Abstract
Background
Antibiotic growth promoters (AGPs) are commonly used within poultry production to improve feed conversion, bird growth, and reduce morbidity and mortality from clinical and subclinical diseases. Due to the association between AGP usage and rising antimicrobial resistance, the industry has explored new strategies including the use of probiotics and other microbial-based interventions to promote the development of a healthy microbiome in birds and mitigate against infections associated with food safety and food security. While previous studies have largely focused on the ability of probiotics to protect against Clostridium perfringens and Salmonella enterica, much less is known concerning their impact on Campylobacter jejuni, a near commensal of the chicken gut microbiome that nevertheless is a major cause of food poisoning in humans.
Results
Here we compare the efficacy of four microbial interventions (two single strain probiotics, the bacterium—Pediococcus acidilactici, and the yeast—Saccharomyces cerevisiae boulardii; and two complex, competitive exclusion, consortia—Aviguard and CEL) to bacitracin, a commonly used AGP, to modulate chicken gut microbiota and subsequently impact C. jejuni infection in poultry. Cecal samples were harvested at 30- and 39-days post hatch to assess Campylobacter burden and examine their impact on the gut microbiota. While the different treatments did not significantly decrease C. jejuni burden relative to the untreated controls, both complex consortia resulted in significant decreases relative to treatment with bacitracin. Analysis of 16S rDNA profiles revealed a distinct microbial signature associated with each microbial intervention. For example, treatment with Aviguard and CEL increased the relative abundance of Bacteroidaceae and Rikenellaceae respectively. Furthermore, Aviguard promoted a less complex microbial community compared to other treatments.
Conclusions
Depending upon the individual needs of the producer, our results illustrate the potential of each microbial interventions to serve flock-specific requirements.
Supplementary Information
The online version contains supplementary material available at 10.1186/s42523-021-00157-6.
Keywords: Probiotics, Campylobacter jejuni, Microbiome, Infection challenge
Introduction
Subtherapeutic doses of antimicrobial growth promoters (AGPs) are commonly used within poultry production to supplement bird diets to improve feed conversion, bird growth, and reduce morbidity and mortality from clinical and subclinical diseases [1]. Since AGPs are not absorbed from the intestines, they do not act therapeutically and have no systematic effects [1]. While the exact mechanism by which AGPs promote growth remains unknown, it has been proposed that they operate through modulating the gut microbiota [2]. One hypothesis is that AGPs exert an overall antibacterial effect, which may protect nutrients from bacterial destruction, decrease production of toxins by intestinal bacteria, and/or reduce incidence of subclinical intestinal infection [1]. Consequently, AGPs have been seen as offering prophylactic benefits by mitigating against infectious agents such as Clostridium perfringens, the causative agent of necrotic enteritis, capable of decimating entire flocks [3–5]. Although AGPs have been shown to be effective against C. perfringens, their impact against other pathogens, particularly those associated with food safety such as Campylobacter jejuni is less documented [4, 6–8]. C. jejuni is considered to possesses a near commensal relationship with chickens and is typically acquired after three weeks of age resulting in asymptomatic infections [9, 10]. In contrast, in humans C. jejuni is a leading source of foodborne illness in industrialized countries [11]. With links between the use of AGPs and the ability of these pathogens to acquire novel mechanisms of antimicrobial resistance there is an urgent need to identify efficacious alternatives, capable of promoting the development of a healthy microbiome [1, 3].
To address global bans on the use of AGPs, the poultry industry has explored the use of probiotics, natural microbes that confer a beneficial effect on their host [12, 13]. Single strain probiotic species including species of Streptococcus, Bacillus, Bifidobacterium, Enterococcus, Lactobacillus, Pediococcus and Saccharomyces, have been shown to have beneficial effects on broiler performance, laying hens, modulation of intestinal gut microbiota and pathogen inhibition [13–16]. However, probiotics have shown varying results between products and farms [17], limiting our understanding of their specific impact on the composition of the gut microbiome. In the case of probiotics this may be in part explained by recent findings of person-, region- and strain-specific patterns of sensitivity to colonization by probiotics in humans [18]. In attempts to overcome these limitations, the use of more complex communities that engraft within the gastro-intestinal (GI) tract, stabilized through mutualistic interactions that rely on metabolic cooperation to exploit specific substrates, is receiving renewed attention [19, 20]. Such interest has been driven by recent successes of fecal microbiota transplantation in humans (FMT) [21, 22]. Applied to poultry, FMTs have been shown to increase feed efficiency, growth performance, immune function and to reduce bacterial infections such as Salmonella [23, 24]. In challenge experiments, transfer of a whole mature microbiome to newly-hatched chicks also reduced transmission and colonisation of C. jejuni [25], reduced mortality from necrotizing enteritis (NE) [26] and decreased colonization by pathogenic and antibiotic resistant E. coli [27].
Given the range of microbial based products available and limited data on their impact on C. jejuni infections, we sought to compare a set of four industrially-relevant microbial interventions to bacitracin, an AGP that has been widely used, on their efficacy against a model of C. jejuni infection in poultry [28]. Among these products were two single strain probiotics: Pediococcus acidilactici and Saccharomyces boulardii; a commercial competition exclusion product, Aviguard; and another experimental competition exclusion, CEL. In addition, we performed 16S rRNA sequence surveys to explore the ability of each product to engraft within the GI tract, alter the composition of the GI microbiome and identify taxa that might be associated with reduction of C. jejuni burden. We further investigated the impact of C. jejuni on gut microbiome in the presence and absence of AGPs.
Results
Two of four microbial solutions reduced colonization of C. jejuni in mature broilers relative to bacitracin
The purpose of this study was to monitor the impact of select microbial solutions on the chicken gut microbiome and their ability to reduce colonization by C. jejuni. To that end, we performed a challenge trial involving a total of 160 broiler chickens (Ross 708; Fig. 1A). One day old chicks were split into 8 groups and subjected to different treatments (see Methods). Four groups received one of the following products: Pediococcus acidilactici (a probiotic bacteria); Saccharomyces cerevisiae boulardii (a probiotic yeast); Aviguard (a commercially available probiotic consortium); and CEL (an experimental probiotic consortium). Two groups received bacitracin (AGP) and two groups were left untreated. At day 14, all groups were orally challenged with C. jejuni apart from two groups to act as control (one bacitracin and one untreated group). At days 30 and 39 post hatch (timepoints associated with peak C. jejuni colonization and broiler slaughter), birds were sacrificed and cecal contents recovered for 16S rDNA sequencing.
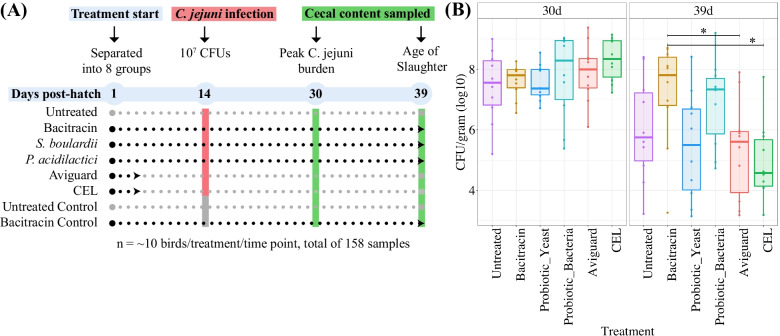
Fig. 1.
Schematic of Experimental Design and C. jejuni burden. A Experimental design. Chickens were divided randomly into 8 groups and assigned to either one of 6 treatments or left untreated at day one of age (Aviguard, CEL, probiotic bacteria (Pediococcus acidilactici CNCMI-4622), probiotic yeast (Saccharomyces cerevisiae boulardii CNCMI-1079), AGP (bacitracin)). On day 14, all groups, except for two (one bacitracin and one untreated group) were orally challenged with 107 CFUs of C. jejuni (strain 81–176). Samples of cecal contents were collected at day 30 and 39 of age (n = 10 birds per time point and treatment group, total of 158 samples for CFU enumeration and 16S rRNA sequencing). After sample collection, tenfold serial dilutions in PBS were plated onto Muller Himton agar containing Preston Campylobacter Selective Supplement. Plates were incubated in microaerophilic conditions at 41 °C and CFUs of C. jejuni were enumerated after 40–48 h and expressed as log10 Campylobacter /g of cecal content. B Boxplots representing the median of CFUs of C. jejuni in each treatment group. Statistical analysis was performed using Kruskal–Wallis test followed by pairwise Dunn's tests. *P < 0.05
Based on cecal colony forming units (CFUs), we found that at 30 days post-hatch none of the probiotic products or bacitracin resulted in significant differences in C. jejuni burden relative to the untreated control (Fig. 1B). At 39 days post hatch, we found that treatment with Aviguard and CEL exhibited a significant decrease in C. jejuni burden relative to birds treated with bacitracin (P < 0.05, Post-hoc pairwise Dunn’s Test). While S. cerevisiae boulardii resulted in a notable decrease in C. jejuni burden relative to bacitracin, this decrease was not found to be statistically significant. We observed no significant differences between any of these treatments and the untreated control group. Aviguard’s ability to lower C. jejuni burden in our study mirrors the success found in other studies where it was able to inhibit intestinal colonization of Clostridium perfringens and Salmonella typhimurium in chickens [29–32]. As such, these results suggest replacing dietary bacitracin with Aviguard, in addition to protecting against necrotic enteritis induced by C. perfringens, as well as colonization by S. typhimurium, may additionally result in a significant reduction in C. jejuni burden.
Alpha diversity in the cecal microbial community increases with age, C. jejuni challenge, and bacitracin AGP treatment
Given the decrease in C. jejuni burden associated with the microbial consortia, we were interested in understanding how the cecal microbiota responds to each treatment and challenge. From the cecal samples harvested from each bird at day 30 and day 39 post hatch, we extracted DNA and performed 16S rDNA surveys. Overall, 4,519,255 paired-end reads were generated. After quality filtering, we retained 2,195,510 paired-end reads with a median of 13,562 reads (5,753–29,542 reads; Additional file 12: Figure S1A) per sample. Sequences were clustered into 1,305 Operational Taxonomic Units (OTUs) based on a de novo assignment with similarity set at 97% (Additional file 12: Figure S1B). Of these, 17 OTUs, represented by 75 reads (0.003% of retained reads) assigned as bacteria, could not be assigned to a phylum. A further 259 OTUs (represented by 72,748 or 3.3% of reads) could not be assigned to a family and 928 OTUs (represented by 961,941 or 43.8% of reads) could not be assigned to a genus. Across all samples, our results confirmed Firmicutes as the major phyla in the adult chicken gut microbiome, represented by 89% ± 14% (mean ± std dev) of assigned reads, compared to Bacteroidetes at 7.1% ± 13%. The most abundant families within the Firmicutes phylum were Ruminococcaceae represented by 34% ± 11% of assigned reads, followed by Clostridiales vadinBB60 group at 24% ± 12% and Lachnospiraceae at 24% ± 11%. The Bacteroidetes phylum consisted of Bacteroidaceae at 3.6% ± 11% and Rikenellaceae at 3.5% ± 8.4%. C. jejuni a member of Epsilonproteobacteria was represented by 0.14% ± 0.30% of all assigned reads. C. jejuni sequence counts exhibited significant correlation with the CFUs of C. jejuni obtained above at both time points (p < 0.001; Spearman’s rank correlation coefficient; Additional file 12: Figure S2A and B). Consistent with the negative CFU results reported for the two unchallenged control groups, we did not find any reads mapping to C. jejuni in either of these samples (Additional file 13: Figure S3). Given that 16S rDNA data provide only information on relative and not absolute abundance, we rely on CFUs as a more reliable indicator of C. jejuni burden to compare treatment effect on pathogen clearance. Furthermore, since only the 39-day samples yielded significant differences in C. jejuni burden, we focus subsequent analyses on samples associated with this later timepoint.
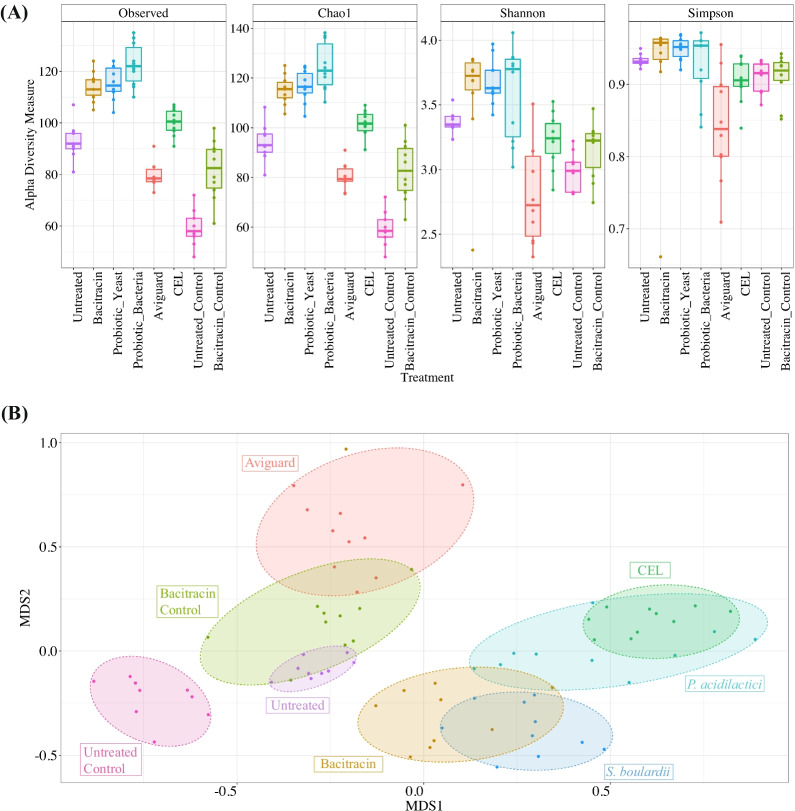
Using four measures of alpha diversity, two measures of species richness (observed number of OTUs and Chao1) and two measures of species evenness (Shannon and Simpson indices), we identified a spectrum of diversity across treatments at day 39 (Fig. 2A). Across all treatment groups, the unchallenged and untreated exhibited the lowest species richness, while the P. acidilactici group had the highest (post-hoc pairwise Wilcoxon of Observed and Chao1; Table 1; Fig. 2A). The former finding potentially highlights the lack of any influence from either treatment and/or challenge with C. jejuni. Samples from birds treated with P. acidilactici also exhibited the highest species evenness (post-hoc pairwise Wilcoxon of Shannon and Simpson; Table 1; Fig. 2A). Interestingly, we found that Aviguard samples, which exhibited the lowest species richness across the challenged groups, were also associated with notably lower species evenness, suggesting the promotion of a relatively less complex microbiome that is less evenly distributed relative to the other treatments (post-hoc pairwise Wilcoxon of Observed, Chao1, Shannon and Simpson; Table 1; Fig. 2A).
Fig. 2.
Alpha and beta diversity of cecal microbial communities. A Boxplots representing alpha diversity metrics of richness (observed number of OTUs and Chao1) and evenness (Shannon and Simpson) for 39-day samples grouped according to treatment (no. of OTUs at 97% similarity). Each point represents the diversity score for a sample, colour-coded according to treatment. B Non-metric multidimensional scaling (nMDS) plot based on Bray–Curtis dissimilarity matrix on relative abundance data in 39-day samples. Colours indicate treatment group
Table 1.
Significant differences in alpha diversity indices in pairwise comparisons between treatments
| Aviguard | Bacitracin | Bacitracin_Control | CEL | Probiotic_Bacteria | Probiotic_Yeast | Untreated | |
|---|---|---|---|---|---|---|---|
| 30-day samples | |||||||
| Observed | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | – | * | |||||
| CEL | ** | – | ** | ||||
| Probiotic_Bacteria | *** | ** | *** | ** | |||
| Probiotic_Yeast | ** | – | ** | – | *** | ||
| Untreated | ** | – | * | – | *** | – | |
| Untreated_Control | ** | ** | ** | *** | *** | *** | *** |
| Chao1 | |||||||
| Bacitracin | * | ||||||
| Bacitracin_Control | – | * | |||||
| CEL | ** | – | ** | ||||
| Probiotic_Bacteria | *** | ** | *** | *** | |||
| Probiotic_Yeast | * | – | ** | – | *** | ||
| Untreated | * | – | * | * | *** | – | |
| Untreated_Control | ** | *** | ** | *** | *** | *** | *** |
| Shannon | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | *** | – | |||||
| CEL | *** | – | – | ||||
| Probiotic_Bacteria | *** | *** | *** | *** | |||
| Probiotic_Yeast | *** | – | – | – | ** | ||
| Untreated | *** | – | ** | – | ** | – | |
| Untreated_Control | *** | – | – | – | *** | – | ** |
| Simpson | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | *** | – | |||||
| CEL | *** | – | – | ||||
| Probiotic_Bacteria | *** | ** | ** | ** | |||
| Probiotic_Yeast | *** | – | – | – | * | ||
| Untreated | *** | – | * | * | ** | – | |
| Untreated_Control | ** | – | – | – | ** | – | – |
| 39-day samples | |||||||
| Observed | |||||||
| Bacitracin | *** | ||||||
| Bacitracin_Control | – | *** | |||||
| CEL | *** | *** | *** | ||||
| Probiotic_Bacteria | *** | * | *** | *** | |||
| Probiotic_Yeast | *** | – | *** | *** | – | ||
| Untreated | ** | *** | * | * | *** | *** | |
| Untreated_Control | *** | *** | *** | *** | *** | *** | *** |
| Chao1 | |||||||
| Bacitracin | *** | ||||||
| Bacitracin_Control | – | *** | |||||
| CEL | *** | *** | ** | ||||
| Probiotic_Bacteria | *** | * | *** | *** | |||
| Probiotic_Yeast | *** | – | *** | *** | – | ||
| Untreated | *** | *** | * | * | *** | *** | |
| Untreated_Control | *** | *** | ** | *** | *** | *** | *** |
| Shannon | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | * | ** | |||||
| CEL | * | ** | – | ||||
| Probiotic_Bacteria | ** | – | * | – | |||
| Probiotic_Yeast | *** | – | *** | *** | – | ||
| Untreated | ** | * | ** | - | – | *** | |
| Untreated_Control | – | ** | – | * | ** | *** | *** |
| Simpson | |||||||
| Bacitracin | * | ||||||
| Bacitracin_Control | – | * | |||||
| CEL | – | * | – | ||||
| Probiotic_Bacteria | * | – | – | – | |||
| Probiotic_Yeast | ** | – | ** | ** | – | ||
| Untreated | * | – | – | * | – | * | |
| Untreated_Control | – | * | – | – | – | ** | * |
P-values were calculated using post-hoc pairwise Wilcoxon rank sum tests for four indices (Observed, Chao1, Shannon, Simpson). Samples are grouped by time point
*p < 0.05; **p < 0.01; ***p < 0.001
Comparing the two unchallenged control groups (untreated and bacitracin) with their respective challenged groups, we found that C. jejuni challenge resulted in increased species richness and increased species evenness at 39 days (post-hoc pairwise Wilcoxon of Observed, Chao1, Shannon and Simpson; Table 1; Fig. 2A). Comparing 39-day samples between the bacitracin and untreated group, we also found that bacitracin treatment increased species richness, in the absence and presence of C. jejuni (post-hoc pairwise Wilcoxon of Observed and Chao1; Table 1; Fig. 2A). In contrast, bacitracin treatment had no impact on species evenness relative to the untreated group.
Each treatment results in a unique microbial signature
We were next interested in comparing the impact of each treatment on the makeup of the microbial communities in the ceca. Using the Bray–Curtis metric as a measure of beta-diversity, we conducted permutational multivariate analysis of variance (PERMANOVA) and permutational analysis of multivariate dispersions (PERMDISP) to measure whether there is a significant separation of samples by treatment or time point due to differences in microbiome structure. PERMANOVA evaluates the significance of separation between centroids of samples grouped by treatment or time point but is sensitive to multivariate dispersion. PERMDISP assesses if the distribution or spread of two sample groups are significantly different and is conducted in parallel to PERMANOVA. For example, a non-significant PERMDISP result would reveal that two sample groups have similar dispersions, which places confidence in a significant PERMANOVA result as it is likely not due to differences in group dispersions. A significant PERMDISP result would mean that two sample groups have different dispersions, therefore a significant PERMANOVA result cannot be interpreted to sample groups having significant separation [33]. To assess if different treatment groups were significantly associated with microbial community structure, we conducted pairwise PERMANOVA and PERMDISP on samples grouped by time point and treatment. Results from these analyses showed that, although most treatment groups at 39 days had significant separation from each other, untreated birds challenged with C. jejuni did not exhibit a significantly different community structure from challenged birds treated with bacitracin, P. acidilactici or S. cerevisiae boulardii (Table 2; Fig. 2B). Consistent with the comparisons of alpha diversity, C. jejuni challenge resulted in a significantly different community structure in both the untreated and bacitracin groups (Table 2; Fig. 2B).
Table 2.
Significant differences in microbial community structure in pairwise comparisons between treatments
| Aviguard | Bacitracin | Bacitracin_Control | CEL | Probiotic_Bacteria | Probiotic_Yeast | Untreated | |
|---|---|---|---|---|---|---|---|
| 30-day samples | |||||||
| PERMANOVA | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | ** | ** | |||||
| CEL | ** | ** | ** | ||||
| Probiotic_Bacteria | ** | ** | ** | ** | |||
| Probiotic_Yeast | ** | ** | ** | ** | ** | ||
| Untreated | ** | ** | ** | ** | ** | ** | |
| Untreated_Control | ** | ** | ** | ** | ** | ** | ** |
| PERMDISP | |||||||
| Bacitracin | – | ||||||
| Bacitracin_Control | – | – | |||||
| CEL | – | – | – | ||||
| Probiotic_Bacteria | – | – | – | – | |||
| Probiotic_Yeast | – | – | – | – | – | ||
| Untreated | – | – | – | – | – | – | |
| Untreated_Control | – | – | – | – | – | – | – |
| PERMANOVA | |||||||
| 39-day samples | |||||||
| Bacitracin | ** | ||||||
| Bacitracin_Control | ** | ** | |||||
| CEL | ** | ** | ** | ||||
| Probiotic_Bacteria | ** | ** | ** | ** | |||
| Probiotic_Yeast | ** | ** | ** | ** | ** | ||
| Untreated | ** | ** | ** | ** | ** | ** | |
| Untreated_Control | ** | ** | ** | ** | ** | ** | ** |
| PERMDISP | |||||||
| Bacitracin | – | ||||||
| Bacitracin_Control | – | – | |||||
| CEL | – | – | – | ||||
| Probiotic_Bacteria | – | – | – | – | |||
| Probiotic_Yeast | – | – | – | – | – | ||
| Untreated | – | * | – | – | * | * | |
| Untreated_Control | – | – | – | – | – | – | – |
P values measured by pairwise PERMANOVA and PERMDISP tests of the Bray–Curtis distances conducted in parallel
*p < 0.05; **p < 0.01; ***p < 0.001
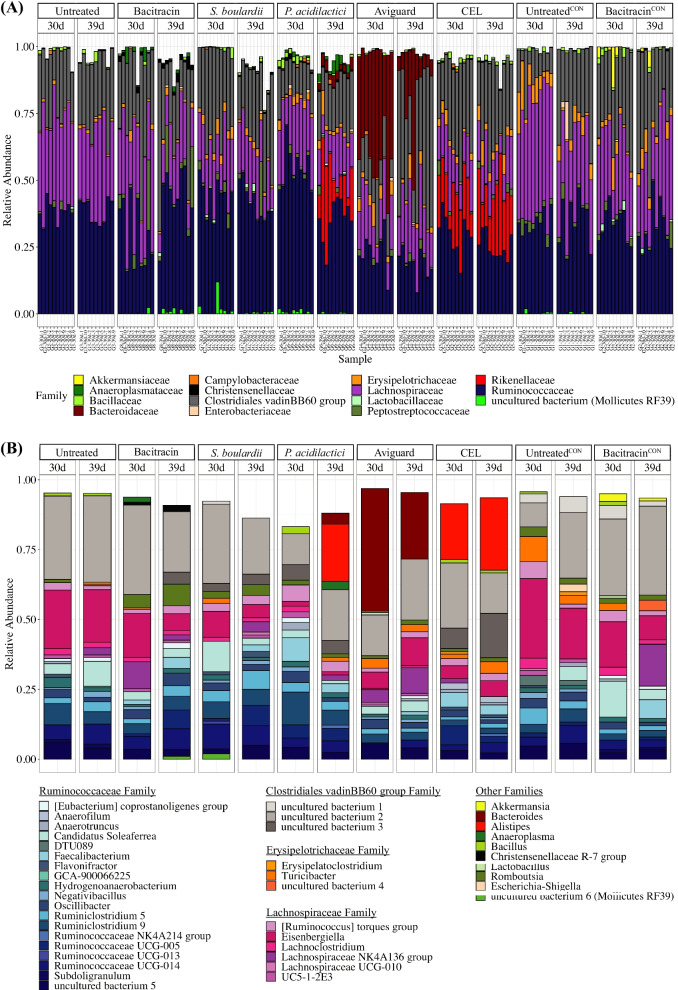
Consistent with observations from the beta diversity analyses, assigning the 1305 OTUs to discrete taxa revealed unique microbial profiles for each treatment and time point (Fig. 3A, B). For example, at both timepoints, treatment with CEL (and P. acidilactici at 39 days), resulted in relatively high abundances of Rikenellaceae; in contrast Aviguard resulted in a high abundance of Bacteroidaceae; while bacitracin yielded signatures associated with Akkermansiaceae among unchallenged birds.
Fig. 3.
Microbial community composition of chicken cecal content. A Stacked bar plots representing relative abundances of the top 15 families in all samples, grouped by treatment and age. Legend lists taxonomic families in order of appearance B Stacked bar plots representing relative abundances of the top 20 genera in each treatment and timepoint
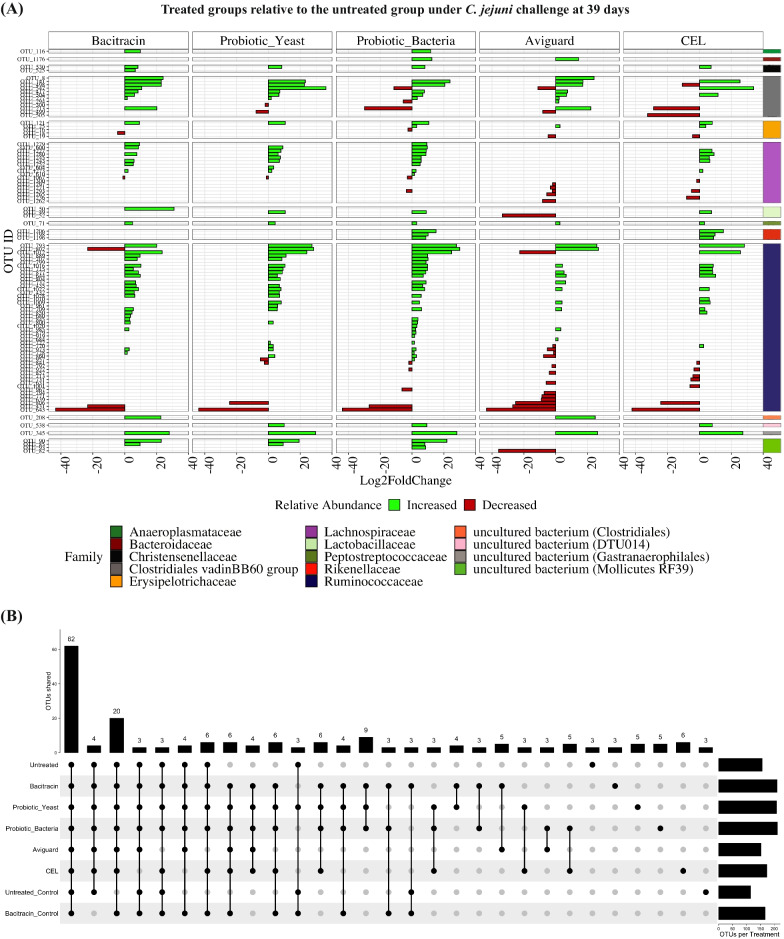
Applying DESeq2 [34] to the 39 day samples, 103 OTUs exhibited differential abundance between treated and untreated groups under C. jejuni challenge (Fig. 4A). Most were associated with a greater relative abundance in the treated groups. A total of 10 OTUs were consistently increased across all treated groups and OTU643 (assigned to Ruminococcaceae) was decreased across all treated groups. OTU19 (Erysipelotrichaceae) was found to be decreased only in groups treated with microbial consortia, CEL and Aviguard. Treatment groups that did not significantly lower C. jejuni burden relative to the untreated group (bacitracin, P. acidilactici and S. cerevisiae boulardii) exclusively shared 9 OTUs that were increased and OTU1067 (assigned to Lachnospiraceae) was commonly decreased. Treatment groups that resulted in a non-significant increase of C. jejuni burden (bacitracin and P. acidilactici) shared 5 differential OTUs that were increased. Furthermore, 5 OTUs (3 increased, 2 decreased) were specific to samples from the challenged bacitracin group. A total of 10 OTUs (7 increased, 3 decreased) were specific to P. acidilactici while 5 OTUs (3 increased, 2 decreased) were exclusive to S. cerevisiae boulardii. Specific to CEL and Aviguard treatment groups, we identified 8 OTUs (1 increased, 7 decreased) and 19 OTUs (2 increased, 17 decreased) respectively.
Fig. 4.
Changes in abundance of OTUs across treatments. A Logarithmic fold changes of differential OTUs from differential abundance analyses with DESeq2 at 39 days post-hatch, comparing treated groups to the untreated group under C. jejuni challenge. OTUs that are in significantly greater or lesser abundance are represented in green and red coloured bars respectively. OTUs are further grouped by their assigned taxonomic families. Legend lists taxonomic families in order of appearance B Vertical bars in the UpSet plot visualizes the number of OTUs unique to each treatment group and the number of OTUs shared between treatment groups at 39 days post-hatch. Horizontal bars represent the total number of OTUs found in each treatment group
Overall, there were significant changes in the abundance of OTUs assigned to Clostridiales vadinBB60 group, Erysipelotrichaceae Lachnospiraceae, Lactobacillaceae, Ruminococcaceae and Peptostreptococcaceae in all samples from challenged birds relative to the challenged untreated birds at both timepoints (Additional file 1: Table S1 and Additional file 2: Table S2; Fig. 4A and Additional file 1: Figure S5A). Of note, all challenged and treated groups at 39 days showed increased relative abundances of a single OTU (OTU71) assigned to Peptostreptococcaceae. Further, we noted that OTU116, assigned to Anaeroplasma, was consistently found in increased abundance in samples from birds treated with bacitracin or P. acidilactici, two of the three treatments that did not mitigate against C. jejuni challenge.
Differential OTUs unassigned at the family level but assigned to either Clostridiales, DTU014 (Clostridia), Gastranaerophilales and Mollicutes RF39 were found among samples from treated groups relative to the challenged, untreated group. Clostridiales was found at an increased relative abundance in 39-day samples from bacitracin and Aviguard groups. OTU538 assigned to DTU014 (Clostridia) was at an increased differential abundance in 39-day samples from birds treated with either S. cerevisiae boulardii, P. acidilactici or CEL. Gastranaerophilales (OTU345) was found at increased relative abundance in all challenged, treatment groups at 39-days. At 39 days, Mollicutes RF39 were at increased relative abundances among samples from bacitracin, S. cerevisiae boulardii and P. acidilactici groups and showed decreased relative abundance in Aviguard samples. No differential OTUs assigned to Mollicutes RF39 were found in CEL samples at 39 days.
Beta diversity analyses and review of taxonomic profiles suggest that each treatment overall resulted in a unique microbial signature. The following sections provide an in-depth review on the key findings associated with each treatment.
Bacitracin treatment yields signatures of Akkermansiaceae in unchallenged conditions
As the impact of AGP-use on C. jejuni in poultry is not well known, we compared the effect of bacitracin on the challenged chickens relative to the unchallenged controls [4, 6, 7]. From the alpha diversity studies above, we noted that bacitracin increased the number of bacterial OTUs in the cecal microbiome but did not promote a more evenly distributed community (Table 1; Fig. 2A and Additional file 13: Figure S4A). Beta diversity plots coupled with PERMANOVA and PERMDISP analyses showed that communities treated with bacitracin significantly differed from those associated with other treatments at 30 days and that community composition is affected by C. jejuni challenge at both time points (Table 2; Fig. 2B and Additional file 13: Figure S4B). However, we found that 39-day samples taken from challenged birds treated with bacitracin do not differ in community composition from 39-day samples taken from challenged and untreated birds (Table 2; Fig. 2B). This is consistent with the alpha diversity analyses that suggested that the combined effect of C. jejuni challenge and bacitracin treatment influenced the cecal microbiome composition. However, both alpha and beta analyses showed that the cecal microbiome was more sensitive to C. jejuni challenge than bacitracin treatment.
As noted above, differential abundance testing between bacitracin groups and the untreated controls that were challenged with C. jejuni consistently identified an increased relative abundance of the OTU116 (Anaeroplasma) (Additional file 1: Table S1 and Additional file 2: Table S2; Fig. 4A and Additional file 14: Figure S5A). While for the day 39 samples, members of Clostridiales vadinBB60 group differential at this time point showed only increased relative abundance whereas other challenged, treatment groups exhibited both increased and decreased relative abundance. At 39 days, differential abundance testing identified 50 OTUs, with 45 increased and 5 decreased. On the other hand, differential abundance testing comparing bacitracin to untreated groups in the absence of C. jejuni challenge identified increased relative abundances of Streptococcaceae and decreased relative abundances in members of Enterobacteriaceae among the 39-day samples. Overall, 70 OTUs were identified to be differential over samples taken from untreated and unchallenged birds, where 44 OTUs were increased and 26 OTUs were decreased.
An absence of differential OTUs assigned to Lactobacillaceae is noted in 30-day samples taken from challenged birds treated with bacitracin as all other samples taken from challenged birds observed significant changes in OTUs assigned to this family. Reads assigned to Akkermansiaceae (Akkermansia genus) were notably unique to samples taken from unchallenged birds treated with bacitracin (Fig. 3A and B). Differential abundance testing confirmed Akkermansiaceae (Akkermansia genus) to be at an increased differential abundance among day 30 samples from unchallenged bacitracin-treated birds over unchallenged and untreated birds (Additional file 3: Table S3).
Single-strain probiotics have a transient impact on the cecal microbiome during C. jejuni challenge
We next focus on the impact of the two single strain probiotics S. cerevisiae boulardii and P. acidilactici. Alpha diversity analyses suggested that P. acidilactici promoted a rich and diverse cecal microbiome (Table 1; Fig. 2A and Additional file 13: Figure S4A). PERMANOVA and PERMDISP analyses indicated that communities at 39 days treated with either single-strain probiotic were similar to those communities obtained from the challenged, untreated birds (Table 2; Fig. 2B). This suggests that over the course of infection by C. jejuni, the influence of the probiotics on the communities may have dissipated. Reflecting this possible transient impact, we were unable to associate any reads to P. acidilactici in samples taken from P. acidilactici treated birds, making it difficult to conclude that the probiotic engrafted under the conditions of this study despite continued administration. Since our sequencing targeted only the 16S rRNA gene, we were unable to monitor the ability of S. cerevisiae boulardii to engraft.
Relative to the untreated but challenged controls, differential OTUs found in samples taken from S. cerevisiae boulardii treated birds at both time points were not found to be specific to its experimental conditions and were common in other treatment groups (Additional file 1: Table S1 and Additional file 2: Table S2; Fig. 4A and Additional file 14: Figure S5A). A total of 50 OTUs were found differential among the S. cerevisiae boulardii group at 39 days, with 43 OTUs increased and 7 OTUs decreased.
The 39-day samples from the P. acidilactici group yielded signatures associated with Bacteroidaceae and Rikenellaceae, absent in the 30-day samples (Fig. 3A, B). Differential abundance testing between samples from the P. acidilactici group and samples from the untreated, challenged group confirmed increased differential abundances of Bacteroidaceae (Bacteroides genus; OTU1176) and Rikenellaceae (Alistipes genus; OTU1206, OTU1196, OTU1198) at 39 days (Additional file 1: Table S1; Fig. 4A). Along with samples taken from challenged birds treated with bacitracin, samples from the P. acidilactici group identified increased relative abundance of OTU116 (assigned to Anaeroplasma) across time points (Additional file 1: Table S1 and Additional file 2: Table S2; Fig. 4A and Additional file 14: Figure S5A). Differential abundance testing identified 67 differential OTUs in P. acidilactici samples over samples taken from challenged and untreated birds at day 39 where 56 strains are increased, and 11 strains are decreased. Of note, 30-day P. acidilactici samples yielded a differential abundance of Bacillaceae (OTU46) not found significant in other samples.
Microbial consortia, Aviguard and CEL promote colonization by Bacteroidaceae and Rikenellaceae respectively
From the alpha diversity studies above, we noted that treatment with Aviguard resulted in a less complex and less even cecal microbiome than the other challenged groups (Table 1; Fig. 2A and Additional file 13: Figure S4A). Beta diversity analyses further revealed that both Aviguard and CEL promoted distinct microbiome structures (Table 2; Fig. 2B and Additional file 13: Figure S4B).
CEL for example, promoted colonization by Rikenellaceae, specifically of the genus Alistipes, that was only otherwise observed in 39-day samples taken from birds treated with P. acidilactici (Additional file 1: Table S1 and Additional file 1: Table S2; Figs. 3A, B, Fig. 4A and Additional file 14: Figure S5A). From the 44 differential OTUs detected among CEL-treated birds over untreated, challenged birds at 39 days, 30 OTUs are increased and 14 OTUs are decreased. Relative to other treatments, samples from both CEL and Aviguard groups yielded a greater proportion of OTUs assigned to Ruminococcaceae that exhibited reduced abundance compared to samples from other treated and challenged groups, potentially reflecting the lower diversity and greater unevenness associated with these samples.
Furthermore, Aviguard promoted a microbiome featuring an abundance of Bacteroidaceae (Bacteroides genus). Indeed, only samples from Aviguard treated birds and the 39-day samples from birds treated with P. acidilactici were found to contain sequence reads assigned to Bacteroidaceae (Bacteroides genus) (Fig. 3A, B). Aside from Bacteroidaceae, samples from the Aviguard group notably observed differential abundances of other taxa that made up its unique microbial signature (Additional file 1: Table S1 and Additional file 2: S2; Fig. 4A and Additional file 14: Figure S5A). The 39-day samples exhibited decreased relative abundances from members of Lachnospiraceae, Lactobacillaceae and Mollicutes RF39 that was not observed in other samples at this time point. OTUs assigned to Christensenellaceae showed differential abundance among all treatment groups except for samples from the Aviguard group. The decrease or absence of differential taxa mentioned may point to lower diversity and greater unevenness in the microbiome. Furthermore, 39-day samples from birds treated with Aviguard consistently had the least number of differentially abundant strains when compared to samples from other treatments with 24 increased strains and 24 decreased strains. Consistent with alpha diversity analyses, these findings suggested that treatment with Aviguard resulted in a less complex microbial community.
Chicken cecal microbial communities share a core microbiome embellished with treatment-specific taxa
Our findings from beta diversity analyses, review of taxonomic profiles and differential abundance testing suggest that each treatment resulted in the establishment of a distinct microbial community. To further investigate the uniqueness of each community, we identified OTUs shared between different treatment groups at each time point (Fig. 4B and Additional file 14: Figure S5B). Across all treatments, we identified a core microbiome of 51 and 62 OTUs for the 30-day and 39-day samples respectively. This increase might be associated with an increase in microbial complexity as the birds age [35, 36]. Also notable is a set of nine taxa common to all 30-day samples with the exception of those samples collected from untreated and unchallenged birds. These increased to 20 taxa for the 39-day samples. This again highlights the effect of C. jejuni challenge and dietary additives on the cecal microbial community.
At the 30-day timepoint, samples from challenged birds that either received bacitracin or P. acidilactici shared 12 OTUs that were not found in any other sample, including members of Mollicutes RF39, Anaeroplasmataceae, Erysipelotrichaceae, Ruminococcaceae, Christensenellaceae and Lachnospiraceae. However, this dropped to just three OTUs at 39 days, which were assigned to Anaeroplasmataceae and Ruminococcaceae. Coincidentally, bacitracin and P. acidilactici were treatments that failed to lower CFUs of C. jejuni. This suggests that failure to alleviate C. jejuni burden may prevent the former OTUs from persisting within the ceca. To investigate further, we calculated Spearman’s rank correlation coefficients for each of the 12 OTUs against C. jejuni sequence counts at 30 days. Only one OTU, assigned to Lachnospiraceae, was significantly (negatively) correlated with C. jejuni sequence counts (R = −0.89, p = 0.033; Spearman’s rank correlation coefficient).
CEL was notable in having the highest number of exclusive OTUs (13 and 6 OTUs at day 30 and 39 respectively), potentially representing components of the CEL consortium itself. For the 30-day samples, these included members of Ruminococcaceae, Clostridiales vadinBB60 group, Lachnospiraceae, Peptococcaceae and Rikenellaceae, while for the 39-day samples, these included members of Christensenellaceae, Peptococcaceae, Ruminococcaceae and Lachnospiraceae. This highlights the unique ability of CEL to promote the persistence of taxa not associated with any other treatment.
Discussion
Foodborne illness in humans caused by C. jejuni through contaminated meat products has directed efforts to reduce avian colonization [37]. In the present study, we assessed and compared the efficacy of four industrially relevant microbial interventions (Aviguard, CEL, S. cerevisiae boulardii and P. acidilactici) and bacitracin to reduce C. jejuni burden. We further explored the impact of each treatment on the composition of the cecal microbiota. Here we focused on the cecum as an important site for C. jejuni colonization and the role of the cecal microbiota on host immunity and restricting colonization by pathogens [38]. Moreover, the cecum plays a key role in nutrient absorption and the production of short chain fatty acids [38]. Examining the impact of different treatments under standardized experimental conditions (housing, genotype, geography, time points and diet) allows the identification of meaningful differences in terms of taxonomic composition.
Bacitracin consists of high molecular weight polypeptides with demonstrated antimicrobial activity against Gram-positive microorganisms by interfering with the formation of the bacterial cell wall [1]. Administered at sub-therapeutic quantities, bacitracin promotes a healthy gut microbiome, resulting in enhanced growth and preventing infections involving food safety pathogens, as well as those impacting flock health [3–5, 8]. For example, dietary supplementation with bacitracin can reduce C. perfringens count in the chicken microbiota and prevent necrotic enteritis [1, 6]. However, bacitracin’s impact on C. jejuni, a Gram-negative bacterium that causes campylobacteriosis, is not well defined [4, 6, 7, 29, 39, 40]. Consistent with reports of an absence of appropriate targets and/or low affinity of binding of bacitracin [41–43], we found that treatment with bacitracin did not lower C. jejuni burden. Further, through the preferential targeting of Gram-positive bacteria, bacitracin treatment may have opened a niche in the gut environment and allowed greater C. jejuni colonization. In addition, it is not recommended to reduce C. jejuni burden in poultry with AGP administration since birds are largely asymptomatic and C. jejuni has been found to readily acquire antibiotic resistance mechanisms [43]. For example, use of fluoroquinolones as an AGP has been linked to the development of ciprofloxacin-resistant Campylobacter in humans [37]. Along with previous literature, results from this study have highlighted that AGP use in poultry production is not ideal and calls to the need for alternatives.
Outside of its ability to mitigate C. jejuni infection, in the absence of this pathogen, we found bacitracin treatment promoted colonization of the ceca by Akkermansia. Although we were unable to define the Akkermansia OTU to the species level, it has been suggested that Akkermansia muciniphila may improve host metabolic functions and immune responses [44]. A. muciniphila is an intestinal symbiont that colonizes the outer mucosal layer, an anaerobe that degrades the mucin for carbon and nitrogen elements and other beneficial by-products that also exerts competitive inhibition on other pathogenic bacteria that degrade mucin [44]. Current evidence shows that A. muciniphila is a promising therapeutic target in microbiome-related diseases such as colitis, metabolic syndrome, immune diseases and cancer [44] and may therefore represent one mechanism by which bacitracin exerts a positive influence on gut health. Interestingly, several studies have reported antibiotic treatment promotes A. muciniphila to be the most abundant member of the gut microbiome [45–47]. For example, vancomycin treatment in mice reduced the abundance of Firmicutes and Bacteroides, while promoting A. muciniphila, and was associated with reducing cumulative diabetes incidence [46]. A separate study of two human patients unexpectedly identified a high proportion (> 40% of all taxa) of A. muciniphila in the gut microbiota after receiving broad-spectrum antibiotic treatments [47].
Supplementation with S. cerevisiae boulardii has been reported to reduce C. jejuni abundance in fecal samples, lead to a higher abundance of beneficial microorganisms and positively influence intestinal mucosa architecture in broiler chickens, with an overall improvement on growth performance [48]. While we did note a slight reduction in C. jejuni burden due to treatment with S. cerevisiae boulardii relative to the bacitracin-treated controls, this decrease was not statistically significant. Sample size may need to be increased in follow-up studies to determine that S. cerevisiae boulardii is effective in reducing C. jejuni burden relative to bacitracin treatment. The aforementioned study also reported higher abundances of Lactobacillaceae following S. cerevisiae boulardii treatment, which reflects other reports of this family to be negatively correlated with C. jejuni [48–51]. Although we found Lactobacillaceae groups to be differentially abundant among communities treated with S. cerevisiae boulardii in this study, it was not specific to this treatment. Further, samples from birds treated with S. cerevisiae boulardii in our study did not indicate colonization of other bacteria unique to this treatment. Similar to our findings, the prior study reported no significant difference in alpha and beta diversity between control samples and treated samples [48]. The surface of S. cerevisiae boulardii has also been found to directly bind to Salmonella potentially preventing invasion in the host and lowering its colonization in the poultry gut [49, 52, 53]. Based on results from this study and previous reports, S. cerevisiae boulardii is a potentially promising alternative to bacitracin for foodborne pathogens of concern in poultry production.
The other single strain probiotic used in this study, P. acidilactici, has been shown to significantly decrease counts of Campylobacter in free-range finishing pigs before slaughter [54]. Moreover, a small cocktail of probiotics that included P. acidilactici isolated from a healthy chicken gut significantly inhibited C. jejuni growth in broiler chickens [39]. Another study in broiler chickens reported a significant reduction of C. jejuni burden after administration of probiotic preparations that included S. cerevisiae boulardii and P. acidilactici [55]. In contrast, we did not see any reduction of pathogen burden through P. acidilactici supplementation, relative to treatment with bacitracin. One explanation for this inconsistency with previous reports of lowered burden of C. jejuni may be the limited ability of P. acidilactici to colonize the birds in our study, as indicated by the inability of 16S rDNA surveys to detect P. acidilactici among samples. At the same time, it should be appreciated that these surveys rely on measures of relative abundance rather than absolute abundance and that other abundant taxa may simply be masking the presence of P. acidilactici. Hence an alternative explanation may reflect P. acidilactici’s ability to secrete pediocins, antimicrobial peptides that target and inhibit the growth of gram-positive bacteria [56]. Thus, P. acidilactici may be acting indirectly on C. jejuni through removing taxa that may otherwise restrict the growth of the gram-negative pathogen. This positive influence on C. jejuni might counterbalance any direct impact of P. acidilactici on the pathogen itself. Beyond any impact on C. jejuni, treatment with P. acidilactici was associated with changes in the relative abundance of other taxa, notably a single OTU assigned to Bacillus, a commonly used single-strain probiotic used in livestock, reported to produce bacteriocins and antimicrobial peptides [5, 12]. We also identified a single OTU assigned to Bacteroidaceae and several OTUs assigned to Rikenellaceae, illustrating the distinct impact of this probiotic on the cecal microbiome.
Aviguard is a competitive exclusion (CE) product administered to newly hatched chicks to promote the formation of a stable and diverse microbiome that exerts colonization resistance. It is a partially characterized, freeze-dried mixture of live commensal bacteria derived from the gut microbiota of specific-pathogen-free (SPF) adult chickens [57]. In our study, Aviguard supplementation mirrored the success found in other studies where it was able to inhibit intestinal colonization of other enteric pathogens such as Clostridium perfringens and Salmonella typhimurium in chickens [29–32]. Aviguard treatment was associated with reduced alpha diversity and reduction in the number of differentially abundant taxa. This suggests that Aviguard, which is cultured from intestinal samples obtained from healthy adult birds, may have rapidly engrafted in the ceca of day old chicks, establishing a microbiome of reduced complexity, potentially dominated by a few highly abundant taxa [32]. This results in a distinct microbiome (as evidenced through beta diversity analyses) and a unique ability to promote colonization by Bacteroidaceae.
Despite also representing a complex microbial consortium, CEL did not elicit the same effect on alpha and beta diversity as Aviguard. However, it was notable that beta diversity calculations of challenged 39-day samples from bacitracin, S. cerevisiae boulardii, P. acidilactici and untreated groups did not have a significant difference from each other. This suggests that microbial consortia treatments result in very distinct microbiomes. This is further reflected by differential abundance of members of Rikenellaceae being associated only in CEL groups, as well as 39-day samples from the P. acidilactici group.
Both Bacteroidaceae and Rikenellaceae belong to the Bacteroidetes phylum, which includes bacteria considered to be a stable part of the GI microbiota and are major producers of short-chain fatty acids (SCFAs), key molecules involved in host homeostasis and disease state [58–60]. Further studies, perhaps through metabolomics, are required to determine the impact of shifts of diversity on the production of SCFAs by the high relative abundance of Bacteroidaceae and Rikenellaceae in the cecal microbiome under Aviguard and CEL treatment respectively. Rikenellaceae (Alistipes genus) is implied to play a critical role in inflammation and disease but its exact mechanisms in the microbiome are yet to be completely elucidated [58, 59, 61]. The observed differential abundance of Rikenellaceae was consistent with previous reports of this family being sensitive to fluctuations in the gut microbiome caused by supplementation with antibiotics or probiotics [62, 63].
Differential abundances of Lactobacillaceae (Lactobacillus genus) were found among samples taken from challenged birds. Members from the Lactobacillaceae are widely used as probiotics to help recover a healthy microbial community after dysbiosis [49, 64–66] and are known to be successful in reducing enteric diseases and maintaining a healthy microbiota in poultry [13]. Lactobacillus strains can lower pathogen burden by decreasing the gut pH via lactic acid secretion and are known to reduce the incidence of Salmonella spp., Clostridium perfringens and Campylobacter infections [48, 67, 68]. Moreover, other studies have associated decreases in Lactobacillaceae populations with increased Campylobacter abundance or stress [12, 48, 50, 51]. Although strains of Lactobacillus have been suggested as probiotics that can mitigate against colonization by pathogens, we found no association between differential abundance of OTUs assigned to this genus with C. jejuni burden in the present study.
Interestingly, higher alpha diversity measures were observed in challenged groups and samples from either bacitracin or P. acidilactici groups while lower alpha diversity measures were seen in the Aviguard group. Coincidentally, treatments of bacitracin and P. acidilactici failed to reduce C. jejuni burden compared to other challenged groups while Aviguard treatment succeeded. In contrast to our findings, previous studies have reported that C. jejuni does not affect the alpha diversity in the cecal microbiota [69–71]. Also in contrast to our study’s findings, bacitracin has also been reported to decrease alpha diversity [72]. Traditionally, increased diversity in the microbiome is considered beneficial to host health while decreased diversity points to a decline [73, 74]. The current study presents contrasting results where microbiomes with lower pathogen burden observe lower diversity, highlighting a need to broaden our understanding of diversity scores to better define the microbial community’s role in host health.
Our data shows that the Firmicutes phylum is predominant in the chicken cecal microbiome, largely consisting of Ruminococcaceae, Lachnospiraceae and Clostridiales vadin BB60 group, with Christensenellaceae, Peptostreptococcaceae and Erysipelotrichaceae at lower percentages. Indeed, bacteria belonging to the phylum, Firmicutes, are the dominant taxa in older birds [35, 38]. Ruminococcaceae and Lachnospiraceae are major SCFA-producing bacterial members of the gut microbiome [38, 49, 75]. Ruminococcaceae and Lachnospiraceae are both obligate anaerobes that metabolise energy by anaerobic respiration and fermentation and are major producers of fermentation products such as the SCFAs: butyrate, acetate, lactate and formate [49, 75]. The Clostridiales vadin BB60 group is largely unclassified and uncharacterized in its role in metabolism and in the microbiome whereas Christensenellaceae has been linked to lower BMI and overall gut health in humans [38]. Samples in this study were taken near the age of slaughter and yield relative abundances of Tenericutes groups such as Mollicutes RF39 and Anaeroplasmataceae (Anaeroplasma), consistent with other studies investigating the mature chicken cecal microbiome [38, 51]. Members of the phylum, Anaeroplasma have also been reported to increase in cecal microbiome of heat-stressed broilers [76]. Interestingly, we noted the increased relative abundance of Anaeroplasma in samples taken from birds treated with bacitracin or P. acidilactici, treatments that did not mitigate against C. jejuni challenge. We therefore speculate that the increased abundance in Anaeroplasma may have resulted from the inability of the bacitracin and P. acidilactici treatments to reduce stressors to the chicken ceca. Moreover, Gastranaerophilales was also seen at differential abundances among samples in this study. Although the role of Gastranaerophilales in the microbiome is uncharacterized, this group is thought to aid host digestion by fermentation and are a source of vitamins B and K [77]. Overall, the present study showed that the core chicken microbiome is dominated by Firmicutes in older birds with other phyla such as Bacteroidetes, Tenericutes, Cyanobacteria and Verrucomicrobia in lower proportions. This reflects previous studies that have shown that Firmicutes, Bacterioidetes, and Proteobacteria make up the core cecal microbiota of birds, with Firmicutes dominating in older birds [9, 35, 78, 79].
Conclusions
Given the varying effects of the microbial interventions, with each conferring a specific benefit (e.g. colonization by Bacteroidetes or Rikenellaceae), our findings suggest that the application of each probiotic may depend on the individual requirements of the farm and/or flock receiving the additives. Overall, microbial-based products are a promising alternative to AGPs addressing the problem of rising antibiotic resistance mechanisms and reducing colonization by C. jejuni in poultry to help mitigate the incidence of human campylobacteriosis.
Methods
Animal trials
All experimental procedures were approved by the University of Guelph Animal Care Committee and conducted according to specifications of the Canadian Council on Animal Care. One hundred and sixty broiler (Ross 708, Aviagen Inc. Huntsville, AL) chickens were divided randomly into 8 groups and assigned to one of 6 treatments at day one of age. On day one, group 1 and 2 received complex microbial consortia Aviguard and CEL respectively in their drinking water as one shot, for less than 24 h. Group 3 received probiotic bacteria (Pediococcus acidilactici CNCM I-4622), group 4 received probiotic yeast (Saccharomyces cerevisiae boulardii CNCMI-1079), and group 5 and 6 received AGP (bacitracin). Group 3, 4, 5 and 6 received their supplements in their feed throughout the entire experimental period. Group 7 and 8 did not receive any dietary additives and were fed a normal diet.
Prior to administration, each complex consortium sachet was dissolved in 500 mL deionized water and provided with 1 L of deionized water to avoid the effects of chlorine. P. acidilactici and S. cerevisiae boulardii supplementation was added to 150 kg of standard feed according to commercial guidelines, resulting in a dose of 1 × 109 CFU/kg of feed for each product. All probiotics were sourced from Lallemand SAS, Blagnac, France. An amount of 110 g of AGP (bacitracin) was added to 220 kg of standard feed for AGP treatment groups.
On day 14, all groups, except for groups 6 and 8 received an oral gavage of 107 CFUs of C. jejuni (strain 81–176) in 500 µl PBS. Fecal droppings were collected every 5 days (starting from day 1 post-hatch to day 39 post-hatch). Approximately 0.5–1 g of cecal contents were collected at day 30 and 39 of age (n = 10 birds per time point and treatment group, total of 158 samples for sequencing). After sample collection, tenfold serial dilutions in PBS were plated onto Muller Hinton agar containing Preston Campylobacter Selective Supplement, an antibiotic cocktail that prevents growth of other bacteria. Plates were incubated in microaerophilic conditions at 41 °C and CFUs of C. jejuni were enumerated after 40–48 h and expressed as log10 Campylobacter /g of cecal content.
Next-generation sequencing (NGS) and sequence processing
Chicken cecal samples were submitted to the Integrated Microbiome Resource at Dalhousie University, Nova Scotia, Canada for 16S rRNA gene high-throughput Illumina sequencing. Microbial communities were analyzed through the V4/V5 hypervariable region of the 16S rRNA gene to capture bacterial taxonomic groups (Primer sequences: 515f (5’-GTGYCAGCMGCCGCGGTAA-3’) and 926r (5’-CCGYCAATTYMTTTRAGTTT-3’)) [80]. The initial quality filtering steps were performed on demultiplexed paired-end reads using the QIIME2 platform [81]. Primers were trimmed using the Cutadapt plugin within QIIME2 [82]. Reads were denoised, dereplicated, filtered for chimeras and merged using the DADA2 plugin within QIIME2 [83]. Sequences were clustered into Operational Taxonomic Units (OTUs) according to a similarity set at 97% based on a de novo assignment [84]. Taxonomic assignment was performed against the SILVA 132 database using the Scikit-learn plugin within QIIME2 at a confidence threshold of 70%, and a phylogenetic tree was produced [85–87].
Statistical analyses
Downstream analyses and graphical outputs were generated in R v.3.6.2 with a combination of R packages including Phyloseq v.1.30.0, Vegan v.2.5.6, PairwiseAdonis v.0.0.1, DESeq2 v.1.26.0 and ComplexHeatmap v. 2.5.3 [34, 88–92]. The Phyloseq package was used to perform normalization, visualize abundance of microbial taxonomic composition and estimate both alpha and beta diversity metrics. P < 0.05 was considered significant for all statistical tests. The effects of microbial-based treatments on colony counts (CFUs) relative to control groups was analyzed with non-parametric Kruskal–Wallis tests, followed by pairwise comparisons with Dunn’s test adjusted with the Benjamini–Hochberg (B–H) method.
Reads were normalized by rarefying to even sequencing depths before calculating species richness (Observed amplicon sequence variants (ASVs) and Chao1) and evenness (Shannon and Simpson index) for alpha diversity estimations. A minimum sequence depth of 5753 was kept and taxa not seen more than 5 times in at least 5 samples were removed. Non-parametric Kruskal–Wallis tests were conducted, followed by subsequent pairwise comparisons with Wilcoxon rank sum test adjusted with the Benjamini–Hochberg (B–H) method to compare alpha diversity metrics among groups.
Total sum scaling was used to normalize raw counts prior to beta diversity analyses and visualizing taxonomic composition. For beta diversity analyses, dissimilarity matrices between samples were calculated with the Bray Curtis method and was visualized with Non-Metric Multidimensional Scaling (nMDS) ordination technique. The nMDS plots in this study were visualized on two dimensions as it exhibited the most separation between treatment groups as opposed to four dimensions. nMDS calculations do not maximize the variability associated with individual axes of the ordination unlike eigenvector-based methods such as principal component analysis, principal coordinates analysis or correspondence analysis [93]. Permutational multivariate analysis of variance (PERMANOVA) with the adonis function from R’s Vegan package was conducted to evaluate the significance of separation between centroids of samples grouped by treatment or time point but is sensitive to multivariate dispersion. Conducted in parallel to PERMANOVA, permutation multivariate analysis of dispersion (PERMDISP) was computed with the function betadisper and permutest from the Vegan package to assess if the distribution or spread of two sample groups are significantly different. Pairwise PERMANOVA and PERMDISP tests were also calculated and adjusted with B–H method.
Differential abundance tests of raw counts were generated with DESeq2 to identify differential taxa among samples. Normalized counts were extracted from DESeq2 to conduct Spearman’s rank tests. Spearman’s rank test was conducted on challenged samples to find taxa negatively/positively correlated with C. jejuni and to compare sequence counts to CFUs of C. jejuni. UpSet plots were created to visualize OTUs shared between or unique among treatment groups, which was implemented with ComplexHeatmap. Prior to building UpSet plots, a filtering step was applied to raw counts where taxa not seen more than 5 times in at least 5 samples were removed.
Supplementary Information
Additional file 1. Table S1. Results from differential abundance analyses with DESeq2 of 39-day samples, comparing treated groups to the untreated group under C. jejuni challenge.
Additional file 2. Table S2. Results from differential abundance analyses with DESeq2 of 30-day samples, comparing treated groups to the untreated group under C. jejuni challenge.
Additional file 3. Table S3. Results from differential abundance analyses with DESeq2, comparing the bacitracin group to the untreated group in the absence of C. jejuni challenge.
Additional file 4. Table S4. Results from differential abundance analyses with DESeq2, comparing samples at 39 days to samples at 30 days post-hatch within each of the 8 treatment groups.
Additional file 5. Table S5. Results from Kruskal–Wallis rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to treatment at each time point.
Additional file 6. Table S6. P-values from post-hoc pairwise Wilcoxon rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to treatment at each time point.
Additional file 7. Table S7. P-values from Wilcoxon rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to days post-hatch within each treatment group.
Additional file 8. Table S8. Influence of treatment on microbiome structure at each time point. Results from PERMANOVA and PERMDISP analyses of the Bray–Curtis distances conducted in parallel.
Additional file 9. Table S9. Significance of separation between centroids of samples grouped by treatment. Results from pairwise PERMANOVA analyses of the Bray–Curtis distances.
Additional file 10. Table S10. Significance of the distribution of samples grouped by treatment. Results from pairwise PERMDISP analyses of the Bray–Curtis distances. Conducted in parallel to pairwise PERMANOVA analyses to place confidence in a significant result.
Additional file 11. Table S11. Influence of days post-hatch on microbiome structure within each treatment group. Results from PERMANOVA and PERMDISP analyses of the Bray–Curtis distances conducted in parallel.
Additional file 12. Figure S1. Samples collection yielded 158 cecal content samples, which were sent for 16S rRNA sequencing. (A) After quality filtering, 2,195,510 paired-end reads were retained with a median of 13,562 reads (5753–29,542 reads) per sample. (B) Sequences were clustered into 1305 Operational Taxonomic Units (OTUs) based on a de novo assignment with similarity set at 97%. Figure S2. Scatter plots showing the Spearman correlation between enumerated CFUs of C. jejuni and sequence counts assigned to C. jejuni in samples at (A) 30 and (B) 39 days post-hatch
Additional file 13. Figure S3. Bar plots representing the relative abundance of C. jejuni in all samples, grouped by treatment and age. Figure S4. Alpha and beta diversity of cecal microbial communities. (A) Boxplots representing alpha diversity metrics of richness (observed number of OTUs and Chao1) and evenness (Shannon and Simpson) for 30-day samples grouped according to treatment (no. of OTUs at 97% similarity). Each point represents the diversity score for a sample, colour-coded according to treatment. (B) Non-metric multidimensional scaling (nMDS) plot based on Bray–Curtis dissimilarity matrix on relative abundance data in 30-day samples. Colours indicate treatment group
Additional file 14. Figure S5. (A) Logarithmic fold changes of differential OTUs from differential abundance analyses with DESeq2 at 30 days post-hatch, comparing treated groups to the untreated group under C. jejuni challenge. OTUs that are in significantly greater or lesser abundance are represented in green and red coloured bars respectively. OTUs are further grouped by their assigned taxonomic families. Legend lists taxonomic families in order of appearance. (B) Vertical bars in the UpSet plot visualizes the number of OTUs unique to each treatment group and the number of OTUs shared between treatment groups at 30 days post-hatch. Horizontal bars represent the total number of OTUs found in each treatment group
Acknowledgements
The authors wish to acknowledge the staff at the Animal Isolation Unit, University of Guelph. We would also like to thank David Reynolds and Caroline Achard (Lallemand SAS) for helpful comments in drafting of the manuscript.
Abbreviations
- AGP
Antibiotic growth promoter
- ASV
amplicon sequence variant
- B–H
Benjamini–Hochberg
- CE
Competitive exclusion
- CFU
Colony forming unit
- FMT
Fecal microbiota transplantation
- GI
Gastro-intestinal
- NE
Necrotizing enteritis
- nMDS
Non-metric multidimensional scaling
- OTU
Operational taxonomic unit
- PERMANOVA
Permutational multivariate analysis of variance
- PERMDISP
Permutational analysis of multivariate dispersions
- SCFA
Short-chain fatty acid
- SPF
Specific-pathogen-free
Authors' contributions
J.P., S.S., M.C. and V.D. conceived and designed the study. K.T-A. performed animal trials, collected samples and analysed C. jejuni counts. M.T. processed and analyzed the sequencing data. J.P. and M.T. wrote the paper and all authors reviewed and/or edited the paper. All authors read and approved the final manuscript.
Funding
This work was funded by grants from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019–06852), Alberta Livestock and Meat Agency, Ontario Ministry of Agriculture, Food and Rural Affairs, the Canadian Poultry Research Council and Lallemand SAS. High performance computing was provided by the Centre for Computational Medicine at the SickKids’ Research Institute.
Availability of data and materials
The sequence data generated during the current study are available from the NCBI Sequence Read Archive with the BioProject identifier PRJNA767870.
Declarations
Ethics approval and consent to participate
All experimental procedures were approved by the University of Guelph Animal Care Committee and conducted according to specifications of the Canadian Council on Animal Care.
Consent for publication
Not applicable.
Competing interests
V.D. and M.C. are employed by Lallemand SAS. The remaining authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Butaye P, Devriese LA, Haesebrouck F. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin Microbiol Rev. 2003;16(2):175–188. doi: 10.1128/CMR.16.2.175-188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson K, et al. Differential impact of subtherapeutic antibiotics and ionophores on intestinal microbiota of broilers. Microorganisms. 2019;7(9):282. doi: 10.3390/microorganisms7090282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou A, Sharif S, Parkinson J. Lactobacillus elicits a 'Marmite effect' on the chicken cecal microbiome. npj Biofilms Microbiomes, 2018. 4(1): 27. [DOI] [PMC free article] [PubMed]
- 4.Neumann AP, Suen G. Differences in major bacterial populations in the intestines of mature broilers after feeding virginiamycin or bacitracin methylene disalicylate. J. Appl Microbiol. 2015;119(6):1515–1526. doi: 10.1111/jam.12960. [DOI] [PubMed] [Google Scholar]
- 5.Caly DL, et al. Alternatives to antibiotics to prevent necrotic enteritis in broiler chickens: a microbiologist's perspective. Front Microbiol. 2015. 6(1336). [DOI] [PMC free article] [PubMed]
- 6.Geier MS, et al. Comparison of alternatives to in-feed antimicrobials for the prevention of clinical necrotic enteritis. J Appl Microbiol. 2010;109(4):1329–1338. doi: 10.1111/j.1365-2672.2010.04758.x. [DOI] [PubMed] [Google Scholar]
- 7.Mwangi S, et al. Characterization of Clostridium perfringens recovered from broiler chicken affected by necrotic enteritis. Poultry Sci. 2019;98(1):128–135. doi: 10.3382/ps/pey332. [DOI] [PubMed] [Google Scholar]
- 8.Craven S., et al. Determination of the Incidence of Salmonella spp., Campylobacter jejuni, and Clostridium perfringens in Wild Birds near Broiler Chicken Houses by Sampling Intestinal Droppings. Avian Dis., 2000;44: 715–20. [PubMed]
- 9.Sergeant MJ, et al. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014;9(3): e91941. [DOI] [PMC free article] [PubMed]
- 10.Sahin O, Morishita TY, Zhang Q. Campylobacter colonization in poultry: sources of infection and modes of transmission. Anim Health Res Rev. 2002;3(2):95–105. doi: 10.1079/ahrr200244. [DOI] [PubMed] [Google Scholar]
- 11.Hermans D, et al. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis. 2012;12(2):89–98. doi: 10.1089/vbz.2011.0676. [DOI] [PubMed] [Google Scholar]
- 12.Patterson JA, Burkholder KM. Application of prebiotics and probiotics in poultry production. Poult Sci. 2003;82(4):627–31. doi: 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- 13.Kabir SML. The Role of Probiotics in the Poultry Industry. Int J Mol Sci. 2009;10(8):3531–3546. doi: 10.3390/ijms10083531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krysiak K, Konkol D, Korczyński M. Overview of the use of probiotics in poultry production. Animals. 2021;11(6):1620. doi: 10.3390/ani11061620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedroso AA, Batal AB, Lee MD. Effect of in ovo administration of an adult-derived microbiota on establishment of the intestinal microbiome in chickens. Am J Vet Res. 2016;77(5):514–26. doi: 10.2460/ajvr.77.5.514. [DOI] [PubMed] [Google Scholar]
- 16.Mikulski D, et al. Effects of dietary probiotic (Pediococcus acidilactici) supplementation on productive performance, egg quality, and body composition in laying hens fed diets varying in energy density. Poultry Sci. 2020;99(4):2275–2285. doi: 10.1016/j.psj.2019.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadde U, et al. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: a review. Anim Health Res Rev. 2017;18(1):26–45. doi: 10.1017/S1466252316000207. [DOI] [PubMed] [Google Scholar]
- 18.Zmora N, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174(6):1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 19.Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annual Review of Microbiology. 2012;66(1):429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 20.McInerney MJ, et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci. 2007;104(18):7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Groot PF, et al. Fecal microbiota transplantation in metabolic syndrome: History, present and future. Gut Microbes. 2017;8(3):253–267. doi: 10.1080/19490976.2017.1293224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vemuri RC, et al. Therapeutic interventions for gut dysbiosis and related disorders in the elderly: antibiotics, probiotics or faecal microbiota transplantation? Benef Microbes. 2017;8(2):179–192. doi: 10.3920/BM2016.0115. [DOI] [PubMed] [Google Scholar]
- 23.Siegerstetter, S.C. et al. Fecal microbiota transplant from highly feed-efficient donors shows little effect on age-related changes in feed-efficiency-associated fecal microbiota from chickens. Appl Environ Microbiol, 2018. 84(2). [DOI] [PMC free article] [PubMed]
- 24.Methner U, Berndt A, Locke M. Salmonella Enteritidis with double deletion in phoP fliC and a competitive exclusion culture elicit substantial additive protective effects against Salmonella exposure in newly hatched chicks. Vaccine. 2017;35(45):6076–6082. doi: 10.1016/j.vaccine.2017.09.071. [DOI] [PubMed] [Google Scholar]
- 25.Gilroy R., et al. Campylobacter jejuni transmission and colonisation in broiler chickens is inhibited by Faecal Microbiota Transplantation. bioRxiv, 2018: p. 476119.
- 26.Hofacre, C.L., et al. Effect of a competitive exclusion culture in a necrotic enteritis challenge model in broilers. J Appl Poultry Res. 2019.
- 27.Methner U, Friese A, Rosler U. Competitive exclusion: a tool to combat extended-spectrum beta-lactamase-producing Escherichia coli strains in chickens. Res Vet Sci. 2019;123:124–128. doi: 10.1016/j.rvsc.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Huyghebaert G, De Groote G. The bioefficacy of zinc bacitracin in practical diets for broilers and laying hens. Poultry Sci. 1997;76(6):849–856. doi: 10.1093/ps/76.6.849. [DOI] [PubMed] [Google Scholar]
- 29.Heimesaat MM, Mrazek K, Bereswill S. Murine fecal microbiota transplantation lowers gastrointestinal pathogen loads and dampens pro-inflammatory immune responses in Campylobacter jejuni infected secondary abiotic mice. Sci Rep. 2019;9(1):19797. doi: 10.1038/s41598-019-56442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofacre CL, et al. Use of aviguard and other intestinal bioproducts in experimental clostridium perfringens-associated necrotizing enteritis in broiler chickens. Avian Diseases. 1998;42(3):579–584. [PubMed] [Google Scholar]
- 31.Nakamura A, et al. Evaluation of aviguard, a commercial competitive exclusion product for efficacy and after-effect on the antibody response of chicks to Salmonella. Poultry Sci. 2002;81(11):1653–1660. doi: 10.1093/ps/81.11.1653. [DOI] [PubMed] [Google Scholar]
- 32.Abudabos AM. Use of a competitive exclusion product (Aviguard(r)) to prevent clostridium perfringens colonization in broiler chicken under induced challenge. Pak J Zool. 2013. 45.
- 33.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monographs. 2013;83(4):557–574. [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Awad WA., et al. Age-related differences in the luminal and mucosa-associated gut microbiome of broiler chickens and shifts associated with campylobacter jejuni infection. Front Cell Infect Microbiol. 2016. 6(154). [DOI] [PMC free article] [PubMed]
- 36.Donaldson EE., et al. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ, 2017. 5: e3587. [DOI] [PMC free article] [PubMed]
- 37.Johnson TJ, Shank JM, Johnson JG. Current and potential treatments for reducing campylobacter colonization in animal hosts and disease in humans. Front Microbiol. 2017. 8(487). [DOI] [PMC free article] [PubMed]
- 38.Richards P, et al. Development of the Caecal Microbiota in three broiler breeds. Front Veterinary Sci. 2019;6:201–201. doi: 10.3389/fvets.2019.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghareeb K, et al. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poultry Sci. 2012;91(8):1825–1832. doi: 10.3382/ps.2012-02168. [DOI] [PubMed] [Google Scholar]
- 40.Grant AQ, Hashem F, Parveen S. Salmonella and campylobacter: antimicrobial resistance and bacteriophage control in poultry. Food Microbiol. 2016;53:104–109. doi: 10.1016/j.fm.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Thibodeau A. et al. Presence and characterization of Campylobacter jejuni in organically raised chickens in Quebec. Can J Vet Res., 2011. 75(4): 298–307. [PMC free article] [PubMed]
- 42.Iovine NM. Resistance mechanisms inCampylobacter jejuni. Virulence. 2013;4(3):230–240. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luangtongkum T, et al. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 2009;4(2):189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T, et al. Akkermansia muciniphila is a promising probiotic. Microbial Biotechnol. 2019;12(6):1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J. Funct. Foods. 2017;33:194–201. doi: 10.1016/j.jff.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen CHF, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 47.Dubourg G, et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. Antimicrobial Agents. 2013;41(2):149–155. doi: 10.1016/j.ijantimicag.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Massacci FR, et al. Dietary Saccharomyces cerevisiae boulardii CNCM I-1079 positively affects performance and intestinal ecosystem in broilers during a Campylobacter jejuni infection. Microorganisms. 2019;7(12):596. doi: 10.3390/microorganisms7120596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moré MI, Swidsinski A. Saccharomyces boulardii CNCM I-745 supports regeneration of the intestinal microbiota after diarrheic dysbiosis – a review. Clin Exp Gastroenterol. 2015: p. 237. [DOI] [PMC free article] [PubMed]
- 50.Sakaridis I. et al. Investigating the association between the caecal microbiomes of broilers and campylobacter burden. Front Microbiol. 2018. 9. [DOI] [PMC free article] [PubMed]
- 51.Ocejo M, Oporto B, Hurtado A. 16S rRNA amplicon sequencing characterization of caecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci Rep. 2019;9(1):2506. doi: 10.1038/s41598-019-39323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Line JE, et al. Yeast treatment to reduce Salmonella and Campylobacter populations associated with broiler chickens subjected to transport stress. Poult Sci. 1997;76(9):1227–31. doi: 10.1093/ps/76.9.1227. [DOI] [PubMed] [Google Scholar]
- 53.Line JE, et al. Effect of yeast-supplemented feed on Salmonella and Campylobacter populations in broilers. Poult Sci. 1998;77(3):405–10. doi: 10.1093/ps/77.3.405. [DOI] [PubMed] [Google Scholar]
- 54.Morales-Partera, Á., et al. Supplementing feed with Pediococcus acidilactici reduces Campylobacter load in finishing pigs. Veterinary Rec. 2020. 187: p. vetrec-2019. [DOI] [PubMed]
- 55.Abd El-Ghany W, Awaad M, Nagwa S. Efficacy of certain feed additives for the prevention of campylobacter jejuni Infection in Broiler Chickens. Asian J Anim Sci. 2015. 9: 427–33.
- 56.Papagianni M, Anastasiadou S. Pediocins: The bacteriocins of Pediococci. Sources, production, properties and applications. Microbial Cell Factor. 2009. 8(1): p. 3. [DOI] [PMC free article] [PubMed]
- 57.Chantziaras, I., et al. The effect of a commercial competitive exclusion product on the selection of enrofloxacin resistance in commensal E. coli in broilers. Avian Pathol. 2018. 47(5): 443–54. [DOI] [PubMed]
- 58.Rautio, M., et al. Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a New Genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and Description of Alistipes finegoldii sp. nov., from Human Sources. Syst Appl Microbiol. 2003. 26(2): 182–88. [DOI] [PubMed]
- 59.Abe, K., et al., Anaerocella delicata gen. nov., sp. nov., a strictly anaerobic bacterium in the phylum Bacteroidetes isolated from a methanogenic reactor of cattle farms. J Gen Appl Microbiol. 2012. 58(6): 405–12. [DOI] [PubMed]
- 60.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol. Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker BJ, et al. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front Immunol. 2020;11:906–906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erb Downward JR, et al. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013. 3(1). [DOI] [PMC free article] [PubMed]
- 63.Clarke SF, et al. Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS ONE, 2013. 8(6): p. e65790. [DOI] [PMC free article] [PubMed]
- 64.Kumari A, Catanzaro R, Marotta F. Clinical importance of lactic acid bacteria: a short review. Acta Biomed. 2011;82(3):177–80. [PubMed] [Google Scholar]
- 65.Turpin W, et al. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int J Food Microbiol. 2010;143(3):87–102. doi: 10.1016/j.ijfoodmicro.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 66.Iqbal MZ, et al. Review: probiotics and their beneficial effects against various diseases. Pak J Pharm Sci. 2014;27(2):405–15. [PubMed] [Google Scholar]
- 67.Baldwin S. et al. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PLOS ONE, 2018. 13(3): p. e0194825. [DOI] [PMC free article] [PubMed]
- 68.Gusils C, et al. Adhesion of probiotic lactobacilli to chick intestinal mucus. Can J Microbiol. 2003;49(7):472–8. doi: 10.1139/w03-055. [DOI] [PubMed] [Google Scholar]
- 69.Hankel J, et al. Caecal Microbiota of experimentally Campylobacter jejuni-infected chickens at different ages. Front Microbiol. 2019;10:2303–2303. doi: 10.3389/fmicb.2019.02303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thibodeau, A. et al. Chicken Caecal microbiome modifications induced by campylobacter jejuni colonization and by a non-antibiotic feed additive. PLOS ONE, 2015. 10(7): p. e0131978. [DOI] [PMC free article] [PubMed]
- 71.Connerton, P.L., et al., The effect of the timing of exposure to Campylobacter jejuni on the gut microbiome and inflammatory responses of broiler chickens. Microbiome, 2018. 6(1). [DOI] [PMC free article] [PubMed]
- 72.Díaz Carrasco JM, et al. Tannins and bacitracin differentially modulate gut microbiota of broiler chickens. BioMed Res Int. 2018;2018:1–11. doi: 10.1155/2018/1879168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Degruttola AK, et al. Current understanding of dysbiosis in disease in human and animal models. Inflammatory Bowel Dis. 2016;22(5):1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsen OFA, Claassen E. The mechanistic link between health and gut microbiota diversity. Sci Rep. 2018. 8(1). [DOI] [PMC free article] [PubMed]
- 75.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio, 2014. 5(2): p. e00889–14-e00889. [DOI] [PMC free article] [PubMed]
- 76.Shi D, et al. Impact of gut microbiota structure in heat-stressed broilers. Poultry Sci. 2019;98(6):2405–2413. doi: 10.3382/ps/pez026. [DOI] [PubMed] [Google Scholar]
- 77.Monchamp M-E, Spaak P, Pomati F. Long term diversity and distribution of non-photosynthetic cyanobacteria in Peri-Alpine Lakes. Front Microbiol. 2019. 9(3344). [DOI] [PMC free article] [PubMed]
- 78.Wei S, Morrison M, Yu Z. Bacterial census of poultry intestinal microbiome. Poultry Sci. 2013;92(3):671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- 79.Oakley BB, et al. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014;360(2):100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- 80.Walters, W. et al., Improved bacterial 16S rRNA Gene (V4 and V4–5) and Fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems, 2016. 1(1): e00009–15. [DOI] [PMC free article] [PubMed]
- 81.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin, M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011. 17(1): p. 10.
- 83.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rognes, T., et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ, 2016. 4: e2584. [DOI] [PMC free article] [PubMed]
- 85.Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quast, C., et al., The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013. 41(Database issue): p. D590–6. [DOI] [PMC free article] [PubMed]
- 87.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12: 2825–30.
- 88.R Core Team. R: A Language and Environment for Statistical Computing. 2019, R Foundation for Statistical Computing: Vienna, Austria.
- 89.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE, 2013;8(4): p. e61217. [DOI] [PMC free article] [PubMed]
- 90.Oksanen, J., et al. vegan: Community Ecology Package. 2019.
- 91.Martinez Arbizu, P. pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. 2017.
- 92.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 93.Legendre P, Legendre L. Numerical ecology. 2012: Elsevier.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Results from differential abundance analyses with DESeq2 of 39-day samples, comparing treated groups to the untreated group under C. jejuni challenge.
Additional file 2. Table S2. Results from differential abundance analyses with DESeq2 of 30-day samples, comparing treated groups to the untreated group under C. jejuni challenge.
Additional file 3. Table S3. Results from differential abundance analyses with DESeq2, comparing the bacitracin group to the untreated group in the absence of C. jejuni challenge.
Additional file 4. Table S4. Results from differential abundance analyses with DESeq2, comparing samples at 39 days to samples at 30 days post-hatch within each of the 8 treatment groups.
Additional file 5. Table S5. Results from Kruskal–Wallis rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to treatment at each time point.
Additional file 6. Table S6. P-values from post-hoc pairwise Wilcoxon rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to treatment at each time point.
Additional file 7. Table S7. P-values from Wilcoxon rank sum tests comparing the alpha diversity indices (Observed, Chao1, Shannon, Simpson) of samples grouped according to days post-hatch within each treatment group.
Additional file 8. Table S8. Influence of treatment on microbiome structure at each time point. Results from PERMANOVA and PERMDISP analyses of the Bray–Curtis distances conducted in parallel.
Additional file 9. Table S9. Significance of separation between centroids of samples grouped by treatment. Results from pairwise PERMANOVA analyses of the Bray–Curtis distances.
Additional file 10. Table S10. Significance of the distribution of samples grouped by treatment. Results from pairwise PERMDISP analyses of the Bray–Curtis distances. Conducted in parallel to pairwise PERMANOVA analyses to place confidence in a significant result.
Additional file 11. Table S11. Influence of days post-hatch on microbiome structure within each treatment group. Results from PERMANOVA and PERMDISP analyses of the Bray–Curtis distances conducted in parallel.
Additional file 12. Figure S1. Samples collection yielded 158 cecal content samples, which were sent for 16S rRNA sequencing. (A) After quality filtering, 2,195,510 paired-end reads were retained with a median of 13,562 reads (5753–29,542 reads) per sample. (B) Sequences were clustered into 1305 Operational Taxonomic Units (OTUs) based on a de novo assignment with similarity set at 97%. Figure S2. Scatter plots showing the Spearman correlation between enumerated CFUs of C. jejuni and sequence counts assigned to C. jejuni in samples at (A) 30 and (B) 39 days post-hatch
Additional file 13. Figure S3. Bar plots representing the relative abundance of C. jejuni in all samples, grouped by treatment and age. Figure S4. Alpha and beta diversity of cecal microbial communities. (A) Boxplots representing alpha diversity metrics of richness (observed number of OTUs and Chao1) and evenness (Shannon and Simpson) for 30-day samples grouped according to treatment (no. of OTUs at 97% similarity). Each point represents the diversity score for a sample, colour-coded according to treatment. (B) Non-metric multidimensional scaling (nMDS) plot based on Bray–Curtis dissimilarity matrix on relative abundance data in 30-day samples. Colours indicate treatment group
Additional file 14. Figure S5. (A) Logarithmic fold changes of differential OTUs from differential abundance analyses with DESeq2 at 30 days post-hatch, comparing treated groups to the untreated group under C. jejuni challenge. OTUs that are in significantly greater or lesser abundance are represented in green and red coloured bars respectively. OTUs are further grouped by their assigned taxonomic families. Legend lists taxonomic families in order of appearance. (B) Vertical bars in the UpSet plot visualizes the number of OTUs unique to each treatment group and the number of OTUs shared between treatment groups at 30 days post-hatch. Horizontal bars represent the total number of OTUs found in each treatment group
Data Availability Statement
The sequence data generated during the current study are available from the NCBI Sequence Read Archive with the BioProject identifier PRJNA767870.