Abstract
Protein phosphatase 2A (PP2A), a serine/threonine phosphatase involved in the regulation of apoptosis, proliferation, and DNA-damage response, is overexpressed in many cancers, including small cell lung cancer (SCLC). Here we report that LB100, a small molecule inhibitor of PP2A, when combined with platinum-based chemotherapy, synergistically elicited an antitumor response both in vitro and in vivo with no apparent toxicity. Using inductively coupled plasma mass spectrometry, we determined quantitatively that sensitization via LB100 was mediated by increased uptake of carboplatin in SCLC cells. Treatment with LB100 alone or in combination resulted in inhibition of cell viability in two-dimensional culture and three-dimensional spheroid models of SCLC, reduced glucose uptake, and attenuated mitochondrial and glycolytic ATP production. Combining LB100 with atezolizumab increased the capacity of T cells to infiltrate and kill tumor spheroids, and combining LB100 with carboplatin caused hyperphosphorylation of the DNA repair marker γH2AX and enhanced apoptosis while attenuating MET signaling and invasion through an endothelial cell monolayer. Taken together, these data highlight the translational potential of inhibiting PP2A with LB100 in combination with platinum-based chemotherapy and immunotherapy in SCLC.
Introduction
Lung cancer is a leading cause of death worldwide with estimated 135,720 deaths and 228,820 new cases in the United States in 2020 alone. Small cell lung cancer (SCLC) accounts for approximately 15% of all lung cancer cases and is an aggressive neuroendocrine tumor, with rapid tumor growth and early development of multiple organ metastases. In addition, the clinical outcome of patients with SCLC is poor due to early relapse and acquired resistance to standard chemotherapy treatments. Unfortunately, treatment options for recurrent tumors are limited and largely ineffective (1–3).
Protein phosphatase 2A (PP2A) is a ubiquitously expressed member of a large family of trimeric Ser/Thr phosphatases, consisting of a catalytic C subunit, a structural A subunit, and a regulatory B-type subunit (4). PP2A regulates a vast portion of the phosphoproteome including pathways involved in apoptosis, proliferation, and DNA-damage response. As a result, PP2A inactivation is a vital step in malignant transformation (5), making it an attractive cancer therapeutic target. Indeed, the therapeutic effects of combining PP2A reactivation along with kinase inhibition to counteract the changes in tumor suppressors and oncogenes that lead to cancer development have been extensively explored (5–7). Conversely, inhibition of PP2A to complement chemotherapy- and radiation-induced cancer cell death is also an area of active investigation.
Recent work on B cells (8) showed that PP2A regulates glycolysis rate and balances energy supply against antioxidant protection through the pentose-phosphate pathway (PPP). The PPP which branches from glycolysis at the first committed step of glucose metabolism, is required for the synthesis of ribonucleotides and is a major source of NADPH. NADPH is required for, and is consumed, during fatty acid synthesis and the scavenging of reactive oxygen species. Thus, the PPP plays a pivotal role in helping glycolytic cancer cells to meet their anabolic demands and combat oxidative stress. Furthermore, in human B acute lymphoblastic leukemia cells, treatment with the PP2A inhibitor LB100 or genetic ablation of PPP2R1A resulted in decreased cancer cell growth and increased survival in a mouse model. A similar mechanism was also observed in SCLC (8); furthermore, PP2A is highly upregulated in SCLC suggesting that it plays an important role in driving tumorigenesis in this cancer (9, 10).
LB100 is a small molecule inhibitor of PP2A that binds to PP2A subunit C. It is derived from the natural compound cantharidin but has a significantly more favorable toxicity profile (11). In preclinical studies, LB100 inhibited proliferation of cell lines from a variety of human solid tumors at low micromolar concentrations (12). More strikingly, it potentiated the activity without significantly increasing the toxicity of cisplatin, doxorubicin, and temozolomide against xenografts of a variety of solid tumors (11, 13–19). Furthermore, inhibition of PP2A by LB100 sensitized human pancreatic cell lines to cisplatin treatment in culture; in pancreatic and ovarian cancer xenograft models LB100 sensitized the tumors to radiation and to cisplatin treatment, respectively (13, 17, 20, 21), underscoring the potential of PP2A as a therapeutic target.
Several lines of evidence suggest that PP2A also plays an important role in regulating the immune system. First, PP2A acts as a negative regulator of cytotoxic T-cell effector function—inhibiting PP2A results in enhanced antigen-specific cytotoxicity of lymphocytes (22). More specifically, PP2A mediates the inhibitory signaling of CTLA-4 by dephosphorylating Akt in activated T cells (23). Second, an in vivo short hairpin RNA screen for immunotherapy targets, identified Ppp2r2d, a regulatory subunit of PP2A. Inhibiting Ppp2r2d enhances the cytotoxic function of tumor-infiltrating lymphocytes (TIL; ref. 23) and silencing of Ppp2r2d results in increased TILs proliferation and cytokine production. Third, a decreased tumor burden and increased survival was observed using adoptive transfer of Ppp2r2d-silenced OT-1 lymphocytes in a B16-ova melanoma model (24). Considered together, these observations strongly suggest that inhibition of PP2A can also be a promising strategy to enhance anticancer immunity.
In the current study, we investigated the effect of pharmacologically inhibiting PP2A with LB100, and LB100/carboplatin in SCLC, employing in vitro and in vivo models. Furthermore, we also examined the effect of LB100 in combination with immunotherapy on the morphology and integrity of three-dimensional (3D) spheroids generated using SCLC cells. Taken together, our results demonstrate that the antitumor effect of chemotherapeutic drugs can be enhanced by blocking PP2A with LB100 by itself or in combination with chemotherapy and immunotherapy in SCLC.
Materials and Methods
General materials for biological experiments
Biological reagents were purchased from Sigma-Aldrich or Thermo Fisher Scientific. Electrophoresis and Western blotting reagents were purchased from Bio-Rad. LB100 was purchased from SelleckChem, cantharidin from Tocris, carboplatin was purchased from Teva Pharmaceuticals, and etoposide was purchased from Fresenius Kabi.
Bioinformatic analysis of PP2A mRNA expression in SCLC
RNA sequencing (RNA-seq) expression data from a previously published study (9) containing 79 SCLC and seven normal control samples were used to study mRNA expression differences in PPP2R1A. The RNA-seq data were previously processed using Illumina HiSeq 2000 and processed using Bowtie (25). Normalized read counts data from these 79 SCLC and seven normal control samples were downloaded from Gene Expression Omnibus (GEO) datasets (GSE60052) and assessed for specific genes of interest. Box plot representation of PPP2R1A shows an upregulation of the subunit the tumor samples. A Mann–Whitney U test was used for a comparison between the normal and SCLC samples.
Tissue microarray
Small cell lung cancer tissue microarrays (TMA) were from US Biomax Inc. (LC818). IHC staining was performed using standard techniques described previously (26) with antibodies against PP2A A (Cell Signaling Technology) in the Pathology/Solid tumor core, The City of Hope. Briefly, each TMA was reviewed and scored by two independent pathologists on a scale of 0 to 3: 0+, no staining, no expression; 1+, weak staining, low expression; 2+, moderate staining, moderate expression; and 3+, strong staining, high expression. The IHC staining of PP2A A was examined by a pathologist who assigned a score 0+ (no staining), 1+ (5%–80% of weak stained cells), 2+ (50%–90% of moderate stained cells), 3+ (70%–100% of strong stained cells). The number of tumor cores with stage I disease (I-IA-IB) was 39 and the pathologic score assigned was between 0+ and 3+. A total of 28 tumor cores were stage II (II-IIA-IIB) and 12 were stage III/IV (IIIA-IIIB-IV). Both groups had the same range of pathologic scores as the stage I group. The TMA slide specification sheet is included in Supplementary Table S1.
Cell culture reagents
Suspension SCLC H524, H526, H82, H446, H69, H146, and HBEC 3KT normal epithelial cell lines were purchased from ATCC and maintained in RPMI1640 (Corning Life Science) supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin (Corning Life Science) and l-glutamine at 37°C with 5% CO2. The morphology of the cell lines was monitored routinely, and the cell lines were routinely tested for Mycoplasma with a mycoplasma detection kit (InvivoGen). For cell line authentication, short tandem repeat analysis was performed with COH Integrative Genomic Core.
Immunoblotting
Whole-cell lysates were prepared using RIPA lysis buffer and proteins were detected by immunoblotting using antibodies specific against PP2A A, PP2A C, Phospho-Histone H2AX (S139), MET, pMet (Tyr 1234/1235), Cleaved Caspase 3, and pan-Actin antibodies from Cell Signaling Technology, Cleaved PARP1 (Santa Cruz Biotechnology), and pMET (Ser985; Thermo Fisher Scientific) were used as described previously (27).
Cell viability assay
To determine specific cytotoxicity, we used Cell Counting Kit-8 (Dojindo Molecular Technologies) as described previously (28). Cells were seeded in 96-well plates at a density 20 × 103 cells per well. Cells were treated with increasing concentrations for 72 hours. Absorbance was measured at 450 nm using a Tecan Spark 10 mol/L multimode microplate reader. IC50 values were generated for each cell line using GraphPad Prism 8. The synergy assays were done as described previously (29). The analysis of the synergy assay was done by the isobologram and combination index (CI) methods. Differences in the inhibition levels of drug combinations were calculated using “Compusyn” software where CI < 1, = 1, and > 1 indicate synergy, additive effect, and antagonism, respectively (30).
Colony formation
Approximately, 1 × 103 cells in 0.3% agarose were seeded in a 96-well plate onto a layer of 0.6% agarose. Cells were grown in the present of LB100, carboplatin, or LB100/carboplatin for 3 weeks to observe colony formation. The colonies were fixed in 4% formaldehyde and stained with crystal violet. Z-stacks of tiled bright-field (BF) images were taken using a 5× objective with a step size of 200 μm on a Zeiss Observer 7 inverted microscope (Carl Zeiss). Using Zen Blue v2.5 (Carl Zeiss Microimaging), stacks were processed by first stitching a reference slice, and then the Extended Depth of Focus module, with default settings, was used to compress the Z-stack information into a single image. Manual counting was conducted on the resulting tiled image using the points tool, and summary measurements generated, in QuPath 0.1.3 (31).
PP2A activity measurement
PP2A immunoprecipitation Ser/Tre Phosphatase Assay Kit (Millipore) was used for measuring PP2A activity following manufacturer's protocol. Briefly, 8 × 106 H524 cells were treated with LB100 for 24 hours. The data are presented as the percentage of relative PP2A activity compared with control.
siPP2A subAα transfection
Ser/Thr phosphatase 2A regulatory subunit A alpha isoform siRNA was purchased from MyBioSource (https://www.mybiosource.com/search/PPP2R1A-siRNA). Cells were transfected with 100 nmol/L siRNA using jetPRIME reagent (Polyplus-transfection). siRNA transient transfection was verified with anti-PPP2R1A abs (MyBioSource).
Transendothelial extravasation assay
The ability of SCLC cells to invade though a layer of endothelial cells was quantified using transendothelial monolayer resistance measurements using an electrical substrate-impedance sensing system (Applied Biophysics), as we have described previously (32).
Monitoring of spheroid growth and cytotoxicity with the IncuCyte live-cell analysis system and IncuCyte cytotox reagent
H446 cells were plated at a density of 10,000 cells per well and spheroid allowed to form (72 hours). Cells were then treated with LB100, carboplatin or LB100/carboplatin and kinetics of spheroid growth were obtained. Spheroids were imaged every 4 hours for 6 days and analyzed using the IncuCyte ZOOM software.
Inductively coupled plasma mass spectrometry assay
Samples were prepared and analyzed for platinum (Pt) concentrations at the Isotoparium (California Institute of Technology, Pasadena, CA), using precleaned Teflon beakers (PFA), Optima grade reagents (Fisher Chemical), and 18.2 MΩ Milli-Q water. Cell pellets were first digested in 500 μL of concentrated HNO3 for 30 minutes at 160°C, before complete dry down. Mouse tumors were digested in 1 mL of concentrated HNO3 for 30–45 minutes at 120°C with periodic degassing, before complete dry down. Samples were cooled to room temperature, placed in 50:50 v/v concentrated HNO3:H2O2 (1 mL for cell pellets, 2 mL for tumors) to burn off organic matter. Cell pellets were placed on a hot plate overnight at 160°C. Tumors were heated at 120°C for 8 hours with periodic degassing. All samples were then evaporated completely and reconstituted in 5 mL 3% v/v HNO3. Holmium (Spex Certiprep Assurance, lot no. 24-80HOM) was used as the internal standard. A stock solution of 3% v/v HNO3 with 2 ppb Ho was used for all sample and standard dilutions. Aliquots of cell lines were diluted 20× using the HNO3 + Ho stock solution, while tumor aliquots were diluted 100× using the same stock solution. Three technical replicates were measured per biological replicate to demonstrate reproducibility. All samples were analyzed using an iCAP RQ (Thermo Fisher Scientific) ICP-MS and an SC-2 DX autosampler (Elemental Scientific). Instrumental tuning parameters (e.g., nebulizer gas flow, torch alignment, and sample uptake rate, quadrupole ion deflector) were optimized to pass the standard performance check prior to analysis. A Pt standard curve (0.001, 0.01, 0.1, 1.0 ppb, Spex Certiprep Assurance, lot no. 24-140PTM) was created using the HNO3 stock solution and measured for sample calibration. For each analysis, both 194Pt and 195Pt as well as 165Ho were measured. Each measurement used five main runs of five sweeps, and each sweep used a dwell time of 50 ms per isotope. To ensure that residual organics did not affect the concentration estimates, each sample was measured in two independent sessions (different days) using two different cone inserts (the High Matrix insert, typically used for geological samples, and the Robust insert, recommended for biological matrices). Both datasets are identical within uncertainty (<± 2%). Pt mass was normalized to total protein mass for cell pellets and tumor mass for mouse samples.
Kinase activity profiling using PamGene's microarray assay
H524 cells were treated with LB100 for 5 hours, to test the effects of the drug on protein tyrosine and serine/threonine kinase activity. PamChips were used to capture the activity of upstream kinases from either the tyrosine kinome (protein tyrosine kinase—PTK) or the serine/threonine kinome (serine/threonine kinase—STK). Both PamChips contain 144 peptides, each composed of 12–15 amino acids, with one or more phosphorylation sites. PTK and STK PamGene assays were performed according to the manufacturer's instructions. Samples were run in triplicate on the PamStation 12 (PamGene) by the High Throughput Screening Core. Image quantification and data processing were conducted with the Evolve and BioNavigator software package (PamGene). The peptides on each chip that had a significant (t test, P < 0.05) log fold change versus the untreated control for at least one drug concentration were analyzed using pathway enrichment analysis (http://reactome.org).
BiOLOG metabolic assay
Phenotype microarrays use a patented redox chemistry, employing cell respiration as a universal reporter. These assays potentially provide a natural fit to support data obtained from metabolomics screens. The redox assay provides for both amplification and precise quantitation of phenotypes. Redox dye mixes contain a water-soluble nontoxic tetrazolium reagent that can be used with virtually any type of animal cell line or primary cell (33).The dyes used in BiOLOG assays measure output of nicotinamide adenine dinucleotide reduced form (NADH) production from various catabolic pathways present in the cells being tested. If cell growth is supported by the medium in an assay well, the actively metabolizing cells reduce the tetrazolium dye. Reduction of the dye results in color formation in the well, and the phenotype is considered “positive.” If metabolism is hampered or growth is poor, then the phenotype is “weakly positive” or “negative,” and little or no color is formed in the well. This colorimetric redox assay allows examination of the effect of treatment on the metabolic rate produced by different substrates and thus is an excellent technique to combine with examination of metabolic output via metabolomics screens.
Glucose uptake assay
Glucose consumption was determined by using a colorimetric glucose assay (Invitrogen) following the manufacturer's instructions. Briefly, cells were seeded into 100 mm plates at a density 2 × 106 cells per well. After 48 hours of cell culture, supernatant of the medium was collected subjected into glucose detection. The uptake of glucose was determined compared with initial glucose concentration in the cell culture medium, which was taken as 100%.
Cell energy phenotype and real-time ATP rate
A Seahorse XF96 instrument (Agilent) was used for cell energy phenotype and real-time ATP assay. Cell energy phenotype assay measures mitochondrial respiration and glycolysis in basal and stressed levels. Real-time ATP measurement detects the rate of ATP production from glycolysis and mitochondria. Before experiment cells were treated for 18 hours with LB100. The day after being treated cells, were washed and seeded at a density 5 × 104 per well in 96-well plates treated with Cell-Tak. The plate was centrifuged to facilitate cell attachment and incubated at 37°C for 60 minutes. Both assays were performed per manufacturer's instructions. Data analysis was done with Wave Desktop 2.6 software (Agilent).
Live imaging of spheroids with drugs and T cells
H446 were generated as described in Materials and Methods (Monitoring of spheroid growth and cytotoxicity with the IncuCyte Live-Cell Analysis System and IncuCyte Cytotox reagent) following incubation with T cells and drugs. The effect of LB100 and atezolizumab in the presence of T cells was monitored with IncuCyte 3D Multi-Tumor Spheroid assay.
Effect of LB100 on tumor growth in subcutaneous H69 cells using a mouse xenograft
Animal studies were performed according to an Institutional Animal Care and Use Committee protocol approved by City of Hope National Medical Center Animal Care and Use Committee. Athymic nude mice (5–6 weeks of age) were purchased from NCI (Frederick, MD). Mice were injected subcutaneously on their right flank with H69 cells suspended (2 × 106) in 100 μL of PBS and 100 μL of matrigel (BD Biosciences). Tumor growth was measured in two dimensions with caliper and when surface tumor was visible (45–50 mm2) mice were randomized in four groups as follow: vehicle (PBS, intraperitoneal three times a week), LB100 (0.25 mg/kg, i.p. three times a week), carboplatin (50 mg/kg, i.p. two times a week), and drug combination (LB100/carboplatin intraperitoneal) for 30 days. At the end of the study, the mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Tumor tissues were excised, weighed, and subsequently fixed in 10% buffered formalin and embedded in paraffin for histologic analysis.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 8. Two sample groups were compared by unpaired, two‐sided Student t tests. Data of more than two groups were analyzed by one-way ANOVA followed by Tukey multiple comparison tests. Values of P < 0.05 were considered significant and indicated as: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Graphs represent the mean ± SE.
Results
PP2A is upregulated in SCLC tumor tissue and cell lines, and knocking down PP2A significantly attenuates proliferation of these cells
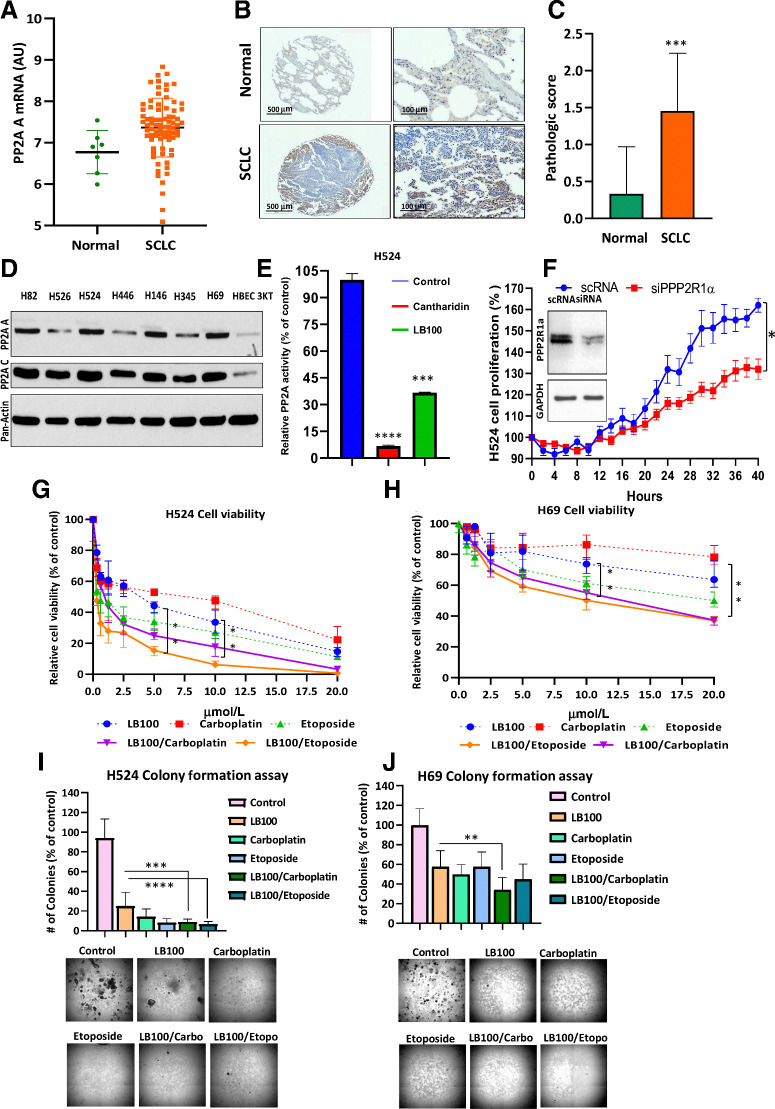
We previously reported that PP2A and its subunits A (PP2A-A) and C (PP2A-C) are overexpressed in several SCLC cell lines (8). This was further confirmed by a bioinformatics analysis of a GEO (https://www.ncbi.nlm.nih.gov/pubmed/27093186) dataset (GSE60052), wherein PP2A-A was significantly overexpressed (P = 0.0144) in SCLC as compared with normal lung (Fig. 1A).
Figure 1.
PP2A A expression in SCLC tumors and cells, effect LB100. A, Scatter plot shows an upregulation of the PP2A-A subunit in the tumor samples (P = 0.0144). A Mann–Whitney U test was used for comparison between the normal and SCLC samples. B, IHC for PP2A was conducted on TMA tissue sections, and images were captured at 4× or 20× using a 3D-Histech PANNORAMIC SCAN whole slide scanner (3D-Histech). PP2A subunit A positively immunostained the cytoplasm and nucleus of normal lung and tumor tissue but was highly upregulated in tumor tissue. TMAs were scored in normal (n = 24) and tumor (n = 79) cores on a scale from 0 (no staining/no protein expression) to 3+ (strong staining/high protein expression). C, Summary bar graph of the average PP2A subunit staining. IHC staining intensity of normal and tumor cores. There was a statistically significant difference between normal and tumor tissue (***, P < 0.001). Student t test was used for comparison between the normal and SCLC samples. D, To compare the expression of PP2A subunits A and C, cell lysates from seven SCLC cell lines and HBEC 3KT (nonmalignant cell line) were subjected to Western blotting (n = 3 biological replicates). E, PP2A activity was determined using a serine/threonine phosphatase activity assay (Millipore) after 24 hours exposure to cantharidin (10 μmol/L) and LB100 (5 μmol/L; n = 3 biological replicates).***, P < 0.001, results were analyzed by ANOVA with Tukey post-test. F, The inset showed reduction of PP2A subunit Aα in H524 cells as well as inhibition of cell proliferation due to PP2A subunit Aα knockdown (n = 3 biological replicates). P < 0.05, Student t test was used for comparison between the groups. LB100 alone or in combination with carboplatin inhibited proliferation and colony formation in SCLC cells. The Cell Counting Kit-8 assay detected cell H524 and H69 cell viability. (n = 3 biological replicates). G and H, Cells were treated with LB100, carboplatin and etoposide, as a single treatment or in combination, at constant ratio. The CI was calculated using Chou-Talalay method to find synergism between LB100 with carboplatin and etoposide (CompuSyn software: www.combosyn.com). **, P < 0.01, ANOVA with Tukey post test was used for comparison between LB100, LB100/carboplatin, and LB100/etoposide. Colony formation assays were used to count the ability of H524 (I) and H69 (J) cells to form colonies. Drug concentrations are listed for two assays with H524 and H69, respectively: LB100 (2.5 μmol/L; 20 μmol/L), carboplatin (4 μmol/L; 20 μmol/L), etoposide (3 μmol/L; 30 μmol/L), LB100/carboplatin (2.5 and 4 μmol/L; 20 and 20 μmol/L), and LB100/etoposide (2.5 and 3 μmol/L; 20 and 30 μmol/L). Representative images of colonies at 4× are shown under the graph (n = 2). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results were analyzed by ANOVA with Tukey post test.
To evaluate the expression levels of PP2A in SCLC, we compared adjacent normal (n = 24) and primary SCLC tumor (n = 79) cores contained within TMAs subjected to IHC using an antibody specific to PP2A-A (Fig. 1B). Each tumor and normal core contained in the TMA was scored independently by a pathologist who was blinded to the identity of the tissue (34, 35). PP2A-A protein was undetectable in most normal cores (0 = 79.17%, 1 = 16.67%, 2 = 4.16%) but was significantly upregulated in tumor tissue (0 = 8.86%, 1 = 41.77%, 2 = 40.5%, 3 = 8.87%; Fig. 1C). The mean pathologic score for PP2A in tumor tissues (1.45 ± 0.088) was significantly higher (P = 0.001) than that of normal tissues (0.333 ± 0.13). Both the publicly available datasets and TMA results showed that PP2A-A expression was significantly upregulated in the SCLC tumor tissue (Fig. 1A–C). Having confirmed overexpression in tumor tissue, we next determined their expression in various SCLC cell lines by immunoblotting, as described previously (27). Both subunits were upregulated in SCLC cell lines including H82, H526, H524, H446, H146, H345, and H69 compared with control HBEC 3KT cells (Fig. 1D).
Cantharidin is the parent compound of LB100 that is known to inhibit PP2A. Therefore, we used cantharidin as a positive control to demonstrate that inhibiting PP2A results in the observed effects in SCLC cells. Indeed, cantharidin treatment reduced PP2A activity by almost 90% while LB100 significantly inhibited phosphatase activity to 65%. (Fig. 1E). Finally, we knocked down PP2A subunit Aα using a specific siRNA in H524 SCLC cells. A scrambled version (scRNA) was used as control. As expected, knocking down PP2A significantly decreased PP2A subunit Aα level and attenuated cellular proliferation in these cells (Fig. 1F, inset).
Combining chemotherapy with LB100 results in synergy
To test the cytotoxicity effect of LB100, carboplatin and etoposide, we treated six SCLC cell lines with various concentrations of each drug for 72 hours. In four cell lines H82, H526, H524, and H446 that were sensitive to cisplatin, LB100 induced cell death more effectively with an IC50 of < 8 μmol/L (Supplementary Table S2) compared with the two other cell lines H146 and H69 that were resistant to cisplatin in which cell death was observed at relatively higher doses of LB100 (IC50 ∼20 μmol/L).
Next, we determined the effect of treating SCLC cell lines with combinations of LB100 and the chemotherapeutic drugs agents, carboplatin and etoposide. Either drug alone was effective in killing H524 SCLC cells that are sensitive to LB100 (Fig. 1G). Cell viability did not change in the control during the 72-hour testing time, remaining at 100%. H524 cell viability decreased to 57.2 ± 3.1% with 2.5 μmol/L LB100 and 5 and 10 μmol/L LB100 reduced cell viability to 44.2 ± 2% and 33.6 ± 4.2%, respectively. However, cell death was significantly higher when LB100 was used in combination with carboplatin or etoposide. LB100/carboplatin or LB100/etoposide with equal concentrations of both drugs dropped the level of viable cells to 32.4 ± 3.2% (2.5 μmol/L LB100/carboplatin); 25 ± 2% (5 μmol/L LB100/carboplatin); 17.8 ± 2.9% (10 μmol/L LB100/carboplatin); 26.8 ± 4.4% (2.5 μmol/L LB100/etoposide); 15.4 ± 1.5% (5 μmol/L LB100/etoposide); and 6.3 ± 1.1% (10 μmol/L LB100/etoposide). CI values at the 50% inhibition of cell proliferation with LB100/carboplatin and LB100/etoposide were below 1, which indicates synergy (0.534 and 0.532, respectively; Fig. 1G; P < 0.01). A similar synergy was seen in the case of H69 SCLC cells. LB100/carboplatin and LB100/etoposide killed LB100-resistant H69 cells with CI values of 0.311 and CI = 0.646, respectively (Fig. 1H). Significant differences in cell viability between single and combination treatments of H69 cells were obtained using 10 and 20 μmol/L as well. Cell viability was decreased for 10 μmol/L LB100 (73.7 ± 2.7%), 10 μmol/L LB100/carboplatin (55.2 ± 3.3%), 10 μmol/L LB100/etoposide (50.1 ± 5.5%). Meanwhile, at 20 μmol/L LB100 viability was found to be (63.6 ± 4.8%), LB100/carboplatin (37.3 ± 1.2%), and LB100/etoposide (37.1 ± 1.6%).
To determine the cytotoxicity effect of LB100 alone and in combination with carboplatin and etoposide on H524 and H69 cells, we also performed colony formation assays. Treatment with single drug (LB100, carboplatin, or etoposide) or in combination (LB100/carboplatin and LB100/etoposide) significantly reduced colony formation in both cell lines (P < 0.0001; P < 0.01; Fig. 1I and J). While colony formation by H524 cells was dramatically reduced compared with LB100 single treatment in both drug combination groups (LB100/carboplatin and LB100/etoposide). However, in the case of the H69 cells, a significant difference was observed only between LB100 and LB100/carboplatin treated cells (Fig. 1J). Therefore, we investigated the effect of LB100 using a 3D cell culture model that resembles the tumor microenvironment more closely.
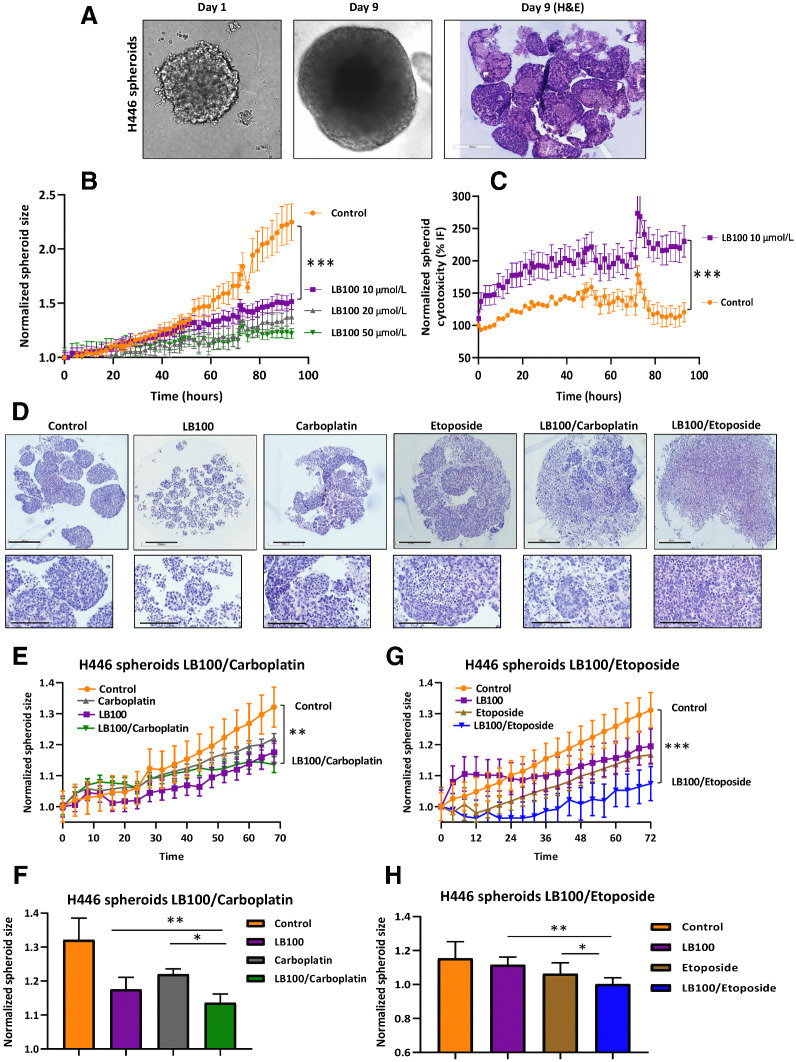
Effect of LB100 on H446 spheroid growth
We further investigated the effect of LB100 and the chemotherapy drugs on spheroids formed by SCLC cells. Three cell lines H524, H69, and H446 were tested. The H524 and H69 cells formed large soft clumps in low-attachment 96-well plates. H446 cells that formed dense spheroids overnight without the addition of extracellular matrix components or matrigel were used for imaging and histologic analysis. Spheroids of 300–500 μm formed in 9 days (Fig. 2A) and the size of the spheroids formed in vitro was comparable with the tumors formed in metastatic sites where the cells experience conditions of hypoxia, inflammation, changes in pH levels and often, nutrient deprivation (36). To test the effect of LB100 on H446 spheroids, we used the IncuCyte Live-Cell Analysis System to record functional changes in real time. H446 spheroids treated with or without 20 μmol/L LB100 were imaged in BF and using green fluorescence over 72 hours. The size of the spheroids was measured using an automated software algorithm that masked the largest BF in the field of view (label-free, real-time live cell assay for spheroids: IncuCyte BF analysis). BF analysis illustrated spheroid shrinkage and increase in the cytotoxicity dye fluorescence after LB100 treatment (Fig. 2B and C). Hematoxylin and eosin (H&E) staining was performed on spheroids treated with LB100, carboplatin alone, and in combination. Before treatment, spheroids had a dense, round shape (Fig. 2D—Control) with very well-defined contours. However, 72 hours of treatment with LB100, carboplatin, etoposide or combination of chemotherapeutic drugs with LB100 significantly changed the morphology of spheroids. Spheroids decreased in size and lost their round shape with LB100 treatment. Carboplatin and etoposide treatments dissociated cells from spheroids, forming diffuse clouds of cells around them. Drug combination of carboplatin or etoposide with LB100 abolished spheroid growth and notably decreased the number of mature spheroids (Fig. 2D). IncuCyte BF analysis on H446 spheroid growth demonstrated that LB100 in combination with carboplatin significantly reduced single spheroid size compared with control or LB100 treatment (Fig. 2E and F). Similar results were obtained with LB100 and etoposide (Fig. 2G and H). These results confirmed the efficacy of LB100 alone and in combination with carboplatin or etoposide in the 3D spheroid model, similar to our observations in 2D cultures.
Figure 2.
Effect of LB-100 on H446 spheroid growth. A, Morphology of a single spheroid of H446 cells on days 1 and 9. Spheroids grow continuously, and H&E staining is represented. B, Spheroid's growth in response to LB100 treatment was recorded with IncuCyte Live-Cell Analysis System. C, Cytotoxicity effect of LB100 was recorded with IncuCyte Live-Cell Analysis System in the presence of LB100 and IncuCyte Cytotox reagent in green fluorescence. A total of 10 μmol/L of LB 100 significantly affected growth and viability of the cells (n = 2, six technical replicates). ***, P < 0.001. Student t test was used for comparison between control and LB100 treatment. D, Effect of LB100, carboplatin, etoposide, and drug combination on H446 spheroid morphology and growth. Representative images of H&E-stained H446 spheroids with LB100, carboplatin, etoposide, and combination treatment. Scale bar, 500 μm. The inserts are enlarged images of spheroids. Scale bar, 200 μm. E and F, Effect of LB100 and carboplatin alone or in combination was monitored using IncuCyte Live Cell system for 70 hours. Maximal significant inhibitory effect of LB100, carboplatin, or drug combination on spheroid's size was observed at timepoint 70 hours. LB100/carboplatin significantly inhibited spheroid's growth compared with control. **, P < 0.01. Results were analyzed with Student t test. G and H, Effect of LB100 and etoposide alone or in combination was monitored using IncuCyte Live Cell system for 72 hours. LB100/etoposide significantly inhibited spheroid's growth compared with control. ***, P < 0.001. Results were analyzed with Student t test. Maximal significant inhibitory effect of LB100, carboplatin, or drug combination on spheroid's size was observed at timepoint 70 and 72 hours (n = 3 biological replicates). *, P < 0.05; **, P < 0.01 results were analyzed by ANOVA with Tukey post test.
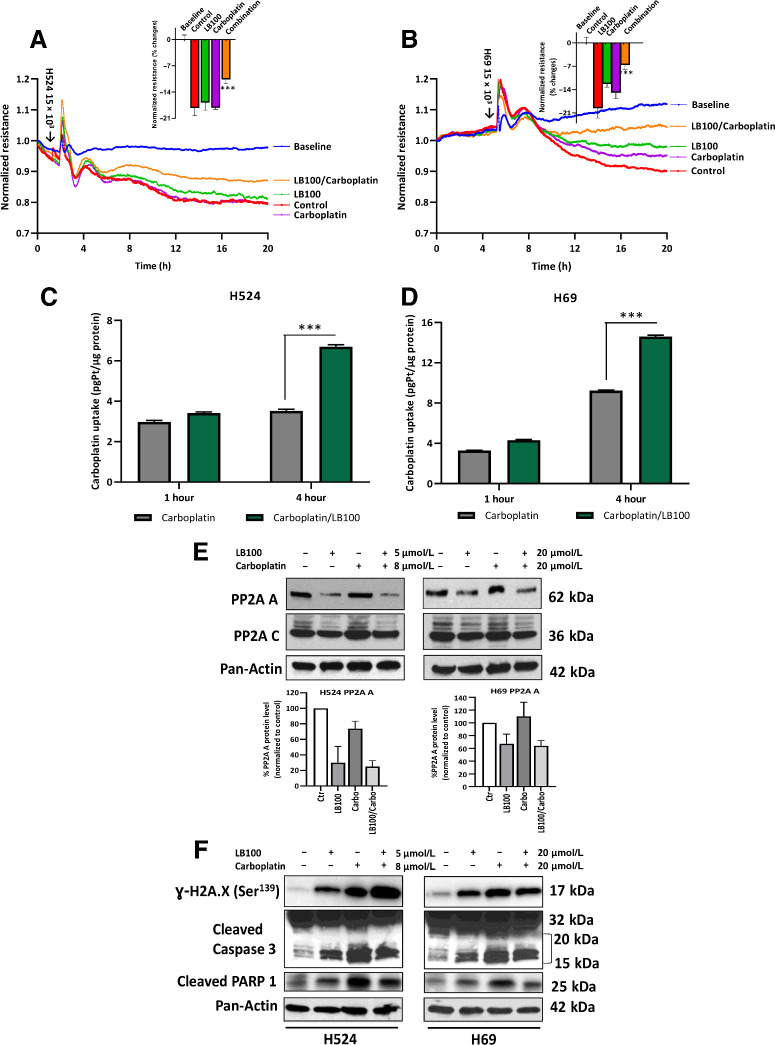
Drug combination inhibits SCLC cell invasion, increases carboplatin uptake, and affects PP2A, DNA damage, and apoptosis regulatory proteins
To discern the effect of LB100 on cell invasion, we tested the ability of SCLC cells to invade monolayer resistance using an electrical substrate-impedance sensing system (Applied Biophysics), as described previously (32). This system continuously measures endothelial monolayer resistance as SCLC cells attach and begin to invade into the monolayer. A decrease in resistance indicates a disrupted endothelial monolayer barrier via transendothelial extravasation of tumor cells. Untreated control cells highly invaded through HUVEC monolayer. After single drug treatments (LB100 or carboplatin), H524 cells showed no changes in transmigration ability (% change control = 18.2 ± 2; LB100 = 16.9 ± 2; carboplatin = 18.2 ± 0.4) and for H69 cells the corresponding values were control = 19.6 ± 1.7; LB100 = 12.3 ± 0.92; carboplatin = 14.9 ± 1.24 (Fig. 3A and B). However, drug combination treatment significantly reduced cell transmigration ability through HUVEC monolayer as compared to untreated control cells (P < 0.001). Inserts indicate a lower percent change of HUVEC barrier disruption for H524 (10.6 ± 1.2%) and H69 (6.6 ± 1.2%) after 20 hours of LB100 + carboplatin treatment (P < 0.001). This suggests that combinatory inhibition of PP2A with chemotherapy could potentially disrupt cell motility through vessels and prevent invasion.
Figure 3.
SCLC cell invasion through HUVEC monolayer. A and B, Graphical representation of H524/H69 cell ability to disrupt a confluent HUVEC monolayer using an electrical substrate-impedance sensing system. Arrows indicate timepoint when cells were added. Inserts show mean values and SD for each group after 20 hours of drug treatment. After treatment, cell viability was counted using an Auto T4 Cell Counter (Nexcelom Cellometer). Cell viability was 90%–95% for drug-treated groups (n = 2). ***, P < 0.001 for control (untreated cells) versus drug combination (LB100/carboplatin) results were analyzed by ANOVA with Tukey post test. Whole-cell Pt accumulation. Graphical representation of LB100 effect on Pt uptake by SCLC cells. Cells were pretreated with LB100 (H524, 5 μmol/L; H69, 20 μmol/L) overnight, then treated with carboplatin for 1 or 4 hours (H524, 10 μmol/L; H69, 20 μmol/L). Whole-cell pellet was used for Pt measurement. Values are normalized to total protein concentration. C and D, Mean values and SD of Pt accumulation for each group. Drug combination significantly increased Pt concentrations in H524 and H69 cells. Pt concentrations in control and LB100 samples were below detection limit (n = 3, technical replicates). P < 0.001 results were analyzed by ANOVA with Tukey post test. Effect of LB100 on PP2A expression and apoptosis regulatory proteins in H524 and H69 cells. Cells were treated with indicated concentrations of LB100, carboplatin, and combination for 72 hours. E, Representative Western blot (WB) panels of the expression of PP2A subunits in H524 and H69 cells. Densitometry analysis shows quantification of PP2A A level in H524 and H69 cells (n = 3 biological replicates). F, Protein phosphorylation of γ-H2AX, caspase 3, and PARP1 cleavage activity was analyzed by WB in H524 and H69 cells after drug treatments. Representative WB panels showed significant increase in γ-H2AX phosphorylation and enhancement of caspase 3 and PARP 1 cleavage activity in H524 and H69 cells after treatment. Pan-actin was used as loading control (n = 3 biological replicates).
Because a combination of LB100 and carboplatin or etoposide showed a synergistic effect, we wished to discern the mechanism by which the drugs worked synergistically. To this end, Pt levels were measured in H524 and H69 cells using ICP-MS. Cells were pretreated with LB100 for 24 hours with subsequent 5 μmol/L (H524 cells) and 20 μmol/L (H69 cells) treatments of carboplatin for 1 or 4 hours. Treating cells for 1 hour with carboplatin only mildly elevated level of Pt in both cell lines relative to the control (Fig. 3C and D). The 4-hour treatment with drug combination significantly increased the level of Pt in both cell lines compared with single treatment of carboplatin alone, suggesting that LB100 enhanced the uptake of Pt in SCLC cells and thus, promoted the pro-apoptotic effect of carboplatin.
Several studies have found that PP2A regulates a large portion of the phosphoproteome, including pathways involved in apoptosis, proliferation, and DNA damage response (11, 37, 38). Thus, we examined the effects of LB100 alone and in combination with carboplatin on the expression of PP2A. The drug treatment drastically reduced the expression of PP2A subunit A in H524 cells (Fig. 3E, top left). But in the case of the H69 cells, subunit A expression was reduced moderately (Fig. 3E, top right). These results were confirmed by densitometry analysis (Fig. 3E, low). The expression of subunit C was unchanged in the control and treated H524 and H69 cells (Fig. 3E, middle). Moreover, LB100, carboplatin, and combination therapy significantly affected phosphorylation of histone γ-H2AX, the marker that correlates with DNA damage and induction of apoptosis in H524 and H69 cells (Fig. 3F). In addition, caspase 3 was activated in H524 and H69 cells after single treatment with LB100 or carboplatin, as well as in combination, as seen by cleavage of the preform (Fig 3F). Moreover, the dysregulation of PP2A induced PARP activity, leading to cell death. Together, these data demonstrated that inhibition of PP2A by LB100 in combination with Pt drugs induced apoptotic signaling in SCLC cells.
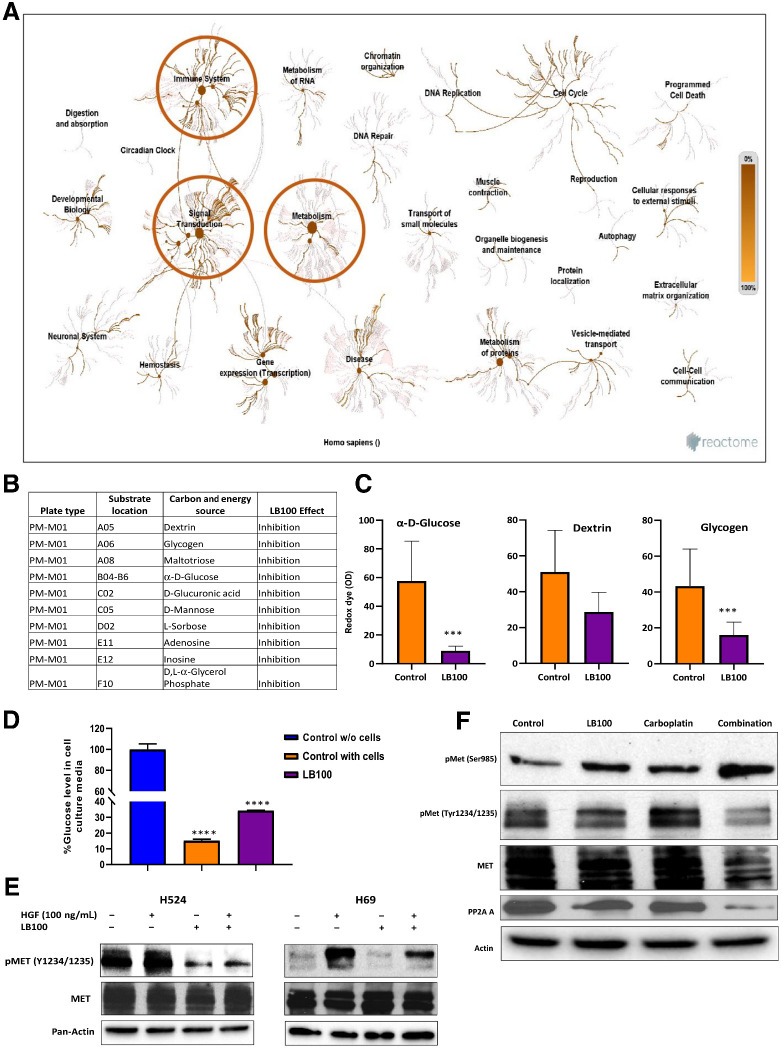
Effect of LB100 on the kinomics profile of H524 cells
Because LB100 selectively inhibits PP2A, we used PamGene technology to detect the phosphorylation of peptides as a functional readout of the cellular STKs. This analysis allowed us to interrogate the inhibitory effect of LB100 on protein phosphorylation throughout a variety of cellular pathways. As expected, LB100, at 5 and 10 μmol/L concentrations significantly increased the phosphorylation of STKs listed in Supplementary Table S3 (n = 20). Surprisingly, treatment of H524 cells with 5 and 10 μmol/L of LB100 significantly reduced the tyrosine kinase peptide phosphorylation (n = 52). The peptides tested are listed in Supplementary Table S4.
A bioinformatics analysis using the Reactome software for enrichment analysis revealed that several pathways were selected as particularly interesting based on a priori knowledge of the effect of LB100 on tumorigenesis (39–42). LB100-mediated inhibition of PP2A strongly influenced both signal transduction and metabolic pathways (Fig. 4A). A closer analysis of the signal transduction pathway showed that, consistent with previous reports (43, 44), LB100 affected HGF-MET signaling. In addition, LB100 also targeted metabolic signaling in SCLC cells.
Figure 4.
Reactome pathway analysis of PamGene PTKs and STKs after LB100 treatment of H524 cells and Biolog phenotype MicroArray. A, Significant changes were observed for signal transduction and metabolic pathways. B, MicroArray analysis showed that overnight treatment with 20 μmol/L LB100 inhibited utilization of carbon substrate sources. Table includes 10 carbon sources affected by LB100 (n = 3). C, LB100 significantly inhibited two carbon substrates utilization by H69 cells. ***, P < 0.001 for control (untreated cells) versus LB100. Results were analyzed by ANOVA with Tukey post test. D, Amplex Red Glucose/Oxidase assay kit was used to measure glucose level in cell culture media. Glucose level was significantly higher in cell culture medium from cells treated with LB100 (20 μmol/L). Glucose concentration detected in initial medium and counted as 100%. Subtracting final medium level of glucose from initial glucose medium concentration yielded % glucose in the medium with cells. Level of glucose dropped in control with cells and in LB100-treated groups (****, P < 0.0001). Results were analyzed by ANOVA with Tukey post test (n = 3 biological replicates). Effect of LB100 on MET phosphorylation. E, H524 and H69 cells were treated overnight with LB100 (H524, 5 μmol/L and H69, 20 μmol/L) following by stimulation with 100 ng/mL HGF in 10 minutes. Cells were collected and lysed for WB analysis with pMET and total MET antibody. Pan-actin was used as loading control (n = 3 biological replicates). F, H524 cell lysates (control, LB100, carboplatin, and combination (LB100/carboplatin) were analyzed by Western blots to check phosphorylation status of MET at Ser985 and Tyr1234/1235. Actin was used as a loading control (n = 3 biological replicates).
Effect of LB100 on metabolic pathways in H69 cells
To discern the effect of LB100 on metabolic signaling, we examined the utilization of carbon sources by H69 employing BiOLOG Phenotype Microarray technology. Using this assay, we examined 94 carbon sources and the redox dye tetrazolium to detect substrate utilization. LB100 inhibited the utilization of 11 carbon substrates compared with control (untreated) H69 cells (Fig. 4B) that could be divided into five groups: sugars (l-sorbose, α-d-Glucose, d-Mannose), polysaccharides (glycogen, D-Glucuronic acid), carbohydrates (dextrin, maltotriose), phosphorylated compounds (D, L-a-Glycerol Phosphate), and amines (adenosine, inosine). Of these, the consumption of three substrates important for anabolic biosynthetic reactions namely, α-d-Glucose (more than 6-fold) and glycogen (more than 2.7-fold) was significantly reduced after LB100 treatment in H69 cells (Fig. 4C). In addition, LB100 inhibited adenosine and inosine substrate utilization in these cells that could have a significant effect on purinergic signaling in SCLC. Finally, glucose uptake by H69 cells from media which contained 11 mmol/L of glucose was measured directly using a Glucose Oxidase Assay and, as expected, was found to be reduced upon treatment with LB100. Levels of glucose in control media remained close to 100% (98.2 ± 3.5%). The percentage of glucose in media containing H69 cells dropped to 15.1 ± 0.42%. After LB100 treatment, the level of glucose reduction was significantly lower, 34.2 ± 0.2% (Fig. 4D; P < 0.0001).
Effect of LB100 on MET phosphorylation in H524 and H69 cells
The PamGene kinomic data showed decreased MET peptide phosphorylation between residues 1227 and 1239. To validate this finding, we performed Western blotting experiments with H524 and H69 cell extracts, following treatment with LB100 (5 and 20 μmol/L, respectively), and stimulation with HGF for 10 minutes using a phospho-MET (pMET) antibody that specifically detects phosphorylated tyrosine 1234/1235. Pretreatment of the H524 cells with LB100 almost abrogated MET basal and HGF activated phosphorylation of MET (Fig. 4E, left). In H69 cells, the level of HGF phosphorylation significantly decreased (Fig. 4E, right) suggesting that inhibiting PP2A with LB100 the affects HGF/MET signaling responsible for cell viability, proliferation, and motility.
Previous studies demonstrated that Ser985 phosphorylation of MET negatively regulated MET kinase activity (45–47). Our results also showed that treatment of H524 cells with LB100 or in combination with carboplatin induced increase in Ser985 phosphorylation and was related with inhibition of MET tyrosine phosphorylation. Moreover, LB100 reduced the expression of PP2A A in LB100/carboplatin samples (Fig. 4F). This finding correlates with PamGene kinomic data that LB100 reduced the Tyr 1234/1235 MET phosphorylation and can be key effect of LB100 on SCLC cells.
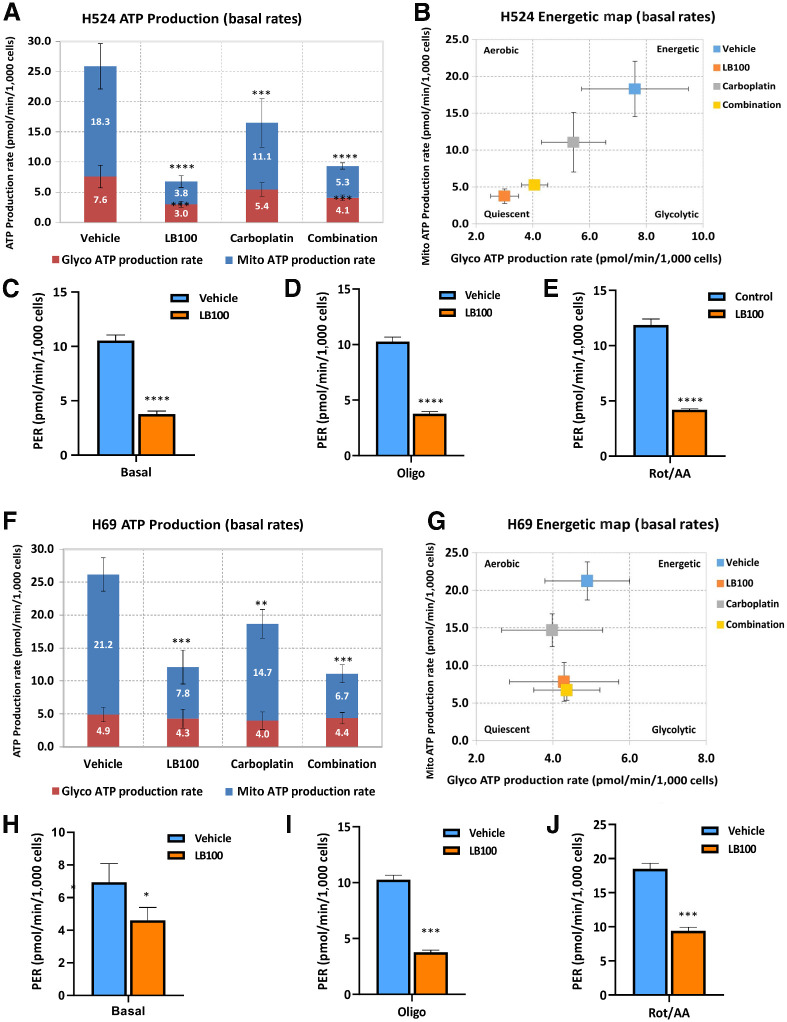
Effect of LB100 on mitochondrial and glycolytic function of SCLC cells
Next, we determined the effect of LB100 on ATP production in SCLC cells employing the Seahorse XF Cell Energy Phenotype Test. H524 and H69 cells were pretreated with half the IC50 dose of LB100 (2.5 and 10 μmol/L, respectively). After drug treatment, we counted the number of cells and examined them for viability using exclusion of trypan blue as a readout. Cellular basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements were determined on a Seahorse XF96 analyzer. H524 and H69 cells were then stressed with a combination of 1 μmol/L of oligomycin (inhibitor of oxidative phosphorylation (OxPhos) and 1 μmol/L carbonyl cyanide p-trifluoromethoxy-phenylhydrazone (FCCP; an uncoupler of OxPhos). Because oligomycin inhibits mitochondrial ATP production and FCCP induces maximum oxygen consumption by uncoupling the H+ gradient in mitochondria, the experimental conditions examined with these two stressed methods reflect the maximum glycolytic capacity and OxPhos capacity of SCLC cells, respectively. Cellular metabolic capacity includes both events and characterizes the limit of cell to acute increases in energy demands. LB100 severely affected energy metabolism of H524 cells; and their basal OCR was 4-fold lower compared with untreated cells (Supplementary Fig. S1A). LB100 treatment also induced inhibition of stressed OCR as well as basal and stressed ECAR (Supplementary Fig. S1B and S1C). These results demonstrated a significant repressive effect of LB100 on glycolytic and OxPhos pathways, the major sources of ATP production in these cells. A significant decrease in basal OCR and ECAR was also observed in H69 cells (Supplementary Fig. S1D). However, there was no significant reduction of stressed OCR and ECAR in these cells upon treatment with LB100 (Supplementary Fig. S1E and S1F).
To determine the role of LB100 alone or in combination with carboplatin on ATP production from mitochondrial respiration and glycolysis, we performed an Agilent Seahorse XF-96 Real-Time ATP rate assay. In H524 cells, total ATP production rate was significantly reduced in all three groups compared with untreated cells by 73.7% (LB100), 36.3% (carboplatin), and 63.7% (LB100/carboplatin; Fig. 5A). Mitochondrial and glycolytic ATP production rates were also significantly lower in drug-treated cells. Importantly, LB100 and LB100/carboplatin were more effective in inhibiting mitochondrial ATP and glycolytic ATP production than carboplatin alone and changed energetic phenotype of H524 cells. The cells tended to become less energetic and glycolytic (Fig. 5B).
Figure 5.
ATP production rate in SCLC cells. A, H524 cells were treated with LB100 (2.5 μmol/L), carboplatin (4 μmol/L), or a combination, and ATP production rate was measured using the Agilent Seahorse XF Real-Time ATP rate assay. mitoATP (mitochondrial) and glycoATP (glycolityc) rates were evaluated in H524 cells without and with drug treatments. All drug treatments significantly reduced mitoATP (top, blue) and glycoATP (bottom, red) production rates. B, Energetic map of H524 cells. After LB100 and drug combination, cells became less glycolytic. C–E, The Agilent Seahorse XF pH sensor probe measures changes in the concentration of free protons, which corresponds to ECAR. Real-Time ATP rate assay includes an improved metric, PER, which detects extracellular acidification from all sources. LB100 drastically reduced PER under basal conditions and after two injections of specific inhibitors of oxidative phosphorylation oligomycin (1.5 μmol/L) and antimycin (0.5 μmol/L)/rotenone (0.5 μmol/L). F, H69 cells were treated with LB100 (10 μmol/L), carboplatin (10 μmol/L), or a combination with LB100/carboplatin. ATP level in cells was measured using the Agilent Seahorse XF Real-Time ATP rate assay. LB100, carboplatin, and combination significantly reduced mitoATP. G, Energetic map of H69 cells. H–J, H69 cellular PER after LB100 treatment from glycolysis of basal and olygomycin and antimycin/rotenone injections. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Results were analyzed by ANOVA with Tukey post test (n = 2, six technical replicates).
To elucidate the effect of the drugs on the glycolytic metabolism of H524 cells, we analyzed the proton efflux rate (PER). PER is calculated by subtracting acidification produced from mitochondrial CO2 production (mitochondrial-derived CO2 can partially hydrate in the extracellular medium, resulting in additional extracellular acidification beyond that contributed by glycolysis) from total acidification or protons efflux (from both glycolysis and mitochondrial) into the extracellular medium. Basal values of the PER were reduced by >50% upon drug treatment compared to untreated cells (Fig. 5C). Measurement of the PER in the presence of oligomycin, an inhibitor of OxPhos, and a second acute injection of antimycin/rotenon (inhibitors of mitochondrial electron transport), showed a significant decrease in LB100-treated group. LB100 treatment also impaired glycolysis and reduced compensatory glycolysis (the ability of the cells to increase glycolysis after OxPhos inhibition with antimycin/rotenone; Fig. 5D and E). In addition, measurements of ATP production in H69 cells. H69 cells showed the same trend as H524 cells in that, the total ATP production rate dropped by 54% in LB100 group, by 12% in carboplatin group and 57% in the LB100/carboplatin group (Fig. 5F). Moreover, LB100 and LB100/carboplatin significantly reduced mitochondrial ATP production rate in H69 cells and the energetic map of H69 cells showed that the glycolytic ATP production rate dropped slightly in comparison with untreated cells (Fig. 5G). To confirm that LB100 also affected glycolytic pathway in LB100-resistant cells, we measured PER in these cells. Basal level of PER was significantly inhibited in LB100 group (Fig. 5H). In addition, LB100 treatment significantly inhibited PER in the presence of mitochondrial electron transport inhibitors (Fig. 5I and J). LB100 alone or in combination with carboplatin led to compromised glycolytic metabolic activity and limited oxidative capacity in H69 cells. Collectively, these results showed that LB100, alone or in combination with carboplatin effectively targeted the metabolic function of SCLC cells, thereby decreasing cell proliferation and migration, rendering them sensitive to chemotherapy.
LB100 and atezolizumab increase the recognition of tumor cells in 3D by CD8+ T cells
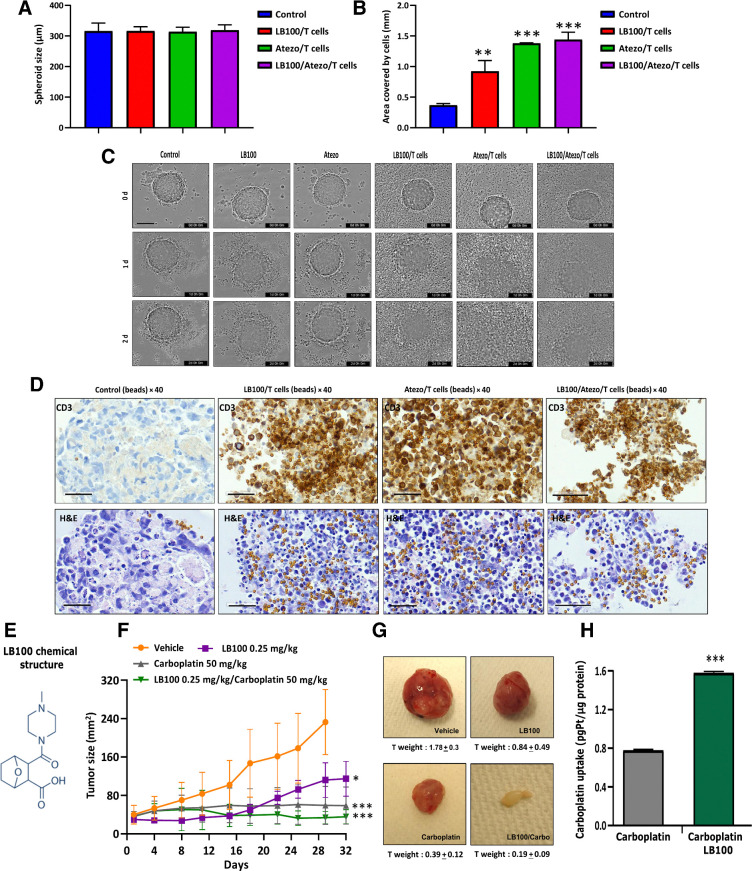
Checkpoint inhibitors can induce an anticancer immune response and PP2A inhibition has been shown to enhance anticancer immunity in several cancers. Therefore, we evaluated the combination of LB100 and atezolizumab together with a humanized IgG antibody that targets PD-L1 using a 3D spheroid of H446 cells formed in the presence of T cells. Cytotoxic CD8+ cells were isolated from whole blood, buffy coat of healthy donors following the protocol described in the Materials and Methods. Supplementary Figure S2A contains a schematic showing the treatment protocol. H446 spheroids were placed in a round bottom 96-well plate with T cells and activated beads and LB100, atezolizumab or a combination of LB100 and atezolizumab and the spheroids were visualized with time-lapse imaging. The average spheroid diameter was between 300 and 350 μm (Fig. 6A) and they had the same morphology at 0 hours (Supplementary Fig. S2B). We investigated the effect of T cells on size and tightness of the spheroids. Results indicated that spheroid shape did not change, and tumor cells did not dissociate from control or control/T cells spheroids after 2 days (Supplementary Fig. S2C). Next, spheroids were monitored for 48 hours after drug treatments and their diameters were measured from phase contrast images (Supplementary Fig. S2B). Cell dissociation diameters significantly (P < 0.001) increased in atezolizumab/T cells and LB100/atezolizumab/T cells groups compared with control (Fig. 6B). LB100 alone had moderate effect (P < 0.01) on spheroid degeneration. T cells in combination with LB100 or atezolizumab affected spheroid integrity. BF images from IncuCyte time-lapse microscopy showed that from day 0 spheroids had a round shape and well-represented spheroid structure (Fig. 6C). LB100 without T cells began disintegrating the spheroids after day 1 and atezolizumab without T cells had no effect on the spheroids. Activated T cells in combination with LB100, atezolizumab or drug combination induced shedding of dead cells, accumulation T cells in spheroid core and at day 2 only spheroid fragments were observed in the images (Fig. 6C). IHC using a CD3 antibody showed T-cell clusters among the tumor cells in three groups: LB100/T cells, atezolizumab/T cells, and LB100/atezolizumab/T cells. Combination treatment induced the destruction of spheroids, led to infiltration of the activated T cells in the spheroids resulting in the dissociation of cells, loss of spheroid morphology, and increased cell cytotoxicity. Clusters of T cells + beads on the H&E staining matched the brown spots of CD3 staining (Fig. 6D). Three-dimensional spheroid tumor cultures are used to study small molecule inhibitors in various cancer types. Use of a combination of small molecule inhibitors with PD-1/PD-L1 inhibitors, T cells, and tumor spheroids will accelerate the development of novel treatments for patients with SCLC. In our study, we employed an allogenic model exploring the effect of LB100, carboplatin, and atezolizumab, in the presence of healthy donor T cells, on SCLC spheroid fate. In the future, it will be important to test combinations of LB100/carboplatin/atezolizumab in autologous conditions using primary tumor-derived and tumor infiltrating lymphocytes from the same patients with SCLC.
Figure 6.
T-Cell infiltration in H446 spheroids in the presence of LB100 and atezolizumab. A, At timepoint 0, single spheroids in 96-well plate were treated with LB100, atezolizumab, and T cells. Column bars present mean values of spheroids at 0 hour. B, Measurement of H446 spheroidal cell distribution after 48 hours treatment with LB100 and atezolizumab in the presence of T cells. LB100/T cells (**, P < 0.01), Atezo/T cells (***, P < 0.001), and LB100/Atezo/T cells (***, P < 0.001). Results were analyzed by ANOVA with Tukey post test (n = 2; 6 technical replicates). C, Sequential images of the same H446 spheroids in control and treated groups. Scale bar, 400 μm. D, H&E and IHC staining with CD3 antibody of H446 spheroids after 48 hours of treatments. Scale bar, 50 μm. Before treatment, 5 × 103 cells were seeded in round bottom 96-well plate and grown for 3 days. LB100 activity alone and with carboplatin against H69 cells subcutaneous mouse xenograft. E, Chemical structure of LB100 (https://www.selleckchem.com/products/lb-100.html). F, Tumor size was measured. Inhibition of tumor growth after LB100 (*, P < 0.05), carboplatin (***, P < 0.001), and their combination (***, P < 0.001) were delivered via intraperitoneal injections. P values show significant differences compared with vehicle group. G, Tumor images from vehicle- and drug-treated groups. H, Columns show total Pt concentration in mouse tumors with carboplatin and LB100/carboplatin treatments (n = 3 as technical replicates). Pt mass was normalized to tumor total mass. Statistical analysis was performed using an ANOVA with Tukey post test (*, P < 0.05), carboplatin (**, P < 0.01).
Effect of LB100 on tumor growth in a mouse model of SCLC
Having demonstrated the potency of LB100 as a main active compound (Fig. 6E) in vitro, we next examined in vivo effect of LB100/carboplatin using a subcutaneous xenograft of human SCLC. Treatment with LB100 or a combination of LB100 and carboplatin resulted in a statistically significant reduction in tumor size (Fig. 6F). Notably, the drugs did not exhibit significant toxicity, nor did they significantly affect the body weight (Supplementary Fig. S3A). However, treatment with LB100, carboplatin, and their combination, caused a significant reduction in tumor weight compared with the vehicle-treated group. LB100/carboplatin inhibited primary tumor growth by 89% compared with vehicle group. The results demonstrated that drug combination maximally suppressed tumor growth (Supplementary Fig. S3B). Tumor weights were greatly reduced in LB100/carboplatin group (vehicle 1.78 ± 0.3 vs. LB100/carboplatin 0.19 ± 0.09; Fig. 6G). Measurement of Pt in mouse tumors after 30 days of treatment with carboplatin and LB100/carboplatin showed a significant increase in intratumoral Pt levels upon combination treatment (Fig. 6H). IHC of the tumors confirmed that pMET, pp2A A, CD31, and Ki67 markers stained low in drug combination group (Supplementary Fig. S4).
Discussion
LB100, a first-in-class small molecule inhibitor of PP2A, has emerged as a promising new drug for solid tumors. A recent study (48) aimed at determining the maximum tolerated dose and the safety, tolerability, and potential activity of LB100 in adult patients with progressive solid tumors, reported positive results and supported its continued development alone and in combination with other therapies. However, as far as we are aware, the drug has not been evaluated for SCLC either by itself or in combination with other standard treatments.
LB-100 is an experimental small molecule inhibitor of PP2A with cytotoxic activity against cancer cells in culture and antitumor activity in animals. Preliminary data suggest that safety and efficacy parameters warrant further clinical testing. Although LB-100 is widely reported as a specific inhibitor of serine/threonine phosphatase 2A (PP2AC/PPP2CA:PPP2CB), recently, D'Arcy and colleagues (2019; ref. 49) using purified enzymes reported that LB-100 is a catalytic inhibitor of both PP2AC and PPP5C. On the basis of these observations together with structural studies and cell-based studies revealing that the effects of LB-100 are mimicked by the genetic disruption of PPP5C, the authors concluded that LB-100 is a catalytic inhibitor of both PP2AC and PPP5C and suggested that the observed antitumor activity might be due to an additive effect achieved by suppressing both PP2A and PPP5C. Nonetheless, it is possible that in vivo particularly in the diseased state, the effects of LB-100 are mediated through the subunit that is relatively upregulated/overexpressed such as in the case of SCLC where the PP2A subunit is overexpressed.
In the current study, we showed that LB100 alone or in combination with chemotherapeutic drugs inhibited cell proliferation and colony formation in SCLC. The maximum inhibitory effect on cell proliferation was observed with a combination of LB100 and carboplatin. Furthermore, the combination was effective in a spheroid model of SCLC that resembles the tumor microenvironment more closely. This drug combination also significantly inhibited invasion of the SCLC cells through HUVEC monolayer compared with the control untreated cells. These results, along with the fact that LB100/carboplatin combination was efficacious in significantly reducing tumor size and weight in a SCLC xenograft mouse model, underscore the potential of this innovative therapeutic option for SCLC.
In addition, LB100 treatment inhibited HGF-induced MET phosphorylation in SCLC cells. Consistent with our results, PP2A is known to regulate MET activation via dephosphorylation of S895 that leads to autophosphorylation of Y1234 and Y1235, resulting in activation of the receptor (46). HGF-induced phosphorylation of MET appears to play an important role in epithelial-to-mesenchymal transition in SCLC (27). In addition, the MET/HGF axis plays a major role in the development of chemoresistance in multiple tumor types, including lung cancer. In NSCLC, the activation of the MET receptor induced chemoresistance by inhibiting apoptosis via activation of PI3K-AKT pathway and downregulation of apoptosis-inducing factor (50). Blockade of this process with a MET inhibitor resensitized these cells to chemotherapy in vitro and in vivo (51). The fact that LB100 can subvert ligand activation of MET suggests that LB100 can also attenuate chemoresistance, a major impediment in treating SCLC. c-MET is also known to be involved in metabolic reprograming in several cancers (52–55).
Cellular metabolic reprogramming is a hallmark of cancer, and aerobic glycolysis is recognized as the dominant metabolic phenotype in cancer cells. However, emerging evidence suggests that cancer cells can acquire a hybrid glycolysis/OxPhos phenotype in which both glycolysis and OxPhos can be utilized for energy production and biomass synthesis. Of note, the hybrid phenotype can facilitate metabolic plasticity of cancer cells and may be specifically associated with metastasis and chemoresistance (56–58).
We had previously shown that, PP2A plays a critical role in regulating the glycolysis rate and balancing energy supply against antioxidant protection through the PPP in B-cell lymphoma. PP2A also plays a critical role in regulating these processes in SCLC cells (8). We observed significant reduction of glucose uptake, as well as glycolytic and OxPhos upon inhibiting PP2A activity with LB100 alone or in combination with carboplatin. Furthermore, the glycolytic capacity and oxidative capacity of these cells were reduced after these treatments. These results suggest that the LB100 and carboplatin treatments lead to the reversal of the hybrid glycolysis/OxPhos phenotype, thus sensitizing the SCLC cells to the chemo drugs. Increased ATP production is associated with increased activity of the ATP-binding cassette (ABC) transporters resulting in chemoresistance (58) which is consistent with the fact that elevated ATP levels directly influence the activity of ABC transporters. The inhibition of glycolysis, OxPhos and deprivation of ATP by LB100 probably led to attenuating the function of the efflux pump, thereby increasing the toxicity of the drug, and reversing drug resistance.
From a mechanistic perspective, our mass spectrometry data suggest that the Pt concentration in SCLC cells and tumor tissue was significantly increased after LB100 treatment. Copper influx/efflux transporters have been suggested to play an important role in Pt-based drug uptake and resistance (59) in cancer. A decrease in Copper transporter 1 (CTR1) expression and increase in ABC transporters, ATP 7A/7B efflux transporters, and multi-drug resistance protein MTB1 is observed in many cancers (60). We believe that the observed increased uptake of Pt in SCLC could be due to the altered expression of one or more of the copper influx/efflux transporters in response to LB100. Consistent with this conjecture, a combination of LB100 and carboplatin acted synergistically to induce DNA damage and apoptosis in SCLC cells.
Tumor cells can evade immunosurveillance by activating immune checkpoint pathways that suppress antitumor immune responses. Immune checkpoint inhibitors restore antitumor immune responses by interrupting co-inhibitory signaling pathways and promote immune-mediated destruction of tumor cells. Emerging evidence suggests that PP2A acts as a negative regulator of cytotoxic T-cell effector function and mediates the inhibitory signaling of CTLA-4 by dephosphorylating Akt in activated T cells. We have demonstrated that PD-L1 is overexpressed in neuroendocrine cells derived from a Rbf/f/Trp53f/f mouse model of SCLC (Supplementary Fig. S5) and combination of atezolizumab and LB100 in the presence of activated T cells induced the destruction of spheroids, led to infiltration of the activated T cells in the spheroids resulting in the dissociation of cells, loss of spheroid morphology, and increased cell cytotoxicity. While additional work needs to be done, we believe that combining immunotherapy with LB100 can prove to be greatly beneficial for the patients with SCLC who have limited options for treatment of this recalcitrant cancer.
While this article was in preparation, Coles and colleagues reported that PKA/CBEP pathway was activated in about 17% of the human SCLC TMA samples (10). The authors of the above article observed that inhibition of PP2A by cantharidin enhanced the phosphorylation of PKA substrates suggesting that PP2A activity counteracts PKA activity in SCLC cells (NJH29 and NCI-H82) and treatment of SCLC cells with two small molecule activators (SMAP) of PP2A, which act via binding to PPP2R1A, suppresses PKA substrate phosphorylation and induce apoptosis in these cells. However, in our study we found that inhibiting PP2A with LB100 resulted in reduced cell viability of SCLC cells and decreased tumor growth in a SCLC mouse xenograft model. Surprisingly however, the authors of the above article acknowledge that PP2A subunit PPP2R1A is highly expressed PP2A a subunit in human SCLC and SCLC cells are dependent on PPP2R1A expression for their growth (corroborating our observation). They further note that PP2A has multiple targets in cells and that they found that SMAP treatment also had potent inhibitory effects on c-MYC expression and mTOR activity in cells. They have not determined the effect of PP2A inhibition by cantharidin or another PP2A inhibitor on cell growth, cell viability, and tumor growth. It is likely that in their study, the cell growth inhibitory effect of the SMAPs via c-MYC and mTOR inhibition is larger than the cell growth promoting effects via activation of PP2A. It is also possible that these molecules are also acting on other protein phosphatases and causing off-target effects. In any case, additional work in the future will be required to discern the controversial role of PP2A in SCLC.
The current data indicate that abrogation of PP2A with LB100 inhibits cell proliferation, tumor growth, and metastasis by asserting its pleotropic effects on, the activity of the oncogene MET, energy production, and drug uptake via altering the expression of transporters thus increasing chemosensitivity. Furthermore, the current data also indicate that combining LB100 with carboplatin and etoposide can enhance these pleotropic effects of LB100 and that, combining immunotherapy with LB100 treatment led to increased T-cell infiltration of H446 spheroids resulting in the disintegration of these spheroids. Taken together, the results from the current study suggest that pharmacologically targeting PP2A appears to be a viable strategy for SCLC.
Authors' Disclosures
J. Miser reports other support from Lixte Biotechnology Holdings, Inc., outside the submitted work. J.S. Kovach reports other support from Lixte Biotechnology Holdings, Inc., outside the submitted work; in addition, J.S. Kovach has a patent for Kovach JS, Johnson; Lixte Biotechnology, Inc., assignee, Oxabicycloheptanes and oxabicycloheptenes, their preparation and use, United States patent US 7998957. 2011 Aug 16. issued; and J.S. Kovach is CEO, President, and Chairman of Lixte Biotechnology Holdings, Inc., and receives compensation as an employee of the company, as disclosed in publicly available filings with the SEC. R. Salgia reports a patent pending for LB 100. No disclosures were reported by the other authors.
Acknowledgments
The work was supported by the NCI of the NIH under award numbers P30CA033572, U54CA209978, R01CA247471, and R01CA218545.
We thank Andrea Bild and Pierre Wallet, Department of Medical Oncology and Therapeutics Research, City of Hope National Medical Center, for experimental advice and for the reagents, and the City of Hope Light Microscopy Core, High Throughput Screening Core and Pathology, Solid Tumor Core, for help with sample processing. The murine SCLC cell lines C2.04, C896.04, and C22.03 were a generous gift from Dr. Anton Berns' lab.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
T. Mirzapoiazova: Conceptualization, investigation, visualization, methodology, writing–original draft, writing–review and editing. G. Xiao: Conceptualization, investigation, writing–original draft, writing–review and editing. B. Mambetsariev: Investigation, writing–review and editing. M.W. Nasser: Methodology, writing–review and editing. E. Miaou: Investigation, writing–original draft, writing–review and editing. S.S. Singhal: Investigation, writing–review and editing. S. Srivastava: Investigation, writing–review and editing. I. Mambetsariev: Investigation, methodology, writing–original draft, writing–review and editing. M.S. Nelson: Investigation, visualization, writing–review and editing. A. Nam: Investigation, writing–original draft, writing–review and editing. A. Behal: Methodology, writing–original draft, writing–review and editing. P. Atri: Conceptualization, formal analysis, investigation, methodology, writing–review and editing. M. Muschen: Conceptualization, investigation, writing–original draft, writing–review and editing. F.L.H. Tissot: Investigation, writing–original draft, writing–review and editing. J. Miser: Conceptualization, investigation, writing–original draft, writing–review and editing. J.S. Kovach: Conceptualization, writing–review and editing. M. Sattler: Conceptualization, writing–review and editing. S.K. Batra: Conceptualization, writing–original draft, project administration, writing–review and editing. P. Kulkarni: Conceptualization, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. R. Salgia: Conceptualization, formal analysis, supervision, funding acquisition, investigation, methodology, writing–review and editing.
References
- 1. Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep 2016;16:644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Khan P, Siddiqui JA, Maurya SK, Lakshmanan I, Jain M, Ganti AK, et al. Epigenetic landscape of small cell lung cancer: small image of a giant recalcitrant disease. Semin Cancer Biol 2020;S1044-579X(20)30240-6 [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Subbiah S, Nam A, Garg N, Behal A, Kulkarni P, Salgia R. Small cell lung cancer from traditional to innovative therapeutics: building a comprehensive network to optimize clinical and translational research. J Clin Med 2020;9:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reynhout S, Janssens V. Physiologic functions of PP2A: Lessons from genetically modified mice. Biochim Biophys Acta Mol Cell Res 2019;1866:31–50. [DOI] [PubMed] [Google Scholar]
- 5. Mazhar S, Taylor SE, Sangodkar J, Narla G. Targeting PP2A in cancer: Combination therapies. Biochim Biophys Acta Mol Cell Res 2019;1866:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thompson JJ, Williams CS. Protein phosphatase 2A in the regulation of Wnt signaling, stem cells, and cancer. Genes 2018;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kauko O, Westermarck J. Non-genomic mechanisms of protein phosphatase 2A (PP2A) regulation in cancer. Int J Biochem Cell Biol 2018;96:157–64. [DOI] [PubMed] [Google Scholar]
- 8. Xiao G, Chan LN, Klemm L, Braas D, Chen Z, Geng H, et al. B-cell-specific diversion of glucose carbon utilization reveals a unique vulnerability in B cell malignancies. Cell 2018;173:470–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLos Genet 2016;12:e1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coles GL, Cristea S, Webber JT, Levin RS, Moss SM, He A, et al. Unbiased proteomic profiling uncovers a targetable GNAS/PKA/PP2A axis in small cell lung cancer stem cells. Cancer Cell 2020;38:129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu J, Kovach JS, Johnson F, Chiang J, Hodes R, Lonser R, et al. Inhibition of serine/threonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage induced defense mechanisms. Proc Natl Acad Sci U S A 2009;106:11697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong CS, Ho W, Zhang C, Yang C, Elder JB, Zhuang Z. LB100, a small molecule inhibitor of PP2A with potent chemo- and radio-sensitizing potential. Cancer Biol Ther 2015;16:821–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai X, Zhi X, Zhang Q, Liang F, Chen W, Liang C, et al. Inhibition of protein phosphatase 2A sensitizes pancreatic cancer to chemotherapy by increasing drug perfusion via HIF-1α-VEGF mediated angiogenesis. Cancer Lett 2014;355:281–7. [DOI] [PubMed] [Google Scholar]
- 14. Hu C, Yu M, Ren Y, Li K, Maggio DM, Mei C, et al. PP2A inhibition from LB100 therapy enhances daunorubicin cytotoxicity in secondary acute myeloid leukemia via miR-181b-1 upregulation. Sci Rep 2017;7:2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Peng Y, Wang F, Tan X, Liu N, Fan S, et al. A synthetic cantharidin analog for the enhancement of doxorubicin suppression of stem cell-derived aggressive sarcoma. Biomaterials 2010;31:9535–43. [DOI] [PubMed] [Google Scholar]
- 16. Martiniova L, Lu J, Chiang J, Bernardo M, Lonser R, Zhuang Z, et al. Pharmacologic modulation of serine/threonine phosphorylation highly sensitizes PHEO in a MPC cell and mouse model to conventional chemotherapy. PLoS One 2011;6:e14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei D, Parsels LA, Karnak D, Davis MA, Parsels JD, Marsh AC, et al. Inhibition of protein phosphatase 2A radiosensitizes pancreatic cancers by modulating CDC25C/CDK1 and homologous recombination repair. Clin Cancer Res 2013;19:4422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lv P, Wang Y, Ma J, Wang Z, Li JL, Hong CS, et al. Inhibition of protein phosphatase 2A with a small molecule LB100 radiosensitizes nasopharyngeal carcinoma xenografts by inducing mitotic catastrophe and blocking DNA damage repair. Oncotarget 2014;5:7512–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon IK, Lu J, Graves CA, Huntoon K, Frerich JM, Hanson RH, et al. Protein phosphatase 2A inhibition with LB100 enhances radiation-induced mitotic catastrophe and tumor growth delay in glioblastoma. Mol Cancer Ther 2015;14:1540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang KE, Wei BR, Madigan JP, Hall MD, Simpson RM, Zhuang Z, et al. The protein phosphatase 2A inhibitor LB100 sensitizes ovarian carcinoma cells to cisplatin-mediated cytotoxicity. Mol Cancer Ther 2015;14:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ho WS, Feldman MJ, Maric D, Amable L, Hall MD, Feldman GM, et al. PP2A inhibition with LB100 enhances cisplatin cytotoxicity and overcomes cisplatin resistance in medulloblastoma cells. Oncotarget 2016;7:12447–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho WS, Wang H, Maggio D, Kovach JS, Zhang Q, Song Q, et al. Pharmacologic inhibition of protein phosphatase-2A achieves durable immune-mediated antitumor activity when combined with PD-1 blockade. Nat Commun 2018;9:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parry RV, Chemnitz JM, Frauwirth KA, Lanfranco AR, Braunstein I, Kobayashi SV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol 2005;25:9543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou P, Shaffer DR, Alvarez Arias DA, Nakazaki Y, Pos W, Torres AJ, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 2014;506:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 2008;47:1025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanteti R, Dhanasingh I, Kawada I, Lennon FE, Arif Q, Bueno R, et al. MET and PI3K/mTOR as a potential combinatorial therapeutic target in malignant pleural mesothelioma. PLoS One 2014;9:e105919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J, Mirzapoiazova T, Tan YHC, Pang KM, Pozhitkov A, Wang Y, et al. Inhibiting crosstalk between MET signaling and mitochondrial dynamics and morphology: a novel therapeutic approach for lung cancer and mesothelioma. Cancer Biol Ther 2018;19:1023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanteti R, Riehm JJ, Dhanasingh I, Lennon FE, Mirzapoiazova T, Mambetsariev B, et al. PI3 kinase pathway and MET inhibition is efficacious in malignant pleural mesothelioma. Sci Rep 2016;6:32992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006;58:621–81. [DOI] [PubMed] [Google Scholar]
- 31. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mirzapoiazova T, Mambetsariev N, Lennon FE, Mambetsariev B, Berlind JE, Salgia R, et al. HABP2 is a novel regulator of hyaluronan-mediated human lung cancer progression. Front Oncol 2015;5:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bochner BR, Siri M, Huang RH, Noble S, Lei XH, Clemons PA, et al. Assay of the multiple energy-producing pathways of mammalian cells. PLoS One 2011;6:e18147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawada I, Hasina R, Lennon FE, Bindokas VP, Usatyuk P, Tan YH, et al. Paxillin mutations affect focal adhesions and lead to altered mitochondrial dynamics: relevance to lung cancer. Cancer Biol Ther 2013;14:679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Faoro L, Singleton PA, Cervantes GM, Lennon FE, Choong NW, Kanteti R, et al. EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mammalian target of rapamycin activation. J Biol Chem 2010;285:18575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finger EC, Giaccia AJ. Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev 2010;29:285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Yang R, Gu J, Yin X, Jin N, Xie S, et al. Cross talk between PI3K-AKT-GSK-3β and PP2A pathways determines tau hyperphosphorylation. Neurobiol Aging 2015;36:188–200. [DOI] [PubMed] [Google Scholar]
- 38. Liu GP, Wei W, Zhou X, Shi HR, Liu XH, Chai GS, et al. Silencing PP2A inhibitor by lenti-shRNA interference ameliorates neuropathologies and memory deficits in tg2576 mice. Mol Ther 2013;21:2247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perrotti D, Neviani P. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol 2013;14:e229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duong FH, Dill MT, Matter MS, Makowska Z, Calabrese D, Dietsche T, et al. Protein phosphatase 2A promotes hepatocellular carcinogenesis in the diethylnitrosamine mouse model through inhibition of p53. Carcinogenesis 2014;35:114–22. [DOI] [PubMed] [Google Scholar]
- 41. Duong FH, Filipowicz M, Tripodi M, Monica NL, Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology 2004;126:263–77. [DOI] [PubMed] [Google Scholar]
- 42. Liu L, Wang H, Cui J, Zhang Q, Zhang W, Xu W, et al. Inhibition of protein phosphatase 2A sensitizes mucoepidermoid carcinoma to chemotherapy via the PI3K-AKT pathway in response to insulin stimulus. Cell Physiol Biochem 2018;50:317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salgia R. Role of c-Met in cancer: emphasis on lung cancer. Semin Oncol 2009;36:S52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hardy-Werbin M, Del Rey-Vergara R, Galindo-Campos MA, Moliner L, Arriola E. MET inhibitors in small cell lung cancer: from the bench to the bedside. Cancers 2019;11:1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gandino L, Longati P, Medico E, Prat M, Comoglio PM. Phosphorylation of serine 985 negatively regulates the hepatocyte growth factor receptor kinase. J Biol Chem 1994;269:1815–20. [PubMed] [Google Scholar]
- 46. Hashigasako A, Machide M, Nakamura T, Matsumoto K, Nakamura T. Bi-directional regulation of Ser-985 phosphorylation of c-met via protein kinase C and protein phosphatase 2A involves c-Met activation and cellular responsiveness to hepatocyte growth factor. J Biol Chem 2004;279:26445–52. [DOI] [PubMed] [Google Scholar]
- 47. Virzì AR, Gentile A, Benvenuti S, Comoglio PM. Reviving oncogenic addiction to MET bypassed by BRAF (G469A) mutation. Proc Natl Acad Sci U S A 2018;115:10058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chung V, Mansfield AS, Braiteh F, Richards D, Durivage H, Ungerleider RS, et al. Safety, tolerability, and preliminary activity of LB-100, an inhibitor of protein phosphatase 2A, in patients with relapsed solid tumors: an open-label, dose escalation, first-in-human, phase I trial. Clin Cancer Res 2017;23:3277–84. [DOI] [PubMed] [Google Scholar]
- 49. D'Arcy BM, Swingle MR, Papke CM, Abney KA, Bouska ES, Prakash A, et al. The antitumor drug LB-100 is a catalytic inhibitor of protein phosphatase 2A (PPP2CA) and 5 (PPP5C) coordinating with the active-site catalytic metals in PPP5C. Mol Cancer Ther 2019;18:556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen JT, Huang CY, Chiang YY, Chen WH, Chiou SH, Chen CY, et al. HGF increases cisplatin resistance via down-regulation of AIF in lung cancer cells. Am J Respir Cell Mol Biol 2008;38:559–65. [DOI] [PubMed] [Google Scholar]
- 51. Cañadas I, Rojo F, Taus Á, Arpí O, Arumí-Uría M, Pijuan L, et al. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clin Cancer Res 2014;20:938–50. [DOI] [PubMed] [Google Scholar]
- 52. Boschert V, Klenk N, Abt A, Raman SJ, Fischer M, Brands RC, et al. The influence of Met receptor level on HGF-induced glycolytic reprogramming in head and neck squamous cell carcinoma. Int J Mol Sci 2020;21:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nguyen TTT, Shang E, Karpel-Massler G, Siegelin MD. Metabolic reprogramming by c-MET inhibition as a targetable vulnerability in glioblastoma. Oncoscience 2020;7:14–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meng F, Wu L, Dong L, Mitchell AV, James Block C, Liu J, et al. EGFL9 promotes breast cancer metastasis by inducing cMET activation and metabolic reprogramming. Nat Commun 2019;10:5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jin N, Bi A, Lan X, Xu J, Wang X, Liu Y, et al. Identification of metabolic vulnerabilities of receptor tyrosine kinases-driven cancer. Nat Commun 2019;10:2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu L, Lu M, Jia D, Ma J, Ben-Jacob E, Levine H, et al. Modeling the genetic regulation of cancer metabolism: interplay between glycolysis and oxidative phosphorylation. Cancer Res 2017;77:1564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jia D, Park JH, Jung KH, Levine H, Kaipparettu BA. Elucidating the metabolic plasticity of cancer: mitochondrial reprogramming and hybrid metabolic states. Cells 2018;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma L, Zong X. Metabolic symbiosis in chemoresistance: refocusing the role of aerobic glycolysis. Front Oncol 2020;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kilari D, Guancial E, Kim ES. Role of copper transporters in platinum resistance. World J Clin Oncol 2016;7:106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 2007;33:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]