cycle relative to bud growth in these cells suggested that mitosis had become uncoupled from bud formation.
FIG. 1.
Cells overexpressing CDC5 or cdc5N209A are multinucleate. To induce Cdc5 (A) or cdc5-N209A (B), 2% galactose was added to mid-logarithmic-phase cultures of strains JC256 (GAL-CDC5) and C1039 (GAL-cdc5N209A GFP-TUB1) for the indicated times. To induce GFP-Tub1 expression, the same cells were then resuspended in SC-methionine plus galactose for 2 h. The cells were fixed and stained with DAPI to visualize nuclei. It should be noted that the frequency of multinucleate cells as visualized here does not reflect the true distribution in these cultures. Rather, these particular fields were chosen in order to demonstrate the variety of multinucleate cells in the culture. Bars, 10 μm.
TABLE 2.
Multinucleate formation in CDC5 and cdc5-N209A overexpression strains
| Strain | Distribution (%)a
|
|
|---|---|---|
| 1 nucleus/mother | 2 nuclei/mother | |
| WT | 99.94 | 0.06 |
| WT + Gal (4) | 99.94 | 0.06 |
| GAL-CDC5 + Gal (0) | 99.7 | 0.3 |
| GAL-CDC5 + Gal (4) | 97.5 | 2.5 |
| GAL-cdc5-N209A + Gal (0) | 99.9 | 0.1 |
| GAL-cdc5-N209A + Gal (4) | 99.1 | 0.9 |
| cdh1/hct1 + Gal (0) | 98.2 | 1.8 |
| cdh1/hct1 + Gal (4) | 98.8 | 1.2 |
| GAL-CDC5 cdh1/hct1 + Gal (0) | 98.9 | 1.1 |
| GAL-CDC5 cdh1/hct1 + Gal (4) | 78.6 | 21.4 |
| GAL-cdc5-N209A cdh1/hct1 + Gal (0) | 99.0 | 1.0 |
| GAL-cdc5-N209A cdh1/hct1 + Gal (4) | 85.0 | 15.0 |
The number of nuclei per mother cell in haploid cells was determined by DAPI staining of asynchronous cultures at 30°C in mid-log phase. Greater than 1,000 cells were counted for each strain. All strains used were derivatives of W303 (wild type [WT]). All cells were grown in rich medium containing raffinose; where indicated, galactose was added for the time periods (hours) shown in parentheses.
Cdh1/Hct1 is a cofactor of the anaphase-promoting complex (APC) and is required for the ubiquitin-mediated destruction of the mitotic cyclins Clb1 and Clb2 at the end of mitosis (39, 47). In cdh1/hct1 null cells, Clb1 and Clb2 inappropriately accumulate during G1 and S (39, 47). We discovered that 1.8% of cdh1/hct1 mutant cells are multinucleate (Table 2). Strikingly, the frequency of multinucleate cells in a culture of cdh1/hct1 cells significantly increased in response to Cdc5 (21.4%) or cdc5N209A (15.0%) overproduction (Table 2).
Cdc5 and Swe1 interact.
Because overexpression of either catalytically active or inactive Cdc5 resulted in multinucleate cells, we predicted that this phenotype might be due to titration of a Cdc5-interacting factor. We carried out a two-hybrid screen using Cdc5 as bait to isolate potential interacting factors (20). One of the positives contained an in-frame fusion to an open reading frame encoding the C-terminal region of Swe1 (amino acid residues 173 to 819) (Fig. 2A) (6). Further two-hybrid experiments showed that a C-terminal region of Cdc5 (between amino acids 340 and 705) is sufficient for interaction with Swe1 (Fig. 2A). This Swe1 interaction region in Cdc5 is distinct from the Cdc5 kinase domain (residues 82 to 337). Likewise, the region of Swe1 (between amino acids 173 and 400) that is sufficient for interaction with Cdc5 is distinct from the Swe1 kinase domain (residues 444 to 789). The Cdc5-Swe1 protein-protein interaction was confirmed by an affinity chromatography strategy (Fig. 2B). Cdc5 has also been independently isolated by another group in a two-hybrid screen using Swe1 as the bait (J. Harrison and D. Lew, personal communication).
FIG. 2.
Cdc5 interacts with Swe1. (A) Two-hybrid assay performed with pDB-Cdc5(1–705), pDB-Cdc5(340–705), pAD-Swe1(173–819), and pAD-Swe1(400–819). pDB (Ga14 DNA binding domain [DB] fusion vector) and pAD (Ga14 activation domain [AD] fusion vector) were used as negative controls. The HIS3 and ADE2 reporter strain PJ69-4a was used, and transformants were tested for growth on medium lacking uracil, leucine, histidine, and adenine. + and − indicates growth and no growth, respectively, after 5 days at 30°C. (B) GST and GST-Swe1 were incubated with yeast extract containing Cdc5 fused to ProA, derived from strain C199 (CDC5-ProA). After 2 h of incubation, beads were washed extensively and the bound proteins (lanes B) were resolved by SDS-PAGE and analyzed by immunoblotting; 10% of the extract added to each incubation was loaded in the input lane (I).
cdh1/hct1 swe1 double mutants have a high frequency of multinucleate cells resulting from an uncoupling of bud formation and mitosis.
Previous studies have concluded that Swe1 does not play a role during an unperturbed cell cycle (6). In agreement with these results, the frequency of multinucleate swe1 cells was very similar to that observed in wild-type cells (Table 3). However, altering the levels of Clb/Cdk affected the accumulation of multinucleate swe1 cells. Examination of cdh1/hct1 swe1 double-mutant cells by DAPI staining revealed greater than 17% with multiple nuclei (Table 3). A representative image is shown in Fig. 3A. In contrast, swe1 and cdh1/hct1 single-mutant cultures were only 0.06 and 1.8% multinucleate, respectively (Table 3). Interestingly, a genetic interaction between CDH1/HCT1 and SWE1 was established based on the slower growth of the cdh1/hct1 swe1 double mutant compared to either single mutant at 37°C (Fig. 3B). Furthermore, overexpression of the mitotic cyclin Clb2 (GAL-CLB2) in swe1 null cells resulted in 10.8% multinucleate cells, compared to only 0.08% multinucleate GAL-CLB2 SWE1 cells (Table 3). The overexpression of the Clb/Cdc28 inhibitor Sic1 (2μm-SIC1 cdh1/hct1 swe1) or deletion of CLB2 (cdh1/hct1 swe1 clb2) partially suppressed the cdh1/hct1 swe1 multinucleate phenotype (Table 3).
TABLE 3.
Multinucleate formation in swe1 mutant strains
| Strain | Distribution (%)a
|
|
|---|---|---|
| 1 nucleus per mother | 2 nuclei per mother | |
| Wild type | 99.94 | 0.06 |
| swe1 | 99.94 | 0.06 |
| cdh1/hct1 | 98.2 | 1.8 |
| cdh1/hct1 swe1 | 82.8 | 17.2 |
| cdh1/hct1 swe1 clb2 | 96.5 | 3.5 |
| cdh1/hct1 swe1 2μm | 84.7 | 15.3 |
| cdh1/hct1 swe1 2μm-SIC1 | 95.7 | 4.3 |
| GAL-CLB2 SWE1 + Gal (0) | 99.93 | 0.07 |
| GAL-CLB2 SWE1 + Gal (4) | 99.92 | 0.08 |
| GAL-CLB2 swe1 + Gal (0) | 99.94 | 0.06 |
| GAL-CLB2 swe1 + Gal (4) | 89.2 | 10.8 |
See Table 2 for full description. Cells were grown in rich medium with two exceptions. Strains transformed with the 2μm or 2μm-SIC1 plasmids were grown in synthetic medium lacking the amino acid tryptophan. The GAL-CLB2 strains were grown in rich medium containing raffinose; where indicated, galactose was added for the time periods (hours) shown (in parentheses). The swe1, cdh1/hct1, and clb2 mutations represent deletions of their respective open reading frames.
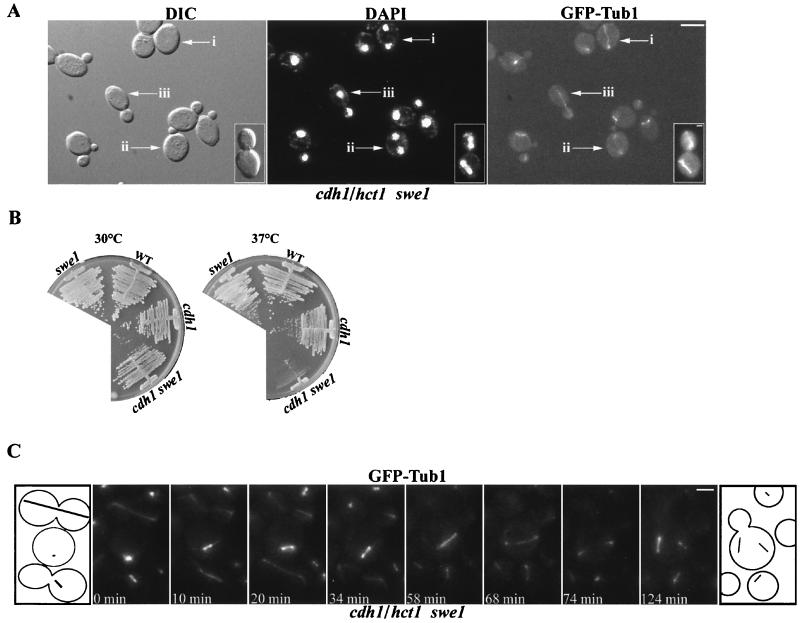
FIG. 3.
Mitosis is unlinked from bud formation in cdh1/hct1 swe1 cells. (A) Visualization of elongated and disassembled spindles in unbudded and small-budded multinucleate cdh1/hct1 swe1 cells. Cultures of C968 (cdh1/hct1 swe1 MET3-GFP-TUB1) were grown to mid-log phase in YPD and transferred to SC lacking methionine for 1 h to induce GFP-Tub1. Arrows mark a multinucleate unbudded cell with an elongated spindle (i), a multinucleate small-budded cell with a disassembled spindle (ii), and a cell undergoing a normal mitosis (iii). (Inset) A rare cell type (0.1% of cdh1/hct1 swe1 GFP-TUB1 cells) in which the mother and its attached daughter are both undergoing mitosis. (B) The cdh1/hct1 swe1 double mutant grows slower than the single mutants at 37°C. Strains C895 (cdh1/hct1 swe1), C798 (swe1), C810 (cdh1/hct1), and W303a (wild type [WT]) were streaked on YPD plates and incubated at 30 or 37°C for 5 days. (C) Video sequence of spindle elongation and disassembly during mitosis in strain C968 (cdh1/hct1 swe1 MET3-GFP-TUB1). Cultures of C968 were grown to mid-log phase in YPD and transferred to SC lacking methionine for 2 h to induce GFP-Tub1. Following induction, they were placed on a slide with a thin agarose pad. A Z series of eight focal planes was collected over 8 s and projected onto a single two-dimensional image. Z series were collected every 2 min for 3 h. An unbudded cell undergoing spindle elongation (interval from 34 to 68 min) and disassembly (interval from 68 to 74 min) is shown (middle cell in the cartoon). The cell above and the cell below the middle cell in the cartoon elongated and aligned their spindles through the bud neck in a wild-type manner. Bars: (A) 10 μm; (C) 2.5 μm.
To test whether mitosis was unlinked from bud formation in cdh1/hct1 swe1 cells, spindle elongation was observed. In cdh1/hct1 swe1 GFP-TUB1 cells, we found unbudded cells with nuclei positioned at the ends of an elongated spindle (Fig. 3A, arrow i) and small-budded cells with nuclei at the end of a disassembling spindle (Fig. 3A, arrow ii). Large-budded cells with an elongated spindle undergoing a normal mitosis were also present (Fig. 3A, arrow iii). These results were similar to the phenotype for cells overexpressing cdc5N209A or Cdc5. Live-cell video microscopy was used to monitor spindle formation in cdh1/hct1 swe1 GFP-TUB1 cells. In Fig. 3C, an unbudded cell undergoing spindle elongation (interval from 34 to 68 min) and disassembly (interval from 68 to 74 min) is shown. This cell completed mitosis, as indicated by spindle breakdown at the 74-min time point, and formed a bud at the 100-min mark (data not shown). In contrast, in most cells that had formed a bud before elongating their spindle, the spindle was properly oriented through the neck into the bud (Fig. 3C, cell above and cell below the center cell in the cartoon). We did observe large-budded cells with misoriented spindles. However, in these rare cases, both the mother and daughter cells were undergoing mitosis and each had a separate spindle (Fig. 3A, inset).
cdc5N209A overproduction suppresses Swe1-dependent phenotypes.
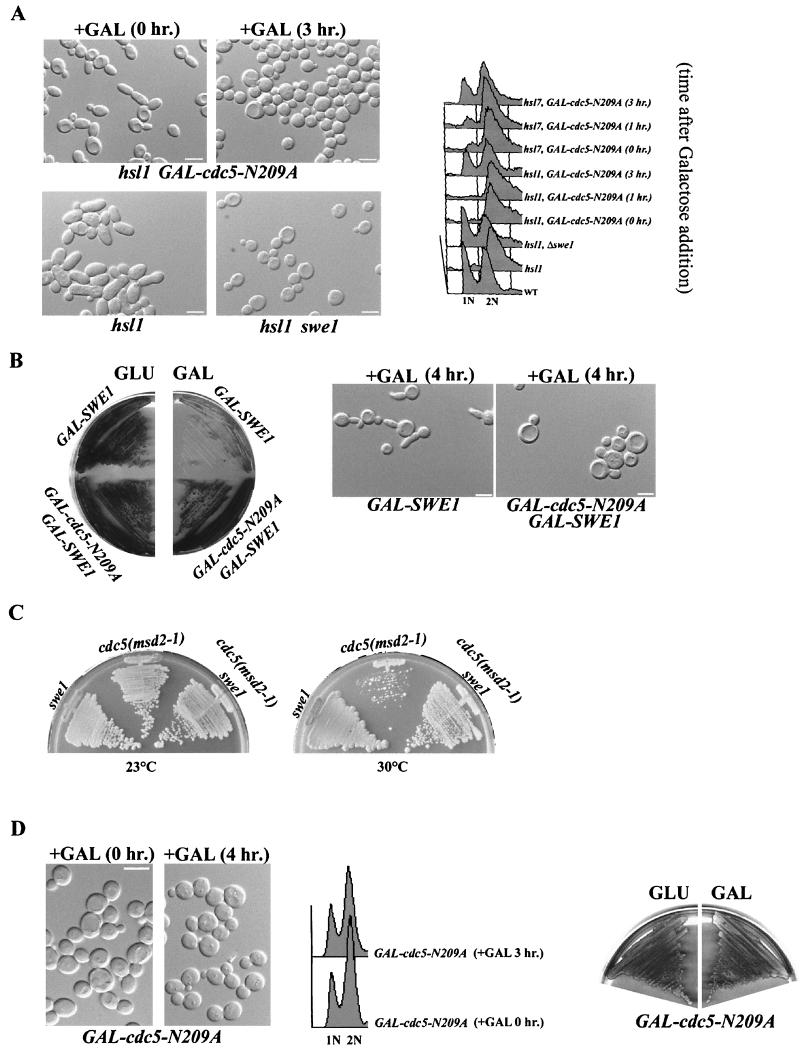
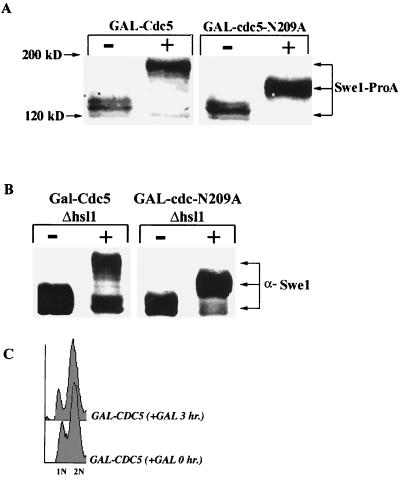
To test whether cdc5N209A overexpression inactivates Swe1 function, we investigated whether overexpression of cdc5N209A in hsl1 or hsl7 mutant cells would suppress the SWE1-dependent defects of these mutants. The hsl1 and hsl7 mutants undergo a prolonged period of apical bud growth during G2 and have Swe1-dependent elongated buds (31). We found that overexpression of cdc5-N209A suppressed the Swe1-dependent elongated bud formation in hsl1 (Fig. 4A) and hsl7 cells. The hsl1 cells overexpressing cdc5-N209A were distinctly round. When the hsl1 and hsl7 cells were examined for DNA content by fluorescence-activated cell sorting (FACS), less than 10% of the hsl1 (Fig. 4A) and hsl7 cells were 1N. These results confirmed a G2 delay for hsl1 and hsl7 cells, as has been previously reported (31). However, a significant fraction of the hsl1 and hsl7 cells overexpressing cdc5-N209A were 1N (Fig. 4A, 3 h after the addition of galactose). The FACS profiles for the hsl1 cells overexpressing cdc5-N209A and the hsl1 swe1 mutant cells were very similar (Fig. 4A). We observed that cdc5-N209A overexpression suppressed the lethality and the formation of elongated buds in response to Swe1 overproduction (Fig. 4B). Taken together, these results supported our model that cdc5-N209A was acting as an inhibitor of Swe1 function. As further evidence, we also found that the extreme slow growth or lethality of cdc5(msd2-1) at 30°C was suppressed in a cdc5(msd2-1) swe1 double mutant (Fig. 4C).
FIG. 4.
cdc5N209A overproduction suppresses Swe1-dependent phenotypes. (A) Strains C508 (hsl1 GAL-cdc5N209A [DIC and FACS]) and C618 (hsl7 GAL-cdc5N209A [FACS only]) were grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for the times indicated. Strains MAY1 (hsl1 [DIC and FACS]), MAY2 (hsl1 swe1 [DIC and FACS]), and W303a (wild type [WT] [FACS only]) were included for comparison. The cells were viewed by DIC (top); they were also stained with propidium iodide and analyzed by FACS (bottom) to evaluate cellular DNA contents. (B) Constitutive expression of SWE1 is toxic and induces elongated buds. Both of these phenotypes were suppressed by cdc5-N209A overproduction. Strains C474 (GAL-Swe1) and C499 (GAL-SWE1 GAL-cdc5-N209A) were streaked on rich plates containing either 2% glucose (repressing conditions) or 2% galactose (inducing conditions) for 5 days (left). Galactose was added to mid-logarithmic-phase cultures of the same strains for 4 h to induce Swe1 (GAL-SWE1) or Swe1 and Cdc5 (GAL-SWE1 GAL-cdc5-N209A) and viewed by DIC (right). (C) Strains C798 (swe1), C745 [cdc5(msd2-1)] and C667 [swe1 cdc5(msd2-1)] were incubated on rich medium at 23 or 30°C for 5 days. (D) Strain JC278 (GAL-cdc5N209A) was grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for the times indicated. The cells were viewed by DIC (left); they were also stained with propidium iodide and analyzed by FACS (middle) to evaluate cellular DNA contents. The same strain was also streaked on rich plates containing either 2% glucose (repressing conditions) or 2% galactose (inducing conditions) for 5 days (right). Bars: 10 μm.
Swe1 is modified and localized to SPBs in response to Cdc5 or cdc5N209A overproduction.
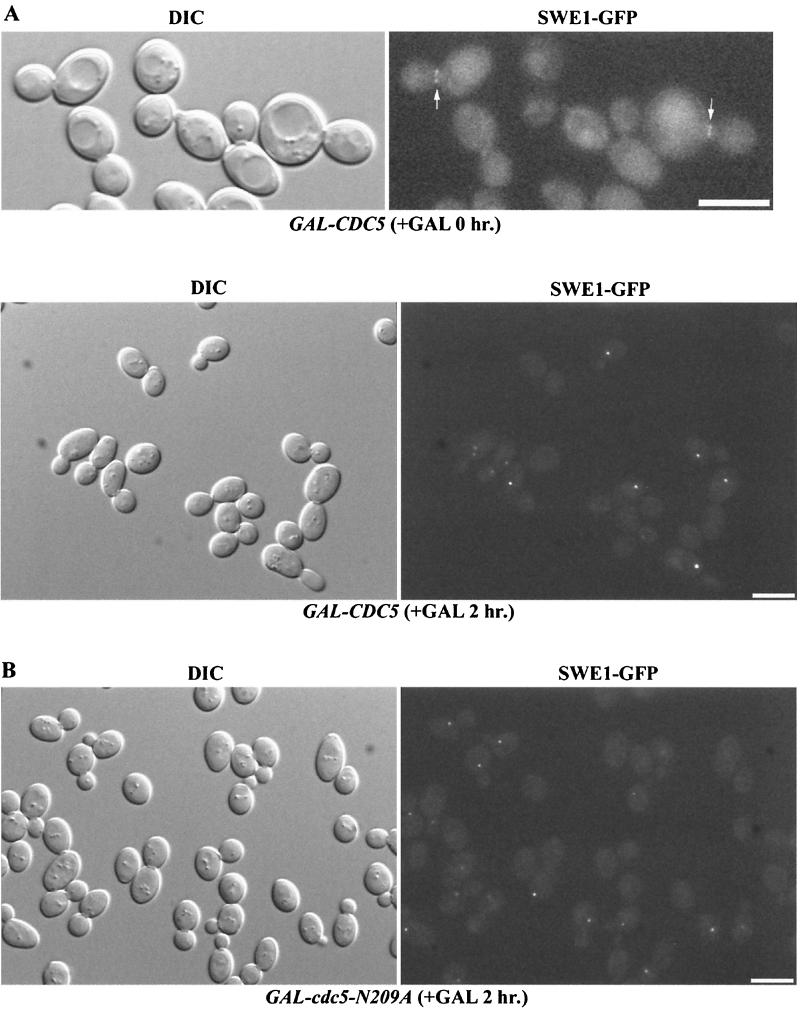
We found that Swe1 is localized to the mother bud neck in a strain expressing Swe1-GFP from the endogenous Swe1 promoter (SWE1-GFP) (Woo and Hardy, unpublished) (Fig. 5A). In contrast, overexpression of Swe1-GFP from the GAL1 promoter (GAL1-SWE1-GFP) resulted in Swe1-GFP localization in the nucleus, at SPBs as well as to the bud neck (Woo and Hardy, unpublished). Interestingly, we observed that endogenous Swe1-GFP localized to one or two bright spots in response to overproduction of Cdc5 (GAL1-CDC5-HA SWE1-GFP) or cdc5-N209A (GAL1-cdc5N209A-HA SWE1-GFP) (Fig. 5). In addition, Swe1-GFP signal was not observed at the neck in cells overexpressing CDC5 or cdc5-N209A. DAPI staining revealed that these spots were on the nuclear periphery of cells (data not shown).
FIG. 5.
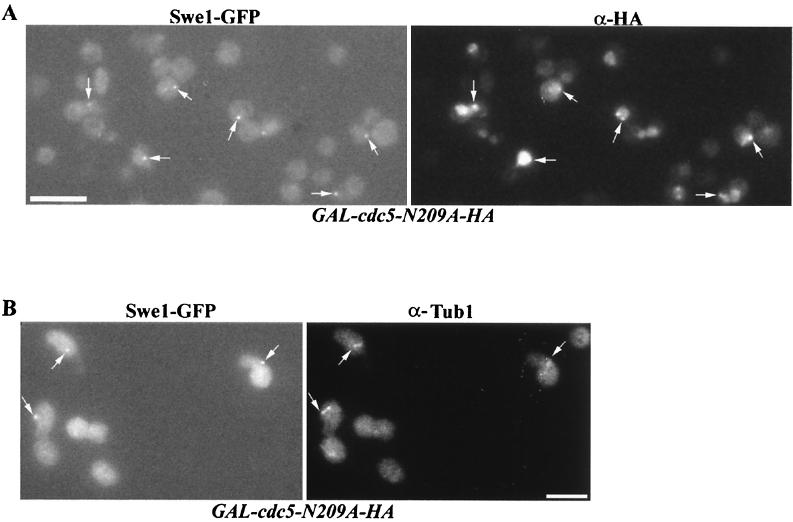
Swe1 is found at one or two distinct spots in cells overexpressing CDC5 or cdc5N209A. (A) Strain C573 (GAL-CDC5-HA SWE1-GFP) was grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for 2 h. (B) Strain C572 (GAL-cdc5-N209A-HA SWE1-GFP) was grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for 2 h. Bars: 10 μm.
The colocalization of Swe1 and Cdc5 in perinuclear spots suggested that this locale might correspond to SPBs. Cdc5 is present at SPBs when expressed under control of its endogenous promoter (41; Woo and Hardy, unpublished) or when overexpressed from the GAL1 promoter (43). Indirect immunofluorescence staining showed that the Swe1-GFP spots colocalized with cdc5-N209A-HA (Fig. 6A) or Cdc5 (data not shown) in response to overproduction of cdc5-N209A or Cdc5, respectively. Combined with anti-Tub1 indirect immunofluorescence microscopy, we determined that Swe1-GFP colocalized to SPBs in response to cdc5N209A overproduction (Fig. 6B). Swe1-GFP also colocalized with SPBs in response to Cdc5 overproduction (data not shown).
FIG. 6.
Swe1 is found associated with cdc5-N209A at the SPBs of cells overexpressing cdc5-N209A. Strain CH572 (GAL-cdc5N209A-HA SWE1-GFP) was grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for 2 h. The cells were fixed and stained for cdc5-N209A-HA (A) or Tub1 (B). Swe1-GFP was visualized by direct fluorescence. Bars: 10 μm.
To examine whether the level or state of Swe1 was altered in cells overexpressing Cdc5 or cdc5N209A, cell extracts were analyzed by immunoblotting. In these experiments, Swe1 was epitope tagged by a chromosomal in-frame integration of the immunoglobulin G binding domain of ProA at the sequence for the Swe1 C terminus. Overexpression of Cdc5 or cdc5N209A resulted in slower electrophoretic mobility for Swe1-ProA (Fig. 7A). The effect was less pronounced in cdc5N209A cells. Because localization of Cdc5 and Swe1 to the mother bud neck is dependent on Hsl1 and Hsl7, Hsl1 and Hsl7 might serve as potential adapters between Cdc5 and Swe1 (30; Woo and Hardy, unpublished). However, electrophoretic shift, and presumed modification, of Swe1-ProA in response to Cdc5 or cdc5N209A overproduction was not dependent on Hsl1 or Hsl7 (Fig. 7B). Furthermore, the localization of Swe1 to SPBs in response to Cdc5 overproduction was also not dependent on Hsl1 or Hsl7 (data not shown).
FIG. 7.
Swe1 is modified in response to Cdc5 or cdc5N209A overproduction. (A) Strains CH473 (GAL-CDC5 SWE1-ProA) and CH459 (GAL-cdc5N209A SWE1-ProA) were grown in raffinose and shifted to 2% galactose. Samples for immunoblot analysis were taken just before (−) and 3 h after (+) addition of galactose. (B) Strains CH507 (hsl1 GAL-CDC5) and CH508 (hsl1 GAL-cdc5N209A) were grown in raffinose and shifted to 2% galactose. Samples for immunoblot analysis were taken just before (−) and 3 h after (+) addition of galactose. (C) Strain JC256 (GAL-CDC5) was grown to mid-logarithmic phase in raffinose. Galactose was added to 2% for the times indicated. The cells were stained with propidium iodide and analyzed by FACS to evaluate cellular DNA contents.
DISCUSSION
Multinucleate formation in response to Cdc5 overexpression is linked to Swe1.
In budding yeast, the Clb/Cdk inhibitor Swe1 does not play a role in mitotic progression in an unperturbed cell cycle and is not required to link bud formation and mitosis (6). In contrast, Swe1 is part of the morphogenesis checkpoint and, in response to perturbations that prevent bud formation, inhibits mitotic progression through negative regulation of Clb/Cdk (24). The results in this paper suggest that the Polo kinase Cdc5 functionally interacts with Swe1. This conclusion is based on multiple pieces of evidence. Most interestingly, a population of cells overproducing Cdc5 has an increased, although low (2.5%), frequency of multinucleate cells. Mitosis appears to be unlinked from bud formation in response to Cdc5 overproduction (Fig. 1). Bud formation requires the action of G1 or Cln cyclins and is inhibited by the mitotic cyclins Clb1 and Clb2, in combination with Cdk (2, 25, 26, 40). Mutants lacking the APC cofactor Cdh1/Hct1 accumulate Clb1/Cdk and Clb2/Cdk activity in G1 (39, 47). We find that a population of cdh1/hct1 null cells has a low frequency of multinucleate cells (1.8%). Strikingly, Cdc5 overproduction or deletion of SWE1 in combination with a deletion in CDH1/HCT1 results in the formation of multinucleate cells at a dramatically higher frequency (18 and 20%, respectively). The cdh1/hct1 swe1 cells undergo mitosis in the absence of bud formation. Therefore, Swe1 prevents the mitotic Cdk from triggering mitotic progression in the absence of bud formation.
Interestingly, Wee1 plays a similar role in S. pombe, where it functions to prevent premature mitosis in cells lacking Ste9, the homolog of Cdh1/Hct1 (21). The comparable responses of cdh1/hct1 cells to the deletion of SWE1 or the overexpression of Cdc5 suggest that Cdc5 overexpression is titrating Swe1 function in the cdh1/hct1 cells. In agreement with this proposal, Swe1-dependent phenotypes in hsl1 and hsl7 mutant cells are suppressed by overexpression of wild-type Cdc5 or a catalytically inactive form (Fig. 4). Because Cdc5 overproduction by itself triggers the formation of multinucleate cells whereas deletion of Swe1 does not, Cdc5 overproduction most likely affects the function of additional factors. It is also possible that high levels of Cdc5 prevent down-regulation of mitotic Cdks by means other than down-regulation of Swe1 activity. It will be interesting to determine whether the interaction between Wee1 and Polo kinases is conserved in other systems. Intriguingly, overexpression of the wild-type or catalytically inactive form of the mammalian Polo kinase Plk1 in HeLa cells results in the formation of multinucleate cells (35).
Polo is a potential negative regulator of Wee1.
In fission yeast, Wee1 is negatively regulated by the Nim1 kinase (9, 12, 36, 38, 51). The C-terminal region of Wee1 that forms the kinase domain is phosphorylated by Nim1 in vitro, resulting in complete inactivation of the Wee1 kinase activity (9, 36, 51). However, regulation of Wee1 most likely involves additional factors, as fission yeast cells lacking Nim1 activity are only slightly delayed in the G2 phase (38). A kinase activity in mitotic Xenopus extracts produces a 60-kDa change in the apparent molecular mass of recombinant fission yeast Wee1 (44). This hyperphosphorylated form of Wee1 is defective for phosphorylation of Clb/Cdk. In contrast to Nim1, the phosphorylation catalyzed by the mitotic Xenopus extracts is restricted to the N-terminal portion of Wee1 (44).
In budding yeast, Swe1 is negatively regulated by the Nim1-like kinase Hsl1, and there is a decrease in the level of Swe1 phosphorylated forms in hsl1 mutant cells (42). It is not known what region of Swe1 is targeted for phosphorylation by Hsl1. Interestingly, we find that Cdc5 interacts with the N-terminal region of Swe1 (Fig. 2). In addition, in cells overproducing Cdc5 or cdc5N209A, Swe1 is converted to a form with an apparent molecular mass increase of 60 or 30 kDa, respectively (Fig. 7). We have not been able to immunoprecipitate the modified forms of Swe1 to determine whether the changes in mobility are due to direct phosphorylation. The largest Swe1 mobility shift to the slowest-migrating form requires Cdc5 kinase activity. However, a shift is observed in cells overexpressing the catalytically inactive cdc5N209A. As Swe1 is targeted to SPBs in response to Cdc5 or cdc5N209A overproduction, other kinases associated with SPBs may be responsible for modifying Swe1. These include both Dbf2 and Cdc15 (14, 52); however, interactions between Swe1 and either Dbf2 or Cdc15 have not been reported.
The inhibitory phosphorylation of the N terminus of Wee1 by the kinase activity in Xenopus mitotic extracts requires the addition of the phosphatase inhibitor okadaic acid (44). Okadaic acid is specific for protein phosphatase 2A (PP2A)-like phosphatases. Interestingly, purification from recombinant baculovirus systems of active Cdc5 and of Plx1 (the Xenopus Polo kinase) requires the addition of okadaic acid (23; Woo and Hardy, unpublished). This correlates with observations that Polo kinases including Cdc5 are activated by phosphorylation (8, 22, 37, 45). Taken together, these results suggest that Plx maybe the N-terminal inhibitory kinase for Wee1. Intriguingly, budding yeast cells that lack the PP2A regulatory subunit Cdc55 are defective in degradation of Swe1 (53). We speculate that cdc55 mutant cells maybe defective in Swe1 degradation because misregulated PP2A targets Cdc5 for inactivation. Alternatively, the misregulated PP2A may target Swe1 or Hsl1.
Possible role for Cdc5 at SPBs.
Mutant analyses in Drosophila, S. pombe, and mammals indicate that Polo kinases play a key role in the formation of bipolar spindles (15). Does Cdc5 play a role in spindle formation? The characterized mutant cdc5 alleles have no reported defects in bipolar spindle formation. However, the cdc5-1 mutant has been linked to microtubule function. After release of the cdc5-1 mutant from a temperature-induced block, the cdc5-1 cells become insensitive to the microtubule-depolymerizing drug methylbenzimidazole-2-yl carbamate (50). In budding yeast, formation of a short bipolar spindle takes places in S phase, and this step is inhibited by overexpression of Swe1 (28). Swe1 expressed from its endogenous promoter is localized to the mother bud neck (30; Woo and Hardy, unpublished). However, when overproduced, Swe1 is also found in the nucleus and at SPBs (Woo and Hardy, unpublished). In this report, we show that Swe1 expressed from its endogenous promoter is targeted to SPBs in response to Cdc5 or cdc5N209A overproduction (Fig. 6). Cdc5 is localized to SPBs just prior to SPB separation, positioning it for a role in Swe1 regulation (Woo and Hardy, unpublished).
In mammalian cells, Wee1 has been localized to the nucleus but has not been found at the microtubule-organizing centers or centrosomes (4, 17). In addition, Wee1 has not been reported to interact with Polo kinases in other systems. However, it is interesting that in fission yeast cells with a mutation in the SPB component Stf1/Cut12, the requirement for Cdc25 is bypassed and Plo1, the Polo kinase in S. pombe, is prematurely recruited to the SPB (7). Cdc25 is a tyrosine phosphatase that opposes Wee1 function and activates Cdc2. The stf1/cut12 mutant cells may bypass Cdc25 by allowing Plo1 to prematurely access and negatively regulate Wee1.
Polo interacts with multiple regulators of mitotic Cdk activity.
Plx1, the Polo kinase in Xenopus, was isolated as a Cdc25-interacting factor, and studies have suggested that Polo activates Cdc2 by phosphorylating and activating Cdc25 (23, 37). Recently it has been shown that the Polo kinase Plk1 in vertebrate cells regulates the nuclear localization of cyclin B1 (46). Plk1 phosphorylates an essential serine residue in the nuclear export signal sequence of cyclin B1 allowing the Cdc2-cyclin B1 complex to accumulate in the nucleus during prophase. Our work shows that Cdc5, the Polo kinase in budding yeast, interacts with Swe1. Further work will be required to determine the function of such a Polo-Wee1 interaction. However, combined with these earlier results, our results lead us to conclude that Polo kinase is a key coordinator for activation of Cdc2 during G2/M.
ACKNOWLEDGMENTS
We thank D. Morgan, J. Charles, J. Kilmartin, R. Booher, A. Straight, A. Murray, A. Amon, M. Grunstein, K. Nasmyth, and D. Lew for strains and plasmids. We thank S. Wente and H. Piwnica-Worms for critical comments.
This work was supported by a grant from NIH to C.F.J.H.
REFERENCES
- 1.Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 3.Bahler J, Steever A B, Wheatley S, Wang Y, Pringle J R, Gould K L, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldin V, Ducommun B. Subcellular localization of human wee1 kinase is regulated during the cell cycle. J Cell Sci. 1995;108:2425–2432. doi: 10.1242/jcs.108.6.2425. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae Wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge A J, Morphew M, Bartlett R, Hagan I M. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev. 1998;12:927–942. doi: 10.1101/gad.12.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L, Hunke L, Hardy C F J. Cell cycle regulation of the Saccharomyces cerevisiae Polo-like kinase Cdc5p. Mol Cell Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman T R, Tang Z, Dunphy W G. Negative regulation of the wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 10.Cormack B, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson A D, Kilmartin J V. Spc42: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunphy W G. The decision to enter mitosis. Trends Cell Biol. 1994;4:202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 13.Epstein C B, Cross F R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 14.Frenz L, Lee S, Fesquet D, Johnston L. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck late in mitosis. J Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 15.Glover D M, Hagan I, Tavares A A M. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 16.Golsteyn R M, Mundt K E, Fry A M, Nigg E A. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heald R, McLoughlin M, McKeon F. Human Wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 18.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs C W, Adams A E M, Szaniszlo P J, Pringle J R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamura K, Maekawa H, Shimoda C. Fission yeast Ste9, a homolog of Hct1/Cdh1 and fizzy related, is a novel negative regulator of cell cycle progression during G1 phase. Mol Biol Cell. 1998;9:1065–1080. doi: 10.1091/mbc.9.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotani K, Tugendreich S, Fujii M, Jorgensen P, Watanabe N, Hoog C, Hieter P, Todokoro K. PKA and MPF-activated Polo-like kinase regulate anaphase promoting complex activity and mitotic progression. Mol Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai A, Dunphy W. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 24.Lew D J. Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr Opin Genet Dev. 2000;10:47–53. doi: 10.1016/s0959-437x(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 25.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 26.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 607–696. [Google Scholar]
- 28.Lim H H, Goh P-H, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephoshorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logarinho E, Sunkel C E. The Drosophila mitotic kinase Polo localises to the centrosome, centromeres and spindle midzone during mitosis and contributes to the phosphorylation of MPM2 reactive epitopes. J Cell Sci. 1998;111:2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- 30.Longtine M S, Theesfeld C L, McMillan J N, Weaver E, Pringle J R, Lew D J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma X J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 32.McMillan J N, Longtine M S, Sia R A, Theesfeld C L, Bardes E S, Pringle J R, Lew D J. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillan J N, Sia R A L, Lew D J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulvihill D P, Petersen J, Ohkura H, Glover D M, Hagan I M. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol Biol Cell. 1999;10:2771–2785. doi: 10.1091/mbc.10.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundt K E, Golsteyn R M, Lane H A, Nigg E A. On the regulation and function of human Polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239:377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 36.Parker L L, Walter S A, Young P G, Piwnica-Worms H. Phosphorylation and inactivation of the mitotic inhibitor wee1 by the nim1/cdr1 kinase. Nature. 1993;363:736–738. doi: 10.1038/363736a0. [DOI] [PubMed] [Google Scholar]
- 37.Qian Y, Erikson E, Li C, Maller J L. Activated Polo-Like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol Cell Biol. 1998;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell P, Nurse P. The mitotic inducer nim1+ functions in a regulatory network of protein kinase homologs controlling the initiation of mitosis. Cell. 1987;49:569–76. doi: 10.1016/0092-8674(87)90459-4. [DOI] [PubMed] [Google Scholar]
- 39.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 40.Shimada Y, Gulli M, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during cell mating. Nat Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- 41.Shirayama M, Zachariae W, Coisk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20/fizzzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shulewitz M J, Inouye C J, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song S, Grenfell T, Garfield S, Erikson R, Lee K. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Coleman T R, Dunphy W G. Two distinct mechanisms for negative regulation of the Wee1 protein kinase. EMBO J. 1993;12:3427–3436. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavares A, Glover D, Sunkel C. The conserved mitotic kinase polo is regulated by phosphorylation and has preferred microtubule-associated substrates in Drosophila embryo extracts. EMBO J. 1996;15:4873–4883. [PMC free article] [PubMed] [Google Scholar]
- 46.Toyoshima-Morimoto F, Taniguchi E, Shinya N, Iwamatsu A, Nishida E. Polo-like kinases phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. 2001;410:215–220. doi: 10.1038/35065617. [DOI] [PubMed] [Google Scholar]
- 47.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 48.Waddle J A, Karpova T S, Waterston R H, Cooper J A. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wente S R, Rout M P, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood J S, Hartwell L H. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982;94:718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu L, Russell P. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature. 1993;363:738–741. doi: 10.1038/363738a0. [DOI] [PubMed] [Google Scholar]
- 52.Xu S, Huang H, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Jiang W, Gentry M, Hallberg R L. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1 degradation. Mol Cell Biol. 2000;20:8143–8156. doi: 10.1128/mcb.20.21.8143-8156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]