Abstract
Background & Aims:
Sterile inflammation is a major clinical concern during ischemia-reperfusion injury (IRI) triggered by traumatic events including stroke, myocardial infarction, and solid organ transplantation. Despite HMGB1 clearly being involved in sterile inflammation, its role is controversial due to a paucity of patient-focused research.
Approach & Results:
Here, we examined the role of HMGB1 oxidation states in human IRI following liver transplantation. Portal blood immediately following allograft reperfusion (liver flush, LF) had increased total HMGB1, but only LF from patients with histopathological IRI had increased disulfide-HMGB1 and induced TLR4-dependent TNFα production by macrophages. Disulfide HMGB1 levels increased concomitantly with IRI severity. IRI+ pre-reperfusion biopsies contained macrophages with hyperacetylated, lysosomal disulfide-HMGB1 that increased post-reperfusion at sites of injury, paralleling increased histone acetyltransferase GTF3C4 and decreased histone deacetylase HDAC5 expression. Purified disulfide-HMGB1 or IRI+ blood stimulated further production of disulfide-HMGB1 and increased pro-inflammatory molecule and cytokine expression in macrophages via a positive feedback loop.
Conclusions:
These data identify disulfide-HMGB1 as a mechanistic biomarker of, and therapeutic target for, minimizing sterile inflammation during human liver IRI.
INTRODUCTION
Sterile inflammation involves a complex interplay between the innate and adaptive immune systems, thereby playing a key role in the overall outcome of mechanical trauma, chemical and environmental insults, and ischemia-reperfusion injury (IRI). IRI is a common clinical condition triggered by sepsis, cardiogenic shock, vascular surgery or organ retrieval for transplantation. Despite this significant clinical concern, there are currently no therapeutics or patient-specific diagnostics available due to the continuing lack of knowledge of the underlying IRI immune mechanisms.
Damage-associated molecular patterns (DAMPs) were proposed as endogenous danger signals generated in response to cellular stress that could be recognized by germline-encoded pattern recognition receptors (PRRs) typically expressed by innate immune cells (1). With the potential for any intracellular component that is detected in the extracellular space by a PRR-bearing cell to act as a DAMP, the search for one specific to IRI has proven difficult. We previously showed via longitudinal evaluation of cytokine signatures in the human blood of orthotopic liver transplant (OLT) recipients that the innate-adaptive immune switch occurs during transplantation, once the recipient portal vein blood (PV) is reperfused through the donor organ (liver flush; LF) (2). Screening for activation of a panel of 10 PRRs by these two blood samples, we then showed that TLR4 and TLR9 were preferentially activated by OLT patient portal blood with biopsy-proven IRI pathology (3), considerably narrowing the search for IRI-mediating DAMPs.
High mobility group box 1 (HMGB1) is a well-characterized DAMP that can bind to TLR4, TLR9 or other receptors depending on its redox state. Under homeostatic conditions, HMGB1 functions as a DNA scaffolding protein in all nucleated cells. Normal nuclear functions occur under reducing conditions, therefore nuclear HMGB1 is in its fully reduced, “all-thiol” redox state (4, 5). Damaged epithelial cells, including hepatocytes, passively release their intracellular HMGB1 into the extracellular space (6–9). HMGB1 has been shown to undergo oxidation that modulates various aspects of its function, including subcellular localization and pro-inflammatory chemokine or cytokine activity via signaling through TLR4 (11, 12). Active secretion of the partially oxidized, “disulfide” redox form of HMGB1 by monocytes and macrophages has been described, in which shuttling of the protein into the cytosol and/or prevention of nuclear import or re-entry of HMGB1 is required (4), and disulfide-HMGB1 is exclusively responsible for HMGB1-TLR4 signaling leading to further pro-inflammatory cytokine secretion (12). Post-translational modification in the form of hyperacetylation on lysine residues within one or both nuclear localization sequences is responsible for this cytoplasmic accumulation (5). Because HMGB1 does not contain a leader sequence, the protein is released via a nonclassical secretory pathway involving specialized lysosomal-associated membrane glycoprotein 1-positive (LAMP1+) vesicles of the endolysosomal compartment (13). Secreted disulfide-HMGB1 can then potentially mediate a positive feedback loop by binding to TLR4 and initiating downstream pro-inflammatory signaling cascade, which can further induce cell death (14, 15).
Because liver IRI has been shown to trigger HMGB1 release (16) which then signals via TLR4 (17) in mouse models, we hypothesized that disulfide-HMGB1-producing macrophages promote IRI in human OLT via TLR4. We used blood and liver biopsies from a large cohort of 92 OLT recipients with and without biopsy-proven IRI and identified the contribution of macrophage-derived disulfide-HMGB1 to the generation and perpetuation of proinflammatory macrophages in patients with IRI. Notably, the histone acetyltransferase GTF3C4 was significantly increased while histone deacetylase HDAC5 was significantly decreased in biopsies from IRI+ patients compared to IRI- patients, providing a molecular mechanism for therapeutic targeting to mitigate downstream effects of IRI and improve overall OLT clinical outcomes. Additionally, GTF3C4 and/or HDAC5 expression levels could serve as a biomarker of IRI across multiple clinical situations such as stroke, sepsis, myocardial infarction, or vascular surgery as well as solid organ transplantation.
RESULTS
HMGB1 is released from donor hepatic tissue into patient portal blood following reperfusion
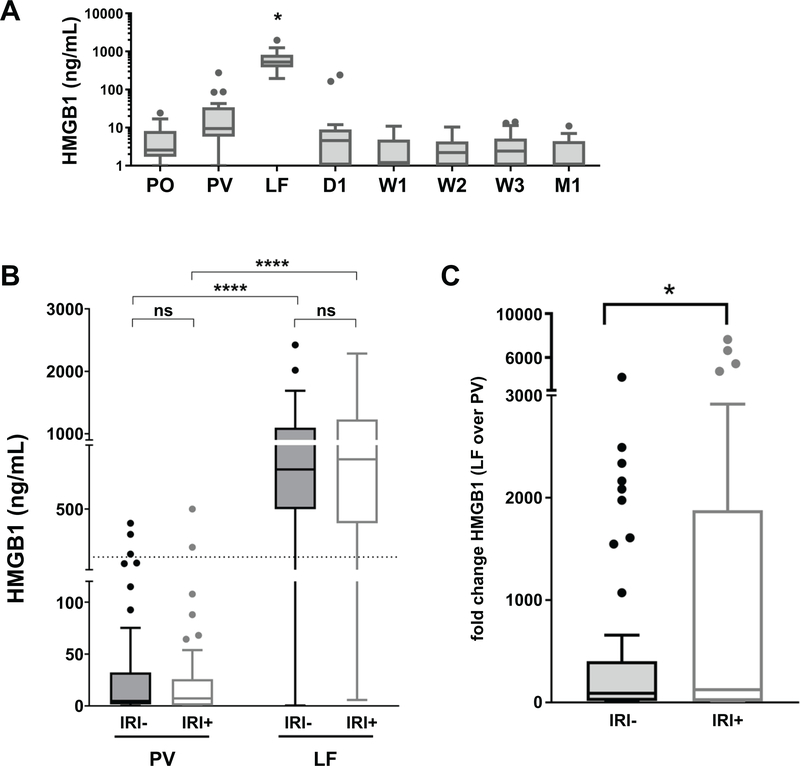
To understand when HMGB1 release occurs in the setting of solid organ transplantation, we performed an ELISA on 10 OLT patient blood samples (5 IRI-, 5 IRI+), obtained at various key time points pre- (pre-operative, PO), intra- (portal vein, PV; and liver flush, LF) and post-transplant (day 1, D1; week 1, W1; week 2, W2; week 3, W3; and month 1, M1) for levels of total HMGB1 (Fig. 1). HMGB1 was found to be below 10 ng/ml in circulating systemic blood at all time points evaluated (Fig. 1A). Recipient portal vein blood obtained just prior to reperfusion (PV) showed slightly elevated levels of HMGB1, which were significantly increased immediately upon reperfusion through the donor allograft (LF) (Fig. 1A). We then performed ELISA for HMGB1 on paired PV and LF samples for the full cohort of 92 OLT recipients (n=92; 46 IRI-, 46 IRI+; Tables 1–3). Although overall HMGB1 levels were not significantly different between IRI+/− groups at either time point (Fig. 1B), HMGB1 levels increased significantly more over time in IRI+ patients than IRI− (Fig. 1C).
Figure 1. Longitudinal orthotopic liver transplant recipient systemic and portal blood samples differ in HMGB1 content by time but not patient IRI status.
Patient plasma samples obtained at key time points pre-, intra- or post-operative time points from either circulating systemic blood (PO, pre-operative; D1, day 1; W1–3, week 1–3; and M1, month 1) or portal blood obtained before (PV, portal vein) or after reperfusion through the donor allograft (LF, liver flush) were assayed by ELISA for levels of HMGB1 present. Results were examined (A) across time, indicating that only PV and LF samples contain HMGB1 (n=10; 5 IRI-, 5 IRI+), or (B) by patient IRI status in PV/LF only (n=92; 46 IRI-, 46 IRI+). Dotted line indicates functional threshold of HMGB1 (200 ng/ml). (C) Paired analysis of fold change of HMGB1 levels in LF over those in PV for each patient by IRI status. Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values and lines represent median values. *P < 0.05. ****P < 0.0001 by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst time and/or IRI status.
Table 1 –
Recipient Characteristics
| Clinical and Demographic Data | (n=92) | IRI− (n=46) | IRI+ (n=46) | |
|---|---|---|---|---|
|
| ||||
| RECIPIENT: | ||||
|
| ||||
| Characteristic | Value | Value | Value | P-value* |
| Age, years [mean±SD] | 56±11 | 57±11 | 54±10 | 0.21 |
| Gender [n (%)] | 0.52 | |||
| Female | 36 (39) | 20 (43) | 16 (35) | |
| Male | 56 (61) | 26 (57) | 30 (65) | |
| Race [n (%)] | 0.97 | |||
| Asian | 8 (9) | 4 (9) | 4 (9) | |
| Black/African American | 5 (5) | 3 (7) | 2 (4) | |
| White/Caucasian | 47 (51) | 24 (52) | 23 (50) | |
| Other | 32 (35) | 15 (33) | 17 (37) | |
| Ethnicity [n (%)] | 0.50 | |||
| Hispanic/Latino | 38 (41) | 16 (35) | 22 (48) | |
| Non-Hispanic/Latino | 51 (55) | 28 (61) | 23 (50) | |
| Unknown/Not Reported | 3 (3) | 2 (4) | 1 (2) | |
| Liver Disease Etiology [n (%)] | 0.12 | |||
| alcoholic | 23 (25) | 7 (15) | 16 (35) | |
| HBV | 6 (7) | 3 (7) | 3 (7) | |
| HCV | 33 (36) | 18 (39) | 15 (33) | |
| NAFLD/NASH | 15 (16) | 10 (22) | 5 (11) | |
| AIH | 4 (4) | 2 (4) | 2 (4) | |
| PBC | 2 (2) | 2 (4) | 0 (0) | |
| PSC | 4 (4) | 2 (4) | 2 (4) | |
| ALF | 2 (2) | 2 (4) | 0 (0) | |
| other | 3 (3) | 0 (0) | 3 (7) | |
| HCC [n (%)] | 26 (28) | 16 (35) | 10 (22) | 0.25 |
| MELD, at list [mean±SD] | 26±12 | 25±12 | 28±11 | 0.21 |
| MELD, at transplant [mean±SD] | 36±6 | 36±5 | 36±6 | 0.67 |
| Transplant(s) [n (%)] | 0.39 | |||
| isolated liver | 78 (85) | 41 (89) | 37 (80) | |
| liver-kidney | 14 (15) | 5 (11) | 9 (20) | |
| Split liver [n (%)] | 1 (1) | 1 (2) | 0 (0) | >0.99 |
| LFTs, opening [mean±SD] | ||||
| ALT | 553±559 | 505±466 | 602±559 | 0.37 |
| AST | 1127±1086 | 1040±980 | 1214±1188 | 0.45 |
| Bilirubin | 8.3±6.5 | 6.9±4 | 9.7±8.1 | 0.04* |
| INR | 1.4±0.2 | 1.4±0.2 | 1.5±0.3 | 0.4 |
Table 3 –
Transplant Characteristics
| Clinical and Demographic Data | (n=92) | IRI− (n=46) | IRI+ (n=46) | |
|---|---|---|---|---|
|
| ||||
| RECIPIENT + DONOR | ||||
|
| ||||
| Characteristic | Value | Value | Value | P-value* |
| ABO [n (%)] | >0.99 | |||
| identical | 81 (88) | 40 (87) | 41 (89) | |
| compatible | 11 (12) | 6 (13) | 5 (11) | |
| Donor Risk Index (DRI) [mean±SD] | 1.5±0.4 | 1.5±0.3 | 1.5±0.4 | 0.58 |
| Sharing [n (%)] | 0.34 | |||
| Local | 52 (57) | 25 (54) | 27 (59) | |
| Regional | 37 (40) | 18 (39) | 19 (41) | |
| National | 3 (3) | 0 (0) | 3 (7) | |
IRI status of OLT patients reflects oxidation state of HMGB1 in portal blood
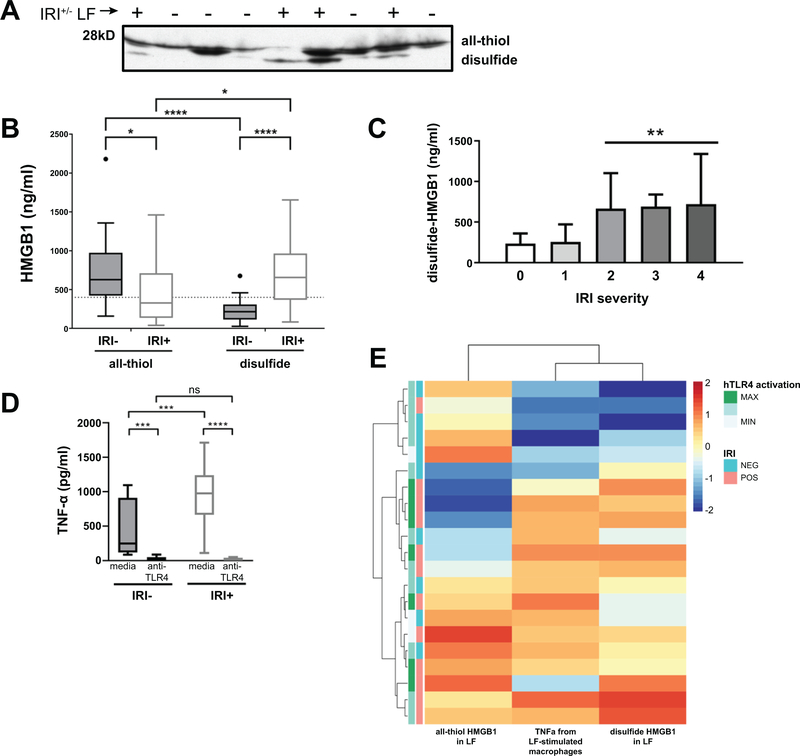
Because HMGB1 presence in LF alone did not predict IRI status, we investigated the redox state of HMGB1, in OLT patient LF samples (Fig. 2). It has been shown that all-thiol HMGB1 is a chemoattractant whereas disulfide-HMGB1 is the form that results in cytokine-inducing activity (12); therefore, we reasoned that IRI+ patients may be differentially stimulating TNFα secretion from monocytes due to varying content of disulfide or all-thiol HMGB1. Gel electrophoresis can separate the two redox forms of HMGB1 due to the disulfide form running slightly faster than the all-thiol form (12). We performed Western blots on LF samples from OLT patients with HMGB1 levels above 400 ng/ml, the detectable limit in our assay (n=77; 43 IRI-, 44 IRI+), to determine the ratio of disulfide to all-thiol HMGB1 in the LF (Fig. 2A). We then combined this information with the concentrations obtained in the HMGB1 ELISA assay from Fig. 1 to extrapolate the concentration of all-thiol and disulfide-HMGB1 in patient LF samples (Fig. 2B). As suspected, IRI+ patients had statistically increased disulfide-HMGB1 than IRI- patients in their LF blood. Additionally, stratifying by IRI severity revealed disulfide-HMGB1 levels steadily increase as histopathological IRI increases (Fig. 2C).
Figure 2. Redox state of HMGB1+ LF is disulfide in IRI+ and all-thiol in IRI- OLT patients.
Patient LF samples obtained just after reperfusion through the donor allograft were assayed by immunoblotting to determine the ratio of disulfide to all-thiol HMGB1 for each patient. (A) Representative western blot showing 28kD bands that correspond to all-thiol HMGB1 and slightly lower bands at ~25kD corresponding to disulfide-HMGB1 for samples from either IRI+ (+) or IRI- (−) patients. (B) Amount of all-thiol and disulfide-HMGB1 was extrapolated by dividing the total HMGB1 content per sample as determined by ELISA in Fig. 1 by the ratio of HMGB1 redox forms determined by Western blotting in 3A. IRI+, n=27; IRI-, n=31 (C) Disulfide-HMGB1 levels by IRI severity. Data shown as mean±SD for each group. 31 IRI-, 27 IRI+; IRI 0 (n=4), 1 (n=27), 2 (n=18), 3 (n=5), 4 (n=4) (D) HMGB1+ LF from 40 OLT patients (20 IRI-, 20 IRI+) was used to stimulate monocytes from healthy third-party donors for 8 hours (as determined in Fig. S5A following pre-treatment for one hour with either media or anti-TLR4 mAb, and supernatant was assayed for secreted TNFα. (E) Comparison of TLR4 activation, disulfide-HMGB1 levels, and TNFα secretion by monocytes as determined by the common cohort of n=21; 9 IRI-, 12 IRI+ from HEK assay, disulfide-HMGB1 levels and TNFα production from macrophages. Shown is a heat map in which the rows represent patients, the columns represent the groups compared, and the colors represent normalized median values per assay (blue = low, red = high). The rows and columns are ordered based on the results of an unsupervised hierarchical clustering, with dendograms on the left side showing groups of IRI- (salmon) or IRI+ (aqua) OLT recipients with or without functional activation of HEK-Blue hTLR4 cells by their LF. *P < 0.05; **P < 0.01; ****P < 0.0001 by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst HMGB1 redox state and/or IRI status.
We added patient LF to monocytes and compared TNFα secretion from cells cultured with samples from 40 OLT patients (20 IRI-, 20 IRI+) (Fig. 2D). LF from IRI+ recipients activated monocytes to secrete significantly more TNFα than IRI- patient LF samples at the 8 hr time point, both of which were abrogated upon pre-treatment with a neutralizing anti-TLR4 mAb, suggesting the HMGB1 found in IRI+ patients is in the disulfide redox state. Additionally, we were able to confirm that LF samples from IRI+ recipients containing high levels of disulfide-HMGB1 stimulated strong TNFα secretion from monocytes via TLR4 activation whereas IRI- patient LF did not (Fig. 2E).
OLT-IRI+ recipient blood activates macrophages towards pro-inflammatory phenotype via TLR4
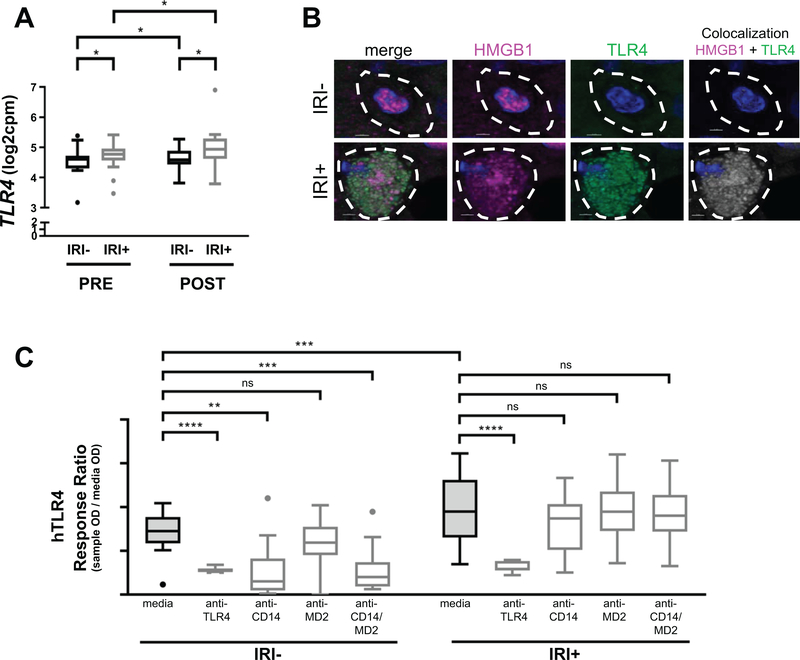
HMGB1 signals via TLR4 only when it is partially oxidized into its disulfide redox form (12). Our previous studies indicated that OLT-IRI+ patient LF samples were significantly more capable of signaling via TLR4 when examining a panel of commercially available human (h)TLR-transfected HEK-293 cell lines (3). Of note, a significant increase in IRI+ patient HMGB1 in LF relative to its PV baseline occurred (Fig. 1C), which mirrored the pattern seen in TLR4 activation in these same samples (3). Therefore, we sought to determine if patient liver macrophages sense HMGB1 through TLR4 similar to peripheral third party monocytes, as macrophages are the primary TLR4+ responders to HMGB1 in the liver via their TNFα production (18). TLR4 transcripts are present in pre-reperfusion donor allografts and increased in post-reperfusion allografts (Fig. 3A), and IRI+ patients have increased transcripts compared to IRI- patients at both time points, despite the number of CD68+ cells in biopsies from OLT patients being similar (Fig. S1A, B). Immunofluorescence (IF) staining of the biopsies for macrophages showed they are positive for TLR4 that colocalizes to HMGB1 (Fig. 3B).
Figure 3. HMGB1+ LF from IRI+, but not IRI-, patients causes TNFα release from monocytes via TLR4.
(A) Transcripts of the TLR4 gene are increased in donor allografts of IRI+ patients and over time. (n=40; 20 IRI- and 20 IRI+) (B) Immunofluorescence staining for HMGB1 (pink) or TLR4 (green) along with the nuclear counterstain DAPI (blue) show TLR4+ macrophages present in IRI+ allografts that are not present in macrophages of IRI- allografts. Importantly, TLR4 and HMGB1 are co-localized in IRI+ macrophages (last panel) Scale bars = 2 um top row, 3 um bottom row as indicated (n=20; 10 IRI- and 10 IRI+.) c, Patient LF was used to stimulate human HEK-Blue TLR4-transfected cell lines (hTLR4) pre-treated with either media, anti-TLR4 mAb, anti-CD14 mAb, anti-MD2 mAb or anti-CD14/anti-MD2 mAbs together, and compared to responses achieved using the natural control ligand LPS or media alone (Fig. S7C) solid (LPS ECmax), dashed (LPS EC50) and dotted (media) lines). (n=27; 14 IRI-, 13 IRI+). Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values and lines represent median values for each time point (A) or patient IRI status (B). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst time, mAb treatment and/or IRI status.
To investigate if HMGB1 requires the same cofactors as LPS to signal via TLR4, we pre-treated HEK-293 cells transfected with hTLR4 with neutralizing antibodies to TLR4, CD14, and/or MD2, then added patient LF and checked for activation of TLR4 (Fig. 3C). As expected, pre-treatment with anti-TLR4 mAb abolished TLR4 activation by patient LF regardless if it was from an IRI- or IRI+ patient. IRI- patient blood was less capable of activating TLR4 when CD14 activity was neutralized, but lack of MD2 signaling did not significantly affect TLR4 activation. Surprisingly, IRI+ patient LF-activation of TLR4 was unaffected by absence of CD14 or MD2. Control disulfide-HMGB1 was only partially affected by CD14 and/or MD2 absence (Fig. S7C), suggesting that although it can signal through TLR4 via these cofactors, they are not required. Overall the effect seen in patients is a combination of ligand and receptor differences.
IRI+ OLT recipients have disulfide-HMGB1-producing macrophages in their allografts that increase after reperfusion
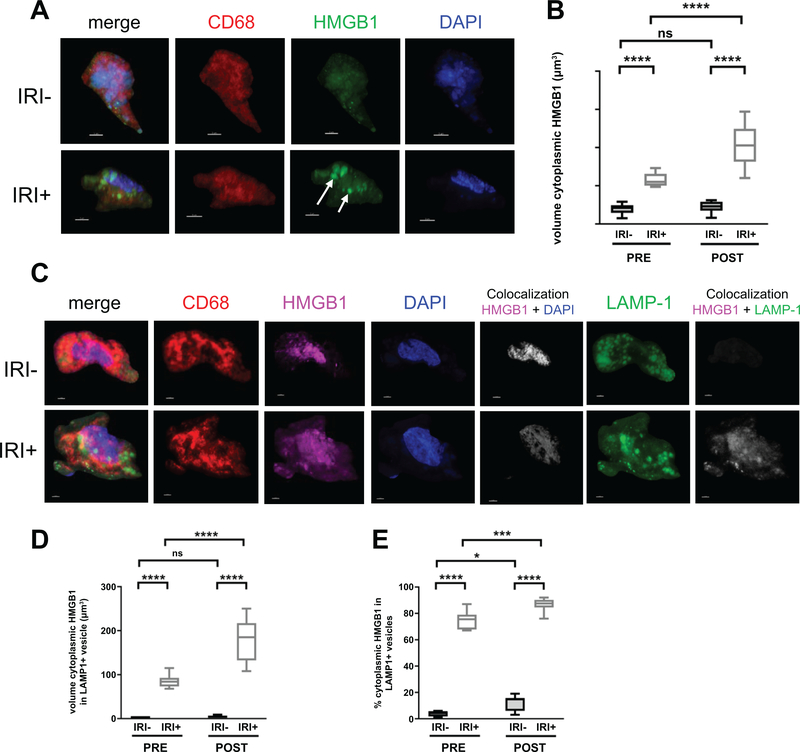
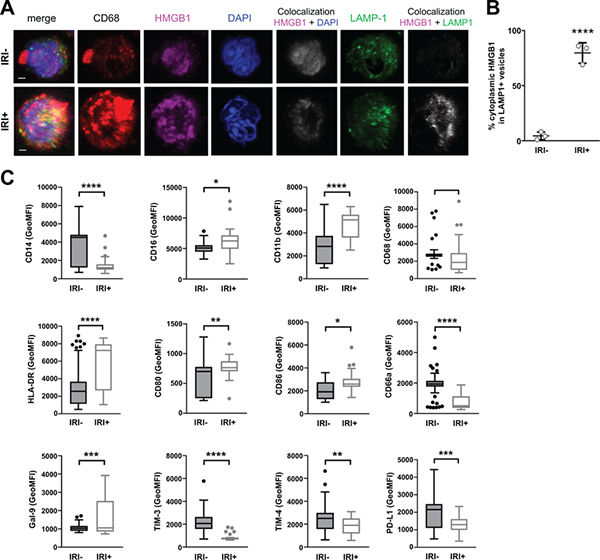
To determine the effect of IRI on the subcellular localization of HMGB1, IF staining was performed. Consistent with other’s observations (17), HMGB1 was almost exclusively localized in the nucleus of hepatocytes in the pre-transplant livers of IRI- patients with no injury (Fig. S2). In patients with IRI, the intensity of pre-transplant HMGB1 staining was increased diffusely throughout the cytoplasm of some hepatocytes, whereas others were depleted of HMGB1 entirely. Active secretion of the disulfide redox form of HMGB1 by monocytes and macrophages requires cytoplasmic translocation as well as loading into LAMP1+ cytoplasmic vesicles (4). IF staining of patient allografts revealed CD68+ macrophages in clusters amongst healthy hepatocytes in both IRI- and IRI+ biopsies (Fig. S2), but appeared to have most of their HMGB1 translocated into cytoplasmic vesicles in only IRI+ patient allografts (Fig. S2, bottom row). Therefore, we characterized the subcellular localization of HMGB1 of macrophages to determine its redox state using three-dimensional reconstruction of high-resolution confocal laser scanning microscopy images of human OLT allografts (Fig. 4), as described in Fig. S3. We found HMGB1 translocated into the cytoplasm of macrophages in the allografts of IRI+ patients (Fig. 4A, arrows), which was significantly different (Fig. 4b) than the macrophages found in IRI- allografts (Video S1, S2). Further, we found the majority of cytoplasmic HMGB1 was restricted within LAMP1+ vesicles of CD68+ macrophages in the allografts of IRI+ patients, but not IRI- recipients at both time points (Fig. 4C–E). These findings suggest that the protective effect occurring in IRI- patients is, at least in part, caused by the prevention of disulfide-HMGB1 formation and release by macrophages.
Figure 4. IRI+ biopsies contain disulfide-producing macrophages that increase over time.
Allograft biopsies were obtained 2 hrs pre-transplant (PRE) or 2 hrs post-reperfusion (POST), fixed in 4% PFA, sucrose-impregnated, embedded in gelatin then OCT blocks for immunofluorescence (IF). (A) 10 or 30um sections were stained for CD68 (red), HMGB1 (green) and counterstained with DAPI to detect cellular nuclei (blue). Scale bars = 3um. (B) Quantification of macrophage-specific cytoplasmic HMGB1 from reconstructed confocal laser-scanning microscopy Z-stacks as described in Fig. S4C, Sections were stained for CD68 (red), HMGB1 (pink), DAPI (blue), and LAMP1 (green). Localization of HMGB1 to the nuclear compartment (white, fifth panel of top and bottom rows) or LAMP1+ cytoplasmic vesicles (white, last panel of top and bottom rows) was determined using Imaris software. (D) Quantification of macrophage-specific cytoplasmic HMGB1 constrained within LAMP1+ vesicles by volume (um3). (E) Quantification of the percent of macrophage-specific cytoplasmic HMGB1 constrained within LAMP1+ vesicles. Numbers shown in B and D represent the average volume of HMGB1 found within the cytoplasm or LAMP1+ cytoplasmic vesicles of ≥3 randomly selected macrophages per patient for n=20 patients; IRI- = 10 and IRI+ = 10, Numbers shown in E represent the percent of cytoplasmic HMGB1 found within LAMP1+ vesicles of ≥3 macrophages per tissue slice for n=20 patients; IRI- = 10 and IRI+ = 10. *P < 0.05; Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values and lines represent median values. *P < 0.05; ***P < 0.001; ****P < 0.0001 by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst HMGB1 redox state and/or IRI status.
Increased GTF3C4 and decreased HDAC5 contribute to hyperacetylation of macrophage-HMGB1 in IRI+ OLT patients
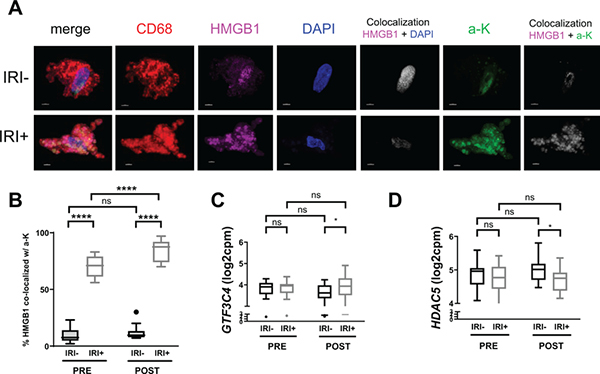
As acetylation of HMGB1 is known to influence its subcellular localization and partial oxidation into secretable disulfide-HMGB1 in macrophages (19), we investigated the acetylation status at lysine residues of HMGB1 in OLT recipient biopsies by IF and confocal microscopy. We found that HMGB1 in IRI+ recipients co-localized predominantly with staining for acetylated lysine residues (Fig. 5A, B), indicating the translocation of HMGB1 observed in macrophages from these patient allografts (Fig. 4) was mediated by hyperacetylation. Along these lines, only a small percentage of acetylated lysine staining co-localized to HMGB1 in macrophages from IRI- biopsies (Fig. 5A, B).
Figure 5. Increased GTF3C4 and decreased HDAC5 contribute to hyperacetylation of macrophage HMGB1 in IRI+ OLT patients.
(A) Allograft biopsies were obtained 2 hrs pre-transplant (PRE) or 2 hrs post-reperfusion (POST), fixed in 4% PFA, sucrose-impregnated, embedded in gelatin then OCT blocks for immunofluorescence (IF). 10 or 30 um sections were stained for CD68 (red), HMGB1 (pink), DAPI (blue), and acetylated lysine residues (green; a-K). Images were acquired by confocal microscopy, and localization of HMGB1 to the nuclear compartment (white, fifth panel of top and bottom rows) or a-K (white, last panel of top and bottom rows) was determined using Imaris software. Scale bars = 3 um (B) Quantification of the percent of macrophage-specific HMGB1 (pink) which co-localized to acetyl-lysine (a-K, green; co-localization, white) from reconstructed confocal laser-scanning microscopy Z-stacks. n = 20; 10 IRI- and 10 IRI+. Gene expression (transcripts per million; log2 CPM) of (C) GTF3C4, a HAT encoding gene, or (D) HDAC5, an HDAC encoding gene, in PRE/POST biopsies from IRI+ and IRI- OLT recipients by RNAseq. n = 40; 20 IRI- and 20 IRI+. Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values, and lines represent median values for each time point. Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values, and lines represent median values for each time point. *P < 0.05 by two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst HMGB1 redox state and/or IRI status.
Hyperacetylation of nuclear proteins, including HMGB1, is performed by histone acetyltransferases (HATs) and balanced by the activity of histone deacetylases (HDACs). Because we observed a difference in hyperacetylation of HMGB1 that corresponded to recipient IRI status, we analyzed the expression of HATs and HDACs via RNAseq (Fig. 5C, D) in OLT recipient biopsies. Out of 12 genes encoding HATs whose expression was detected in OLT patient biopsies (Fig. S4), seven were differentially expressed between pre- and post-reperfusion time points. Interestingly, GTF3C4, a gene encoding a HAT also known as KAT12, had increased transcripts in IRI+ allografts obtained post-reperfusion (Fig. 5C). Further, of the 12 genes encoding HDACs found to be expressed in OLT recipient biopsies (Fig. S5), six were differentially expressed from pre- to post-reperfusion, yet only HDAC5 was decreased in IRI+ patient biopsies as compared to IRI- (Fig. 5D), and this occurred at the same time point that GTF3C4 transcripts were decreased.
Disulfide-HMGB1+ LF triggers monocytes to translocate nuclear HMGB1 into LAMP1+ vesicles and become more pro-inflammatory
It has been reported that HMGB1 is selectively internalized by infiltrating mononuclear phagocytes (20). Further, HMGB1 has been proposed to perpetuate inflammatory amplification loops in other disease settings such as infection (21). We found a significant increase in disulfide-HMGB1-producing macrophages in the post-reperfusion biopsies over the pre-reperfusion samples; therefore, we speculated that this uptake of HMGB1 by newly arriving myeloid cells could stimulate them to become disulfide-producing macrophages within the allograft. To investigate this possibility, we stimulated monocytes with either HMGB1-containing patient LF or commercially-available purified all-thiol HMGB1 or disulfide-HMGB1 as controls. We found that IRI+ LF stimulated monocytes to translocate their own HMGB1 into their cytoplasm and package it into lysosomal vesicles within 2 hours (Fig. 6A, B) similar to results obtained with control disulfide-HMGB1 (Fig. S6A, B) whereas IRI- LF (Fig. 6A, B) and all-thiol HMGB1 (Fig. S6A, B) did not induce this change. Additionally, disulfide-HMGB1-containing IRI+ LF stimulated a change in phenotype of monocytes in culture for 3 days (Fig. 6C) to become more pro-inflammatory and invasive, upregulating HLA-DR, CD80, CD86 and CD11b while simultaneously downregulating molecules that were increased in monocytes cultured with IRI- LF, including CEACAM1, TIM3, TIM4, and PD-L1, molecules involved in anti-inflammatory and pro-resolution as well as T cell exhaustion.
Figure 6. Disulfide-HMGB1+IRI+ patient LF activates monocytes to translocate nuclear HMGB1 into cytoplasmic vesicles and become more pro-inflammatory.
PBMC-derived monocytes from healthy, third-party donors were cultured with patient LF (n=6; 3 IRI-, 3 IRI+) for 2 hours to determine HMGB1 localization by confocal microscopy or 3 days to determine functional phenotype by flow cytometry. (A) Representative images of monocytes stained by multicolor IF for CD68 (red), HMGB1 (pink), DAPI (blue), and LAMP1 (green), and z-stack images were acquired by confocal microscopy and reconstructed in Imaris. Scale bars = 2 um (B) Quantification of monocyte-specific cytoplasmic HMGB1 in LAMP1+ vesicles from reconstructed confocal laser-scanning microscopy Z-stacks. Data are presented as dots for each of three separate experiments with long lines indicating mean. (C) Stimulated monocytes were multiplex stained with a panel of directly-labeled antibodies and run on a flow cytometer to determine surface expression levels of CD14, CD16, CD11b, CD68, HLA-DR, CD80, CD86, CD66a, Gal-9, TIM-3, TIM-4, and PD-L1. Geometric mean fluorescence intensity (GeoMFI) for the respective surface marker on cells stimulated by n=77; 43 IRI- versus 34 IRI- LF are shown. Data are presented as Tukey box-and-whisker plots: whiskers are inner fences reaching 1.5 times the interquartile range and boxes represent the interquartile ranges, dots indicate outlying values and lines represent median values. ns = no significance; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by Student’s T test.
DISCUSSION
IRI is a pathophysiologic process in which hypoxic organ damage is accentuated following return of blood flow and oxygen delivery. Liver IRI consists of direct hepatocellular damage as the result of the ischemic insult, as well as delayed dysfunction and damage that results from activation of innate inflammatory pathways. Innate immune cells are activated after ischemic injury to initiate the tissue repair processes needed to regain homeostasis, and to provide defense against microbial invasion. However, excessive activation can lead to exaggerated local and systemic inflammation that may extend the tissue damage. HMGB1 is an early mediator of inflammation and cellular injury after IRI, and its pro-inflammatory actions have been shown to require TLR4 in liver IRI mouse models (16). Our human studies have confirmed liver IRI to be TLR4-driven (3); therefore, we sought to determine if pro-inflammatory HMGB1 was mobilized in response to IRI in liver transplant patients. Similar to prior reports (22), we found that HMGB1 levels are increased following reperfusion in OLT (LF sample); however, this did not correlate to biopsy-proven patient IRI status. We provide the first evidence that in human OLT, IRI+ recipient LF contained a significantly increased amount of disulfide-HMGB1, which corresponded to the degree of IRI severity, the amount of TNFα secreted by LF-stimulated macrophages as well as activation of TLR4. Therefore, we propose the partially oxidized disulfide-HMGB1 redox form is a more relevant mechanistic biomarker and therapeutic target than total HMGB1 in OLT-IRI, and likely other clinical states involving sterile inflammation.
Our data shows that TLR4 is increased in IRI+ liver grafts and is co-localized with HMGB1 in the cytoplasmic vesicles of macrophages within the allograft. However, the precise mechanism of TLR4 activation by HMGB1 remains to be clarified. Direct binding of HMGB1 to TLR4 (14) or LPS-mediated binding (23) are observed with CD14 (15), or MD2 proposed to be necessary (24, 25) for the latter. Interestingly, in our human studies HMGB1 did not require either MD2 or CD14 or even LPS, as the addition of polymyxin B (PMB) did not alter the ability of patient LF or control disulfide-HMGB1 to activate TLR4 (3) or stimulate TNFα from monocytes. Despite these conflicting results, the affinity between HMGB1 and TLR4 is very low (14), indicating one or more binding partners are required to enhance this association. It is possible that there is an alternate high-affinity receptor (or co-receptor) for HMGB1 on the surface of macrophages for TLR activation, and scavenger receptors have been proposed in this capacity (26), specifically class A scavenger receptors MSR1 and MARCO (20). TLR4 signals that are activated by HMGB1/scavenger receptors or HMGB1 with other co-receptors may be different in OLT-IRI than those activated by LPS, as it was shown that MARCO uptake of HMGB1 led to neuroprotection by the M2 macrophages generated, whereas uptake by TLR4 alone resulted in the generation of M1 macrophages secreting pro-inflammatory cytokines (26). Our study uncovers the role of the disulfide-HMGB1/TLR4/TNFα axis in driving the OLT-IRI-generated sterile inflammation response towards an adaptive immunity, but further characterization of the precise molecules involved in this pathway is warranted.
A limitation of this clinical study is that we were unable to determine the originating cellular source, or specific timing and/or localization of HMGB1 oxidation. It is well established that IR-stressed hepatocytes translocate their nuclear HMGB1 into the cytoplasm (7), and we saw this effect in patient allografts as well. Therefore, it is reasonable to speculate injured hepatocytes are a major cellular source of extracellular HMGB1 found in patient blood. However, we found no evidence to support disulfide-HMGB1 originating from within hepatocytes, as cytoplasmic HMGB1 found in hepatocytes was diffuse rather than localized into LAMP1+ vesicles. We also cannot exclude the possibility that non-disulfide HMGB1 originating from damaged hepatocytes is partially oxidized into disulfide-HMGB1 in the extracellular space by ROS generated under IRI. However, high oxidative environments such as this have been shown to produce a terminally oxidized, sulfonyl, non-immunogenic form of HMGB1 (11), making this more likely to be a mechanism for initiation of IRI rather than a mechanism for driving IRI once it has begun.
We found that pre-transplant donor allografts contained CD68+ myeloid cells with HMGB1 restricted primarily in LAMP1+ cytoplasmic vesicles rather than the nucleus, indicating that these cells were generated initially as a response to ischemic conditions resulting from organ procurement. Disulfide-HMGB1-containing CD68+ cells were significantly increased in IRI+ allografts post-reperfusion, suggesting that IRI+ recipient blood flow into the organ amplified the initial ischemic response. HMGB1 is important in perpetuating inflammatory amplification loops in other disease settings such as infection (21); therefore, it is possible that disulfide-HMGB1, either alone or in concert with other binding partners, induces a self-perpetuating cycle in OLT-IRI by activating macrophages to secrete more disulfide-HMGB1. In agreement with this concept, the addition of exogenous disulfide-HMGB1 to freshly isolated monocytes caused them to translocate hyperacetylated HMGB1 into cytoplasmic vesicles within 2 hours, whereas the addition of all-thiol HMGB1 did not initiate any further translocation out of the nuclear compartment. We also found macrophages in donor allografts of IRI+ patients with translocated HMGB1 in their LAMP1+ vesicular compartment that increased after reperfusion, indicating these macrophages are differentially activated from those found in IRI- patient biopsies who predominantly contained nuclear HMGB1. Because our in vitro experiments with monocytes treated with LF from IRI+ patients resulted in the upregulation of several pro-inflammatory molecules, including HLA-DR, CD80 and CD86, it is reasonable to presume a comparable liver graft macrophage activation profile following stimulation with patient LF over the course of the first week post-transplant. Attenuating the activities of these specific cells may be a preferable therapeutic approach towards mitigating hepatic IRI and improving liver transplant clinical outcomes.
The release of HMGB1 by LPS-challenged macrophages is dependent on the activation of PARP-1 (33), which ultimately regulates the translocation of HMGB1 to the cytoplasm through upregulating the acetylation of HMGB1 by elevating the activity ratio of histone acetyltransferases (HATs) to deacetylases (HDACs), which can catalyze acetylation and deacetylation of HMGB1 (19). GTF3C4 can act as a HAT by interacting with histone H3 (34), a level of epigenetic control which was shown to regulate Th cell differentiation into effector phenotypes (35). Our RNAseq analysis identified increased expression of GTF3C4 in IRI+ patient allografts, making this a potential regulator of IRI and downstream effector differentiation of CD4+ T cells via HMGB1 hyperacetylation in macrophages in which HMGB1 has also been described as having a role in histone H3 activity and these two proteins are likely to be in regular close contact. HDAC4 and 5 (36, 37), have been implicated in ischemic brain injury and our mouse studies have suggested that the HDAC Sirtuin 1 (Sirt1) in mediating liver IRI (38). In our screening of RNAseq results for HDACs related to human OLT-IRI, HDAC5 was found to be increased in IRI+ patients post-reperfusion, whereas HDAC4 was significantly decreased and Sirt1 was similarly expressed between IRI+/− OLT recipients. These data indicate that, in human OLT recipients, HDAC5 is the major HDAC needed to deacetylate HMGB1 and constrain it within the nuclear compartment for regaining cellular homeostasis. A larger cohort with more representative cases across all IRI severities will be necessary to assess correlation with IRI severity. Taken together, these results indicate that the combined effect of increased GTF3C4 and decreased HDAC5 contribute to overall hyperacetylation of HMGB1 in macrophages of IRI+ patients, which drives it into cytoplasmic vesicles for secretion as a pro-inflammatory cytokine capable of self-perpetuating clinical or sub-clinical levels of inflammation and possibly an initiation of adaptive immune responses via TLR4 signaling.
Ischemia-reperfusion injury is not limited to solid organ transplantation, with aspects of the condition occurring in many other surgical situations and health conditions. However, it is currently an unavoidable consequence of the transplantation procedure, allowing us the unique opportunity to investigate the mechanisms of HMGB1-stimulated pro-inflammatory macrophage activation in a powered patient cohort. We find that the disulfide redox state of HMGB1 is responsible for mediating IRI and its subsequent switch from innate to adaptive immune activation via TLR4 signaling in macrophages found in the allograft. Further, this redox state of HMGB1 stems from macrophages activated to hyperacetylate HMGB1, thereby resulting in its translocation from the nucleus into the cytoplasmic vesicles for secretion upon further stimulation. The histone acetyltransferase GTF3C4 and deacetylase HDAC5 appear to work in concert to promote translocation and partial oxidation of HMGB1. Therefore, disulfide-HMGB1, GTF3C4, and HDAC5 comprise a group of molecules that could not only serve as putative mechanistic biomarkers, but as targets for future therapeutic development aimed at decreasing the occurrence of IRI in the human OLT setting as well as other types of sterile inflammatory conditions.
METHODS
Study design and sample collection
Adult primary orthotopic liver transplant (OLT) recipients were recruited between May 10, 2013 and July 28, 2018 (Tables 1–3). Routine standard-of-care and immunosuppressive therapy were administered as specified by UCLA liver transplant protocols. All studies described were reviewed and approved by the UCLA Institutional Research Board (IRB #13–000143). Patients gave written informed consent prior to their participation in the study. Study data were collected and managed using REDCap electronic data capture tools hosted at UCLA (39). Donor organs were procured from both donation after brain death (DBD) and donation after circulatory death (DCD) donors using standardized techniques. No donor organs were obtained from executed prisoners or other institutionalized persons. Organs were perfused with and stored in cold University of Wisconsin solution (ViaSpan; Bristol-Meyers Squibb Pharma, Garden City, NY). Cold ischemia time was defined as the time from the perfusion of the donor with preservation solution to the removal of the liver from cold storage. Tru-Cut needle biopsies were taken of donor allografts approximately two hours prior to transplantation (PRE biopsies). Recipient venous blood was collected with acid-citrate-dextrose (ACD) anti-coagulant during two main phases relative to the transplant: pre-operative (PO), and post-operative at 1 day (D1), 1 week (W1) and 1 month (M1). Intra-operative portal blood was collected from the recipient portal vein prior to reperfusion (PV) and as it was first flushed through the vena cava of the donor liver during reperfusion (LF). Protocol Tru-Cut needle biopsies were taken from the left lobe intra-operatively after complete revascularization of the allograft (two hours post-reperfusion; POST biopsies) prior to surgical closing of the abdomen and graded for IRI as previously described (2).
OLT recipient and donor characteristics
Ninety-two (92) OLT recipients were scored for IRI by histopathology using intra-operative post-reperfusion biopsies of the liver allograft. The demographic data and clinical parameters of the recipients are shown in Table 1, donors are shown in Table 2, and combined recipient/donor parameters for the transplant are shown in Table 3. Subcohort selection was necessary in some assays as described in the figure legends, due to limited patient sample quantities. For correlation between demographic data and IRI, the student’s T-test was used for continuous variables, and the Fisher’s exact test was used for categorical variables. There was no correlation between IRI status and any of the parameters for the overall cohort or any subcohorts used except bilirubin levels at the opening of the post-transplant phase, approximately 8 hours post-reperfusion, which were significantly increased in IRI+ patients (Table 1).
Table 2 –
Donor Characteristics
| Clinical and Demographic Data | (n=92) | IRI− (n=46) | IRI+ (n=46) | |
|---|---|---|---|---|
|
| ||||
| DONOR: | ||||
|
| ||||
| Characteristic | Value | Value | Value | P-value* |
| Age, years [mean±SD] | 40±17 | 40±17 | 40±16 | 0.97 |
| Gender [n (%)] | 0.68 | |||
| Female | 41 (45) | 22 (48) | 19 (41) | |
| Male | 51 (55) | 24 (52) | 27 (59) | |
| Race [n (%)] | 0.27 | |||
| Asian | 8 (8.7) | 2 (4.4) | 6 (13) | |
| Black/African American | 8 (9) | 4 (9) | 4 (9) | |
| White/Caucasian | 46 (50) | 27 (59) | 19 (41) | |
| Other | 30 (33) | 13 (28) | 17 (37) | |
| Ethnicity [n (%)] | 0.37 | |||
| Hispanic/Latino | 29 (31.5) | 12 (26) | 17 (37) | |
| Non-Hispanic/Latino | 62 (67) | 33 (72) | 29 (63) | |
| Other | 1 (1) | 1 (2) | 0 (0) | |
| Status [n (%)] | 0.40 | |||
| DBD | 67 (73) | 31 (67) | 36 (78) | |
| DCD | 4 (4) | 3 (7) | 1 (2) | |
| Unknown | 21 (23) | 12 (26) | 9 (20) | |
| Warm Ischemia, minutes [mean±SD] | 53±14 | 52±12 | 54±17 | 0.40 |
| Cold Ischemia, hours [mean±SD] | 7.4±1.8 | 7.2±1.6 | 7.6±2.0 | 0.24 |
| Cause of Death [n (%)] | 0.61 | |||
| trauma | 34 (37) | 18 (39) | 16 (35) | |
| CVS | 29 (32) | 13 (28) | 16 (35) | |
| anoxia | 27 (29) | 13 (28) | 14 (30) | |
| other | 2 (2) | 2 (4) | 0 (0) | |
| DM [n (%)] | 9 (10) | 4 (9) | 5(11) | 1.00 |
| HTN [n (%)] | 23 (25) | 10 (22) | 13 (29) | 0.48 |
| CAD [n (%)] | 4 (4) | 2 (4) | 2 (4) | 1.00 |
| LFTs, at procurement [mean±SD] | ||||
| ALT | 64±116 | 42±54 | 86±152 | 0.07 |
| Bilirubin | 1.0±0.7 | 0.9±0.6 | 1.1±0.8 | 0.12 |
HMGB1 detection in patient plasma samples
HMGB1 ELISA (IBL International) was performed according to manufacturer’s instructions. To determine the redox state of HMGB1, OLT patient LF samples were run undiluted using gel electrophoresis and detected with anti-HMGB1 polyclonal Ab (Abcam). Ratio of 25kD band (bottom band, disulfide-HMGB1) 28 kD band (top band, all-thiol HMGB1) was quantified using ImageJ (NIH). The ratio of all-thiol:disulfide-HMGB1 determined by Western blot analysis was used to extrapolate the amount of each redox form of HMGB1 present in patient LF samples by dividing the total HMGB1 content (as determined by ELISA) by each redox form’s relative amount for each patient sample.
Human primary monocyte isolation, immune phenotyping and functional cytokine assay
Monocytes were enriched from healthy donor blood using negative selection with RosetteSep technology (StemCell Tech) at a final purity of >92.6%. For monocyte stimulation, 30,000 cells were exposed to either RPMI-1640 media with 10% FBS alone or 10ng/ml M-CSF (R&D Systems), 50 ng/ml LPS (Invivogen), 200 ng/ml all-thiol HMGB1 (HMGBiotech), 200 ng/ml disulfide-HMGB1 (HMGBiotech), or with 10% patient LF samples and incubated at 37° for 2, 4, 8, or 24 hours or 3 days.
TNFα ELISA (R&D systems) was performed according to manufacturer’s instructions on supernatants from monocytes stimulated with purified HMGB1 in either the all-thiol or disulfide redox state, and compared to the well-characterized canonical TLR4-ligand lipopolysaccharide (LPS) to determine the HMGB1-mediated timing of monocyte activation and TNFα secretion (Fig. S5A), and the functional threshold of disulfide-HMGB1 (Fig. S5B).
For confocal microscopy, cells were incubated with primary antibodies for CD68 (BioLegend), HMGB1, LAMP1, or acetyl-lysine (Abcam), then appropriate fluorescence-conjugated secondary antibodies and DAPI. Cells were imaged in optically-clear wells (Ibidi, Fitchburg, WI), with liquid mounting media (Ibidi).
Monocyte phenotypes were assessed by flow cytometry using fluorochrome-labeled detection antibodies: CD14-BV785 (clone M5E2),CD16-BV510 (clone 3G8), CD11b-PCP/Cy5 (clone ICRF44), CD68-PE/Cy7 (clone Y1/82A), CD86-BV605 (clone IT2.2), HLA-DR-BV650 (clone L243), Gal-9-FITC (clone 9M1–3), TIM-3-APC (clone F38–2E2), and TIM-4-PE (clone 9F4) from Biolegend; CD66a-AF700 (clone COL-1) from Novus Biologicals; and CD80-APC/H7 (clone L307.4) and PD-L1-PECF594 (clone MIH1) from BD Biosciences. Samples were run on an LSR Fortessa (BD Biosciences, San Jose, CA) within 7 days of fixation. Data were acquired with FACSDiva software (BD Biosciences, San Jose, CA), and analyzed using FlowJo software (TreeStar).
TLR4-activation functional assay
Human (h)TLR4-specific HEK-Blue™ reporter cells (InvivoGen, San Diego, CA), were grown, maintained and utilized as previously described (3). Neutralizing antibodies (InvivoGen) were added for one hour prior to cell stimulation with patient LF to block the function of TLR4, CD14, and/or MD2.
Hierarchical clustering analysis
Abundances irrespective of conditions were normalized using robust Z-scaling with median and median absolute deviation. Then, medians of scaled values for each group were color-coded and plotted in heat maps. Unsupervised hierarchical clustering was performed on rows and columns using Euclidean distance as the similarity measure with Ward’s linkage.
Immunohistochemical and immunofluorescence staining of allograft biopsies
For immunohistochemical (IHC) staining, Formalin-fixed, paraffin-embedded biopsy sections were stained with primary antibody to CD68, detected using labelled HRP (Agilent), and visualized with diaminobenzidine (DAB), then counterstained with hematoxylin. Whole stained slides were converted to high resolution digital brightfield images using an Aperio ScanScope AT high throughput scanning system (Leica Biosystems). Images were then acquired using Aperio ImageScope software (Leica Biosystems) and analysis was performed using Tissue Studio software (Definiens).
For immunofluorescence (IF) staining, allograft biopsies were fixed in phosphate-buffered 4% paraformaldehyde (PFA), then impregnated with 30% sucrose. Samples were embedded in gelatin blocks then OCT-embedded and cryosectioned. Immunofluorescence of tissue sections was performed as previously described (40, 41). Sections were then incubated with primary antibodies for CD68 (Biolegend), HMGB1 (Abcam), LAMP1 (Abcam), or acetyl-lysine (a-K; Abcam), then incubated with appropriate fluorescence-conjugated secondary antibodies and DAPI. Sections were mounted on a glass slide with Prolong Gold Antifade, coverslipped and allowed to dry/cure overnight.
Confocal laser scanning microscopy of immunofluorescence staining
Samples were observed and imaged using three-dimensional confocal laser scanning microscopy as previously described (40, 41). In brief, immunofluorescence images were acquired using a scanning confocal laser scanning microscope (LSM 880, Zeiss) using Zen Black 2009 software.
Reconstructing HMGB1 subcellular localization within macrophages in three dimensions
The three-dimensional structure of macrophages in the allograft was reconstructed from confocal z-stacks using Imaris 9.2 (Bitplane). Digital isolation of macrophages present in the tissue sample was performed for each image as previously described (40, 41). To verify the localization of labeled proteins at the subcellular level, liver biopsy sections were counterstained with the nuclear dye DAPI and/or the lysosomal marker LAMP1 and nuclear or cytoplasmic/lysosomal localization of HMGB1 was determined based on the colocalization with contents of the DAPI+ and/or LAMP1+ compartment remodeled in three dimensions as described in Fig. S3.
RNAseq analysis
RNAseq testing was performed by the UCLA Technology Center for Genomics and Bioinformatics (TCGB) and analyzed as previously described (2). Transcript abundances are expressed as transcripts per million (TPM) (42) log2 scale. Data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE87487.
Statistics
Two-way analysis of variance (ANOVA) with Sidak’s multiple comparisons test was used to determine differences amongst time and/or IRI status unless otherwise indicated. A P-value less than 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the BSCRC Imaging Core, as well as thank Gregg Kunder, Stephanie Younan, and the liver transplant team who go above and beyond to collect patient samples for this project. The authors hereby express their thanks for all of the organ donors and their families, for giving the gift of life and the gift of knowledge, by their generous donation.
This work was supported by:
NIH Ruth L. Kirschstein National Research Service Award T32CA009120 (RAS), NIH L60 MD011903 (RAS), NIH PO1 AI120944 (JWKW and EFR), and UL1TR001881 (UCLA Clinical and Translational Science Institute)
Abbreviations:
- Ab
antibody
- AU
absorbance units
- CD
cluster of differentiation
- DAMP
danger-associated molecular pattern
- ELISA
enzyme-linked immunosorbent assay
- GTF3C4
General Transcription Factor IIIC Subunit 4
- HAT
histone acetyl transferase
- HDAC
histone deacetylase
- HMGB1
high-mobility group box 1
- IFN
interferon
- IL
interleukin
- IRI
ischemia-reperfusion injury
- LF
liver flush
- mAb
monoclonal antibody
- MPO
myeloperoxidase
- mRNA
messenger RNA
- NF-KB
nuclear factor-KB
- OLT
orthotopic liver transplantation
- PRR
pattern recognition receptor
- PBMC
peripheral blood mononuclear cells
- PV
portal vein blood
- TLR
toll-like receptor
- TNFα
tumor necrosis factor
Footnotes
COMPETING INTERESTS STATEMENT
The authors have no competing interests to disclose.
REFERENCES
- 1.Matzinger P Tolerance, danger, and the extended family. Annu Rev Immunol 1994;12:991–1045. [DOI] [PubMed] [Google Scholar]
- 2.Sosa RA, Zarrinpar A, Rossetti M, Lassman CR, Naini BV, Datta N, Rao P, et al. Early cytokine signatures of ischemia/reperfusion injury in human orthotopic liver transplantation. JCI Insight 2016;1:e89679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sosa RA, Rossetti M, Naini BV, Groysberg VM, Kaldas FM, Busuttil RW, Chang YL, et al. Pattern Recognition Receptor-reactivity Screening of Liver Transplant Patients: Potential for Personalized and Precise Organ Matching to Reduce Risks of Ischemia-reperfusion Injury. Ann Surg 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoppe G, Talcott KE, Bhattacharya SK, Crabb JW, Sears JE. Molecular basis for the redox control of nuclear transport of the structural chromatin protein Hmgb1. Exp Cell Res 2006;312:3526–3538. [DOI] [PubMed] [Google Scholar]
- 5.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. Embo j 2003;22:5551–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002;418:191–195. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, Nace GW, McDonald KA, Tai S, Klune JR, Rosborough BR, Ding Q, et al. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology 2014;59:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosakote YM, Brasier AR, Casola A, Garofalo RP, Kurosky A. Respiratory Syncytial Virus Infection Triggers Epithelial HMGB1 Release as a Damage-Associated Molecular Pattern Promoting a Monocytic Inflammatory Response. J Virol 2016;90:9618–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi A, Shimizu M, Shinya E, Takahashi H. HMGB1 released from intestinal epithelia damaged by cholera toxin adjuvant contributes to activation of mucosal dendritic cells and induction of intestinal cytotoxic T lymphocytes and IgA. Cell Death Dis 2018;9:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leavy O HMGB1-mediated inflammatory cell recruitment. Nature Reviews Immunology 2012;12:232. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Lundback P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, et al. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol Med 2012;18:250–259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Venereau E, Casalgrandi M, Schiraldi M, Antoine DJ, Cattaneo A, De Marchis F, Liu J, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med 2012;209:1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardella S, Andrei C, Ferrera D, Lotti LV, Torrisi MR, Bianchi ME, Rubartelli A. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep 2002;3:995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Hreggvidsdottir HS, Palmblad K, Wang H, Ochani M, Li J, Lu B, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A 2010;107:11942–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Kim SY, Pribis JP, Lotze M, Mollen KP, Shapiro R, Loughran P, et al. Signaling of high mobility group box 1 (HMGB1) through toll-like receptor 4 in macrophages requires CD14. Mol Med 2013;19:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsung A, Sahai R, Tanaka H, Nakao A, Fink MP, Lotze MT, Yang H, et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med 2005;201:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsung A, Klune JR, Zhang X, Jeyabalan G, Cao Z, Peng X, Stolz DB, et al. HMGB1 release induced by liver ischemia involves Toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. The Journal of experimental medicine 2007;204:2913–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen XL, Sun L, Guo F, Wang F, Liu S, Liang X, Wang RS, et al. High-mobility group box-1 induces proinflammatory cytokines production of Kupffer cells through TLRs-dependent signaling pathway after burn injury. PLoS One 2012;7:e50668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Li L, Chen L, Yuan W, Dong L, Zhang Y, Wu H, et al. PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation. J Immunol 2014;193:6114–6123. [DOI] [PubMed] [Google Scholar]
- 20.Shichita T, Ito M, Morita R, Komai K, Noguchi Y, Ooboshi H, Koshida R, et al. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat Med 2017;23:723–732. [DOI] [PubMed] [Google Scholar]
- 21.Gougeon ML, Melki MT, Saidi H. HMGB1, an alarmin promoting HIV dissemination and latency in dendritic cells. Cell Death Differ 2012;19:96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilmakunnas M, Tukiainen EM, Rouhiainen A, Rauvala H, Arola J, Nordin A, Makisalo H, et al. High mobility group box 1 protein as a marker of hepatocellular injury in human liver transplantation. Liver Transpl 2008;14:1517–1525. [DOI] [PubMed] [Google Scholar]
- 23.Youn JH, Oh YJ, Kim ES, Choi JE, Shin JS. High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-alpha production in human monocytes. J Immunol 2008;180:5067–5074. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Wang H, Ju Z, Ragab AA, Lundback P, Long W, Valdes-Ferrer SI, et al. MD-2 is required for disulfide HMGB1-dependent TLR4 signaling. J Exp Med 2015;212:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troseid M, Lind A, Nowak P, Barqasho B, Heger B, Lygren I, Pedersen KK, et al. Circulating levels of HMGB1 are correlated strongly with MD2 in HIV-infection: possible implication for TLR4-signalling and chronic immune activation. Innate Immun 2013;19:290–297. [DOI] [PubMed] [Google Scholar]
- 26.Komai K, Shichita T, Ito M, Kanamori M, Chikuma S, Yoshimura A. Role of scavenger receptors as damage-associated molecular pattern receptors in Toll-like receptor activation. Int Immunol 2017;29:59–70. [DOI] [PubMed] [Google Scholar]
- 27.Watson M, Stott K, Fischl H, Cato L, Thomas JO. Characterization of the interaction between HMGB1 and H3-a possible means of positioning HMGB1 in chromatin. Nucleic acids research 2014;42:848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellgaard L Catalysis of disulphide bond formation in the endoplasmic reticulum. Biochem Soc Trans 2004;32:663–667. [DOI] [PubMed] [Google Scholar]
- 29.Sahu D, Debnath P, Takayama Y, Iwahara J. Redox properties of the A-domain of the HMGB1 protein. FEBS Lett 2008;582:3973–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamkanfi M, Sarkar A, Vande Walle L, Vitari AC, Amer AO, Wewers MD, Tracey KJ, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol 2010;185:4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai Y, Shen XD, O’Connell R, Gao F, Lassman C, Busuttil RW, Cheng G, et al. Cutting edge: TLR4 activation mediates liver ischemia/reperfusion inflammatory response via IFN regulatory factor 3-dependent MyD88-independent pathway. J Immunol 2004;173:7115–7119. [DOI] [PubMed] [Google Scholar]
- 32.Oyama J, Blais C Jr., Liu X, Pu M, Kobzik L, Kelly RA, Bourcier T. Reduced myocardial ischemia-reperfusion injury in toll-like receptor 4-deficient mice. Circulation 2004;109:784–789. [DOI] [PubMed] [Google Scholar]
- 33.Davis K, Banerjee S, Friggeri A, Bell C, Abraham E, Zerfaoui M. Poly(ADP-ribosyl)ation of high mobility group box 1 (HMGB1) protein enhances inhibition of efferocytosis. Mol Med 2012;18:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh YJ, Kundu TK, Wang Z, Kovelman R, Roeder RG. The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol Cell Biol 1999;19:7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falvo JV, Jasenosky LD, Kruidenier L, Goldfeld AE. Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv Immunol 2013;118:37–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He M, Zhang B, Wei X, Wang Z, Fan B, Du P, Zhang Y, et al. HDAC4/5-HMGB1 signalling mediated by NADPH oxidase activity contributes to cerebral ischaemia/reperfusion injury. Journal of cellular and molecular medicine 2013;17:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evankovich J, Cho SW, Zhang R, Cardinal J, Dhupar R, Zhang L, Klune JR, et al. High mobility group box 1 release from hepatocytes during ischemia and reperfusion injury is mediated by decreased histone deacetylase activity. J Biol Chem 2010;285:39888–39897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura K, Zhang M, Kageyama S, Ke B, Fujii T, Sosa RA, Reed EF, et al. Macrophage heme oxygenase-1-SIRT1-p53 axis regulates sterile inflammation in liver ischemia-reperfusion injury. J Hepatol 2017;67:1232–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosa RA, Murphey C, Robinson RR, Forsthuber TG. IFN-gamma ameliorates autoimmune encephalomyelitis by limiting myelin lipid peroxidation. Proc Natl Acad Sci U S A 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sosa RA, Murphey C, Ji N, Cardona AE, Forsthuber TG. The Kinetics of Myelin Antigen Uptake by Myeloid Cells in the Central Nervous System during Experimental Autoimmune Encephalomyelitis. Journal of Immunology 2013;191:5848–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2010;26:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.