Abstract
In this study, peptide entry inhibitors against the fusion processes of severe acute respiratory syndrome coronavirus-2 (SCV2) and influenza A virus (IAV) were designed and evaluated. Fusion inhibitor peptides targeting the conformational shift of the viral fusion protein were designed based on the relatively conserved sequence of HR2 from SCV2 spike protein and the conserved fusion peptide from hemagglutinin (HA) of IAV. Helical HR2 peptides bind more efficiently to HR1 trimer, while helical amphipathic anti-IAV peptides have higher cell penetration and endosomal uptake. The initial sequences were mutated by increasing the amphipathicity, using helix favoring residues, and residues likely to form salt- and disulfide-bridges. After docking against their targets, all anti-SCV2 designed peptides bonded with the HR1 3-helical bundle's hydrophobic crevice, while AntiSCV2P1, AntiSCV2P3, AntiSCV2P7, and AntiSCV2P8 expected to form coiled coils with at least one of the HR1 strands. Four of the designed anti-IAV peptides were cell-penetrating (AntiIAVP2, AntiIAVP3, AntiIAVP4, AntiIAVP7). All of them interacted with the fusion peptide of HA and some of the residues in the conserved hydrophobic pocket of HA2 in H1N1, H3N1, and H5N1 subtypes of IAV. AntiIAVP3 and AntiIAVP4 peptides had the best binding to HA2 conserved hydrophobic pocket, while, AntiIAVP2 and AntiIAVP6 showed the best binding to the fusion peptide region. According to analyses for in-vivo administration, AntiSCV2P1, AntiSCV2P7, AntiIAVP2, and AntiIAVP7 were the best candidates. AntiSCV2 and AntiIAV peptides were also conjugated using an in vivo cleavable linker sensitive to TMPRSS2 applicable as a single therapeutic in coinfections or uncertain diagnosis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10989-021-10357-y.
Keywords: Coronavirus, Spike glycoprotein, Influenza A virus, Hemagglutinin, Peptide, Drug design
Introduction
Enveloped viruses are one of the ever-present categories of viruses, responsible for many classic and emerging infections that cause considerable morbidity and mortality each year. Many known pathogens such as hepatitis C, human immunodeficiency virus (HIV), smallpox, influenza, Ebola, SARS, and MERS viruses belong to this family (Badani et al. 2014). Enveloped viruses must first bind to the cell surface and then fuse their membrane with the host cell membrane to start an infection (Behzadipour et al. 2021a). One or more surface glycoproteins mediate the process of viral-cell fusion in enveloped viruses. One of these glycoproteins is usually denoted as the fusion protein. These fusion proteins will ultimately coordinate close contact between cellular and viral membranes and disrupt the cellular membrane's continuity to enable the release of the viral genome into the host cell (Harrison 2008).
Viral fusion proteins have various structural properties and mechanisms of activation. Based on their secondary structure, these fusion proteins can be classified into three different categories. Class I viral fusion proteins can be found in viruses such as influenza, HIV, SARS, and MERS with typically an α-helical trimer conformation that will further fold into a helical hexamer (Schibli and Weissenhorn 2004). Class II fusion proteins have a β-sheet rich structure, while class III fusion proteins are a mix of α-helix and β-sheet conformations (White et al. 2008). Regardless of their category, activation of fusion proteins may occur after binding to cell surface receptors or acidification inside an endosome, resulting in a rearrangement of their conformation. This rearrangement exposes a fusion peptide (class I fusion proteins) or a fusion loop (class II and III fusion proteins) that will disturb the integrity of the cellular membrane and start the process of membrane fusion (Badani et al. 2014). Severe acute respiratory syndrome coronavirus-2 (SCV2) is the virus to blame for the current global pandemic, and influenza A virus (IAV), responsible for many seasonal epidemics and pandemics, are enveloped viruses both with class I fusion proteins. Many bioactive peptides can bind to fusion proteins and therefore inhibit the entry of enveloped viruses into host cells. The FDA approved one of these peptide entry inhibitors (Enfuvirtide) as a therapeutic against HIV-1 (Matthews et al. 2004). We believe fusion proteins, although often dismissed, can potentially be targets of effective antiviral countermeasures.
The surface glycoprotein or S protein of SCV2 has two domains, namely S1 and S2. S1 is responsible for attachment to host cell receptors, and the S2 subunit mediates the membrane fusion. After the receptor binding at the cell surface, the virus can enter cells using both endosomal and non-endosomal pathways (Yang and Shen 2020). Some host proteases such as furin and transmembrane protease serine 2 (TMPRSS2) can cleave the S protein into its two subunits S1 and S2, and then at S2ʹ, therefore activating the S protein for the fusion process (Fig. 1) (Coutard et al. 2020). A conformational rearrangement occurs in S2, resulting in two heptad repeats forming a 6-helix bundle fusion core; the N-terminal heptad repeat (NHR or HR1) region as the homotrimer middle and three C-terminal heptad-repeat (CHR or HR2) regions packed into its surrounding. Peptides designed based on the HR2 act as decoys to the viral CHR, potentially forming a non-functional helical hexamer and antagonizing the conformational shift in the fusion protein. They are, therefore, competitive inhibitors of the virus-cell fusion process (Wang et al. 2018). Previous studies on HR2 inspired peptides to combat SCV2 and its predecessors SARS and MERS confirm the feasibility of employing these peptides against SCV2 (Badani et al. 2014; Wang et al. 2018; Xia et al. 2020; Xiu et al. 2020).
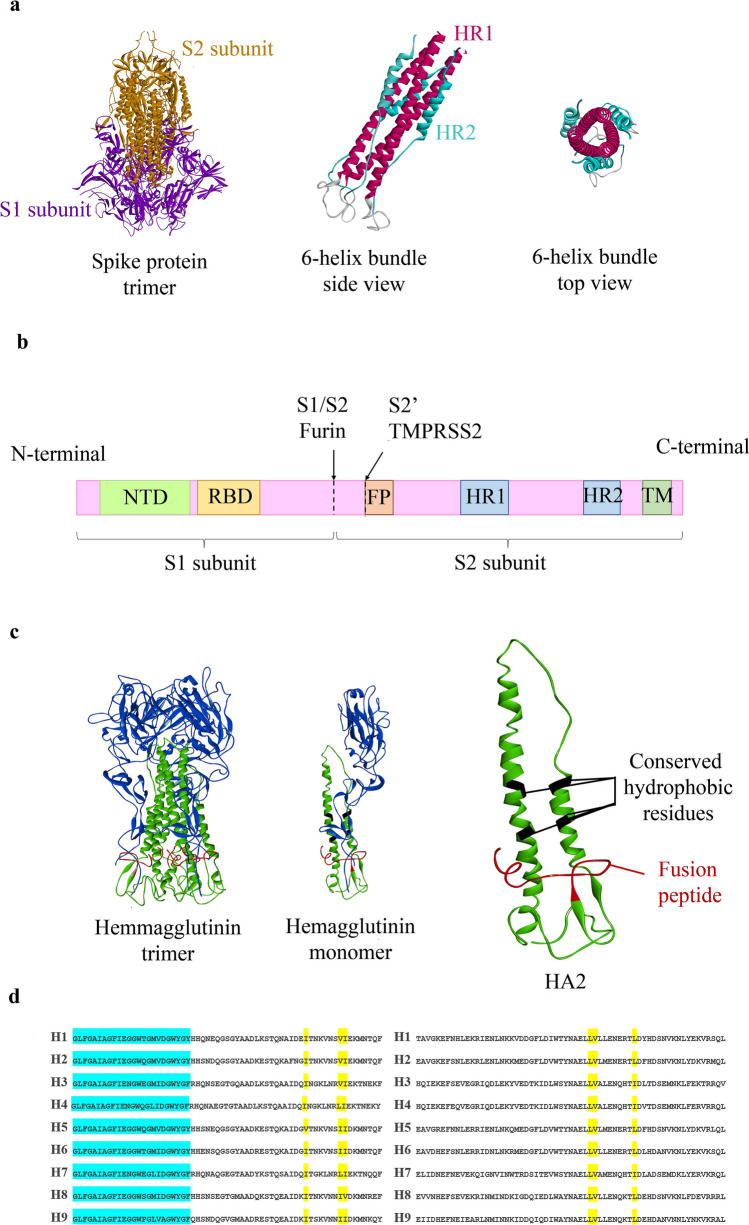
Fig. 1.
Structures of SCV2 S protein and IAV Hemagglutinin. a Models of SCV2's S protein trimer, as well as a schematic 6-helix bundle. b An illustration of S protein primary structure and its cleavage sites, including NTD (N-terminal domain), RBD (receptor binding domain), FP (fusion peptide), HR1 (heptad repeat 1), HR2 (heptad repeat 2), TM (transmembrane domain). The arrows represent the cleavage sites of furin and TMPRSS2 proteases (Schütz et al. 2020). c 3D structure of influenza A Hemagglutinin trimer, monomer, and HA2. d Amino acid alignment of 10 different subtypes of IAV. Blue shaded residues are the conserved fusion peptides, while yellow shaded residues are previously reported binding sites in the conserved hydrophobic pocket of HA2 (Lin et al. 2017)
Hemagglutinin (HA), one of IAV's major glycoproteins, is the fusion protein responsible for binding and entry of the virus into host cells. HA is composed of two subunits HA1 and HA2. Much like SCV2, cleavage of HA into HA1 and HA2 is followed by a low pH-dependent conformational shift of HA2 into a helical hexamer that will result in the fusion of the virus to the endosomal membrane. However, peptides derived from the HR2 region of IAVs have not significantly inhibited the entry of these viruses into host cells (Lin et al. 2017). Therefore, one should consider other targets of IAV entry inhibition, from which the conserved hydrophobic pocket of HA2 is noteworthy. Some of the amino acids present in the hydrophobic pocket of HA2, including residues 48, 55, 56, 99, 100, and 108, are often occupied by hydrophobic residues crucial to the conformational shift of Hemagglutinin (Bullough et al. 1994). These amino acid targets are highly conserved across different influenza A subtypes, making them expedient targets for developing broad anti-IAV molecules (Fig. 1).
Since coexisting infections with SCV2 and IAV have been observed simultaneously with common and sometimes confusing symptoms during the COVID-19 pandemic, the availability of a product capable of controlling both viruses seems essential (Bai et al. 2021; Miatech et al. 2020). As the duration of effectiveness of vaccines remains unclear, inhibitors of virus entry into the host cells will be one of the most effective preventive and therapeutic agents available after infection. This study describes a step-by-step rational design of novel antimicrobial peptide candidates against SCV2 and IAV based on the entry inhibitor peptides.
Methods
Retrieval of Fusion Protein Models and Template Peptide Sequences
SCV2 S Protein Fusion Core and HR2
The HR2 region (residues 1171–1200) of S protein (Uniprot ID: P0DTC2) from SCV2 was used as the template peptide for peptide design. The HR1 trimer in the fusion core of PDB code 6M1V was used as the target (Sun et al. 2020).
Influenza A Virus HA2 and Fusion Peptide
The fusion peptide of IAV was chosen as the template for peptide design against this virus. The sequence was retrieved from the N-terminal of HA2 subunit (residues 345–367) of hemagglutinin protein of influenza A virus (Uniprot ID: Q82774). The HA2 region of PDB codes: 4EDB, 4UO0, and 3S11, which belong to influenza A H1N1, H3N1, and H5N1 strains, respectively, were selected as targets and models of viral fusion protein before the structural shift.
Modeling and Secondary Structure of Designed Peptides
The HR2 region (residues 1171–1200) of S protein and the fusion peptide of IAV that are viral template sequences will be referred to as wild peptides from here on. Peptides designed based on the mentioned wild template sequences with potential entry inhibitor properties will be referred to as designed peptides henceforth. All wild and designed peptides were modeled using the PEPstrMOD program to evaluate the degree of helicity in the designed peptide (http://crdd.osdd.net/raghava/pepstr/) (Singh et al. 2015). Ramachandran plots were used to assess the robustness of models using pdbsum, as previously reported (Rahmatabadi et al. 2019).
Not all helices can remain stable in solution since their backbone hydrogen bonds can be attacked by water molecules (Jarrold 2007). Peptides with higher than 70% predicted helicity were also evaluated for forming helices in water using the FMAP program (https://membranome.org/fmap). FMAP predicts helix formation in different environments, including water, with 95% accuracy.
Docking of the Peptides and Evaluation of Coiled-Coils Formation
Peptides were docked into the target fusion construct by the ClusPro program (https://cluspro.bu.edu/) (Vajda et al. 2017). The fusion construct was uploaded as the receptor and the peptides as ligands. Results were sorted based on their cluster sizes. Furthermore, the result with the largest cluster size was selected. Docked structures were submitted to the Coilcheck+ program (http://caps.ncbs.res.in/coilcheckplus/action/coilcheckplus-form.php) (Sunitha et al. 2012) to investigate the interactions and calculate the energy between the designed anti-SCV2 peptides and HR1 trimer.
Evaluation of Cell Penetration for IAV Entry Inhibitor Peptides
The designed anti-IAV peptides were evaluated for their cell penetration ability using the MLCPP program (http://www.thegleelab.org/MLCPP/) (Manavalan et al. 2018). MLCPP screens the amino acid composition and the sequences' physicochemical properties in its first layer of prediction and uses a highly randomized tree to predict cell penetration.
Evaluation of Hydrophobic Moment, Wimley–White, and Bowman Index
The wild peptides and the designed viral entry inhibitors were evaluated for their amphipathicity by the Heliquest (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) (Gautier et al. 2008), using their hydrophobic moment. All the helical wheel illustrations of this study were drawn using Heliquest. Wimley–White and Bowman index of the peptides were calculated using the APD3 tool (https://aps.unmc.edu/prediction) (Wang et al., 2016).
Lipid-Binding Discrimination Factor of Designed Peptides
Entry inhibitory peptides in possession of membrane-tropic features show a higher affinity to the membrane. They are more likely to interact with fusion proteins of viruses at the cell membrane interfaces (Wang et al. 2018). Each peptide's lipid-binding discrimination factor (D) was calculated using the hydrophobic moment (µH) and the net charges (z) obtained from heliquest as follows:
Evaluation of the Designed Peptides for In Vivo Administration
De novo designed peptides were submitted to ToxinPred (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php) for evaluation of the toxicity (Gupta et al. 2013). Plifepred (https://webs.iiitd.edu.in/raghava/plifepred/batch.php) predicted half-lives of peptides (Mathur et al. 2018). Vaxijen V 2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to evaluate antigenicity (Zaharieva et al. 2017). HemoPI (https://webs.iiitd.edu.in/raghava/hemopi/multiple_test.php) and AllerTOP (https://www.ddg-pharmfac.net/AllerTOP/) were applied to determine allergenicity and hemolytic potency (Chaudhary et al. 2016), respectively (Dimitrov et al. 2013). Prosperous (https://prosperous.erc.monash.edu/) calculated susceptibility to human proteases (Song et al. 2017).
Prediction of the Antimicrobial Activity Using Bioinformatics Tools
The designed anti-IAV and anti-SCV2 peptides were submitted to the iAMPpred program available at (http://cabgrid.res.in:8080/amppred/), an SVM-based tool that predicts the antibacterial, antiviral, and antifungal potential of the peptides (Meher et al. 2017).
Modelling of In Vivo Cleavable Fused Peptides
The FASTA sequence of one of the most promising entry inhibitory peptides against SCV2 and IAV fused by an in vivo cleavable sequence was submitted to the I-TASSER program for modeling. The model with the highest C-score was selected.
Results and Discussion
Rational Design of Viral Entry Inhibitor Peptides
As mentioned earlier, CHR-based designed peptides, also called C-peptides, antagonize the SCV2 entry process by forming faulty fusion cores and hindering the conformational shift. Previous studies on the coiled-coil 6-helix bundle of HIV-1 virus—as one of the best-characterized structures among other class I enveloped viruses—have shown that the degree of helicity and amphipathicity of the designed peptide is of more importance compared to its sequence for an effective attachment to NHR (Su et al. 2017; Wang et al. 2018). The studies have shown that even though C-peptides are usually in random coil conformation in the free state, they will shift into a helical conformation when bound to the NHR helical trimer. C-peptides' bonding to the NHR region involves a large amount of energy penalty due to the loss of entropy by taking the more constrained helix conformation. The smaller the peptide, the harder it will be to make up the energy penalty (Sia et al. 2002). By constraining C-peptides in their unbound state, the binding energy penalty will be reduced. Therefore, peptides with a higher degree of helical conformation have a higher potential of being effective inhibitors of SCV2 S protein.
One of the arguments regarding the unsuccessful trials in developing an entry inhibitor peptide therapeutic against IAV is their inability to enter endosomes alongside the virus since HA only uses the endosomal pathway for host cell entry (Lin et al. 2017). To overcome this obstacle, scientists have tried to increase the peptide's affinity to the host membrane surface by lipidation, which entails conjugating the designed C-peptide to a lipid moiety, thus increasing the probability of peptide and fusion protein interactions (Lee et al. 2011). A class of peptides known for their high affinity to membrane interfaces is the cell-penetrating peptides (CPPs) (Behzadipour and Hemmati 2019; Henriques et al. 2006). Previous reports of successful viral entry inhibition have been reported by conjugating the entry inhibitor peptides to the TAT peptide, one of the most studied CPPs (Behzadipour et al. 2021b; Figueira et al. 2018; Melnik et al. 2011; Sadeghian et al. 2018). A peptide with a high hydrophobicity value (to target the hydrophobic pocket of the IAV HA2 region) and high membrane affinity is needed to follow said strategies. The fusion peptide of HA is s peptide with these characteristics in the IAV sequence. There are also previous positive results available in favor of choosing the fusion peptide of IAV as the scaffold for anti-IAV peptide design (Wu et al. 2015). Since the fusion peptide scaffold is highly hydrophobic, by designing an amphipathic alpha-helical peptide, mutating the originally non-cell penetrating frame towards a peptide with a higher cell-penetration ability is observed (Kalafatovic and Giralt 2017).
To conclude, viral fusion inhibitors that are primarily helical in their unbound state were designed by performing rational mutations on the wild peptides derived from SCV2 and IAV themselves, employing three general strategies and then combining the best features of the most helical peptides:
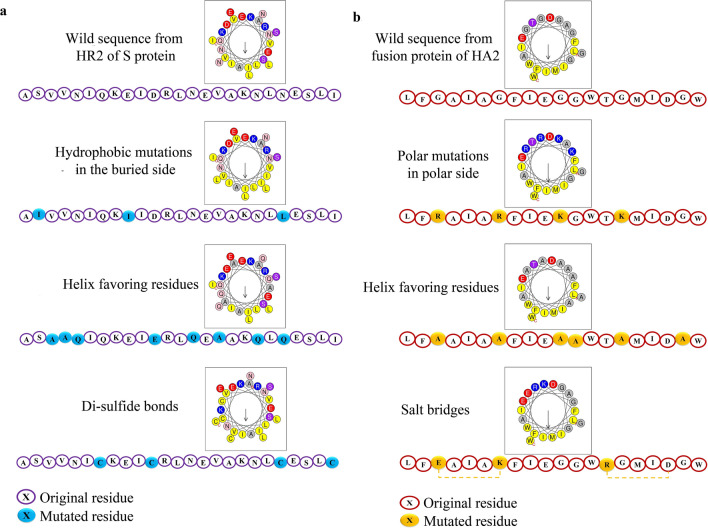
- Increasing the amphiphilicity of the peptides:One of the main driving forces of coiled-coil formation is the amphipathicity of the designed C-peptides in a way that the helix forms hydrophobic and hydrophilic faces. The hydrophobic face of the peptide will be buried in the coiled-coil formation (Wang et al. 2018). Therefore, using the helical wheel representation of HR2 from SCV2, we tried to substitute the residues in the hydrophobic side with more potent hydrophobic amino acids.
- Helix favoring residuesThe peptide must pay an entropic penalty to fold from a random coiled conformation into a more restrained helical conformation. Since different amino acids have diverse propensities to form helices, each residue's helical penalty also differs. Previously PACE et al. (Nick Pace and Martin Scholtz 1998) introduced a system to differentiate the helix favoring amino acids. Therefore, the less helix-favoring viral peptide residues will be substituted with more helix favoring residues, based on the helix propensity table (Table 1).
- Salt bridges and disulfide bondsAnother strategy to stabilize helix formation in short peptides is employing salt and disulfide bridges. The salt-bridge approach puts amino acids with opposite charges in the peptide at a distance from each other. The attraction of opposite charges constrains the peptide in a helical conformation. Based on previous studies (Yin 2012), one of the popular strategies with helix stabilizing effects is placing the amino acids (positively charged Lysine or Arginine and negatively charged Glutamate or Aspartate) in i and i+4 positions. Two cysteine residues in the abovementioned positions can also have a helix stabilizing effect.
Multiple strategies
The best peptides obtained by each strategy will be further mutated according to the other two approaches to get peptides with the highest helicity ratio.
Table 1.
Differences in helical penalties of the twenty natural amino acids (Nick Pace and Martin Scholtz 1998)
| Amino acid | Helical penalty (kcal mol−1) |
|---|---|
| Alanine | 0 |
| Arginine | 0.21 |
| Leucine | 0.21 |
| Methionine | 0.24 |
| Lysine | 0.26 |
| Glutamine | 0.39 |
| Glutamic acid | 0.4 |
| Isoleucine | 0.41 |
| Tryptophan | 0.49 |
| Serine | 0.5 |
| Tyrosine | 0.53 |
| Phenylalanine | 0.54 |
| Histidine | 0.61 |
| Valine | 0.61 |
| Asparagine | 0.65 |
| Threonine | 0.66 |
| Cysteine | 0.68 |
| Aspartic acid | 0.69 |
| Glycine | 1 |
| Proline | 3.16 |
Design of C-Peptides Against SARS-CoV-2
The "IQKEIDRLNEVAKN" sequence from the wild SCV2 HR2 (GINASVVNIQKEIDRLNEVAKNLNESLIDL) was predicted to be helical by the PEPstrMOD program. A twenty-five amino acid sequence was selected from HR2 as the scaffold for peptide design (ASVVNIQKEIDRLNEVAKNLNESLI). Peptides were mutated based on the mentioned strategies in the previous section to enhance the helicity ratio (Table 2, Fig. 2). Designed peptides were modeled using the PEPstrMOD program. Peptides with higher than 70% helicity were mutated again based on all three strategies to achieve newer peptides with a higher degree of helicity (Table 3). For example, the residues in a peptide with hydrophobic mutations on its buried side were further substituted by helix favoring and/or salt bridge forming amino acids. Peptides were modeled again, and 27 designed peptides with more than 70% helicity were submitted to further analyses. In summary, out of the three strategies, hydrophobic mutations in the peptide's buried side and the consequent enhancement in amphipathicity resulted in an increased number of peptides with higher than 70% helicity. As expected, using more than one strategy simultaneously produced peptides with a higher ratio of the helical structure.
Table 2.
SCV2 entry inhibitor peptides designed based on three strategies and their helicity ratio. Mutated residues are underlined
| Design strategy | Mutated sequence | Predicted helical segment | Helicity percentage (%) |
|---|---|---|---|
| Wild sequence | ASVVNIQKEIDRLNEVAKNLNESLI | IQKEIDRLNEVAKN | 56 |
| Hydrophobic mutation in buried site | AIVVNIQKIIDRLNEVAKNLLESLI | IQKIIDRLNEVAKNLLES | 72 |
| AYVVNIQKFIDRLNEVAKNLWESLI | IQKFIDRLNEVAKNLWE | 68 | |
| ALVVNIQKLIDRLNEVAKNLFESLI | IQKLIDRLNEVAKNLFES | 72 | |
| AYVVNIQKLIDRLNEVAKNLMESLI | IQKLIDRLNEVAKNLMES | 72 | |
| AAVVNIQKVIDRLNEVAKNLMESLI | IQKVIDRLNEVAKNLMES | 72 | |
| AIVVNIQKAIDRLNEVAKNLYESLI | IQKAIDRLNEVAKNLYE | 68 | |
| AFVVNIQKAIDRLNEVAKNLMESLI | IQKAIDRLNEVAKNLMES | 72 | |
| ALVVNIQKLIDRLNEVAKNLAESLI | IQKLIDRLNEVAKNLAE | 68 | |
| Helix favoring residues | ASVVQIQKEIERLNEVAKQLQESLI | IQKEIERLNEVAKQLQE | 68 |
| ASVVAIQKEIERLNEVAKALAESLI | AIQKEIERLNEVAKALAE | 72 | |
| ASVVRIQKEIERLNEVAKRLRESLI | IQKEIERLNEVAKRLRE | 68 | |
| ASVVAIQKEIERLAEVAKKLAESLI | AIQKEIERLAEVAKKLAE | 72 | |
| ASVVAIQKEIARLNEVAKALAESLI | AIQKEIARLNEVAKALAE | 72 | |
| ASAANIQKEIDRLNEAAKNLNESLI | NIQKEIDRLNEAAK | 56 | |
| ASAANIQKEIERLNEAAKNLNESLI | NIQKEIERLNEAAK | 56 | |
| ASAAQIQKEIERLQEAAKQLQESLI | AAQIQKEIERLQEAAKQLQE | 80 | |
| Salt bridges and disulfide bond.s | ASVVNIQKEIDELNEVAKNLNESLI | IQKEIDELNEVAKN | 56 |
| ASVVNIEKEIKELNEVAKNLNESLI | IEKEIKELNEVAKN | 56 | |
| ASVVNIQEEIDRLNEVAKNLNESLI | IQEEIDRLNEVAKN | 56 | |
| ASVENIQKEIDELREVAKNLNESLI | VENIQKEIDELREVAKN | 68 | |
| ASVRNIQEEIKRLNEVAKNLNESLI | RNIQEEIKRLNEVAKN | 64 | |
| ASVVNICKEICRLNEVAKNLNESLI | VVNICKEICRLNEVAKN | 68 | |
| ASCVNICkEICRLNEVAKNLNESLI | CVNICKEICRLNEVAKN | 68 | |
| ASVVNICKEICRLNEVAKNLCESLC | VVNICKEICRLNEVAKNLCE | 80 |
Fig. 2.
Helical wheel representations of Anti-SCV2 and Anti-IAV designed peptides. a Helical wheel presentation and sequences of wild HR2 of SCV2 and three peptides, each designed based on one of the design strategies. b Helical wheel representation and sequences of the design scaffold from wild fusion peptide of IAV and three peptides, each designed according to one of the design strategies
Table 3.
The most helical designed SCV2 entry inhibitor peptides using each of the three strategies and multiple strategies, as well as their helicity ratios
| Mutated sequence | Helical segment | Helicity percentage (%) | |
|---|---|---|---|
| Wild sequence | ASVVNIQKEIDRLNEVAKNLNESLI | IQKEIDRLNEVAKN | 56 |
| Most helical peptides obtained from each of three employed strategies | AIVVNIQKIIDRLNEVAKNLLESLI | IQKIIDRLNEVAKNLLES | 72 |
| ALVVNIQKLIDRLNEVAKNLFESLI | IQKLIDRLNEVAKNLFES | 72 | |
| AYVVNIQKLIDRLNEVAKNLMESLI | IQKLIDRLNEVAKNLMES | 72 | |
| AAVVNIQKVIDRLNEVAKNLMESLI | IQKVIDRLNEVAKNLMES | 72 | |
| AFVVNIQKAIDRLNEVAKNLMESLI | IQKAIDRLNEVAKNLMES | 72 | |
| ASVVAIQKEIERLNEVAKALAESLI | AIQKEIERLNEVAKALAE | 72 | |
| ASVVAIQKEIERLAEVAKKLAESLI | AIQKEIERLAEVAKKLAE | 72 | |
| ASVVAIQKEIARLNEVAKALAESLI | AIQKEIARLNEVAKALAE | 72 | |
| ASAAQIQKEIERLQEAAKQLQESLI | AAQIQKEIERLQEAAKQLQE | 80 | |
| ASVVNICKEICRLNEVAKNLCESLC | VVNICKEICRLNEVAKNLCE | 80 | |
| Multiple strategies | AAVEQIQKEIERLQEAAKQLQEQLK | AVEQIQKEIERLQEAAKQLQE | 84 |
| AAVEQIQKEIERLQEAAKNLNEQLI | AVEQIQKEIERLQEAAKNLNE | 84 | |
| AAVEQIQKEIERLQEAAKNLLESLK | AVEQIQKEIERLQEAAKNLLE | 84 | |
| ASVRNIQEEIKRLNEVAKNLLESLK | RNIQEEIKRLNEVAKNLLE | 76 | |
| AAVEQIQKAIERLQEAAKNLAEQLI | AVEQIQKAIERLQEAAKNLAE | 84 | |
| KAVKNIQKAIDRLNEVAKNLAESLK | VKNIQKAIDRLNEVAKNLAE | 80 | |
| ASAAQIQKEIERLQEAAKKLAESLI | AAQIQKEIERLQEAAKKLAE | 80 | |
| ASAAQIQKEIERLQEAAKNLLESLK | AAQIQKEIERLQEAAKNLLE | 80 | |
| AAVEQIQKAIERLQEAAKKLAESLI | AVEQIQKAIERLQEAAKKLAE | 84 | |
| AAVVNICKEICRLNEVAKNLCEALC | AVVNICKEICRLNEVAKNLCE | 84 | |
| ASVRNIQEEIKRLNEVAKNLAEQLK | RNIQEEIKRLNEVAKNLAE | 76 | |
| KAVENICKEICRLNEVAKNLCEALC | AVENICKEICRLNEVAKNLCE | 84 | |
| KAVENICKEICRLNEVAKNLCEKLC | AVENICKEICRLNEVAKNLCE | 84 | |
| KAVENICKEICELNEVAKNLCEKLC | AVENICKEICELNEVAKNLCE | 84 | |
| ASVRNIQEEIKRLNEVAKKLAESLI | RNIQEEIKRLNEVAKKLAE | 76 | |
| AAVEQIQKEIERLQEVAKNLLESLR | AVEQIQKEIERLQEVAKNLLE | 84 | |
| AAVEQIQKEIERLQEAAKNLLESLR | AVEQIQKEIERLQEAAKNLLE | 84 |
Mutated residues are underlined
Design of Entry Inhibitor Peptides Against a Conserved Sequence of IAV
The wild fusion peptide of hemagglutinin was predicted to have random coil conformation by PEPstrMOD. From the fusion peptide sequence of IAV, a twenty amino acid sequence was selected as the scaffold for peptide design (LFGAIAGFIEGGWTGMIDGW). In addition to the strategies used for the design of SCV2 C-peptides, polar mutations were performed on the polar side of the fusion peptide helical wheel representation (Table 4, Fig. 2). As the hydrophobic side of the wild peptide is entirely hydrophobic, additional hydrophobic mutations on the hydrophobic side were not necessary. The four most helical sequences from the 24 initially designed peptides were further submitted to mutations to achieve a higher degree of helicity (Table 5). In the end, the 18 designed peptides with more than 70% helicity were prepared for further analyses. Out of the three strategies using helix favoring residues, primarily Ala in the peptide sequence, it had a more beneficial effect on the designed peptides' helicity ratio.
Table 4.
IAV entry inhibitor peptides designed based on three strategies and their helicity ratio
| Design strategy | Mutated sequence | Helical segment | Helicity percentage (%) |
|---|---|---|---|
| Wild peptide | LFGAIAGFIEGGWTGMIDGW | - | 0 |
| Polar mutation in polar side | LFGAIARFIERGWTRMIDGW | FGAIA | 25 |
| LFGAIAKFIEKGWTKMIDGW | FGAIAKFIE | 45 | |
| LFRAIARFIEKGWTKMIDGW | LFRAIARFIEKGWTKM | 80 | |
| LFGAIADFIEDGWTDMIDGW | FGAIADF | 35 | |
| LFGAIAEFIEEGWTEMIDGW | AIAE | 20 | |
| LFDAIADFIERGWTRMIDGW | FDAIADFI | 40 | |
| LFSAIASFIETGWTHMIDGW | LFSAIASFI | 45 | |
| LFNAIANFIEQGWTHMIDGW | LFNAIANFIE | 50 | |
| Helix favoring residues | LFAAIAAFIEGAWTGMIDAW | FAAIAAF | 35 |
| LFLAIALFIEGLWTGMIDLW | IALFIEGLWTGMI | 65 | |
| LFLAIALFIEGAWTGMIDAW | - | 0 | |
| LFMAIAMFIEGAWTGMIDAW | MAIAMF | 30 | |
| LFGAIARFIEGAWTRMIDAW | FGAIARF–-AWT | 50 | |
| LFGAIAKFIEGLWTKMIDLW | FGAIAKFIEGLWTKMID | 85 | |
| LFGAIAKFIEKAWKGMIDAW | FGAIAKFIEKAWKGM | 75 | |
| LFAAIAAFIEAAWTAMIDAW | LFAAIAAFIEAAWTAMIDA | 95 | |
| Salt bridges and disulfide bonds | LFGAIAGFIEGGWKGMIDGW | GAI | 15 |
| LFEAIAKFIEGGWTGMIDGW | LFEAIAKF | 40 | |
| LFEAIAKFIEGGWRGMIDGW | LFEAIAKFI | 45 | |
| LFCAIACFIEGGWTGMIDGW | CAIACF | 30 | |
| LFGAIACFIECGWTGMIDGW | IACF | 20 | |
| LFCAIACFIEGGWTCMIDCW | - | 0 | |
| LFEAIAKFIECGWTCMIDGW | LFEAIAKF | 40 | |
| LFCAIACFIEGGWKGMIDGW | AIACF | 25 |
Mutated residues are underlined
Table 5.
The most helical designed IAV entry inhibitor peptides using each of the three strategies and multiple strategies, as well as their helicity ratios
| Sequence | Helical segment | Helicity percentage (%) | |
|---|---|---|---|
| Wild peptide | LFGAIAGFIEGGWTGMIDGW | – | 0 |
| The most helical peptides obtained from each of three employed strategies | LFRAIARFIEKGWTKMIDGW | LFRAIARFIEKGWTKM | 75 |
| LFGAIAKFIEGLWTKMIDLW | FGAIAKFIEGLWTKMID | 85 | |
| LFGAIAKFIEKAWKGMIDAW | FGAIAKFIEKAWKGM | 75 | |
| LFAAIAAFIEAAWTAMIDAW | LFAAIAAFIEAAWTAMIDA | 95 | |
| Multiple strategies | LFKAIAKFIEKAWTKMIDAW | FKAIAKFIEKAWTKM | 75 |
| LFKAIAKFIEKLWTKMIDLW | FKAIAKFIEKLWTKM | 75 | |
| LFLAIAKFIELLWTKMIDLW | LAIAKFIELLWTKM | 70 | |
| LFLAIAKFIEKAWKKMIDAW | LAIAKFIEKAWKKM | 70 | |
| LFLAIAKFIEKAWKLMIDAW | LAIAKFIEKAWK | 60 | |
| LFLAIAKFIEKAWKLMIDLW | LAIAKFIEKAWK | 60 | |
| LFAAIAAFIEAAWKAMIDAW | AAIAAFIEAAWKAM | 70 | |
| LFAAIAAFIEALWKAMIDLW | AAIAAFIEALWKAM | 70 | |
| LFAKIAAFIEALWKAMIDLW | AKIAAFIEALWKAM | 70 | |
| LFKAIAKFIEKAWTKMIEAW | FKAIAKFIEKAWTKM | 75 | |
| LFKAIAKFIEKLWTKMIELW | FKAIAKFIEKLWTKMI | 80 | |
| LFLAIAKFIELLWTKMIELW | LAIAKFIELLWTKMI | 75 | |
| LFLAIAKFIEKAWKKMIEAW | LAIAKFIEKAWKKM | 70 | |
| LFAAIAAFIEAAWKAMIEAW | AAIAAFIEAAWKAM | 70 | |
| LFAAIAAFIEALWKAMIELW | AAIAAFIEALWKAMI | 75 | |
| LFAKIAAFIEALWKAMIELW | AKIAAFIEALWKAMI | 75 |
Mutated residues are underlined
Evaluation of the Secondary Structure of Designed Entry Inhibitor Peptides in Water
Most peptides have random coil formation in an aqueous solution. As the backbone hydrogen bonds of α-helices may not necessarily withstand the attack of water molecules, peptides' helical structure may be compromised in aqueous solutions. The Fmap program can evaluate the propensity of helix formation in aqueous media and calculate the energy penalty of helix formation. Designed peptides with 70% and higher helical structure in their models were submitted to the Fmap program to evaluate their secondary structure in an aqueous environment. A total of 27 designed SCV2 entry inhibitor peptides and 14 designed IAV entry inhibitor peptides, as well as the wild scaffolds, were submitted to the Fmap program. Overall, nine anti-SCV2 peptides (AntiSCV2P1-9) and eight Anti-IAV peptides (AntiIAVP1-9) were predicted to have 70% and more helical conformation in water (Table 6). These peptides were selected as potential entry inhibitors against the two viruses submitted to further analyses. Meanwhile, the wild sequence from HR2 was predicted to have only five amino acids with helical conformation in water. The wild sequence from the IAV fusion peptide was predicted to have a 100% coil structure.
Table 6.
Helical domains of the Anti-SCV2 and Anti-IAV designed peptides in an aqueous solution
| Peptide category | Peptide name | Peptide sequence | Helical segment | Energy penalty (kcal mol−1) | Helicity percent (%) |
|---|---|---|---|---|---|
| Scaffold from wild sequence of SCV2 HR2 | ASVVNIQKEIDRLNEVAKNLNESLI | 15–19 | 1.7 | 20 | |
| Designed C-peptides against SCV2 | AntiSCV2P1 | AAVEQIQKEIERLQEAAKNLLESLK | 4–24 | 1.8 | 84 |
| AntiSCV2P2 | ASVRNIQEEIKRLNEVAKNLLESLK | 6–24 | 1.3 | 76 | |
| AntiSCV2P3 | ASAAQIQKEIERLQEAAKKLAESLI | 5–23 | 2.5 | 76 | |
| AntiSCV2P4 | ASAAQIQKEIERLQEAAKNLLESLK | 5–24 | 2.3 | 80 | |
| AntiSCV2P5 | AAVEQIQKAIERLQEAAKKLAESLI | 4–23 | 2.4 | 80 | |
| AntiSCV2P6 | ASVRNIQEEIKRLNEVAKNLAEQLK | 6–22 | 1.8 | 68 | |
| AntiSCV2P7 | ASVRNIQEEIKRLNEVAKKLAESLI | 6–23 | 1 | 72 | |
| AntiSCV2P8 | AAVEQIQKEIERLQEVAKNLLESLR | 4–22 | 3.1 | 76 | |
| AntiSCV2P9 | AAVEQIQKEIERLQEAAKNLLESLR | 4–24 | 1.8 | 84 | |
| Scaffold from wild sequence of IAV fusion peptide | LFGAIAGFIEGGWTGMIDGW | – | – | 0 | |
| Designed IAV entry inhibitory peptides | AntiIAVP1 | LFLAIAKFIELLWTKMIDLW | 3–17 | 1 | 75 |
| AntiIAVP2 | LFLAIAKFIEKAWKKMIDAW | 3–17 | 2.1 | 75 | |
| AntiIAVP3 | LFLAIAKFIEKAWKLMIDAW | 3–18 | 2 | 80 | |
| AntiIAVP4 | LFLAIAKFIEKAWKLMIDLW | 3–18 | 2 | 80 | |
| AntiIAVP5 | LFAAIAAFIEALWKAMIDLW | 4–18 | 2.1 | 75 | |
| AntiIAVP6 | LFLAIAKFIELLWTKMIELW | 3–18 | 1.5 | 80 | |
| AntiIAVP7 | LFLAIAKFIEKAWKKMIEAW | 3–19 | 1.7 | 85 | |
| AntiIAVP8 | LFAAIAAFIEALWKAMIELW | 4–18 | 1.8 | 75 | |
AntiSCV2P7 and AntiSCV2P8 had the lowest and highest helical penalty among anti-SCV2 peptides, respectively. Between Anti-IAV peptides, AntiIAVP1 has the most increased stability, while AntiIAVP2 and AntiIAVP5 had the lowest stability.
Docking of Selected Viral Entry Inhibitor Peptides
To confirm the binding of designed peptides to the target protein sequences, nine AntiSCV2 peptides, eight AntiIAV peptides, and the initial wild peptides were docked against their respective targets (HR1 trimer from S protein of SCV2 and HA2 from IAV) using the ClusPro program.
Docking of Anti-SCV2 HR2 Mimicking Peptides Against SCV2 HR1 Trimer
The PDB file of SCV2 HR1 trimer structure (PDB code 6M1V) was submitted as the fusion core receptor, and the nine AntiSCV2 designed peptides as the ligand. As per the instructions using the program's balanced coefficient, the results for each peptide were ranked according to their cluster sizes. The model with the largest cluster size was selected as the representative result of the docking. All of the designed peptides formed bonds with the fusion core in a similar position to that of HR2 in the original hexamer by connecting to at least two of the helices simultaneously (Fig. 3). The models with the largest cluster sizes belonged to AntiSCV2P3, AntiSCV2P6, and AntiSCV2P2, respectively. The cluster sizes of all the representative docking models are available in Online Resource 1.
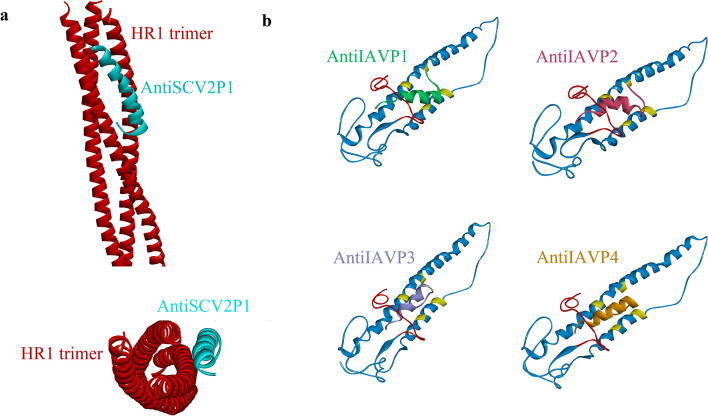
Fig. 3.
Illustrations of Anti-SCV2 and Anti-IAV peptides docking against their targets. a Positioning of AntiSCV2P1 after docking against HR1 trimer. b Docking results of AntiIAVP1-4 against HA2 from H1N1 subtype of IAV (HA2: blue, fusion peptide: red, conserved hydrophobic residues: yellow) (Color figure online)
Docking of Anti-IAV Peptides Against HA Fusion Protein
The PDB file of HA2 (PDB code 4EDB), which belongs to the H1N1 subtype of IAV, was submitted to ClusPro as the receptor protein, and the eight designed AntiIAV peptides as ligands. The resulting models with the largest cluster size using the program's balanced coefficient were regarded as the representatives of the docking procedure. The largest cluster sizes belonged to peptides AntiIAVP8 and AntiIAVP5, respectively. The cluster sizes of all the representative docking models of AntiIAV peptides are available in Online Resource 1.
All eight designed peptides showed favorable binding positions as in all of the representative models. The designed peptide interacted with the fusion peptide of the hemagglutinin protein and some of the residues in the conserved hydrophobic pocket of HA2 (Fig. 3). The binding of the peptides to either of the mentioned sites is considered to hinder the entry of the virus into host cells (Lin et al. 2017; Münch et al. 2007). Furthermore, the peptides were able to interact with the fusion peptide sufficiently and at least one of the mentioned conserved hydrophobic residues when docked against H3N1 (PDB code 4UO0) and H5N1 (PDB code 3S11). This indicates that the designed peptides can be expected to be used as broad-spectrum AntiIAV agents. A list of the peptides' interactions with the critical sites of HA2 from H1N1 is available in Table 7. AntiIAVP3 and AntiIAVP4 peptides were able to form interactions with the highest number of conserved hydrophobic pocket residues of HA2, while, AntiIAVP2 and AntiIAVP6 could bind to the most residues in the fusion peptide region.
Table 7.
A list of the designed AntiIAV peptides' interactions with the fusion peptide and conserved hydrophobic pocket residues of HA2 from H1N1 (4EDB)
| Peptide name | Residues of the HA2 designed peptides interacted with | |
|---|---|---|
| Fusion peptide residues | Conserved hydrophobic pocket residues | |
| Scaffold from wild sequence of IAV fusion peptide | THR15, TRP21, TYR22 | ILE48 |
| AntiIAVP1 | ILE6, ILE10, GLY12, TRP14, MET17, VAL18, ASP19, TRP21 | ILE48 |
| AntiIAVP2 | ILE6, ILE10, GLY12, GLY13, TRP14, GLY16, MET17, VAL18, ASP19, TRP21, TYR22 | ILE48, LEU108 |
| AntiIAVP3 | ILE6, ILE10, GLY13, TRP14, MET17, TRP21, GLY23 | ILE48, VAL100, LEU108 |
| AntiIAVP4 | ILE6, ILE10, GLY12, TRP14, TRP21, TYR22, GLY23 | ILE48, VAL100, LEU108 |
| AntiIAVP5 | ILE6, ILE10, GLY12, TRP14, MET17, TRP21 | ILE48, LEU108 |
| AntiIAVP6 | ILE6, ILE10, GLY12, TRP14, MET17, VAL18, ASP19, GLY20, TRP21, TYR22 | ILE48 |
| AntiIAVP7 | ILE6, ILE10, TRP14, GLY16, MET17, TRP21, TYR22 | LEU108 |
| AntiIAVP8 | ILE6, ILE10, TRP14, MET17, TRP21 | ILE48, LEU108 |
Evaluation of Coiled-Coil Formation and Binding Energy of Anti-SCV2 Designed Peptides
When two or more helices wrap around each other, there is a possibility that they can form highly stable helical structures, known as coiled-coils (McFarlane et al. 2009). When designed peptides form coiled-coil structures with the HR1 trimer, they are expected to have higher efficiency in binding to the fusion core and preventing viral entry into host cells. The docking results collected from ClusPro were submitted to the CoilCheck plus program, and the stabilizing energy of the docked structure was calculated. A more negative stabilizing energy corresponds to a stronger interaction between the designed peptide and the HR1 trimer. Designed peptides and the HR1 trimer will be considered coiled-coil structures if the calculated stabilizing energy per residue between the designed peptides and any HR1 monomers in the docked structure is between − 1.0 to − 4.0 (kJ mol−1) (Sunitha et al. 2012). The stabilizing energy per residue is obtained using the total stabilizing energy, a sum of hydrogen bond, electrostatic, and Vander Waals energies. Since each peptide has bound to two HR1 monomers, the stabilizing energy of binding to each HR1 helix is calculated separately. Among the nine AntiSCV2 peptides, the energy per residue value of AntiSCV2P1, AntiSCV2P3, AntiSCV2P7, and AntiSCV2P8 is within the mentioned interval (Table 8). This means these peptides are expected to form coiled-coil conformation with at least one monomer in the HR1 trimer. AntiSCV2P7 is expected to form coiled-coils with both HR1 monomers. The total binding energy of the designed peptides to the HR1 trimer in this study equals the sum of the two calculated total stabilizing energies. Peptides AntiSCV2P7 and AntiSCV2P3 have the most negative total binding energies.
Table 8.
The calculated stabilizing energies of the designed AntiSCV2 peptides interacting with the HR1 trimer
| Name of the peptide | Total stabilizing energy (kJ mol−1) | Energy per residue (kJ mol−1) | Total binding energy | ||
|---|---|---|---|---|---|
| Binding to the first chain | Binding to the second chain | Binding to the first chain | Binding to the second chain | ||
| Scaffold from the wild sequence of SCV2 HR2 | − 83.7 | − 15.5 | − 0.8 | − 0.15 | − 99.2 |
| AntiSCV2P1 | − 108.9 | − 52.4 | − 1.1 | − 0.5 | − 161.3 |
| AntiSCV2P2 | − 72.5 | − 71.9 | − 0.7 | − 0.7 | − 144.4 |
| AntiSCV2P3 | − 60.7 | − 114.1 | − 0.6 | − 1.1 | − 174.8 |
| AntiSCV2P4 | − 47.2 | − 86.7 | − 0.5 | − 0.9 | − 133.9 |
| AntiSCV2P5 | − 47.5 | − 79.9 | − 0.5 | − 0.8 | − 127.4 |
| AntiSCV2P6 | − 86.0 | − 81.9 | − 0.8 | − 0.8 | − 167.9 |
| AntiSCV2P7 | − 97.7 | − 98.9 | − 1.0 | − 1.0 | − 196.6 |
| AntiSCV2P8 | − 100.6 | − 66.6 | − 1.0 | − 0.7 | − 167.2 |
| AntiSCV2P9 | − 84.9 | − 76.9 | − 0.8 | − 0.8 | − 161.8 |
Evaluation of the Cell Penetration of IAV Entry Inhibitor Designed Peptides
Designed peptide sequences were submitted to the MLCPP program to investigate whether the designed AntiIAV peptides can enter the host cell along with the virus. The probability of effective entry inhibitory peptide and fusion protein interaction increases by enhancing the cell penetration ability of IAV entry inhibitor peptides. Peptides with a probability score of equal or more than 0.50 are considered probable CPPs by the MLCPP algorithm. While the wild sequence was predicted as a non-CPP, peptides AntiIAVP2, AntiIAVP3, AntiIAVP4, and AntiIAVP7 reached a probability score of ≥ 0.5. Although the actual cell penetration of the designed peptide should be evaluated using experimental analyses, it is evident that the strategies undertaken to develop AntiIAV peptides have raised the cell penetration probability compared to the wild sequence.
AntiSCV2 and AntiIAV Peptides Hydrophobic Moment and Lipid Discrimination Factor Calculation
The hydrophobic moment of the helix is a vector pointing to the hydrophobic side of the helix. The size of this vector is a marker of the helices' amphipathicity. Peptides with a higher hydrophobic moment value have a higher degree of amphipathicity. Amphipathicity of the peptides plays a decisive role in their interaction with membrane interfaces.
In connection with viral entry inhibitor peptides, an increased affinity to membrane indicates a higher concentration of these peptides at cell surfaces where they are expected to act against viruses. Regarding C-peptides, the increased amphipathicity implies a higher probability of the hydrophobic side's interaction with the fusion core. Heliquest program projects the hydrophobic moment vector on the helical wheel representation of the helical peptide based on a model developed by Eisenberg (Eisenberg et al. 1982). The calculated hydrophobic moment values are available in Table 9. As can be perceived from the table, all the designed peptides have a larger hydrophobic moment vector than the wild sequences. Out of Anti-IAV peptides, AntiIAVP2 and among Anti-SCV2 peptides, AntiSCV2P2 and AntiSCV2P8 have the highest hydrophobic moment value.
Table 9.
Hydrophobic moment, net charge, lipid-binding discrimination factor, Boman, and Wimley–White index of designed AntiSCV2 and AntiIAV peptides
| Peptide category | Peptide name | Hydrophobic moment | Net charge | Lipid-binding discrimination factor | Boman index (kcal mol−1) | Wimley–White index |
|---|---|---|---|---|---|---|
| Wild HR2 sequence from SCV2 | 0.43 | − 1 | 0.08 | 1.95 | 10.54 | |
| Designed Anti-SCV2 peptides | AntiSCV2P1 | 0.57 | 1 | 0.87 | 2.05 | 14.60 |
| AntiSCV2P2 | 0.58 | 1 | 0.87 | 2.39 | 12.39 | |
| AntiSCV2P3 | 0.48 | 0 | 0.45 | 1.67 | 12.27 | |
| AntiSCV2P4 | 0.50 | 0 | 0.48 | 2.01 | 12.27 | |
| AntiSCV2P5 | 0.56 | 0 | 0.52 | 1.37 | 12.21 | |
| AntiSCV2P6 | 0.55 | 1 | 0.85 | 2.80 | 13.57 | |
| AntiSCV2P7 | 0.54 | 1 | 0.84 | 2.25 | 12.39 | |
| AntiSCV2P8 | 0.58 | − 1 | 0.21 | 2.34 | 13.78 | |
| AntiSCV2P9 | 0.57 | − 1 | 0.21 | 2.43 | 13.88 | |
| Wild fusion peptide sequence from IAV | 0.53 | − 2 | 0.16 | − 1.19 | − 3.89 | |
| Designed Anti-IAV peptides | AntiIAVP1 | 0.56 | 0 | 0.53 | − 1.33 | − 4.21 |
| AntiIAVP2 | 0.67 | 2 | 1.29 | − 0.35 | − 0.35 | |
| AntiIAVP3 | 0.61 | 1 | 0.91 | − 0.87 | − 1.90 | |
| AntiIAVP4 | 0.64 | 1 | 0.94 | − 1.03 | − 2.63 | |
| AntiIAVP5 | 0.64 | − 1 | 0.28 | − 1.61 | − 3.54 | |
| AntiIAVP6 | 0.55 | 0 | 0.52 | − 1.40 | − 3.42 | |
| AntiIAVP7 | 0.67 | 2 | 1.29 | − 0.44 | 0.44 | |
| AntiIAVP8 | 0.64 | − 1 | 0.27 | − 1.70 | − 2.75 | |
Another measure of membrane affinity is the lipid-binding discrimination factor introduced by Heliquest (Keller 2011). The lipid-binding discrimination factor is calculated using hydrophobic moment and the net charge of the peptide. Peptides with a lipid-binding discrimination factor higher than 0.68 are considered to be potentially lipid binding. AntiSCV2P1, AntiSCV2P2, AntiSCV2P6, and AntiSCV2P7 amongst AntiSCV2 and AntiIAVP2, AntiIAVP3, AntiIAVP4, and AntiIAVP7 out of the AntiIAV designed peptides were predicted to be potentially lipid-binding helices (Table 9). These peptides have a higher affinity to membrane surfaces compared to the other designed peptides. This affinity also increases the likelihood of peptide entrance into the endosome alongside the virus, a feat which is usually achieved by conjugating entry inhibitory peptides to lipid chains. None of the wild sequences were evaluated as lipid-binding using this method.
Another method for evaluating a peptide’s membrane affinity and overall hydrophobicity is the Wimley–White whole-residue hydrophobicity (Cunsolo et al. 2020). Peptides with negative Wimley–White index values have a higher interaction with membrane surfaces. The designed IAV peptides show more negative Wimley–White index values than designed anti-SCV2 peptides, which is in line with their higher probability of membrane penetration (Table 9). Boman index is an indicator of the protein binding potential of a peptide; a higher index shows a higher probability of protein binding (Boman 2003). The designed anti-IAV peptides all have negative Boman indexes (Table 9). Usually, values < 1 and negative values indicate a potential antimicrobial peptide with low systemic side effects. The low Boman index values in anti-IAV peptides indicate their scaffold being a fusion peptide and having a high affinity for the membrane compared to proteins. However, the designed anti-SCV2 peptides have a high Boman index value, which is in line with their intended mechanism of binding to the HR1 trimer. Furthermore, the values are all < 3 which should minimize their probable systemic side effects due to binding to host proteins.
Analyses Regarding the In Vivo Administration of the Designed Antiviral Peptides
While conventional methods of in vitro and in vivo evaluation of peptides are the most trustworthy, they can be costly and time-consuming. Using in silico analyses to evaluate potential biotherapeutic candidates before their in vivo trials saves valuable resources. Several computational tools have been developed to facilitate this process. Some of these tools assess the peptide's bioavailability, while others evaluate potential toxicity or side effects.
Bioavailability Evaluation of the Designed Peptides
Bioavailability is one of the significant hurdles in the development of peptide-based therapeutics. Some of the critical factors in determining the bioavailability of a peptide are its half-life and antigenicity. Peptides with longer half-lives have higher bioavailability and more promising drug candidates(Mathur et al. 2016). The Plifepred program showed no significant difference between the half-lives of designed AntiIAV peptides, with values around 1000 s (Table 10). However, the half-life values of AntiSCV2 peptides varied from more than 5000 s (AntiSCV2P9) to about 1200 s for AntiSCV2P6 and AntiSCV2P2.
Table 10.
Bioavailability and toxicity of the designed antiviral peptides using computational tools
| Peptide category | Peptide name | Half-life (s) | Antigenicity | Protease susceptibility | Hemolytic potency (PROB score) | Allergenicity |
|---|---|---|---|---|---|---|
| Wild HR2 sequence from SCV2 | 854 | ✖ | 6 | 0.48 | ✖ | |
| Designed AntiSCV2 peptides | AntiSCV2P1 | 3339 | ✖ | 9 | 0.49 | ✔ |
| AntiSCV2P2 | 1245 | ✖ | 11 | 0.48 | ✔ | |
| AntiSCV2P3 | 2489 | ✖ | 9 | 0.48 | ✔ | |
| AntiSCV2P4 | 2614 | ✖ | 11 | 0.49 | ✔ | |
| AntiSCV2P5 | 3596 | ✖ | 8 | 0.48 | ✔ | |
| AntiSCV2P6 | 1211 | ✖ | 10 | 0.49 | ✖ | |
| AntiSCV2P7 | 1247 | ✖ | 10 | 0.47 | ✖ | |
| AntiSCV2P8 | 3246 | ✖ | 6 | 0.50 | ✔ | |
| AntiSCV2P9 | 5397 | ✖ | 9 | 0.49 | ✔ | |
| Wild fusion peptide sequence from IAV | 938 | ✖ | 3 | 0.48 | ✖ | |
| Designed AntiIAV peptides | AntiIAVP1 | 1056 | ✔ | 0 | 0.50 | ✖ |
| AntiIAVP2 | 1165 | ✖ | 5 | 0.50 | ✖ | |
| AntiIAVP3 | 1182 | ✔ | 4 | 0.50 | ✔ | |
| AntiIAVP4 | 1144 | ✔ | 0 | 0.51 | ✔ | |
| AntiIAVP5 | 1076 | ✔ | 0 | 0.49 | ✖ | |
| AntiIAVP6 | 1206 | ✖ | 0 | 0.49 | ✔ | |
| AntiIAVP7 | 1090 | ✖ | 2 | 0.50 | ✖ | |
| AntiIAVP8 | 1262 | ✖ | 0 | 0.49 | ✔ | |
Antigenicity: antigenic ✔, non-antigenic ✖
Allergenicity: probable allergen ✔, probable non-allergen ✖
Protease susceptibility: number of cleavage sites with positive PROSperous scores, regarding many human proteases such as metallopeptidases, caspases, cathepsins, thrombin, plasmin, and furin.
Proteolytic degradation of therapeutic peptides in the blood is detrimental to the peptides' half-life and circulation time. To predict whether the designed peptides are susceptible to blood proteases, they were submitted to the PROSPERous program. The program predicts the possible cleavage sites in a protein or peptide sequence. In this study, the designed peptides' susceptibility to twenty-one human blood protease, including thrombin, furin, and caspases, was predicted, and any peptide harboring cleavage sites with a positive score was chosen as a potential substrate. Based on the predictions among Anti-SCV2 peptides, AntiSCV2P8 was the most resistant, while AntiSCV2P2 and AntiSCV2P4 were the most susceptible sequences to human blood proteases. Meanwhile, the designed AntiIAV peptides were more resistant to the selected proteases. Five of the designed peptides (AntiIAVP1, AntiIAVP4, AntiIAVP5, AntiIAVP6, and AntiIAVP8) had no cleavage sites for the chosen proteases. Resistant peptides are expected to achieve higher bioavailability after administration.
Except for vaccines, the immunogenicity of peptide and protein therapeutics is often unfavorable. The immune reaction results in the production of anti-drug antibodies. These antibodies can cause inconveniences such as neutralizing the therapeutic, which affects the drug's efficacy or even safety (Fernandez et al. 2018). The possibility of immune responses to the designed peptides was evaluated using Vaxijen. According to the gathered data, all the designed AntiSCV2 peptides and half of the AntiIAV peptides (AntiIAVP2, AntiIAVP6, AntiIAVP7, and AntiIAVP8) were recognized as non-antigenic.
Toxicity Evaluation and Probable Side Effects Prediction of the Designed Peptides
Compared to small molecule therapeutics, peptide-based therapeutics are frequently associated with decreased probability of side effects or drug–drug interactions (Lau and Dunn 2018). However, there are several ways a therapeutic peptide can cause complications (Sisakht et al., 2021). Some peptides are inherently toxic, which can show themselves in the form of inflammation, pain, cardiovascular irregularities, or even neurotoxicity. Therefore, designed peptides were assessed using the ToxinPred program for possible toxic motifs in their sequences. None of the designed AntiSCV2 and AntiIAV peptides were predicted as toxic.
Other examples of peptide-induced complications are RBC lysis and allergenicity. Hemolysis can occur for many reasons, including the toxicity of the peptide, oxidation or ion channel, and pump inhibition (Chaudhary et al. 2016). HemoPI program evaluated the peptides by assigning a PROB score. All the designed peptides had relatively equal PROB scores around 0.5 and a relatively low probability of hemolysis induction.
Allergenicity is one of the concerning side effects of the vaccines against SCV2 (Kleine-Tebbe et al. 2021). To test the allergenicity of the designed peptides, they were submitted to the AllergenFP program. From both categories, peptides AntiSCV2P6 and AntiSCV2P7 as well as AntiIAVP1 AntiIAVP2, AntiIAVP5, and AntiIAVP7 were predicted to be probable non-allergens.
Selection of the Most Promising AntiSCV2 and AntiIAV Peptides
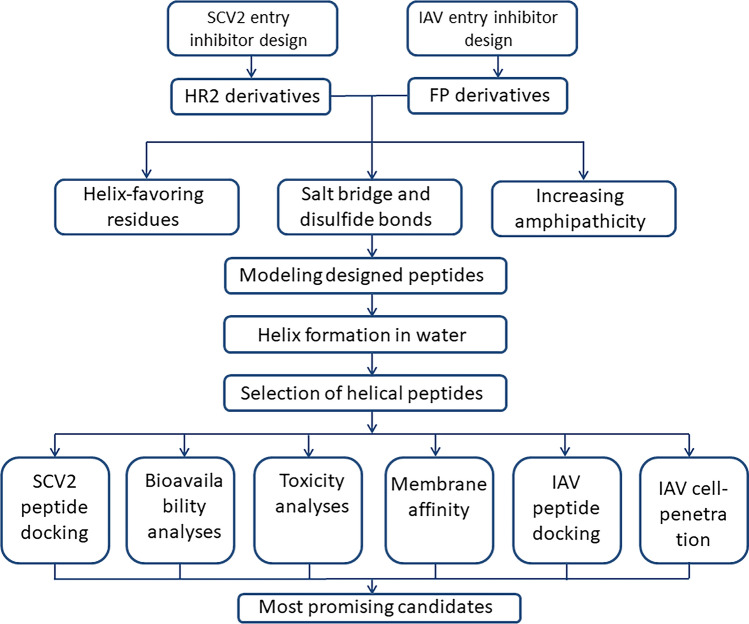
From the initial 40 designed AntiSCV2 and AntiIAV peptides, nine AntiSCV2 and eight AntiIAV peptides could maintain 70% or more helical conformation both in modeling and in the aqueous environment. Hereafter, using several computational tools, the peptides' efficacy, safety, and bioavailability were evaluated. A schematic summary of the design and evaluation workflow is available in Fig. 4. The most promising candidates for further in vitro and in vivo evaluations can be chosen by characterizing the acquired data. To do so, a scoring function, some of which was previously used by Hemmati et al. (Hemmati et al. 2020), is employed. In summary:
AntiSCV2 Peptides that were able to form coiled-coils with HR1 trimer were scored "+ 1", and others scored "0".
AntiIAV peptides that were predicted as cell-penetrating by MLCPP were scored "+ 1", and others scored "0".
Lipid-binding peptides got a score of "+ 1" and non-lipid-binding peptides a score of "0".
Peptides with longer and shorter than 1800s half-life were scored "+ 1" and "0", respectively.
The most protease-resistant peptides in each category were defined as "+ 1", and the other peptides as "0".
Non-antigenic and antigenic peptides were designated as "+ 1" and "0", respectively.
Probable non-allergenic and probable allergenic peptides were assigned "+ 1" and "0", respectively.
Considering their hemolytic potency, peptides with a PROB score of ≥ 0.50 were scored "0", and peptides with a lower prob score were scored by "+ 1".
Any toxic peptides would have been eliminated.
Fig. 4.
A summary of the workflow employed by this study to design and evaluate AntiSCV2 and AntiIAV entry inhibitor peptides
A sum of all the scores designated to each peptide was calculated, and the peptides with the highest overall score were selected as the most promising antiviral candidates. AntiSCV2P1 and AntiSCV2P7 scored highest among AntiSCV2 peptides, whereas AntiIAVP2 and AntiIAVP7 displayed the AntiIAV category highest scores.
Prediction of the Antimicrobial Potential Using iAMPpred
There are bioinformatics tools developed to assist scientists in evaluating the antimicrobial peptides prior to in vitro and in vivo experiments. In this study, we have employed one of the sequence-based bioinformatics tools that can evaluate the antibacterial, antifungal, and antiviral probability of a submitted peptide to further confirm the antimicrobial activity of the designed peptides. Based on the obtained results (Table 11), all of the designed peptides have an antiviral activity with a probability score of > 0.5 and can be considered positive antiviral peptides. Four anti-IAV peptides (AntiIAVP2, AntiIAVP3, AntiIAVP4, AntiIAVP7) were predicted as potential antibacterial and antifungal peptides. All designed peptides had a substantially higher antiviral probability and can be regarded as selective antivirals.
Table 11.
Antibacterial, antifungal, and antiviral probabilities of anti-SCV2 and anti-IAV designed peptides using the iAMPpred program
| Peptide category | Peptide name | Antibacterial probability | Antiviral probability | Antifungal probability |
|---|---|---|---|---|
| 1 | AntiIAVP1 | 0.285 | 0.93 | 0.202 |
| 2 | AntiIAVP2 | 0.868 | 0.96 | 0.786 |
| 3 | AntiIAVP3 | 0.762 | 0.96 | 0.618 |
| 4 | AntiIAVP4 | 0.712 | 0.97 | 0.576 |
| 5 | AntiIAVP5 | 0.475 | 0.84 | 0.292 |
| 6 | AntiIAVP6 | 0.264 | 0.95 | 0.209 |
| 7 | AntiIAVP7 | 0.812 | 0.97 | 0.755 |
| 8 | AntiIAVP8 | 0.603 | 0.91 | 0.326 |
| 9 | AntiSCV2P1 | 0.073 | 0.81 | 0.078 |
| 10 | AntiSCV2P2 | 0.016 | 0.6 | 0.032 |
| 11 | AntiSCV2P3 | 0.219 | 0.89 | 0.442 |
| 12 | AntiSCV2P4 | 0.099 | 0.92 | 0.278 |
| 13 | AntiSCV2P5 | 0.244 | 0.81 | 0.282 |
| 14 | AntiSCV2P6 | 0.029 | 0.52 | 0.032 |
| 15 | AntiSCV2P7 | 0.03 | 0.76 | 0.058 |
| 16 | AntiSCV2P8 | 0.032 | 0.63 | 0.035 |
| 17 | AntiSCV2P9 | 0.047 | 0.78 | 0.076 |
Designing AntiSCV2 and AntiIAV Conjugates with an In Vivo Cleavable Linker
Each of the final candidates (AntiSCV2P1, AntiSCV2P7, AntiIAVP2, and AntiIAVP7) has the potential to be developed as an entry inhibitor therapeutic by itself. We put forth the idea of creating an in vivo cleavable peptide-peptide conjugate from both AntiSCV2 and AntiIAV categories. Inserting an in vivo cleavable linker between an AntiSCV2 and an AntiIAV peptide will prolong the plasma half-life of the complex compared to the individual components and preserve the bioactivity of each domain without hindrance for the other peptide (Chen et al. 2013). One strategy for designing in vivo cleavable linkers are protease-sensitive linkers. These linkers use sequences that are liable to specific proteases in the human body. The oligopeptide nature of the in vivo cleavable protease-sensitive linker makes the whole construct's production as a recombinant peptide in an expression host feasible. At the same time, the complications of the chemical conjugation method can be avoided. The linker can be sensitive to one of the proteases present in the bloodstream. Some of the linkers from this category previously employed are factor XIa/FVIIa, factor Xa, and thrombin-sensitive linkers (Goyal and Batra 2000; Park et al. 2012; Schulte 2009). This leads to the rapid destruction of the linker in the blood circulation.
In a different strategy, the enzymatically cleavable linker is the substrate of a specific protease overexpressed under one particular pathological condition or in a specific organ. Designing conjugates with these linkers can be applied to site activation of biotherapeutics in targeted locations. TMPRSS2 is a transmembrane serine protease expressed in the alveolar epithelium. This enzyme plays an essential role in the SCV2 and influenza fusion process by priming the S protein and hemagglutinin, respectively (Shen et al. 2020). Inhibitors of this enzyme are also considered AntiSCV2 and anti-influenza drug candidates. Using substrates of this enzyme as a conjugate linker will ensure the on-site release of AntiSCV2 and AntiIAV peptides. The linker itself acts as a competitive inhibitor of TMPRSS2.
Herein, an in vivo cleavable conjugate was designed using AntiSCV2P1 and AntiIAVP2, two of the most promising designed peptides from AntiIAV and AntiSCV2 categories. These peptides are linked together by the "DPLKPTKRSFIED" sequence (cleaved by TMPRSS2) in the conjugate (Fig. 5). This sequence is found at the S2' site of the wild-type SCV2 and is cleaved during the SCV2 cellular entry (Hoffmann et al. 2020).
Fig. 5.
An illustration of the AntiSCV2P1 and AntiIAVP2 fused together by a TMPRSS2 cleavable linker
Conclusion
Viral entry inhibitors for respiratory infections are favorable pharmaceuticals for systemic injection or nasal and buccal formulations to prevent early infection. Entry inhibitor peptides against SCV2 and IAV can be designed to target any steps of viral entry into host cells, including the viral cell surface receptor binding, the protease processing of the fusion protein, endocytosis, or the fusion process. In this study, nine AntiSCV2 and eight AntiIAV peptides targeting the fusion process of both viruses are introduced. The AntiSCV2 peptides were designed based on the relatively conserved HR2 region of SCV2 S protein. Peptides were intended to be highly helical to facilitate their binding to the hydrophobic cervices in the HR1 trimer. The AntiIAV peptides, on the other hand, were designed based on the conserved fusion peptide of hemagglutinin protein and were devised as helical amphipathic peptides with cell-penetration potential to increase their endosomal uptake. Peptides from both categories were evaluated for binding to their target fusion proteins, their affinity to membrane interfaces, and their safety in the case of in vivo administration. Our study showed that using different strategies, including amphipathicity enhancement, the addition of helix favoring residues, and insertion of salt and disulfide-bridges simultaneously, is more successful in achieving highly helical peptides. AntiSCV2P1 and AntiSCV2P7 as the most promising AntiSCV2 peptides, as well as, AntiIAVP2 and AntiIAVP7 from the AntiIAV category are candidates for further in vitro and in vivo studies.
The bioavailability of the peptides is a key limitation to their clinical application; however, upper and lower airways, the sites of SCV2 and IAV infections, are accessible using inhalation formulations. As it will result in a higher concentration of these peptides in the lungs and lower the probability of systematic adverse effects, the low uptake from the respiratory epithelium is beneficial. The AntiSCV2 and AntiIAV peptides were also conjugated using a TMPRSS2 sensitive linker, which can be developed as a single therapeutic, with antiviral activity against both viruses used in case of coinfections or uncertain diagnosis.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Shiraz University of Medical Sciences, Shiraz, IRAN for the financial support of this work (Grant Number 23205).
Author contributions
YB: Formal analysis, Investigation, Writing—Original Draft; Formal analysis SH: Conceptualization, Resources, Writing—Review & Editing, Supervision, Project administration.
Availability of Data and Material
Not applicable.
Code Availability
Not applicable.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethics Approval
IR.SUMS.REC.1400.517.
Consent for Publication
Not applicable.
Consent to Participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Badani H, Garry RF, Wimley WC. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim Biophys Acta. 2014;1838:2180. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Zhao Y, Dong J, Liang S, Guo M, Liu X, Wang X, Huang Z, Sun X, Zhang Z, Dong L, Liu Q, Zheng Y, Niu D, Xiang M, Song K, Ye J, Zheng W, Tang Z, Tang M, Zhou Y, Shen C, Dai M, Zhou L, Chen Y, Yan H, Lan K, Xu K. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity. Cell Res. 2021 doi: 10.1038/s41422-021-00473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadipour Y, Hemmati S. Considerations on the rational design of covalently conjugated cell-penetrating peptides (CPPs) for intracellular delivery of proteins: a guide to CPP selection using glucarpidase as the model cargo molecule. Molecules (Basel, Switzerland) 2019;24:4318. doi: 10.3390/molecules24234318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadipour Y, Gholampour M, Pirhadi S, Seradj H, Khoshneviszadeh M, Hemmati S. Viral 3CLpro as a target for antiviral intervention using milk-derived bioactive peptides. Int J Pept Res Ther. 2021;27(4):2703–2716. doi: 10.1007/s10989-021-10284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadipour Y, Sadeghian I, Ghaffarian Bahraman A, Hemmati S. Introducing a delivery system for melanogenesis inhibition in melanoma B16F10 cells mediated by the conjugation of tyrosine ammonia-lyase and a TAT-penetrating peptide. Biotechnol Prog. 2021;37:e3071. doi: 10.1002/btpr.3071. [DOI] [PubMed] [Google Scholar]
- Boman HG. Antibacterial peptides: basic facts and emerging concepts. J Intern Med. 2003;254(3):197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Chaudhary K, Kumar R, Singh S, Tuknait A, Gautam A, Mathur D, Anand P, Varshney GC, Raghava GPS. A web server and mobile app for computing hemolytic potency of peptides. Sci Rep. 2016;6:22843. doi: 10.1038/srep22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zaro JL, Shen W-C. Fusion protein linkers: property, design and functionality. Adv Drug Del Rev. 2013;65:1357. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunsolo V, Schicchi R, Chiaramonte M, Inguglia L, Arizza V, Cusimano MG, Schillaci D, Di Francesco A, Saletti R, Lo Celso F, Barone G. Identification of new antimicrobial peptides from mediterranean medical plant Charybdis pancration (Steinh.) Speta. Antibiotics. 2020;9(11):747. doi: 10.3390/antibiotics9110747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2013;30:846. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Bustos R, Zapata C, Garcia J, Jauregui E, Ashraf G. Immunogenicity in protein and peptide based-therapeutics: an overview. Curr Protein Pept Sci. 2018;19:958. doi: 10.2174/1389203718666170828123449. [DOI] [PubMed] [Google Scholar]
- Figueira TN, Augusto MT, Rybkina K, Stelitano D, Noval MG, Harder OE, Veiga AS, Huey D, Alabi CA, Biswas S, Niewiesk S, Moscona A, Santos NC, Castanho MARB, Porotto M. Effective in vivo targeting of influenza virus through a cell-penetrating/fusion inhibitor tandem peptide anchored to the plasma membrane. Bioconjug Chem. 2018;29:3362. doi: 10.1021/acs.bioconjchem.8b00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008;24:2101. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Goyal A, Batra JK. Inclusion of a furin-sensitive spacer enhances the cytotoxicity of ribotoxin restrictocin containing recombinant single-chain immunotoxins. Biochem J. 2000;345(Pt 2):247. doi: 10.1042/bj3450247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8:e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati S, Behzadipour Y, Haddad M. Decoding the proteome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for cell-penetrating peptides involved in pathogenesis or applicable as drug delivery vectors. Infect Genet Evol. 2020;85:104474. doi: 10.1016/j.meegid.2020.104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques ST, Melo MN, Castanho MARB. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem J. 2006;399:1. doi: 10.1042/BJ20061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrold MF. Helices and sheets in vacuo. Phys Chem Chem Phys. 2007;9:1659. doi: 10.1039/B612615D. [DOI] [PubMed] [Google Scholar]
- Kalafatovic D, Giralt E. Cell-penetrating peptides: design strategies beyond primary structure and amphipathicity. Molecules. 2017;22(11):1929. doi: 10.3390/molecules22111929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RCA. New user-friendly approach to obtain an Eisenberg plot and its use as a practical tool in protein sequence analysis. Int J Mol Sci. 2011;12:5577. doi: 10.3390/ijms12095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Tebbe J, Klimek L, Hamelmann E, Pfaar O, Taube C, Wagenmann M, Werfel T, Worm M. Severe allergic reactions to the COVID-19 vaccine—statement and practical consequences. Allergol Select. 2021;5:26. doi: 10.5414/ALX02215E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JL, Dunn MK. Therapeutic peptides: historical perspectives, current development trends, and future directions. Biorg Med Chem. 2018;26:2700. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Lee KK, Pessi A, Gui L, Santoprete A, Talekar A, Moscona A, Porotto M. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem. 2011;286:42141. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Luo Y, Yang G, Li F, Xie X, Chen D, He L, Wang J, Ye C, Lu S, Lv L, Liu S, He J. Potent influenza A virus entry inhibitors targeting a conserved region of hemagglutinin. Biochem Pharmacol. 2017;144:35. doi: 10.1016/j.bcp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- Manavalan B, Subramaniyam S, Shin TH, Kim MO, Lee G. Machine-learning-based prediction of cell-penetrating peptides and their uptake efficiency with improved accuracy. J Proteome Res. 2018;17:2715. doi: 10.1021/acs.jproteome.8b00148. [DOI] [PubMed] [Google Scholar]
- Mathur D, Prakash S, Anand P, Kaur H, Agrawal P, Mehta A, Kumar R, Singh S, Raghava GPS. PEPlife: a repository of the half-life of peptides. Sci Rep. 2016;6:36617. doi: 10.1038/srep36617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Singh S, Mehta A, Agrawal P, Raghava GPS. In silico approaches for predicting the half-life of natural and modified peptides in blood. PLoS ONE. 2018;13:e0196829. doi: 10.1371/journal.pone.0196829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004;3:215. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- McFarlane AA, Orriss GL, Stetefeld J. The use of coiled-coil proteins in drug delivery systems. Eur J Pharmacol. 2009;625:101. doi: 10.1016/j.ejphar.2009.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meher PK, Sahu TK, Saini V, Rao AR. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci Rep. 2017;7:42362. doi: 10.1038/srep42362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik LI, Garry RF, Morris CA. Peptide inhibition of human cytomegalovirus infection. Virol J. 2011;8:76. doi: 10.1186/1743-422X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miatech JL, Tarte NN, Katragadda S, Polman J, Robichaux SB. A case series of coinfection with SARS-CoV-2 and influenza virus in Louisiana. Respir Med Case Rep. 2020;31:101214. doi: 10.1016/j.rmcr.2020.101214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J, Ständker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pöhlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann W-G, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Nick Pace C, Martin Scholtz J. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422. doi: 10.1016/S0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Yamaguchi Y, Inouye M. Intramolecular regulation of the sequence-specific mRNA interferase activity of MazF fused to a MazE fragment with a linker cleavable by specific proteases. Appl Environ Microbiol. 2012;78:3794. doi: 10.1128/AEM.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmatabadi SS, Sadeghian I, Ghasemi Y, Sakhteman A, Hemmati S. Identification and characterization of a sterically robust phenylalanine ammonia-lyase among 481 natural isoforms through association of in silico and in vitro studies. Enzyme Microb Technol. 2019;122:36. doi: 10.1016/j.enzmictec.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Sadeghian I, Khalvati B, Ghasemi Y, Hemmati S. TAT-mediated intracellular delivery of carboxypeptidase G2 protects against methotrexate-induced cell death in HepG2 cells. Toxicol Appl Pharmacol. 2018;346:9. doi: 10.1016/j.taap.2018.03.023. [DOI] [PubMed] [Google Scholar]
- Schibli DJ, Weissenhorn W. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion (review) Mol Membr Biol. 2004;21:361. doi: 10.1080/09687860400017784. [DOI] [PubMed] [Google Scholar]
- Schulte S. Half-life extension through albumin fusion technologies. Thromb Res. 2009;124(Suppl 2):S6. doi: 10.1016/s0049-3848(09)70157-4. [DOI] [PubMed] [Google Scholar]
- Schütz D, Ruiz-Blanco YB, Münch J, Kirchhoff F, Sanchez-Garcia E, Müller JA. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv Drug Del Rev. 2020;167:47. doi: 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L-W, Qian M-Q, Yu K, Narva S, Yu F, Wu Y-L, Zhang W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci Rep. 2020;10:7635. doi: 10.1038/s41598-020-64368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Short constrained peptides that inhibit HIV-1 entry. Proc Natl Acad Sci. 2002;99:14664. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Singh H, Tuknait A, Chaudhary K, Singh B, Kumaran S, Raghava GPS. PEPstrMOD: structure prediction of peptides containing natural, non-natural and modified residues. Biol Direct. 2015;10:73. doi: 10.1186/s13062-015-0103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisakht M, Solhjoo A, Mahmoodzadeh A, Fathalipour M, Kabiri M, Sakhteman A. Potential inhibitors of the main protease of SARS-CoV-2 and modulators of arachidonic acid pathway: non-steroidal anti-inflammatory drugs against COVID-19. Comput Biol Med. 2021;136:104686. doi: 10.1016/j.compbiomed.2021.104686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Li F, Leier A, Marquez-Lago TT, Akutsu T, Haffari G, Chou K-C, Webb GI, Pike RN. PROSPERous: high-throughput prediction of substrate cleavage sites for 90 proteases with improved accuracy. Bioinformatics. 2017;34:684. doi: 10.1093/bioinformatics/btx670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Wang Q, Xu W, Yu F, Hua C, Zhu Y, Jiang S, Lu L. A novel HIV-1 gp41 tripartite model for rational design of HIV-1 fusion inhibitors with improved antiviral activity. AIDS. 2017;31:885. doi: 10.1097/qad.0000000000001415. [DOI] [PubMed] [Google Scholar]
- Sun H, Li Y, Liu P, Qiao C, Wang X, Wu L, Liu K, Hu Y, Su C, Tan S, Zou S, Wu G, Yan J, Gao GF, Qi J, Wang Q. Structural basis of HCoV-19 fusion core and an effective inhibition peptide against virus entry. Emerg Microbes Infect. 2020;9:1238. doi: 10.1080/22221751.2020.1770631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunitha MS, Nair AG, Charya A, Jadhav K, Mukhopadhyay S, Sowdhamini R. Structural attributes for the recognition of weak and anomalous regions in coiled-coils of myosins and other motor proteins. BMC Res Notes. 2012;5:530. doi: 10.1186/1756-0500-5-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Kozakov D. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85:435. doi: 10.1002/prot.25219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao L, Xia S, Zhang T, Cao R, Liang G, Li Y, Meng G, Wang W, Shi W, Zhong W, Jiang S, Liu K. De novo design of α-helical lipopeptides targeting viral fusion proteins: a promising strategy for relatively broad-spectrum antiviral drug discovery. J Med Chem. 2018;61:8734. doi: 10.1021/acs.jmedchem.8b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lin D, Shen X, Li F, Fang Y, Li K, Xun T, Yang G, Yang J, Liu S, He J. New influenza A virus entry inhibitors derived from the viral fusion peptides. PLoS ONE. 2015;10:e0138426. doi: 10.1371/journal.pone.0138426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu S, Dick A, Ju H, Mirzaie S, Abdi F, Cocklin S, Zhan P, Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J Med Chem. 2020;63:12256. doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Shen H-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci. 2020;16:1724. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. Constrained peptides as miniature protein structures. ISRN Biochem. 2012;2012:692190. doi: 10.5402/2012/692190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharieva N, Dimitrov I, Flower D, Doytchinova I. Immunogenicity prediction by VaxiJen: a ten year overview. J Proteom Bioinf. 2017;10:298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.
Not applicable.