Abstract
Metabolic glycan probes have emerged as an excellent tool to investigate vital questions in biology. Recently, methodology to incorporate metabolic bacterial glycan probes into the cell wall of a variety of bacterial species has been developed. In order to improve this method, a scalable synthesis of the peptidoglycan precursors is developed here allowing for access to essential peptidoglycan immunological fragments and cell wall building blocks. The question was asked if masking polar groups of the glycan probe would increase overall incorporation, a common strategy exploited in mammalian glycobiology. Here, we show through cellular assays that E. coli do not utilize peracetylated peptidoglycan substrates yet do employ methyl esters. The 10-fold improvement of probe utilization indicates that masking the carboxylic acid is favorable for transport and that bacterial esterases are capable of removing the methyl ester for use in peptidoglycan biosynthesis. This investigation advances bacterial cell wall biology, offering a prescription on how to best deliver and utilize bacterial metabolic glycan-probes.
Keywords: bacterial peptidoglycan, cell wall recycling, carbohydrate probes, metabolic labeling, esterases
Graphical Abstract
Cell walls are vital components for bacteria, providing them protection against environmental insults such as changes in osmotic pressure or small molecule threats.1 A major component of the bacterial cell wall envelope is the bacterial peptidoglycan (PG), an essential mesh-like polymer that bacteria assemble with units of N-acetylglucosamine (NAG) and N-acetyl muramic acid (NAM) around their cytosolic membrane.2 This carbohydrate backbone is connected through β-1,4-linkages and further crosslinked through complex d- and l-amino acid-containing peptide chains from the 3-position of NAM (Figure 1).2 This architecture along with modifications around the muropeptide scaffold render the dynamic and protective properties of this essential microbial macromolecule against turgor, the host immune system, and antibiotics.3, 4 To gain insight into the causative mechanism of bacterial-related diseases, inflammatory disorders, and in turn to design novel antibiotics capable of targeting this polymer,5 it is imperative to understand how fragments naturally generated from this molecule interact at the molecular level with the immune system.6 Therefore, the development of labeling strategies for PG on the carbohydrate backbone are invaluable, enabling the study of the PG biosynthetic process and the identification of key structural features via fluorescent imaging and spectrometric techniques.7
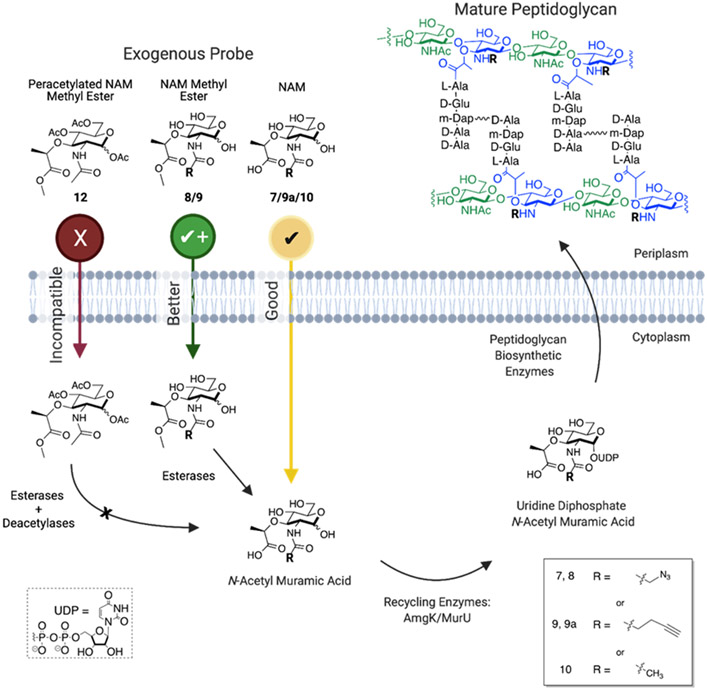
Figure 1: Metabolic and Recycling Pathways of N-acetyl-Muramic Acid Probes.
The NAM methyl ester (azide (8) and alkyne (9)) and peracetylated NAM methyl ester (12) compounds are proposed to incorporate more efficiently than NAM (alkyne (7) and azide (9a)) by masking the polar and negatively charged hydroxyls and carboxylic acid, respectively. Cellular esterases and deacetylases would then deprotect the probe in the cytoplasm. The unmasked NAM probe is converted to the requisite UDP-NAM PG building block. UDP-NAM is then incorporated in the bacterial PG by the biosynthetic enzymes. Bioorthogonal probes are presented within the NAM probe by providing handles such as azide or alkyne on the N-acetyl position of the carbohydrate. Created with BioRender.com
We previously reported methodology for metabolic PG labeling by utilizing glycan-probes bearing a bioorthogonal handle at the 2-amino position of the NAM (7 and 9a, Figure 1).8-10 We found that derivatization at this position of the NAM was an acceptable modification for the biosynthetic and recycling enzymes, permitting NAM containing bioorthogonal tags to be displayed on the surface of both Gram-positive and Gram-negative bacteria (Figure 1). It is worth highlighting that the method uses the enzymes NAM/NAG anomeric kinase (AmgK) and NAM α-1-phosphate uridylyltransferase (MurU) first identified by the Mayer group in Pseudomonas putida.11 These recycling enzymes circumvent the difficult synthetic preparation of uridine diphosphate (UDP)-NAM sugars, which are challenging to isolate and carry two negative charges at physiological pH, making it difficult to cross the bacterial membrane(s) (Figure 1). It is also noteworthy that these glycan probes complement strategies labeling PG via the d-amino acids of the peptide.12-20 E. coli have multiple pathways to uptake PG fragements.21 For example, some bacteria use the AmpG permease to bring anhydro-muropeptides through the membrane while others can take up NAM (10, Figure 1) via MurP, which simultaneously phosphorylates the compound at the 6-position and ultimately produces UDP-MurNAc. Subsequently, MurNAc 6-phosphate is converted to GlcNAc 6-phosphate by MurQ etherase. From here the GlcNAc can be rerouted into PG biosynthesis or channeled into glycolysis. The recombinant E. coli (EQKU) cells utilized here lack the MurQ etherase, which permits processing of the probe solely through the inserted recycling enzymes AmgK and MurU (Figure 1) for incorporation into mature PG.
The previously reported NAM probes (7 and 9a, Figure 1) have been used in a series of biological experiments to illuminate the recently identified bacterial glycosyltransferase, FtsW,22, 23 and define zones of enhanced cell wall synthesis in Helicobacter pylori.24 However, we wondered if the method could be improved by simplifying the synthesis of the critical 2-amino precursor (6, Scheme 1) and enhancing the probe delivery (Figure 1). The latter is commonly addressed in mammalian glycobiology through acetylation of the carbohydrate probe to mask polar groups; these groups are then deprotected by cellular esterases/deacetylases in the cytoplasm.25, 26 Recently, Dube and coworkers, have shown that acetylated glycan probes could be used to assess the glycosylation patterns of secreted colonization factors in H. pylori,27 suggesting that this could be a strategy to improve uptake and subsequent incorporation of the N-acetyl-muramic acid probes. Here, a multigram synthesis of the N-acetyl-muramic acid probe precursor is reported and a detailed study exploring the permissibility of acetylated PG probes is presented. Ultimately, the data show that bacteria have a limited set of esterases/deacetylases and one must be careful about which groups are masked on the glycan, as esterases/deacetylases are not universal across mammalian and bacterial biology. For the N-acetyl-muramic acid probes, it is found that acetate protecting groups on the hydroxyl groups are not tolerated whereas the negative charge on the carboxylic acid can be masked with an ester (Figure 1). Through microscopy and mass spectrometry assays, we established that E. coli and other species do not utilize peracetylated derivatives of N-acetyl-muramic acid, suggesting that the specific cellular esterases/deacetylases are not able to unmask the precursors. Yet, the data show methyl esters are tolerated by the system and removed by an esterase. By simply masking the negative charge on the carboxylic acid on the substrate, similar levels of cell wall labeling were achieved using significantly less probe. In optimizing these critical steps, the best production and delivery strategies for the peptidoglycan probes were realized.
Scheme 1.
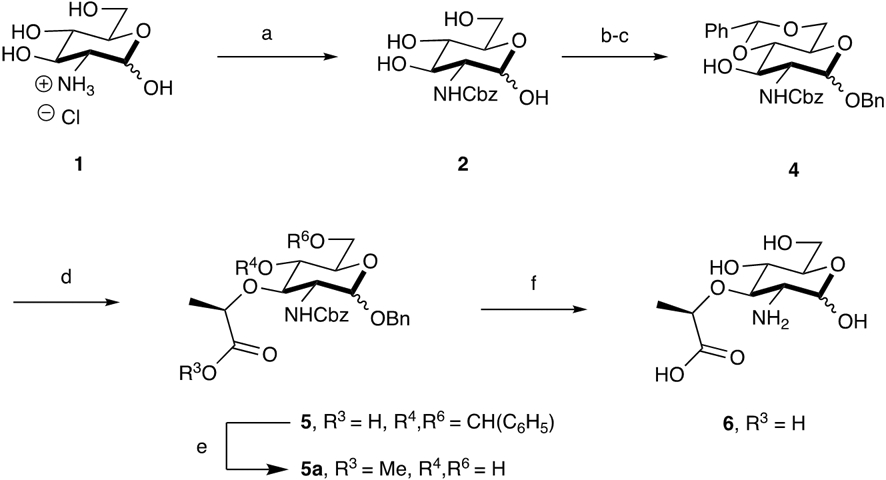
Synthetic Route for the Preparation of 2-amino Muramic Acid: Scalable synthesis of 2-amino-muramic acid reported over 5 synthetic steps in 25% yield on a multigram scale. Reaction Conditions: (a) Cbz-OSu, NaHCO3, 1:1 THF:H2O (80%); (b) BnOH, AcCl, 80°C; (c) PhCH(OMe)2, pTSA, DMF (57% over 2 steps); (d) 60% NaH, (S)-(−)-2-Bromopropionic Acid, THF, −20°C (69%); (e) IRA H+ resin, MeOH, 60°C (58%); (f) H2, Pd(OH)2, 14:5:1 THF:H2O:AcOH (77%)
Results and Discussion:
Modular synthesis of 2-amino-NAM probes
Herein an updated synthetic method that allows modular, scalable production of the 2-amino precursor to the azide and alkyne NAM probes is reported. Previously, an azido transfer reaction was the initial step of the synthesis to access 6 (Scheme 1).8-10 However, the pyrophoric nature of the reactants and intermediates therein limits the scale of this step to 5 grams in the academic setting, which is not adequate for downstream biological applications.28-30 This problem was circumvented by employing a carboxybenzyl (Cbz) protecting group that is labile under palladium catalyzed hydrogenation conditions (Scheme 1). Although others have explored this route of using the Cbz protecting group in combination with a benzyl and benzylidene acetal groups to prepare NAM and muramyl dipeptide (MDP) fragments,31, 32 final products were never isolated nor reported. The route shown in Scheme 1 is an accessible method to produce the Cbz-protected NAM (supporting Information, NMR and high-resolution spectra), with 25% overall yield (all intermediates reported in the Supporting Information).
In this route 2-amino muramic acid, 6, is obtained over five steps. First, commercially available d-(+)-glucosamine hydrochloride (1) and N-(benzyloxycarbonyloxy)succinimide (Cbz-OSu) are used in the initial step to obtain 2. Subsequently, the benzyl ether protecting group is installed (3, Scheme S3) via a Fischer glycosylation where HCl is generated in situ. This is followed by benzylidene acetal protection of the 4,6-diol catalyzed by pTSA (4, Scheme 1). It should be noted that in the lactic acid coupling step, the reaction temperature is essential to acquire 5. If the reaction is carried out at room temperature utilizing sodium hydride, partial removal of the Cbz protecting group is observed. This suggests that the hydride can act as a base and attack the carbonyl carbon of the Cbz on 4, resulting in the removal of the 2-amino protecting group.33 However, by reducing the temperature to −20°C, Cbz removal was prevented and the corresponding lactic acid containing intermediate 5 was obtained in 69% yield. With the lactic acid moiety installed, palladium catalyzed hydrogenation yields the critical intermediate, 2-amino muramic acid, 6, in 77 % yield. 6 can be subsequently derivatized at the 2-amino position using mild acylating conditions. In order to show the utility of the synthesis in preparing PG building blocks/fragments, we pivoted this synthesis at intermediate 5 to prepare muramyl dipeptide (MDP) (19, Scheme S18), a synthetic immunostimulatory fragment ubiquitous in PG from most bacterial species34, as well as N-glycolyl-MDP (20, Scheme S19), a fragment derived from the PG of the pathogen Mycobacterium tuberculosis.35, 36 These fragments are standard compounds when assessing bacterial activation of immune pathways. Recent studies from our lab have shown that it is essential to look beyond the commercially available fragments, as different PG fragments induce the expression of genes distinctively.37 Therefore, the ease of preparing compounds 19 and 20 (Scheme S18 and S19) and the modularity-- deprotection at the 4- and 6-positions for further diversification around the whole sugar scaffold--38 of the synthesis emphasizes the accessibility of preparing an array of bacterial cell wall fragments/subunits, both synthetic and biologically inspired, that are more complex mono- and potentially disaccharides.
While other groups have reported different protecting group strategies at the 2-amino position such as tert-butyloxylcarbonyl (BOC),39 the Cbz offers greater stability and utility in preparation of the NAM (and other PG fragment probes). Moreover, the added hydrophobicity of the Cbz group reduces the number of flash chromatography columns in the purification steps, instead taking advantage of recrystallization and precipitation techniques which allows the 2-amino muramic acid derivative to be produced in an overall 25% yield when starting from 15 grams of d-(+)-glucosamine over 5 steps.
Optimization of NAM probe delivery, utilization and incorporation into bacterial peptidoglycans
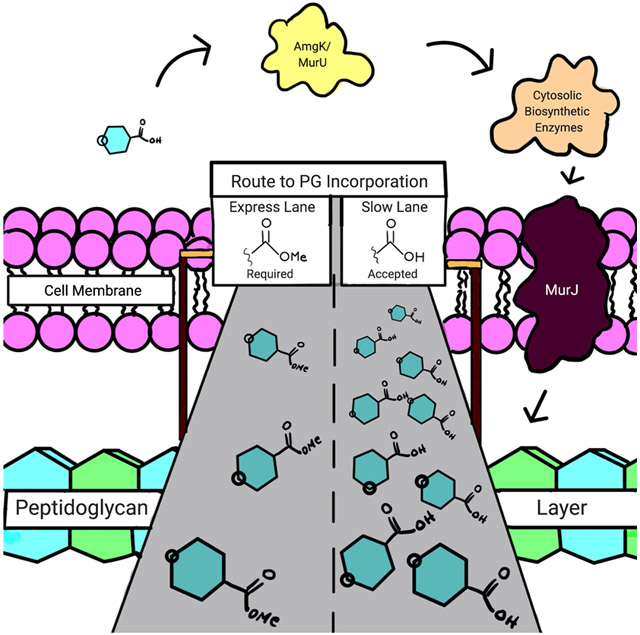
With a robust synthetic route to the NAM probe derivatives in hand, we then sought to improve the metabolic labeling efficiency of the NAM probes in E. coli. In considering mammalian systems (and select bacterial systems) that use glycan metabolic incorporation, the carbohydrate probe’s hydroxyl groups are masked via acetylation to increase membrane permeability.40-42 The polar nature of the hydroxyl containing compounds limits cell uptake across the bilayer in the absence of dedicated transporters. Acetylation and esterification permits the probes to be more readily transported into the cytosol for utilization by biosynthetic enzymes or at least improves metabolic flux of these compounds. Once in the cell, endogenous, cytosolic carboxylic esterases/deacetylases would reveal the hydroxyl groups. We reasoned that this strategy would be useful in reducing the concentration of NAM probe to sufficiently label the PG core structure of bacteria. Previous protocols8 for metabolic labeling of PG utilized 6 mM of glycan probe (azide/alkyne (7 and 9a) though only a fraction of these molecules was incorporated, implying that probe uptake is limiting overall incorporation. The polarity of the N-acetyl-muramic acid probe should be considered in regards to access to the cytoplasm. The probe has three hydroxyl groups and a carboxylic acid group (Figure 1), which at physiological pH will carry a substantial negative charge. In addition, the outer membrane of E. coli adds another barrier through which exogenous probe must traverse. In order to quickly assess if this approach would be useful in E. coli PG remodeling strategies, the peracetylated probe (12, Scheme 2) was prepared. We note that when 10 was subjected to acid catalyzed esterification conditions and then global acetylation with acetic anhydride, a bicyclic ring system formed between the 3-carboxylic acid group and 4-OH (12a, Scheme S13).43 As a result, 10 was converted to an ester at the 3- position and subsequently subjected to acetylating conditions (12, Scheme 2) to avoid obtaining 12a. These compounds were then assessed in a bacterial growth assay using the E. coli ΔMurQ- KU (EQKU cells) model system in which recycling enzymes AmgK and MurU are expressed and can recycle NAM when natural PG biosynthesis is inhibited with the antibiotic fosfomycin (Figure 1);8, 11 in the presence of fosfomycin, bacteria cannot generate de novo NAM (10, Figure 1) and must rely on the exogenous NAM for PG biosynthesis. Previous work shows that the EQKU cell line can rescue growth with 7 and 9a. However, in this work, when the acetylated derivative 12 was used, E. coli growth was stymied (Figure S2); whereas the positive control with non-acetylated NAM 10, E. coli survived under fosfomycin growth conditions (Figure S3). The observed cell death could be attributed to the generated acetate ions as it is known that acetate inhibits bacterial growth at high concentrations. In order to determine if high concentration of acetate ions killed the bacteria, growth curve analyses were performed in the presence of acetate ions. Growth curves were obtained at 2.4, 24 and 240 mM, (the concentration of acetate equivalent to what would be released in the cytosol if all of the acetates were removed at 6 mM of probe and then also at concentrations 10-fold above and 10-fold below). The assay demonstrated that the growth of E. coli was comparable to that of the control at 2.4 and 24 mM, while 240 mM showed a steep decline in growth rate which was expected. Nevertheless, complete death was not observed as was the case with the acetylated probe 12 (Figure S1). It should be noted that the expression of amgk/murU in the E. coli model system (Figure 1) is maintained by chloramphenicol, which is cited to inhibit bacterial esterase function.44 In order to assess if chloramphenicol was inhibiting deacetylation of the probe, P. putida, which harbor amgK and murU in their genomes, were treated with peracetylated compounds. The P. putida cultures did not rebound when incubated overnight with fully acetylated probe (12, Scheme 2). Finally, Helicobacter pylori (an organism that in previous work was amenable to muramic acid probes (7 and 9a) and acetylated NAG probes)27, 45 was assessed for the ability to use the acetylated probe 12; these bacteria also failed to incorporate (data not shown). This led us to propose that the endogenous (cytosolic) carboxylic esterases are able to deacetylate the N-acetyl-glucosamine derivatives but not probe 12. Or if the peracetylated probe is processed, it is unmasked at a rate that is not compliant with the rate of PG turnover. Previous work by Tippmann and coworkers suggested that E. coli do not contain sufficient endogenous esterases to deprotect carbohydrates, supporting the findings that acetylated NAM probes are not utilized in the system.46 To assess if this in the case with acetylated muramic acid, 12 was incubated with E. coli lysates and analyzed via mass spectrometry experiments; in all the experiments only the ester was removed and not the acetate protecting groups (Supporting Information, Table S1, Figures S13-S19). The data suggest that an acetyl protection strategy would not be useful with the N-acetyl-muramic acid PG remodeling method.
Scheme 2:
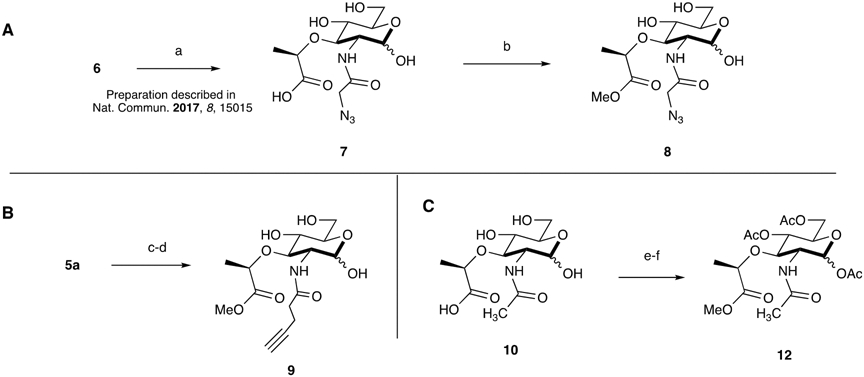
Synthesis of Methyl Ester Azide (A) and Alkyne (B) NAM Probes and Preparation of Peracetylated NAM (C). Reagent conditions: (a) 2-azidoacetic acid NHS, Na2CO3, MeOH; (b) IRA H+ resin, MeOH (18%); (c) 4-pentynoic acid, EDC-HCl, 1:1 H2O:THF (26%); (d) IRA H+ resin, MeOH; (e) Ac2O, Cat. DMAP, pyridine (31% over 2 steps).
Next, a strategy was developed to solely transform the carboxylic acid moiety, which presents as a carboxylate in vivo, to an ester. We have previously shown using a 3-azido muramic ester probe that the system is able to incorporate this substrate into the PG of E. coli.9, 10 Therefore, it appears that there are esterases present within the bacterial cell capable of hydrolyzing the methyl ester functionality from the lactic acid position of NAM. The NAM methyl ester possessing the alkyne functionality at the 2-position was synthesized using ethylcarbodiimide hydrochloride (EDC-HCl) rather than the N-hydroxysuccinimide (NHS) ester, which had been used in the preparation of 9.8 During purification, the residual NHS proved difficult to separate from the final compound, 9; this is evidenced by a singlet peak at ~2.7 ppm in the 1H NMR. Late-stage methylation of 7 and 10 yielded the azide NAM methyl ester, 8, and the peracetylated N-acetyl methyl ester, 12, respectively. These reactions were catalyzed by IRA H+ resin at room temperature. It is essential to carefully monitor the methylation process; addition to the anomeric hemiacetal is also possible if the reaction proceeds for long periods, converting the C1 hydroxyl to a methyl ether. Conversely, the alkyne NAM methyl ester was prepared by refluxing intermediate 5 in methanol with IRA H+ resin at 60 °C, simultaneously removing the benzylidene acetal and installing a methyl group. 5a was subsequently hydrogenated and derivatized with the alkynyl functionality. This strategy was required for the alkyne NAM probe, because the acetal swiftly formed when the anomeric position was unprotected in the presence of IRA H+ resin. The presence of multiple functional groups around the NAM sugar ring makes these simple transformations challenging.
With the two probes in hand, these compounds were then assessed for utilization by the recycling machinery in the E. coli model system. We implemented a 96-well growth curve plate assay to quickly monitor incorporation of probe (Figure 2). Briefly, each probe was monitored using a concentration range from 60 μM to 6 mM for a course of 6 hours. In addition to testing the modified NAM methyl ester probes, the parent compounds (7 (Scheme 2) and 9a (Figure 1)) were assayed for comparison. In both cases, it was found that the methyl ester derivatives incorporated at a lower concentration than the free acid derivatives. The azide NAM (AzNAM) methyl ester (8) showed incorporation at 150 μM, whereas no incorporation at this concentration was observed in the AzNAM (7) probe (Figure 2b, Figure S4). The alkyne NAM (AlkNAM) methyl ester probe (9) incorporated at a lower concentration compared to the parent compound (9a, Scheme S3a) (600 μM vs 1500 μM) but at four-fold higher concentration than the AzNAM (7) (Figure 2C, FigureS5). These data suggest that masking the carboxylic acid is an effective strategy to dampen the negative charge on the NAM and improve uptake, allowing the concentration of probes used in downstream biological experiments to be reduced by up to four-fold. Nonetheless, the bioorthogonal NAM probes are incorporated less efficiently than the natural substrate N-acetyl muramic acid, which can rescue fosfomycin treatment at a concentration as low as 60 μM, suggesting that even the small alkyne and azide modifications hinder utilization by the PG recycling and biosynthetic machinery (Figure 1, Figure S3).
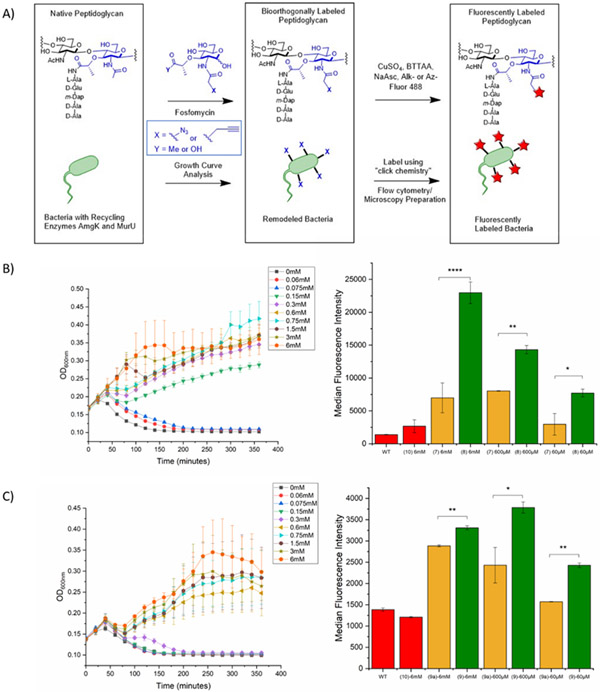
Figure 2. Growth curve and flow cytometry analysis demonstrate that E. coli accept carboxylic acid protected muramic acid probes (8 and 9) and incorporate into cell wall.
A. Overall PG Remodeling Strategy. Fosfomycin treatment selects for bacteria that utilize the recycling and biosynthetic enzymes for the incorporation of bioorthogonal PG building blocks into mature PG. Click chemistry then affords fluorescence modification onto the NAM unit of PG. B. AzNAM methyl ester (8) recovers growth of E. coli cells at concentrations of 150 μM or above. Some incorporation of lower concentration probe is observed over the first hour of growth. Flow cytometry confirms incorporation of probe at 6 mM , 600 μM, and 60 μM ), with a statically higher median fluorescence intensity than the AzNAM free acid probe lacking the methyl ester (7). C. AlkNAM methyl ester (9) recovers growth of E. coli cells at concentrations of 600 μM or above. Some incorporation of lower concentration probe is observed over the first hour of growth. Flow cytometry confirms incorporation of probe at 6 mM,600 μM,and 60 μM, with a statically higher median fluorescence intensity than the AzNAM free acid probe lacking the methyl ester (7). AzNAM (7), AlkNAM (9a) and NAM (10) growth curves and flow cytometry are in the supporting Information. Error bars represent the standard deviation of three biological replicates from one trial. The graphs are representative of the average of triplicates from one of the three biological experiment replicates. Statistical analysis was performed by ANOVA with Tukey’s test (*p < 0.05, **p<0.01, ***p<0.001, ****p<0.0001).
After growth curve analysis, the incorporation of the methyl ester probes was confirmed with a series of experiments. First, mass spectrometry was used to verify the presence of “bioorthogonal” PG products. Briefly, cells were grown and remodeled with the methyl ester probes. Following incorporation, cells were treated with lysozyme to digest and generate the corresponding peptidoglycan fragments. Cells were filtered and mass spectrometry analysis confirmed incorporation into the bacterial PG (Figure S9-S12), as the data show the presence of the major disaccharide compounds as either azide or alkyne modified.
Next, the labeling efficiency of the bioorthogonal containing probes was assessed with flow cytometry by copper (I) catalyzed azide-alkyne cycloaddition (CuAAC) utilizing either an Alk488 or Az488 to the azide and alkyne NAM probes, respectively. In looking at the AzNAM methyl ester (8) data, the median fluorescence intensity was found to be statistically higher than the AzNAM (7) (Figure 2B). For 6 mM of probe, 8 had a median fluorescence intensity value of 22964.5±1639.8 compared to 7’s median fluorescence intensity of 6985±2271 (****p<0.0001). For 600 μM of probe, 8 had a median fluorescence intensity value of 14310±636.40 compared to 7’s median fluorescence intensity of 8037.5±53.0 (**p<0.01). For 60 μM of probe, 8 had a median fluorescence intensity value of 7718.5±593.3 compared to 7’s median fluorescence intensity of 2965.5±92.6 (*p<0.05). This trend was also followed by the AlkNAM methyl ester, 9, probe compared with AlkNAM (9a). For 6 mM of probe, 9 had a median fluorescence intensity value of 3308±48 compared to 9a’s median fluorescence intensity of 2885±23 (**p<0.01). For 600 μM of probe, 9 had a median fluorescence intensity value of 3785±130 compared to 9a’s median fluorescence intensity of 2429.33±418.60 (*p<0.05). For 60 μM of probe, 9 had a median fluorescence intensity value of 2426.5±54.5 compared to 9a’s median fluorescence intensity of 1570±65 (**p<0.01). It should be noted that overall the median fluorescence intensity of the azide modifications is higher than the alkyne modifications, suggesting that the promiscuity of the PG biosynthetic pathway is more amenable to the azide modification than the alkyne modification, specifically at lower concentrations. It also confirms that the use of the methyl ester modification on the lactic acid at the 3-position of the NAM allows the probe to be incorporated at a higher degree when concentration is decreased.
Confocal microscopy was also performed on the samples showing incorporation of the probe into the cell wall. However, we note the quality of fluorescence imaging decreases significantly following a decrease in probe concentration (Figure S6). This could potentially be due to the decreased efficiency of the click reaction or due to incomplete reaction, as the incorporation of the probe is still high as shown by the flow cytometry data. This result suggests that higher concentrations of probe are needed for imaging purposes but lower can be used to remodel for other applications such as utilizing the probe as an affinity handle for mass-spectrometric enrichment studies.
Previous methods to label the carbohydrate backbone of peptidoglycan take advantage of the promiscuity of both the bacterial cell wall recycling and PG biosynthetic machinery (Figure 1).8 While the method has proven illuminating in visualizing the cell wall on defined carbohydrate units, here the method was improved by increasing uptake of the carbohydrate building blocks. A common strategy in mammalian glycobiology is the utilization of acetates and here we asked if masking polar groups of the glycan core would increase overall probe incorporation. Through microscopy and mass spectrometry assays, it was established that E. coli and other species do not utilize acetylated NAM substrates on the alcohols of the carbohydrates, suggesting that the specific cellular esterases are not able to deacetylate the precursors. Alternatively, one could argue that the acetylated sugars might not be substrates for the specific PG sugar transporters, which can be utilized by bacteria to recycle PG.47 It could be reasoned that E. coli do have deacetylases and a N-acetyl-muramic acid transporter is the actually gatekeeper of this process (Figure 1). In order to quickly test this possibility, E. coli cell lysates were assessed for the ability to unmask both the acetate and methyl ester protecting group from 12. Mass spectrometry data revealed only the deprotected carboxylic acid muramic acid derivatives; deprotection of the acetates was not observed (Supporting Information, Table S1). These data support that bacteria are charged with a specific set of esterases: those capable of removing carboxylic acid ester masks from muramic acid but not the acetates from alcohol groups of these peptidoglycan building block. These data go along with a study by Tippmann and coworkers whose work with peracetylated N-acetyl-glucosamine suggested cellular deacetylases for this specific carbon source do not exist.46 Here, this work shows that this phenomenon extends beyond GlcNAc to critical peptidoglycan building blocks, suggesting E. coli lack deacetylases capable of unmasking NAM derivatives. However, Tippmann and coworkers’ data suggest that E. coli do not have methylesterases to deprotect masked carboxylic acid derivatives, which is contrary to the data presented here in which esters of muramic acid derivatives are efficiently removed both in cell lysates and whole cell experiments. Therefore, it can be concluded that methyl ester NAM derivatives are tolerated by the PG labeling strategy (8 and 9, Scheme 2). By simply masking the negative charge of carboxylic acid on the muramic acid, similar levels of cell wall labeling were achieved by using significantly less probe (Figure 2). In parallel, an improved synthesis of the peptidoglycan precursors was developed which allows scalable synthesis of essential building blocks. A new, improved route for the preparation of NAM probes for metabolic labeling as well as immunological privileged PG fragments is described. This rapid, tunable, and high yielding synthesis of PG derivatives allows the production of analogues to interrogate the PG biosynthesis and to develop inhibitors of this process. Moreover, the benefits of the methyl ester transformation extend permissibility to traditional coupling conditions, sometimes eliminating the use of NHS, which have proven difficult to remove from final carbohydrates. The findings presented here will greatly increase the utility of both production of muramic acid probe synthesis, as the improved 2-amino muramic acid synthesis will allow a variety of chemical biological probes to be produced. Additionally, immunostimulatory probes such as the canonical, muramyl dipeptide and more complex PG fragments, the standards used by immunologist when assessing immune activity of PG.48 The improved uptake mechanism will decrease the amount of probe necessary and permit for large scale tracking experiments to be performed to ultimately identify the biologically relevant PG fragments and which innate immune receptors are engaged with them.
Supplementary Material
Acknowledgments
This work was supported by the NIH Glycoscience Commonfund (U01CA221230). C.L.G is a Pew Biomedical Scholar and thanks the Pew Foundation. K.A.W. and S.N.H. thank the NIH for support through a CBI training grant: 5T32GM133395A. We thank C.Fromen for access to the flow cytometer and D.Kerecman for assistance with mass spectrometry. Instrumentation support was provided by the Delaware COBRE and INBRE programs, supported by the National Institute of General Medical Sciences (P30 GM110758-02, P20 GM104316-01A1, and P20 GM103446). We thank N. Salama and J. Taylor (Fred Hutchinson Cancer Research Center and University of Washington Department of Microbiology) for testing the NAM methyl ester compounds in H. pylori.
Abbreviations:
- Ac2O
Acetic anhydride
- AcCl
Acetyl chloride
- PhCH(OMe)2
Benzaldehyde dimethyl acetal
- AcOH
Acetic acid
- AlkNAM
Alkyne N-acetylmuramic acid
- AmgK
NAM/NAG anomeric kinase
- AzNAM
Azide N-acetylmuramic acid
- BnOH
Benzyl alcohol
- BOC
Tert-butyloxylcarbonyl
- Cbz
Carboxybenzyl
- Cbz-OSu
N-(benzyloxycarbonyloxy)succinimide
- CuAAC
Copper (I) catalyzed azide-alkyne cycloaddition
- DMAP
4-Dimethylaminopyridine
- DMF
Dimethylformamide
- EDC-HCl
1-Ethyl-3-(3′-dimethylaminopropyl)carbodiimide · HCl
- GlcNAc
N-acetylglucosamine
- MDP
Muramyl dipeptide
- MeOH
Methanol
- MurU
NAM α-1-phosphate uridylyltransferase
- Na2CO3
Sodium carbonate
- NAG
N-acetylglucosamine
- NaH
Sodium Hydride
- NaHCO3
Sodium bicarbonate
- NAM
N-acetyl muramic acid
- NHS
N-hydroxysuccinimide
- Pd(OH)2
Palladium hydroxide
- PG
Peptidoglycan
- pTSA
p-Toluenesulfonic acid
- THF
Tetrahydrofuran
- UDP
Uridine diphosphate
Footnotes
Supporting Information: Experimental procedures and materials include: growth curve assay conditions; bioorthogonal cell labeling procedures, microscopy, and flow cytometry; mass spectrometry incorporation assay; synthetic procedures; 1H NMR and 13C NMR spectra for reported compounds; and high resolution mass spectrometry spectra for reported compounds.
The Supporting Information is available free of charge on the ACS Publications website.
Competing Financial Interests
The authors declare no conflict of interest.
References
- 1.Silhavy TJ; Kahne D; Walker S, The bacterial cell envelope. Cold Spring Harb. Perspect. Biol 2010, 2 (5), a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmer W; Blanot D; De Pedro MA, Peptidoglycan structure and architecture. FEMS Microbiology Reviews 2008, 32 (2), 149–167. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds PE, Structure, biochemistry and mechanism of action of glycopeptide antibiotics. European Journal of Clinical Microbiology and Infectious Diseases 1989, 8 (11), 943–950. [DOI] [PubMed] [Google Scholar]
- 4.Page JE; Walker S, Natural products that target the cell envelope. Curr. Opin. Microbiol 2021, 61, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher JF; Mobashery S, Constructing and deconstructing the bacterial cell wall. Protein Sci. 2020, 29 (3), 629–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump GM; Zhou J; Mashayekh S; Grimes CL, Revisiting peptidoglycan sensing: interactions with host immunity and beyond. Chem. Commun. (Camb.) 2020, 56 (87), 13313–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taguchi A; Kahne D; Walker S, Chemical tools to characterize peptidoglycan synthases. Curr. Opin. Chem. Biol 2019, 53, 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang H; DeMeester KE; Hou C-W; Parent MA; Caplan JL; Grimes CL, Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nature communications 2017, 8, 15015–15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMeester KE; Liang H; Jensen MR; Jones ZS; D’Ambrosio EA; Scinto SL; Zhou J; Grimes CL, Synthesis of Functionalized N-Acetyl Muramic Acids To Probe Bacterial Cell Wall Recycling and Biosynthesis. Journal of the American Chemical Society 2018, 140 (30), 9458–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMeester KE; Liang H; Zhou J; Wodzanowski KA; Prather BL; Santiago CC; Grimes CL, Metabolic Incorporation of N-Acetyl Muramic Acid Probes into Bacterial Peptidoglycan. Current protocols in chemical biology 2019, 11 (4), e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gisin J; Schneider A; Nägele B; Borisova M; Mayer C, A cell wall recycling shortcut that bypasses peptidoglycan de novo biosynthesis. Nat Chem Biol 2013, 9 (8), 491–3. [DOI] [PubMed] [Google Scholar]
- 12.Liechti GW; Kuru E; Hall E; Kalinda A; Brun YV; VanNieuwenhze M; Maurelli AT, A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 2014, 506 (7489), 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu Y-P; Booher G; Egan A; Vollmer W; VanNieuwenhze MS, d-Amino Acid Derivatives as in Situ Probes for Visualizing Bacterial Peptidoglycan Biosynthesis. Accounts of chemical research 2019, 52 (9), 2713–2722. [DOI] [PubMed] [Google Scholar]
- 14.Siegrist MS; Whiteside S; Jewett JC; Aditham A; Cava F; Bertozzi CR, D-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS chemical biology 2013, 8 (3), 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuru E; Hughes HV; Brown PJ; Hall E; Tekkam S; Cava F; de Pedro MA; Brun YV; VanNieuwenhze MS, In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-Amino Acids. Angewandte Chemie International Edition 2012, 51 (50), 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu Y-P; Hall E; Booher G; Murphy B; Radkov AD; Yablonowski J; Mulcahey C; Alvarez L; Cava F; Brun YV; Kuru E; VanNieuwenhze MS, Fluorogenic d-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nature Chemistry 2019, 11 (4), 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu Y-P; Rittichier J; Kuru E; Yablonowski J; Pasciak E; Tekkam S; Hall E; Murphy B; Lee TK; Garner EC; Huang KC; Brun Yves V.; VanNieuwenhze MS, Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chemical Science 2017, 8 (9), 6313–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shieh P; Siegrist MS; Cullen AJ; Bertozzi CR, Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proceedings of the National Academy of Sciences 2014, 111 (15), 5456–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fura JM; Kearns D; Pires MM, D-Amino Acid Probes for Penicillin Binding Protein-based Bacterial Surface Labeling. Journal of Biological Chemistry 2015, 290 (51), 30540–30550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apostolos AJ; Pidgeon SE; Pires MM, Remodeling of Cross-bridges Controls Peptidoglycan Cross-linking Levels in Bacterial Cell Walls. ACS Chemical Biology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer C; Kluj RM; Mühleck M; Walter A; Unsleber S; Hottmann I; Borisova M, Bacteria's different ways to recycle their own cell wall. International Journal of Medical Microbiology 2019, 309 (7), 151326. [DOI] [PubMed] [Google Scholar]
- 22.Yang X; McQuillen R; Lyu Z; Phillips-Mason P; De La Cruz A; McCausland JW; Liang H; DeMeester KE; Santiago CC; Grimes CL; de Boer P; Xiao J, A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat Microbiol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taguchi A; Welsh MA; Marmont LS; Lee W; Sjodt M; Kruse AC; Kahne D; Bernhardt TG; Walker S, FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol 2019, 4 (4), 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor JA; Bratton BP; Sichel SR; Blair KM; Jacobs HM; DeMeester KE; Kuru E; Gray J; Biboy J; VanNieuwenhze MS; Vollmer W; Grimes CL; Shaevitz JW; Salama NR, Distinct cytoskeletal proteins define zones of enhanced cell wall synthesis in Helicobacter pylori. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nischan N; Kohler JJ, Advances in cell surface glycoengineering reveal biological function. Glycobiology 2016, 26 (8), 789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almaraz RT; Aich U; Khanna HS; Tan E; Bhattacharya R; Shah S; Yarema KJ, Metabolic oligosaccharide engineering with N-Acyl functionalized ManNAc analogs: cytotoxicity, metabolic flux, and glycan-display considerations. Biotechnol. Bioeng 2012, 109 (4), 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champasa K; Longwell SA; Eldridge AM; Stemmler EA; Dube DH, Targeted identification of glycosylated proteins in the gastric pathogen Helicobacter pylori (Hp). Mol Cell Proteomics 2013, 12 (9), 2568–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green SP; Wheelhouse KM; Payne AD; Hallett JP; Miller PW; Bull JA, Thermal Stability and Explosive Hazard Assessment of Diazo Compounds and Diazo Transfer Reagents. Org Process Res Dev 2020, 24 (1), 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer N; Goddard-Borger ED; Greiner R; Klapotke TM; Skelton BW; Stierstorfer J, Sensitivities of some imidazole-1-sulfonyl azide salts. J. Org. Chem. 2012, 77 (4), 1760–4. [DOI] [PubMed] [Google Scholar]
- 30.DeMeester KE; Liang H; Zhou J; Wodzanowski KA; Prather BL; Santiago CC; Grimes CL, Metabolic Incorporation of N-Acetyl Muramic Acid Probes into Bacterial Peptidoglycan. Curr. Protoc. Chem. Biol 2019, 11 (4), e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiso M; Goh Y; Tanahashi E; Hasegawa A; Okumura H; Azuma I, Synthesis and immunoadjuvant activities of novel N-acylmuramoyl dipeptides related to the lipid A constituent of the bacterial lipopolysaccharide. Carbohydrate Research 1981, 90 (2), C8–C11. [Google Scholar]
- 32.Okumura H; Kamisango K.-i.; Saki I; Tanio Y; Azuma I; Kiso M; Hasegawa A; Yamamura Y, Synthesis and Immunoadjuvant Activity of Acylamino Analogs of N-Acetylmuramoyl-L-alanyl-D-isoglutamine. Agricultural and Biological Chemistry 1982, 46 (2), 507–514. [Google Scholar]
- 33.Hesek D; Lee M; Noll BC; Fisher JF; Mobashery S, Complications from Dual Roles of Sodium Hydride as a Base and as a Reducing Agent. The Journal of Organic Chemistry 2009, 74 (6), 2567–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellouz F; Adam A; Ciorbaru R; Lederer E, Minimal structural requirements for adjuvant activity of bacterial peptidoglycan derivatives. Biochem. Biophys. Res. Commun 1974, 59 (4), 1317–25. [DOI] [PubMed] [Google Scholar]
- 35.Jeyanathan M; Alexander DC; Turenne CY; Girard C; Behr MA, Evaluation of in situ methods used to detect Mycobacterium avium subsp. paratuberculosis in samples from patients with Crohn's disease. J. Clin. Microbiol 2006, 44 (8), 2942–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melnyk JE; Mohanan V; Schaefer AK; Hou C-W; Grimes CL, Peptidoglycan Modifications Tune the Stability and Function of the Innate Immune Receptor Nod2. Journal of the American Chemical Society 2015, 137 (22), 6987–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bersch KL; DeMeester KE; Zagani R; Chen S; Wodzanowski KA; Liu S; Mashayekh S; Reinecker H-C; Grimes CL, Bacterial Peptidoglycan Fragments Differentially Regulate Innate Immune Signaling. ACS Central Science 2021, 7 (4), 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaefer AK; Melnyk JE; Baksh M; Lazor KM; Finn MG; Grimes CL, Membrane-Association Dictates Ligand Specificity for the Innate Immune Receptor NOD2. ACS Chem. Biol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K-T; Huang D-Y; Chiu C-H; Lin W-W; Liang P-H; Cheng W-C, Synthesis of Diverse N-Substituted Muramyl Dipeptide Derivatives and Their Use in a Study of Human NOD2 Stimulation Activity. Chemistry – A European Journal 2015, 21 (34), 11984–11988. [DOI] [PubMed] [Google Scholar]
- 40.Luchansky SJ; Hang HC; Saxon E; Grunwell JR; Yu C; Dube DH; Bertozzi CR, Constructing Azide-Labeled Cell Surfaces Using Polysaccharide Biosynthetic Pathways. In Methods in Enzymology, Academic Press: 2003; Vol. 362, pp 249–272. [DOI] [PubMed] [Google Scholar]
- 41.Laughlin ST; Bertozzi CR, Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nature Protocols 2007, 2 (11), 2930–2944. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar AK; Fritz TA; Taylor WH; Esko JD, Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal beta 1-->4GlcNAc beta-O-naphthalenemethanol. Proceedings of the National Academy of Sciences of the United States of America 1995, 92 (8), 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severn WB; Richards JC, A Novel-Approach for Stereochemical Analysis of 1-Carboxyethyl Sugar Ethers by Nmr-Spectroscopy. Journal of the American Chemical Society 1993, 115 (3), 1114–1120. [Google Scholar]
- 44.Smith GN; Worrel CS; Swanson AL, Inhibition of Bacterial Esterases by Chloramphenicol (Chloromycetin). Journal of Bacteriology 1949, 58 (6), 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark EL; Emmadi M; Krupp KL; Podilapu AR; Helble JD; Kulkarni SS; Dube DH, Development of Rare Bacterial Monosaccharide Analogs for Metabolic Glycan Labeling in Pathogenic Bacteria. Acs Chemical Biology 2016, 11 (12), 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonczak AK; Simova Z; Tippmann EM, A critical examination of Escherichia coli esterase activity. J. Biol. Chem 2009, 284 (42), 28795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer C; Kluj RM; Muhleck M; Walter A; Unsleber S; Hottmann I; Borisova M, Bacteria's different ways to recycle their own cell wall. Int. J. Med. Microbiol 2019, 309 (7), 151326. [DOI] [PubMed] [Google Scholar]
- 48.Bersch KL; DeMeester KE; Zagani R; Chen S; Wodzanowski KA; Liu S; Mashayekh S; Reinecker H-C; Grimes CL, Bacterial Peptidoglycan Fragments Differentially Regulate Innate Immune Signaling. ACS Central Science 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.