Abstract
Mutations in the smu-1 gene of Caenorhabditis elegans were previously shown to suppress mutations in the genes mec-8 and unc-52. mec-8 encodes a putative RNA binding protein that affects the accumulation of specific alternatively spliced mRNA isoforms produced by unc-52 and other genes. unc-52 encodes a set of basement membrane proteins, homologs of mammalian perlecan, that are important for body wall muscle assembly and attachment to basement membrane, hypodermis, and cuticle. We show that a presumptive null mutation in smu-1 suppresses nonsense mutations in exon 17 but not exon 18 of unc-52 and enhances the phenotype conferred by an unc-52 splice site mutation in intron 16. We have used reverse transcription-PCR and RNase protection to show that loss-of-function smu-1 mutations enhance accumulation in larvae of an alternatively spliced isoform that skips exon 17 but not exon 18 of unc-52. We have identified smu-1 molecularly; it encodes a nuclearly localized protein that contains five WD motifs and is ubiquitously expressed. The SMU-1 amino acid sequence is more than 60% identical to a predicted human protein of unknown function. We propose that smu-1 encodes a trans-acting factor that regulates the alternative splicing of the pre-mRNA of unc-52 and other genes.
Alternative splicing of pre-mRNA is a complex but common mechanism of regulating gene function in multicellular eukaryotes. Current estimates suggest that at least one-third of all genes in humans (17) are alternatively spliced. Patterns of alternative splicing are in some cases known to be cell type specific or developmentally regulated (16, 24). Regulated alternative splicing of pre-mRNA most often results in the synthesis of multiple (functional) proteins from the same pre-mRNA, but alternative splicing can also function as an on-off switch for gene function (1).
Analysis of a genetic interaction in C. elegans has led to the proposal that the mec-8 gene encodes an RNA binding protein that regulates alternative splicing of unc-52 pre-mRNA (26). The unc-52 gene encodes, via alternative splicing and alternative 3′-end usage, homologs of perlecan, the core protein of the mammalian basement membrane heparan sulfate proteoglycan (39, 40). A subset of UNC-52 isoforms is present in the basement membrane adjoining body wall muscle (12, 30, 33, 39) and is important both for initiation of myofilament lattice assembly and for maintaining anchorages of body wall muscle to basement membrane, hypodermis, and cuticle. Null mutations in unc-52 lead to paralysis and arrest at the twofold stage of embryonic elongation (19, 39, 46); the muscle cells in these mutants have severely disorganized thick and thin filaments. Viable unc-52 mutants exhibit normal embryogenesis but suffer from a progressive disruption of body wall muscle and progressive paralysis during larval development (15). The unc-52 viable mutations are located in a region of unc-52 that when transcribed is alternatively spliced; most are nonsense mutations in alternatively spliced exons that prematurely truncate some of the UNC-52 protein isoforms (40). Null alleles of mec-8, which are viable by themselves, are lethal in combination with viable alleles of unc-52 (25). The accumulation of two alternatively spliced unc-52 mRNA isoforms that skip all of the unc-52(viable) mutations is severely reduced in mec-8 mutants (26). As a consequence, mutations in mec-8 reduce the amount of functional UNC-52 protein detected in embryos of unc-52(viable) mutants (26, 33), and mec-8; unc-52(viable) embryos are arrested in their development, with a phenotype resembling that of unc-52 null mutants (25).
Mutations in mec-8 by themselves lead to mechanosensory (6) and chemosensory (36) defects that appear to be unrelated to unc-52 function (26), suggesting that MEC-8 may affect the processing of multiple pre-mRNAs. Null alleles of mec-8 also display an incompletely penetrant cold-sensitive lethality that is correlated with defects in body muscle attachment to adjacent hypodermis and cuticle (25). This phenotype also seems to be unrelated to unc-52 function. A mutation, unc-52(ra515), that deletes the region of unc-52 in which mec-8-dependent splicing occurs (33) has been identified. Homozygous unc-52(ra515) animals have an essentially wild-type phenotype, and the mec-8; unc-52(ra515) double mutant shows no synthetic lethality but exhibits the same cold-sensitive, incompletely penetrant lethality as the mec-8 single mutant (33).
The interaction between mutations in a gene that regulates splicing (mec-8) and hypomorphic alleles of an essential alternatively spliced gene (unc-52) provides a starting point for powerful genetic screens directed at finding new genes that regulate splicing in vivo. Recessive, loss-of-function mutations in the gene smu-1 were identified as suppressors of the synthetic lethal interaction between mec-8(u218ts) and unc-52(e669su250ts) at a restrictive temperature (25). The smu-1 mutations also promote a weak bypass suppression of the other mec-8 mutant phenes and suppress the muscular dystrophy conferred by specific unc-52(viable) alleles.
We have found that smu-1 mutations affect the accumulation of a specific alternatively spliced unc-52 transcript, resulting in enhanced skipping of unc-52 mutations suppressed by smu-1. smu-1 encodes a highly conserved nuclear protein predicted to have five WD repeats, a motif associated with protein-protein interactions. WD repeats are found in proteins involved in diverse cellular processes, including splicing and other aspects of RNA metabolism (34). We propose that SMU-1 interacts with one or more additional factors to regulate the alternative splicing of unc-52 and other transcripts.
MATERIALS AND METHODS
Strains and culture.
Nematodes were cultured as described previously (5, 43). RW7000 was the source of the dimorphism bnP1, formerly called TCbn2 (47). In this paper, mec-8(ts) refers to mec-8(u218ts) I and unc-52(ts) refers to unc-52(e669su250ts) II. Other alleles unspecified in the text were (18) unc-101(m1) I, unc-59(e261) I, smu-2(mn416) II, unc-36(e251) III, mut-6(st702) IV, and him-5(e1467) V. Using the previously described screen (25), we identified smu-1(mn602), smu-1(mn609), smu-2(mn610), and smu-2(mn611) following gamma irradiation; smu-1(mn615) arose spontaneously in a mut-6 mutator genetic background (31).
Genetic and physical mapping of smu-1.
Unc-101 non-Unc-59 and Unc-59 non-Unc-101 recombinants were picked from the self progeny of unc-101 smu-1 + unc-59/+ + bnP1 + hermaphrodites. Each recombinant chromosome was made homozygous and scored with respect to bnP1 using PCR (47): 41 of 43 unc-101 unc-59(+) and 2 of 35 unc-101(+) unc-59 chromosomes were bnP1. Recombinant chromosomes were tested for the presence of smu-1 by crossing hermaphrodites to smu-1(mn415); unc-52(ts); him-5 males. F1 cross progeny were picked and allowed to self at 25°C. The absence of Unc-52 F2 progeny indicated that the recombinant chromosome was smu-1. Six of 20 unc-101 bnP1 chromosomes and two of nine unc-59 chromosomes were smu-1.
Analysis of genetic interactions between smu-1 and unc-52.
Double-mutant combinations of smu-1(mn415) and viable alleles of unc-52 other than unc-52(ts) were created by picking many non-Unc-52 non-Unc-101 progeny from unc-101 +/+ smu-1; unc-52/+ hermaphrodite parents. Progeny that failed to segregate Unc-101 animals were assumed to be homozygous for smu-1, and Unc-52 progeny from these plates were picked; multiple lines were analyzed for each unc-52 allele. The time at which the larvae became paralyzed was determined by creating a synchronized population of L1 larvae which was observed throughout larval development. The frequencies of embryonic arrest and abnormal larvae in the smu-1; unc-52(e1421) strain were determined 24 h after removal of egg-laying hermaphrodites from a growth plate. After an additional 72 h, 8 of 33 severely abnormal larvae and 21 of 21 mildly abnormal larvae had developed into adults. In control experiments, less than 3% of smu-1(mn415) or unc-52(e1421) self progeny from homozygous parents were arrested during embryogenesis.
Molecular biology and cloning of smu-1.
Standard molecular biology techniques (41) were used. All plasmid subcloning was done with pBluescript SK(−) (Stratagene) unless otherwise indicated.
smu-1(mn415); unc-52(ts); unc-36 hermaphrodites were transformed with yeast artificial chromosomes (YACs) or cosmids containing C. elegans genomic DNA (provided by A. Coulson). DNA was coinjected with R1p16, a clone that rescues unc-36 (obtained from L. Lobel), at 50 ng/μl. Non-Unc-36 F1 or F2 animals were placed at 25°C, and their progeny were inspected for non-Unc-36 Unc-52 animals. smu-1 rescue in a mec-8(ts) background was assayed by similar methods. Yeast genomic DNA containing YACs was prepared as described (9), except that DNA was purified over a Genomic-tip 100/G column (Qiagen) and not by CsCl-ethidium bromide density ultracentrifugation. This DNA was injected at 80 to 100 ng/μl; all other DNAs were injected at 20 ng/μl. Long PCR products were generated from CC4 DNA using the Expand long-template PCR system (Boehringer Mannheim). Primers CC4.3F (TTAAAAAGGGGGACGAATAGAGGTC) and CC4.3R (ATACGAACACAAGTACCGGTGTGC) amplified the predicted gene CC4.3; this PCR product rescued smu-1 in four of four lines and was cloned to create pCS212.
In addition to allele-specific DNA sequence changes, a GC-to-AT base substitution predicted to cause an amino acid substitution was found in all smu-1 mutants and named mn621. We recovered mn621 from the original strain used to generate smu-1 mutants and determined that mn621 does not suppress mec-8(ts) and does not affect the developmental timing of larval paralysis or embryonic development in any of the unc-52 mutants. For clarity, we have omitted mn621 from smu-1 genotypes.
RT-PCR.
RNA was isolated (26) and treated with RNase-free DNase (RQ1; Promega). Reverse transcription (RT) reaction mixtures contained 5 μg of RNA, 100 ng of random hexamers, 40 U of RNasin, 500 μM concentrations of each deoxynucleoside triphosphate, and 10 U of avian myeloblastosis virus reverse transcriptase in 20 μl of avian myeloblastosis virus-RT buffer (Promega). Reaction mixtures were incubated at 42°C for 2 h followed by 5 min at 95°C and diluted with 60 μl of TE buffer (41). Primers for unc-52 PCR included 18/19R (GAGGCTCTTGGTCCTCAG) and others described previously (26). Primers for ama-1 PCR were A3 (CCCGGAGGAGATTAAACGCATG), A2 (CAGTGGCTCATGTCGAGTTTCCAGA), and B2 (CGACCTTCTTTCCATCATTCATCGG). To assess linearity, PCR was performed on 1 μl of template for various numbers of cycles and at 25 cycles for multiple-template dilutions. PCR was performed in 25 μl of PCR buffer A (Fisher) with 250 μM concentrations of each deoxynucleoside triphosphate, 1 U of RedTaq (Sigma), 25 pmol of each unc-52 PCR primer, and 5 pmol of each ama-1 PCR primer. Two combinations of primers were used, with similar results: (i) 16F plus 18/19R and A3 plus B2 or (ii) 16F plus 19R and A2 plus B2. All cDNA samples were tested in two independent RT-PCR experiments, with similar results. 16-18-19 PCR products generated from smu-1(mn415); unc-52(ts) and mec-8(ts) smu-1; unc-52(ts) RNAs were cloned; all 14 of the clones isolated had normal 16-18 splice junctions. Reactions were cycled for 30 s at 94°C, 1 min at 54°C, and 1 min at 72°C. Gels were blotted and hybridized with end-labeled primer 16F-B (TGATCCAGATACCGGAGCGCCAATCG), 18R, or A2. Data were collected and analyzed using a PhosphorImager and ImageQuant software (Molecular Dynamics).
RNase protection.
RNA for RNase protection and Northern analysis was isolated with Trizol (Life Technologies). RNase protection was performed using an RPAIII kit (Ambion) on 4 to 20 μg of total RNA. The probe for RNase protection was generated using XhoI-linearized pCS168, a T7 MAXIscript in vitro transcription kit (Ambion) and 2.5 μl of [32P]CTP (800 Ci/mmol, 20 mCi/ml) in 10 μl. Each RNase protection reaction mixture contained 2.5 × 105 cpm of gel-isolated probe that was hybridized to RNA for at least 14 h at 42°C prior to RNase digestion (37°C for 30 min; supplied enzyme mix at 1:100 to 1:250). Protected fragments were resolved on a 5% polyacrylamide–urea gel. Plasmid pCS168 was generated by cloning the PCR product amplified from N2 cDNA template by the unc-52 16F and 16/18R (TGGAGTGCCTTGAGCTTC) primers into the EcoRV site of pBluescript KS(−) (Stratagene) in the reverse orientation. Positive controls to confirm the identities of the protected 16-18 and 16-17 fragments were performed by conducting RNase protection experiments with unc-52 sense RNAs generated in vitro from the appropriate cDNAs; these RNAs produced fragments of the expected sizes. Quantitative data were collected and analyzed as for RT-PCR. Linearity was verified in each experiment using multiple concentrations of two or more total RNAs.
smu-1 expression constructs.
pCS167 is a full-length translational fusion of smu-1 with the green fluorescent protein (GFP) gene, gfp (7). A PstI site that replaced the endogenous stop codon was created, and a PCR-generated gfp-containing fragment with PstI sites at each end was cloned into this site. The remainder of smu-1 came from pCS212 (3.8-kb XhoI-XbaI fragment) and CC4 (5.5-kb ApaI-XhoI fragment). pCS175, a smu-1 construct tagged with hemagglutinin (HA) epitopes (45), was created in the same manner. pCS167 and pCS175 rescued smu-1 in 7 of 10 and 1 of 6 independently generated lines, respectively. Chromosomal integration of the smu-1::gfp array was induced by gamma irradiation (28). Fixation and antibody staining of embryos and larvae were as described previously (4, 10). Dilutions of the antibodies were 1:50 for anti-GFP (Clontech), 1:500 for anti-HA (Boehringer Mannheim), and 1:500 for fluorescein isothiocyanate-conjugated goat anti-rabbit or goat anti-rat secondary antibodies (Cappel).
Nucleotide sequence accession number.
The sequence of the nearly full-length cDNA yk449d4 has been deposited in GenBank with accession no. AF330595.
RESULTS
smu-1 suppresses unc-52 nonsense mutations in exon 17 but not exon 18.
Loss-of-function mutations in smu-1 were originally shown (25) to be strong suppressors of the adult-onset paralysis seen in the viable and temperature-sensitive allele unc-52(e669su250ts), which was derived as a spontaneous, intragenic suppressor of the stronger viable allele e669 (27) and contains the e669 nonsense mutation in exon 17 as well as a single base change, su250, in the intron immediately upstream of exon 17 (40). We asked if smu-1(mn415), a probable null allele of smu-1, is able to suppress stronger but still viable alleles of unc-52, even weakly. The developmental stage during which body paralysis was seen in a majority of the larvae was determined for unc-52 and smu-1(mn415); unc-52 strains. We found that smu-1(mn415) suppressed unc-52(e669) and unc-52(e1012) in addition to unc-52(e669su250) (Fig. 1A). Paralysis was delayed by approximately 24 h for both e669 and e1012. The suppressed alleles contain nonsense mutations in exon 17 of unc-52 (40). smu-1(mn415) did not suppress nonsense mutations in exon 18, represented by unc-52(e444) and unc-52(e998).
FIG. 1.
smu-1(mn415) suppresses unc-52 alleles e669, e669su250, and e1012 in larvae and enhances unc-52(e1421) in embryos. (A) Developmental stage of phenotypic onset for various strains containing unc-52 mutations and smu-1(+) or smu-1(mn415). (B) A representation of the alternatively spliced region of unc-52. Exons are represented by boxes, introns by horizontal lines. The unc-52 mutations e669, e1012, e444, and e998 are premature stop codons; e1421 changes a splice donor site from GTAAG to GTAAA; and su250 is a single base-pair change within intron 16 (40). Some of the alternatively spliced unc-52 transcripts are shown below the exons as thin lines. The splice form increased by smu-1 (16-18) is shown as a thick line. Arrows above the exons represent the RT-PCR primers used to detect the 16-18 and 16-17-18-19 splice forms. (C) smu-1(mn415); unc-52(e1421) animals with defects in embryonic elongation and development. On the left is an animal that hatched with hypodermal constrictions and bulges; on the right is an arrested embryo that has elongated just over twofold. The arrow points to the well-developed pharynx characteristic of fully developed embryos, and the arrowheads point to hypodermal bulges. (D) Wild-type newly hatched L1 larva and embryo elongated just over twofold for comparison. Bar, 17 μm (magnifications in panels C and D are identical).
smu-1 enhances the phenotype of unc-52(e1421).
The unc-52(e1421) allele alters the exon 16 splice donor site in the alternatively spliced region of unc-52 (40). We found that 25% of the self progeny (n = 464) of smu-1(mn415); unc-52(e1421) hermaphrodites were arrested as embryos, and an additional 28% exhibited hypodermal constrictions and bulges at hatching (Fig. 1C). The arrested embryos had defects in embryonic elongation. Wild-type embryos elongate to approximately 3.5 times the length of the egg before hatching (37); arrested smu-1(mn415); unc-52(e1421) embryos elongated only 2 to 3 times the length of the egg (Fig. 1C). The arrested embryos showed normal pharyngeal development but had hypodermal constrictions and bulges. The most severely affected larvae grew slowly or not at all, but the less affected larvae recovered and often appeared to be relatively normal until the onset of paralysis at the early adult stage, which is characteristic of unc-52(e1421) animals. The strongest defects seen in the smu-1(mn415); unc-52(e1421) embryos and larvae were similar to, but weaker than, those seen in unc-52(null) mutations and in mec-8; unc-52(viable) double mutants (25).
smu-1 mutations cause an increase in the accumulation of the 16-18-19 alternatively spliced transcript of unc-52 in larvae.
The observation that mutations in smu-1 could suppress the adult-onset paralysis of unc-52 alleles with nonsense mutations in exon 17 but not exon 18 suggested that smu-1 mutations might increase the amount of a splice form that skips exon 17 but not exon 18 (Fig. 1). Experiments were therefore designed to monitor the relative level of such unc-52 mRNA in L2-L4 larvae.
Initially, we measured the relative amount of the 16-18-19 mRNA isoform found in unc-52(e669su250ts) and smu-1(mn415); unc-52(e669su250ts) mutants raised at 25°C, where loss of smu-1 function had a very obvious phenotypic effect (Fig. 2A). Semiquantitative RT-PCR was performed on larval RNAs isolated from these strains using primers designed to amplify both unc-52 (diagrammed in Fig. 1) and ama-1 cDNAs. In unc-52(e669) and unc-52(e669su250) larvae, the unc-52 PCR primers amplified both the 16-17-18-19 and 16-18-19 isoforms, but the 16-18-19 isoform was more abundant. ama-1 encodes the large subunit of RNA polymerase II (3) and was used to control for variation in the amount of total cDNA because its message is expressed at a relatively constant level during postembryonic development (20, 21). The data for two independent RNA isolations from these strains are plotted in the left half of Fig. 2A, which shows about a fourfold increase in the relative amount of the 16-18-19 isoform in the strains carrying smu-1(mn415). The magnitude of the relative increase in the 16-18-19 isoform in all repetitions of this experiment ranged between two- and fivefold (Fig. 2 and data not shown).
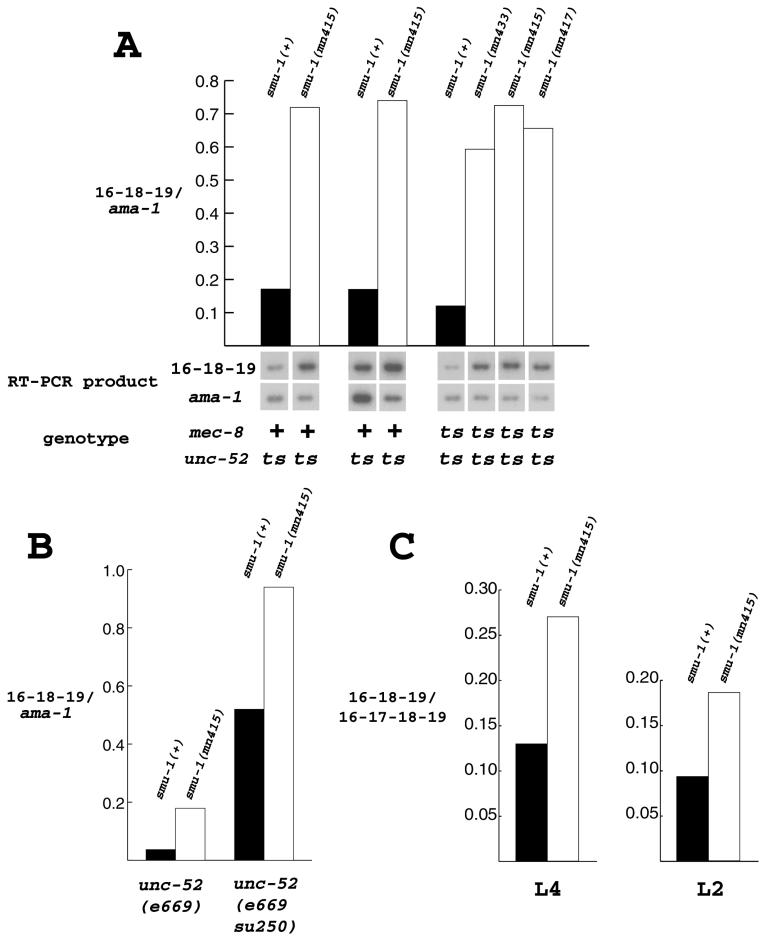
FIG. 2.
RT-PCR analysis of the unc-52 16-18-19 transcript in larvae. (A and B) Animals with the indicated genotypes were raised at 25°C until the L3 larval stage. The amount of the unc-52 16-18-19 isoform detected in each strain, normalized with respect to the amount of ama-1 mRNA amplified in the same RT-PCR, is shown on the y axis. In panel A, film exposures are shown below the graph. The graphs in panel C are similar, but the amount of the unc-52 16-18-19 isoform detected was normalized with respect to the unc-52 16-17-18-19 isoform instead of ama-1 transcript. These strains were mec-8(+); unc-52(+) and were raised at 20°C until either the L2 or L4 larval stage.
The same type of semiquantitative PCR experiment was performed on cDNA prepared from strains containing mec-8(u218ts) in addition to unc-52(e669su250ts). The results (right half of Fig. 2A) were essentially identical to those seen in the previous experiments, suggesting that mec-8 does not affect the level of this particular isoform. This experiment also demonstrated that three different alleles of smu-1 had the same effect on the levels of the 16-18-19 isoform produced by unc-52. To assess the contribution of the su250 mutation on the altered abundance of the 16-18-19 isoform, we compared the relative levels of this isoform among (i) unc-52(e669), (ii) smu-1(mn415); unc-52(e669), (iii) unc-52(e669su250), and (iv) smu-1(mn415); unc-52(e669su250) larvae raised at 25°C (Fig. 2B). This experiment demonstrated that the loss of smu-1 function results in an increase in the amount of the 16-18-19 isoform in an unc-52(e669) mutant background. The magnitude of this increase was approximately 3.5-fold. This experiment also showed that the e669 intragenic suppressor mutation su250 located in intron 16 (Fig. 1) causes a four- to sevenfold increase in the amount of the 16-18-19 isoform (Fig. 2B). When both su250 and a smu-1 mutation are present, the level of the 16-18-19 isoform is apparently high enough to suppress completely the paralysis conferred by e669.
Instead of normalizing the 16-18-19 isoform to ama-1 mRNA, which does not distinguish altered abundance of all unc-52 transcripts from changes in just the specific isoform measured, we compared the ratio of the 16-18-19 unc-52 mRNA to the 16-17-18-19 isoform in wild-type and smu-1 larvae (Fig. 2C). In contrast to the experiments involving unc-52(e669) mutations, we found a high level of the 16-17-18-19 isoform and a relatively low level of the 16-18-19 isoform. This altered proportion is probably due to mRNA surveillance (38), which would be expected to reduce the abundance of the 16-17-18-19 isoform containing the e669 nonsense mutation in exon 17. We compared the 16-18-19/16-17-18-19 ratio in wild-type and smu-1 larvae (Fig. 2C) and found that there is roughly a twofold increase in the ratio in smu-1 mutants. To confirm that there was a similar increase in smu-1; unc-52 strains, we compared the relative proportions of the 16-18-19 and 16-19 isoforms in larvae from unc-52(e669) and unc-52(e669su250) strains with and without smu-1. The ratio of 16-18-19 isoform to 16-19 isoform was about twofold higher in the smu-1 strains and was much higher in unc-52(e669su250) animals than in unc-52(e669) animals (data not shown).
RNase protection experiments were performed to confirm the results of the RT-PCR experiments. We used the antisense unc-52 RNA probe diagrammed in Fig. 3A. In this experiment, the proportions of the 16-18 to 16-17 plus 16-19 unc-52 mRNA isoforms in wild-type and smu-1 larvae were compared. We saw an approximately two- to threefold increase in the ratio of the 16-18 to 16-17 plus 16-19 isoforms in smu-1 mutants, consistent with an overall increase in the level of the 16-18 mRNA isoform (Fig. 3B). The experiment performed on the L4 stage RNA also confirmed that this increase is a common property of three different alleles of smu-1. We repeated this experiment once more with the L4-stage RNA from N2, smu-1(mn415) animals, and smu-1(mn417) animals with essentially identical results.
FIG. 3.
RNase protection analysis of the 16-18 transcript in wild-type and smu-1 mutant larvae. (A) The antisense probe used for RNase protection and the unc-52 RNAs protected by this probe. (B) Data collected from experiments using this probe. The amount of the 16-18 isoform detected in each strain is normalized with respect to the other unc-52 isoforms detected by the antisense probe and is plotted on the y axis. The RNA came from larval populations grown at 20°C to the L2 and L4 stages.
Molecular identification of smu-1.
We identified the smu-1 gene by positional cloning. Genetic mapping placed smu-1 between unc-101 I and an insertional dimorphism due to the transposable element Tc1, called bnP1 (Fig. 4A). We were able to rescue smu-1 by transformation with a YAC and a cosmid located in this physical interval (Fig. 4B), and a small rescuing region was defined using long PCR products (Fig. 4C).
FIG. 4.
Genetic and physical locations of the smu-1 gene. (A) Genetic map position of smu-1. (B) Data from rescue experiments using the YAC Y102H4 and three cosmids in the physical interval between unc-101 and bnP1. The number of rescuing lines is expressed as a proportion of the total number of lines generated. (C) Structure of the smu-1 gene and a partial restriction map of the smu-1 genomic sequence. The structure of CC4.2 was derived from ESTs and partial sequencing of the cDNA clone yk384h4. Clones and PCR products tested for smu-1 rescue are shown in the lower portion of the figure.
We confirmed that smu-1 was the predicted gene CC4.3 by identifying allele-specific lesions in the exons of this gene in five of the six smu-1 alleles (Fig. 5A). Southern blot analysis following HindIII and EcoRI digestion identified a rearrangement associated with the remaining allele (mn615) near exon 4 of CC4.3. The molecular characterizations of mn415 and mn609 indicate that they are probably null mutations. mn415 changes a glutamine codon to a stop codon (CAA to TAA) at amino acid position 255 of the predicted protein. mn609 is a 247-bp deletion in exon 4 that alters the reading frame of the predicted gene at amino acid position 106.
FIG. 5.
smu-1 encodes a highly conserved gene with five predicted WD repeats. (A) Alignment of the amino acid sequence of C. elegans (Ce) SMU-1 with a predicted human (Homo sapiens [Hs]) protein. Amino acid identities are boxed in black, and amino acid similarities are boxed in gray. Predicted WD repeat sequences are underlined, and the positions of amino acid changes found in the mutant alleles are indicated with arrows. (B) Alignment of the cores of the WD repeats with the published consensus sequence (34). Residues in bold match the consensus sequence.
Analysis of the smu-1 transcript.
The predicted exon-intron structure of smu-1 was confirmed by sequencing the nearly full-length cDNA yk449d4 (Fig. 4C). Many cDNA clones derived from smu-1 are represented as expressed sequence tags (ESTs) in sequence databases and confirm this exon-intron structure. A probe made from yk449d4 detects a single transcript of approximately 1.9 kb on Northern blots of total RNA extracted from wild-type embryos or L4 larvae (data not shown). The yk449d4 cDNA does not begin with any of the spliced-leader sequences found in C. elegans, so we performed 5′ rapid amplification of cDNA ends (5′ RACE) (13). Eight clones from two independent 5′ RACE experiments indicated that the smu-1 transcript is not trans spliced and that the transcriptional start site is 16 to 25 nucleotides upstream of the beginning of yk449d4. The 5′ untranslated region of the smu-1 mRNA contains a 3′ splice consensus site (TTTTCAG/C) upstream of the predicted translation initiation site and 24 nucleotides downstream of the first predicted transcriptional start site. We did RT-PCR using the spliced leaders SL1 and SL2 as forward primers and found that this consensus site can be trans spliced to SL1. However, since yk449d4 and the 5′ RACE clones leave this predicted 3′ splice site intact, we conclude that SL1 trans splicing to this site is rare. These data are consistent with the observation that trans splicing to SL1 is inefficient when the length of the AU-rich sequence upstream of the 3′ splice site is 41 nucleotides or fewer (8).
The predicted SMU-1 protein contains five WD repeats and is 62% identical to a predicted mammalian protein.
Database homology searches with the predicted SMU-1 protein revealed similarities with a large family of proteins that contain a repeated motif known as a WD (or WD-40 or β-transducin) repeat (34). SMU-1 contains five predicted WD repeats at its C-terminal end. The WD repeats were identified by a hidden Markov model search against the Pfam database (2) with the predicted SMU-1 amino acid sequence. Each WD motif is underlined in Fig. 5A, and the core of each WD repeat is aligned with the published consensus sequence (34) in Fig. 5B. The last (most C-terminal) WD repeat is a poorer match to the consensus sequences than are the other WD repeats (the logarithm of the odds [LOD] score from the hidden Markov model search is only 3.9, where 15 is generally considered significant). A predicted homolog of smu-1 found in humans encodes a predicted protein with a stronger match to the WD repeat consensus sequence in an equivalent position (Fig. 5; this repeat has a LOD score of 27.7). This protein is predicted to have five WD repeats in exactly the same positions as SMU-1. The amino acid sequence of the predicted human homolog of SMU-1 is based on the sequence of cDNA AK001667. The completely sequenced cDNA was isolated from an NT2 teratocarcinoma cell line, but human EST sequences suggest that the corresponding gene is expressed in many different tissues and organs.
SMU-1 is 62% identical and 78% similar to its predicted human homolog. The amino acid sequence similarity extends throughout the lengths of these proteins (Fig. 5A). We searched EST and protein databases using the unique N-terminal region of SMU-1 to determine what other organisms had potential smu-1 homologs. Highly conserved smu-1 homologs were found in other invertebrates (Drosophila melanogaster) and in vertebrates (Mus musculus, Danio rerio, and Xenopus laevis) and plants (Arabidopsis thaliana) but not in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Rescuing SMU-1::GFP and SMU-1::3HA fusion proteins are nuclearly localized and widely expressed.
To determine the expression and subcellular localization patterns of smu-1 in C. elegans embryos and larvae, we created expression constructs in which smu-1 was fused to either the gene for GFP or sequence encoding three tandem HA tags (Fig. 4C). The expression patterns of these two full-length rescuing transgenes were analyzed by direct visualization of GFP or by antibody staining with anti-GFP or anti-HA antibodies. The transgenes were ubiquitously expressed throughout development, and the protein products were found localized to nuclei (Fig. 6).
FIG. 6.
smu-1 transgenes are widely expressed. (A) L3 worm carrying the smu-1::3HA transgene stained with an anti-HA antibody. (B) GFP expression in the intestine and germ line of an adult hermaphrodite carrying the rescuing smu-1::gfp transgene. (C) Differential interference contrast image of the worm shown in panel B. The large nuclei at the tops of panels B and C are intestinal (black arrows), and the smaller nuclei below (white arrows) are germ line. The punctate pattern in the intestine is autofluorescence unrelated to GFP. (D) Expression of smu-1::gfp in a nine-cell embryo stained with an anti-GFP antibody. The arrow points to a mitosis. (E) DAPI image of the embryo shown in panel D.
In hermaphrodite larvae, we saw expression of the rescuing transgenes in all types of somatic nuclei (Fig. 6A) and in the nuclei of the female germ line (Fig. 6B). Germ line expression was surprising because most transgenic arrays seem to be subject to germ line silencing (22). Occasionally neuronal nuclei identified by DAPI (4′,6′-diamidino-2-phenylindole) failed to stain with anti-GFP antibodies in integrated lines containing the rescuing smu-1::gfp transgene. This was a very small proportion of the neurons, and lack of staining in these cases could have been due either to technical aspects of the antibody staining protocol or to incomplete expression of the integrated transgene (28). Nuclei of the other major tissue types (hypodermis, intestine, and muscle) always stained under the same conditions.
In mid- to late-stage embryos, autofluorescing GFP was visible in all or most nuclei. In embryos of less than about 200 cells, we were unable to detect GFP directly, but fixation and staining with anti-GFP antibodies indicated that GFP was present in all of the nuclei of early embryos that were not in the process of dividing (Fig. 6C). Tiny foci of anti-GFP staining within these nuclei were often seen (Fig. 6C), but it is not clear whether they represent a pattern of subnuclear localization or are due to fixation or staining artifacts. Obvious foci were not seen in the cells of other developmental stages, although in some cases the fluorescence had a rough or uneven appearance when studied closely.
DISCUSSION
We have demonstrated that mutations in smu-1, a highly conserved gene predicted by genetic analysis to be involved in alternative splicing, affect the pattern of transcript accumulation from the unc-52 gene in larvae. Since smu-1 does not encode a protein predicted to have RNA binding activity, we suggest that the wild-type SMU-1 protein affects unc-52 transcript accumulation by interacting with and influencing the activity of other proteins. What are the functions of these other proteins?
SMU-1 probably does not regulate mRNA stability.
We first conclude that smu-1 is not a member of the smg family of genes, which are involved in nonsense-mediated mRNA decay in C. elegans (38). smg mutations confer phenotypes not seen in smu-1 mutants (25), and a smg-1 mutation has no effect on the phenotype of unc-52(e669su250ts) (data not shown), an allele of unc-52 that is completely suppressed by loss-of-function smu-1 mutations. Mutations in the smg genes alter the accumulation of alternatively spliced products with premature stop codons (32, 29). In contrast, mutations in smu-1 influence the accumulation of an alternatively spliced product of unc-52 that does not have a premature stop codon.
It is possible that smu-1 is a member of an uncharacterized complex that influences RNA stability in C. elegans. We have done RT-PCR experiments looking at other mRNA isoforms produced by the unc-52 gene (data not shown) and have found no evidence that the level of any of the other isoforms produced by unc-52 is altered in smu-1 mutants. The observation that smu-1 mutations suppress unc-52 mutations with stop codons only in exon 17 also suggests that the function of smu-1 is specific with respect to unc-52. If the level of the 16-18-19 isoform rises in smu-1 mutants due to increased stability, the complex responsible for destabilizing that isoform in wild-type animals in vivo must be able to distinguish it from the other, very similar, unc-52 mRNA isoforms. It seems more likely that such a specific effect would be due to a small change in the pattern of alternative splicing of the unc-52 pre-mRNA.
SMU-1 affects alternative splicing of unc-52 pre-mRNA.
The physical nature of many intragenic suppressor mutations of unc-52(viable) mutations supports the idea that unc-52(viable) mutations can be readily suppressed by changes in the pattern of alternative splicing. The changes in nucleotide sequence associated with 13 independent intragenic suppressors of the unc-52 alleles e1421, e1012, e669, and e998 have been determined (40). Most (11 of 13) were mutations in splice sites within the alternatively spliced region of unc-52 and were predicted to alter the patterns of alternative splicing of unc-52 transcripts. One of these intragenic suppressors was su250, which was located in the middle of an intron and predicted to suppress e669 by enhancing the skipping of exon 17. Consistent with this prediction, we found that there is an increase in the 16-18-19 unc-52 mRNA isoform in e669su250 larvae grown at 25°C compared to e669 larvae. At 25°C, e669 is partially suppressed by su250. In general, suppression of e669 by su250 at 25°C is phenotypically similar to the suppression of e669 by smu-1, although su250 may be a slightly stronger suppressor of e669 than smu-1. Considering that virtually all of the intragenic suppressor mutations appear to affect the splicing patterns of unc-52 pre-mRNA, it seems most likely that the extragenic suppressor mutation smu-1 acts similarly. In the rest of this discussion, we consider the implications of our hypothesis that the wild-type smu-1 gene plays a role in regulating alternative splicing.
SMU-1 is not an essential RNA splicing factor.
The function of smu-1 is clearly not required for basic splicing activity in C. elegans. Null mutations in smu-1 are viable, and the expected phenotype for null mutations in genes that are absolutely required for splicing is embryonic lethality. There are no characterized mutations in core splicing factors in C. elegans, but the activities of some core splicing factors have been disrupted using the technique of RNA interference (11). These factors include both subunits of U2AF and the factor cSAP49, a subunit of the U2-associated factor SF3b (14, 48). As expected, RNA interference of any of these three genes results in embryonic lethality. It is possible that smu-1 does play a role in basic splicing but that its role is masked by the function of one or more genes that overlap it in function. However, the idea that smu-1 does not encode a core component of the spliceosome is supported by the observation that no homolog of smu-1 exists in S. cerevisiae, an organism that has been widely used to study basic splicing reactions. A biochemical purification of human spliceosomes did not identify a human homolog of SMU-1 as a component (35).
From these observations, we propose that SMU-1 is a member of a complex that plays a role in splice site selection during regulated alternative splicing and that removing smu-1 function causes subtle alterations in the ratios of alternatively spliced products produced by several genes. We propose that the sum of these small changes in alternative splicing patterns causes the slightly smaller brood sizes and the other subtle phenotypes that have been observed in smu-1 mutant animals (25). Since mec-8 is also predicted to affect the alternative splicing of multiple genes (9, 26), this model accounts for our observation that smu-1 suppresses not only certain alleles of unc-52 but also the phenotypes associated with mutations in mec-8. smu-1 is a weak suppressor of mec-8, suggesting that smu-1 may have smaller effects on alternative splicing than mec-8. This is the case for unc-52, the only identified target of both smu-1 and mec-8.
SMU-1 is highly conserved among multicellular eukaryotes, suggesting that its function is also conserved. Excellent in vitro systems for studying splicing have been developed for Drosophila and mammalian tissue culture cells and could be used to define SMU-1's role in splice site selection. Members of the SR family of RNA binding proteins are involved in splice site selection in these systems (24), and SR proteins are candidates for proteins that interact with SMU-1. Functional characterization of SR proteins using RNA interference has demonstrated that removing the individual functions of these proteins often results in a mild or nonobvious phenotype in C. elegans, possibly due to overlapping or redundant gene function (23). Like smu-1, no SR protein counterparts are found in S. cerevisiae (44), and their presence is correlated with complex alternative splicing. Members of this protein family also were not recovered in the biochemical purification of spliceosomes mentioned previously (35), although SR proteins did copurify with spliceosomes under less stringent purification conditions. Although the rescuing SMU-1::GFP fusion protein did not obviously localize to subnuclear structures, as has been observed for mammalian pre-mRNA splicing factors such as SR proteins (42), it is not clear whether this type of localization is easily visualized in C. elegans. C. elegans transformation techniques generally result in multiple transgene copies (28) and protein overexpression, which might obscure a pattern of subnuclear localization, particularly if SMU-1 is only transiently associated with the splicing machinery.
SMU-1 function in larvae.
We suggest that the wild-type function of smu-1 in splicing unc-52 transcripts in larvae is either to promote the 16-17 splice or to repress the 16-18 splice. The best way to distinguish these two hypotheses will be to determine what other proteins SMU-1 interacts with and whether any of these other proteins are able to bind to specific sequences in the alternatively spliced unc-52 pre-mRNA. Candidate target binding sequences would be the relevant splice sites and nearby sequences that regulate splice site usage. It seems likely that an intron sequence encompassing the su250 mutation modulates splice site choice, since this mutation affects the level of the 16-18 splice form. Loss-of function mutations in the gene smu-2 strongly resemble mutations in smu-1 (25; A. Spartz, unpublished data), suggesting that the product of the smu-2 gene may interact with SMU-1 or function in the same process. We have observed that GFP fluorescence from the rescuing smu-1::gfp transgene is greatly diminished in homozygous smu-2(mn416) mutants, suggesting that smu-2 may be required for the production or stability of SMU-1 (data not shown).
SMU-1 function in embryos.
The observation that a loss-of-function mutation in smu-1 enhances the embryonic arrest phenotype but not the larval onset-of-paralysis phenotype conferred by unc-52(e1421) suggests that smu-1 regulates the splicing of unc-52 transcripts in embryos and larvae differently. This difference could be related to differences between embryos and larvae in the alternative splicing of unc-52 transcripts controlled by other factors, such as MEC-8. Recent experiments suggest that there is both temporal and spatial regulation of the production of UNC-52 protein isoforms generated by alternative splicing (33). If MEC-8's promotion of the 15-19 and 16-19 unc-52 isoforms occurs predominantly in embryos (as suggested by our unpublished experiments), for example, then an increase in the 16-19 isoform in smu-1 embryos at the expense of the 15-19 isoform could account for the enhancement of the embryonic arrest phenotype of unc-52(e1421) by smu-1 mutation; an increase in the exon 16-containing isoform would lead to increased dependence on the e1421 mutant splice site. A change in the relative amounts of the mec-8-dependent 15-19 and 16-19 isoforms by smu-1 mutation would not be expected to affect the embryonic phenotypes of other viable unc-52 mutations, which affect exons 17 and 18.
ACKNOWLEDGMENTS
We thank Todd Starich for much good advice, Claire Kari for able technical help, Heather Gardner and Angela Spartz for useful discussions about RT-PCR, and Y. Kohara for cDNA clones. Some nematode strains were supplied by the Caenorhabditis Genetics Center.
This work was supported by U.S. National Institutes of Health (NIH) research grants GM56367 (J.E.S.) and GM22387 (R.K.H.). C.A.S. was a recipient of a doctoral dissertation fellowship from the University of Minnesota and a fellowship from NIH training grant HD07480.
REFERENCES
- 1.Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 2.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird D M, Riddle D L. Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol Cell Biol. 1989;9:4119–4130. doi: 10.1128/mcb.9.10.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowerman B, Draper B W, Mello C C, Priess J R. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Conrad R, Lea K, Blumenthal T. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- 9.Davies A G, Spike C A, Shaw J E, Herman R K. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics. 1999;153:117–134. doi: 10.1093/genetics/153.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Francis R, Waterston R H. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frohman M. RACE: rapid amplification of cDNA ends. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 28–38. [Google Scholar]
- 14.Fujita M, Kawano T, Terashima K, Tanaka T, Sakamoto H. Expression of spliceosome-associated protein 49 is required for early embryogenesis in Caenorhabditis elegans. Biochem Biophys Res Commun. 1998;253:80–84. doi: 10.1006/bbrc.1998.9747. [DOI] [PubMed] [Google Scholar]
- 15.Gilchrist E J, Moerman D G. Mutations in the sup-38 gene of Caenorhabditis elegans suppress muscle-attachment defects in unc-52 mutants. Genetics. 1992;132:431–442. doi: 10.1093/genetics/132.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabowski P J. Splicing regulation in neurons: tinkering with cell-specific control. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- 17.Hanke J, Brett D, Zastrow I, Aydin A, Delbruck S, Lehmann G, Luft F, Reich J, Bork P. Alternative splicing of human genes: more the rule than the exception? Trends Genet. 1999;15:389–390. doi: 10.1016/s0168-9525(99)01830-2. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkin J. Genetics. In: Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. C. elegans II. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 881–1047. [Google Scholar]
- 19.Hresko M C, Williams B D, Waterston R H. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon M, Gardner H F, Miller E A, Deshler J, Rougvie A E. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone I L, Barry J D. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly W G, Xu S, Montgomery M K, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langman D, Johnstone I L, Caceres J F. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 25.Lundquist E A, Herman R K. The mec-8 gene of Caenorhabditis elegans affects muscle and sensory neuron function and interacts with three other genes: unc-52, smu-1 and smu-2. Genetics. 1994;138:83–101. doi: 10.1093/genetics/138.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundquist E A, Herman R K, Rogalski T M, Mullen G P, Moerman D G, Shaw J E. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122:1601–1610. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- 27.Mackenzie J M, Jr, Garcea R L, Zengel J M, Epstein H F. Muscle development in C. elegans mutants exhibiting retarded sarcomere construction. Cell. 1978;15:751–762. doi: 10.1016/0092-8674(78)90261-1. [DOI] [PubMed] [Google Scholar]
- 28.Mello C C, Fire A. DNA transformation. In: Epstein H F, Shakes D C, editors. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, Calif: Academic Press; 1995. pp. 451–482. [Google Scholar]
- 29.Mitrovitch Q M, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moerman D G, Hutter H, Mullen G P, Schnabel R. Cell autonomous expression of perlecan and plasticity of cell shape in embryonic muscle of Caenorhabditis elegans. Dev Biol. 1996;173:228–242. doi: 10.1006/dbio.1996.0019. [DOI] [PubMed] [Google Scholar]
- 31.Mori I, Moerman D G, Waterston R H. Analysis of a mutator activity necessary for germline transposition and excision of Tc1 transposable elements in Caenorhabditis elegans. Genetics. 1988;120:397–407. doi: 10.1093/genetics/120.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison M, Harris K S, Roth M B. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:9782–9785. doi: 10.1073/pnas.94.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullen G P, Rogalski T M, Bush J A, Gorji P R, Moerman D G. Complex patterns of alternative splicing mediate the spatial and temporal distribution of perlecan/UNC-52 in Caenorhabditis elegans. Mol Biol Cell. 1999;10:3205–3221. doi: 10.1091/mbc.10.10.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 35.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 36.Perkins L A, Hedgecock E M, Thomson J N, Culotti J G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 37.Priess J R, Hirsh D I. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 38.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 39.Rogalski T M, Williams B D, Mullen G P, Moerman D G. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 1993;7:1471–1484. doi: 10.1101/gad.7.8.1471. [DOI] [PubMed] [Google Scholar]
- 40.Rogalski T M, Gilchrist E J, Mullen G P, Moerman D G. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–169. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sleeman J E, Lamond A I. Nuclear organization of pre-mRNA splicing factors. Curr Opin Cell Biol. 1999;11:372–377. doi: 10.1016/S0955-0674(99)80052-0. [DOI] [PubMed] [Google Scholar]
- 43.Sulston J, Hodgkin J. Methods. In: Wood W B, editor. The nematode Caenorhabditis elegans. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- 44.Tacke R, Manley J L. Determinants of SR protein specificity. Curr Opin Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 45.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams B D, Waterston R H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams B D, Schrank B, Huynh C, Shownkeen R, Waterston R H. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zorio D A, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA. 1999;5:487–494. doi: 10.1017/s1355838299982225. [DOI] [PMC free article] [PubMed] [Google Scholar]