Abstract
Summary
The combination, analysis and evaluation of different studies which try to answer or solve the same scientific question, also known as a meta-analysis, plays a crucial role in answering relevant clinical relevant questions. Unfortunately, metabolomics studies rarely disclose all the statistical information needed to perform a meta-analysis. Here, we present a meta-analysis approach using only the most reported statistical parameters in this field: P-value and fold-change. The P-values are combined via Fisher’s method and fold-changes by averaging, both weighted by the study size (n). The amanida package includes several visualization options: a volcano plot for quantitative results, a vote plot for total regulation behaviours (up/down regulations) for each compound, and a explore plot of the vote-counting results with the number of times a compound is found upregulated or downregulated. In this way, it is very easy to detect discrepancies between studies at a first glance.
Availability and implementation
Amanida code and documentation are at CRAN and https://github.com/mariallr/amanida.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The widespread of metabolomics as a potential tool for clinical diagnosis has increased the systematic reviews and meta-analysis on this topic. Meta-analysis is the statistical combination in a single estimate for results from primary studies answering the same question, which is a common practice in medical research. Typical protocols for meta-analysis only consider one metric to conduct the analysis, whether statistical significance or relative change. They require raw data [Metaboanalyst’ (Chong et al., 2018)] or statistics parameters like standard error, standard deviation or variance [R package ‘meta’ (Balduzzi et al., 2019)], rarely disclosed in metabolomics studies (Lee et al., 2020; Tofte et al., 2020). This causes that in some cases about half of the studies on systematic reviews are not suitable to be included in the meta-analysis (Guasch-Ferré et al., 2016; Pang et al., 2021). Although some standardizations have been proposed for the chemical analysis of metabolomics experiments (sample preparation, experimental analysis, quality control, metabolite identification and data pre-processing) (Sumner et al., 2007), clinical metabolomics needs standardized statistical reporting (Mutter et al., 2019) including how to process meta-analysis. Also, metabolomics public repositories, such as MetaboLights (Haug et al., 2020), do not disclose the statistics data for all studies. On this matter, some approaches have been developed when the studies to be included for meta-analysis do not disclose statistical data, such as vote-counting (Bushman and Wang, 2009), a qualitative estimate that takes into account only the trend of the compound. Some omics have high established protocols for meta-analysis, i.e. microarray in genomics, although the last improvements these methods cannot be adapted to metabolomics as they require the same genes or probes between studies and/or mean per group with standard deviation (Huo et al., 2020; Marot et al., 2009), which is not the case for metabolomics, due to the different analytical techniques used. In this omics, standards protocols only recommend to disclose the overall result of the univariate analysis, P-value and fold-change, without the need of reporting deviation or other metrics (Viant et al., 2019).
Public meta-analysis tools can only by applied to data with standard deviation or directly to raw data (see Supplementary Table S1). Currently there is no available methodology to do a meta-analysis based on studies that only disclose overall results. Amanida addresses the issue of combining overall results to perform meta-analysis based on statistical significance (P-value), relative change (fold-change) and study size. This approach increases the power of meta-analysis in metabolomics where the relative change is as important as the statistical significance (Sinclair and Dudley, 2019; Tolstikov et al., 2020). Estimates are weighted by study size to give more value to big studies where results are less overfitted or spurious (Rattray et al., 2018). Amanida R package also includes the option of performing a qualitative vote-counting plot, as many metabolomics studies will only report the relative change trend, and in this case, only a systematic review can be performed.
2 Statistical information
For significance evaluation using the statistic result P-value, we use the weighted P-values combination (Yoon et al., 2021), which is a variant of Fisher’s method (Fisher, 1925). A gamma distribution is used to assign non-integral weights proportional to study size to each P-value (1).
| (1) |
The fold-change is logarithmically transformed (base 2) to reduce skewness due to methodology (Curran-Everett, 2018), so that the variability is more homogeneous, and the distribution of the sample mean is consistent with a normal distribution. The logarithmically transformed fold-change values are averaged with weighting by study size (2).
| (2) |
Missing data are ignored, and negative values fold-change which stands for inverse comparison (control/case), are reversed (-1/value).
A qualitative analysis of the data can be performed with a vote-counting approach. Vote-counting comprises the general behaviour of the metabolites per study. As previously described (Bushman and Wang, 2009), votes are assigned as follows: value of +1 for compounds up-regulated, value of -1 for down-regulated, and 0 if no change in the behaviour is reported. Then, the total vote per compound is obtained summing the votes.
3 Software description
The Amanida R package allows a meta-analysis of metabolomics data, combining the results of different studies addressing the same question. The user provides the input data through text files (in txt, csv or xlsx format) containing the following information: identifier (previously curated by the user), P-value, fold-change, study size (N) and reference. Then amanida computes the quantitative and qualitative meta-analysis. Results are disclosed in two tables, one for the quantitative meta-analysis, with the global P-value and fold-change obtained, and one for the qualitative meta-analysis, with the vote-counting and number of articles.
Results can be graphically inspected via different plots: the volcano_plot where P-value and fold-change are plotted in logarithmic scale (see Fig. 1), for which the user can select the cut-off thresholds, labelling the selected compounds with their identifiers; the vote_plot where the vote-counting results are plotted per each compound; and the explore_plot where the vote-counting results are plotted against the total number of articles in which each compound is reported as upregulated or downregulated (see Fig. 2). All analysis can be obtained in a completely automatic manner using amanida_report function. To illustrate the package we have used a dataset from a urinary metabolomics meta-analysis study of colorectal cancer (Mallafré et al., 2021).
Fig. 1.
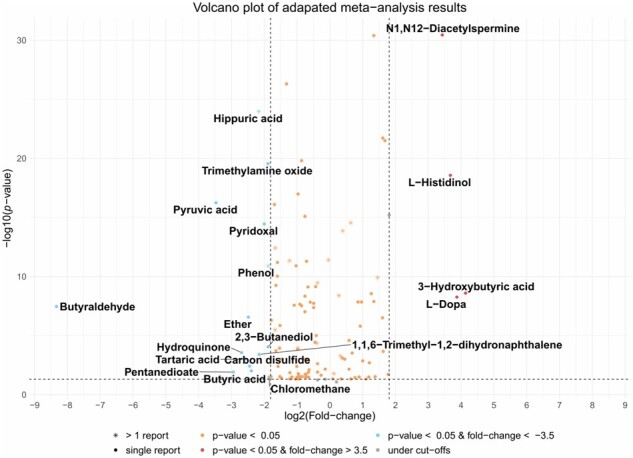
Amanida volcano plot. Quantitative meta-analysis results with a cut-off of 0.05 for P-value and 3 for fold-change. Data obtained from (Mallafré et al., 2021)
Fig. 2.
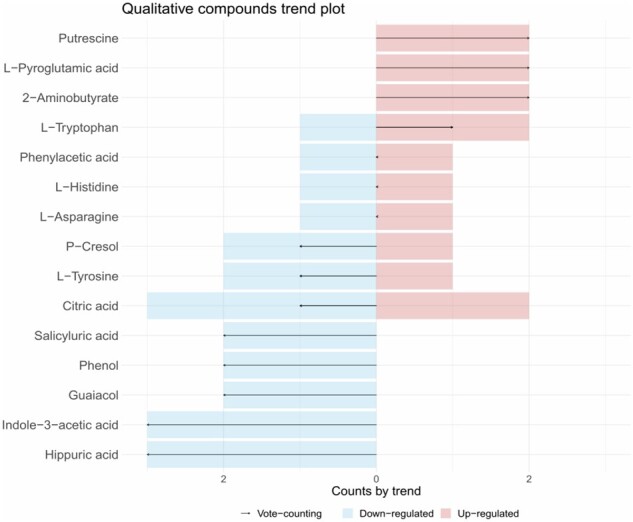
Amanida explore plot. Vote-counting results plotted against total number of articles divided by trend. Data obtained from (Mallafré et al., 2021)
4 Validation
To evaluate the package, we selected a metabolomics meta-analysis with data disclosed (Lee et al., 2020) which describes the association of metabolites with lung cancer risk. We could only use the information of amino acids concentration disclosed in six studies to compare both methodologies, as it was the only fully available (Supplementary Table S2). We must mention that to include the original six studies as in Lee et al. from one study the numeric values have been extracted directly from box-plots (Pietzke et al., 2019). The meta-analysis used on the reviews compares the weighted means between groups using the fixed model or random model if there is high heterogeneity (I2 > 40%) between studies. The principal difference is that the fixed model assumes the effect in all studies is the same and the random model assumes that the effects between studies vary according to a normal distribution, used for convention. When there is high heterogeneity between studies with random models the studies are weighted more equally, this gives higher weight to small studies which is an unwanted approximation in metabolomics. Regarding results, Lee et al. found three statistically significant compounds associated with lung cancer risk: Methionine (I2 = 86.0%), Proline (I2 = 87.1%) and Tryptophan (I2 = 83.6%). Following the same procedure, we have repeated the meta-analysis using the ‘Meta’ R package (Balduzzi et al., 2019). We obtained significant results (see Supplementary Figs) for Proline [random model (I^2 = 87.1%)]. Differences in the results are due to the difference in the box-plot estimations from (Pietzke et al., 2019).
We applied the amanida approach to the same data as Lee et al. for amino acids, where 20 of the 21 amino acids achieve statistical significance (P-value combined < 0.05, Supplementary Table S3), however the fold-change combined in all cases is smaller than 1.5, far from the threshold of 2 to consider a biological change. This means it might be a pattern in the amino acids of lung cancer patients, but the biological effect is small.
Evaluation of meta-analysis results includes multiple metrics including the multiple steps of processing applied compound by compound, while amanida results are obtained straightforward in a detailed report. As mentioned before, amanida has the advantage to work with P-values and fold-change, metrics more disclosed in metabolomics studies than the compound concentrations with their deviations. Also, it increases the readability of the results, classical meta-analysis includes multiple parameters to consider, such as heterogeneity, ranks or multiple models to obtain relevant results. We must say that Lee et al. considered as good metabolites, those with extremely high heterogeneity, and that Cochrane recommends not to do meta-analysis when considerable variation of results, i.e. I2 is 75–100% (Higgins et al., 2019). The validation study used, was the only one that the authors found that reported both mean differences and standard deviations along with P-values and fold-changes.
5 Limitations
Applying amanida meta-analysis directly to the statistical estimates reduces the combination accuracy since without the raw data is not possible to know how much dispersion there is or how many outliers are included. Another drawback is found when the studies only disclose the statistical information for the significant compounds whereas preventing the correct combination of all results. Good practices in metabolomics suggest using a minimum of 50 samples per group to obtain relevant results, in a meta-analysis, these small studies are not recommended to be included due to the variability introduced, as we can observe with Proenza et al.’s (2003) and Yue et al.’s (2018) studies which have 14 and 20 participants per group, respectively. A common criticism for all meta-analyses described in Cochrane Handbook is that they ‘combine apples with oranges’ (Higgins et al., 2019), which have been also said for metabolomics as there is a wide range of techniques for compound detection or extraction methods with different sensibility that are combined. This limitation can be restricted on the systematic search, selecting only studies with the same technique and protocol, but it will reduce substantially the number of studies to combine.
6 Summary
\Amanida has been developed to deal with two issues, the few data disclosed in metabolomics and attribute different weights for the studies according to sample size. Standards metabolomics protocols basically recommend to disclose the overall result of the analysis, P-value and fold-change, without the need of reporting their means or deviation or other metrics (used in classical meta-analysis). In amanida, both overall statistical results are weighted according to the sample size, where larger studies will have more importance. Classical meta-analysis does not measure the strength of the combined result, only shows if the data has a pattern. To look for the strength we combine the statistical significance (P-value) with the effect (fold-change), which measures the quantity of change between groups. Amanida is the first approach to a meta-analysis with non-integral data.
Supplementary Material
Acknowledgement
The authors thank Dr Mariona Vinaixa (Universitat Rovira i Virgili, Spain) for the many useful discussions and helpful suggestions.
Funding
This work was supported by Spanish MINECO project Total2DChrom [RTI2018-098577-B-C21] and Catalan AGAUR project [2018LLAV00072]. This project received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No [798038]. M.LL. is thankful for her graduate fellowship from URV PMF-PIPF program [ref. 2019PMF-PIPF-37]. They acknowledge the AGAUR consolidated group [2017 SGR 1119]. IISPV is a member of the CERCA Programme/Generalitat de Catalunya. This article is based upon work from COST Action 805 CA17118, supported by COST (European Cooperation in Science and Technology).
Conflict of Interest: none declared.
Contributor Information
Maria Llambrich, Department of Electrical Electronic Engineering and Automation, Universitat Rovira i Virgili, IISPV, 43007 Tarragona, Spain; Metabolomics Interdisciplinary Group, Department of Nutrition and Metabolism, Institut d’Investigació Sanitària Pere Virgili, 43201 Reus, Catalonia, Spain; Biomedical Research Centre in Diabetes and Associated Metabolic Disorders (CIBERDEM), ISCIII, 28029 Madrid, Spain.
Eudald Correig, Department of Biostatistics, Universitat Rovira i Virgili, 43201 Reus, Catalonia, Spain.
Josep Gumà, Oncology Department, Hospital Universitari Sant Joan de Reus, Institut d’Investigació Sanitària Pere Virgili, Universitat Rovira i Virgili, 43204 Reus, Spain.
Jesús Brezmes, Department of Electrical Electronic Engineering and Automation, Universitat Rovira i Virgili, IISPV, 43007 Tarragona, Spain; Metabolomics Interdisciplinary Group, Department of Nutrition and Metabolism, Institut d’Investigació Sanitària Pere Virgili, 43201 Reus, Catalonia, Spain; Biomedical Research Centre in Diabetes and Associated Metabolic Disorders (CIBERDEM), ISCIII, 28029 Madrid, Spain.
Raquel Cumeras, Department of Electrical Electronic Engineering and Automation, Universitat Rovira i Virgili, IISPV, 43007 Tarragona, Spain; Metabolomics Interdisciplinary Group, Department of Nutrition and Metabolism, Institut d’Investigació Sanitària Pere Virgili, 43201 Reus, Catalonia, Spain; Biomedical Research Centre in Diabetes and Associated Metabolic Disorders (CIBERDEM), ISCIII, 28029 Madrid, Spain; West Coast Metabolomics Center, University of California Davis, CA 95616, USA.
References
- Balduzzi S. et al. (2019) How to perform a meta-analysis with R: a practical tutorial. Evid. Based Ment. Health, 22, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman B.J., Wang M.C. (2009) Vote-counting procedures in meta-analysis. In: The Handbook of Research Synthesis and Meta-Analysis. New York, Russel Sage Foundation. pp. 207–220. [Google Scholar]
- Chong J. et al. (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res., 46, W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran-Everett D. (2018) Explorations in statistics: the log transformation. Adv. Physiol. Educ., 42, 343–347. [DOI] [PubMed] [Google Scholar]
- Fisher R. (1925) Statistical Methods for Research Workers, 1st edn. Oliver and Boyd, Edinburgh, Scotland.
- Guasch-Ferré M. et al. (2016) Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care, 39, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K. et al. (2020) MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res., 48, D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P.T. et al. (2019) Cochrane Handbook for Systematic Reviews of Interventions, 2nd edn. Wiley.
- Huo Z. et al. (2020) P-value evaluation, variability index and biomarker categorization for adaptively weighted Fisher’s meta-analysis method in omics applications. Bioinformatics, 36, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.B. et al. (2020) Association between metabolites and the risk of lung cancer: a systematic literature review and meta-analysis of observational studies. Metabolites, 10, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallafré C. et al. (2021) Comprehensive volatilome and metabolome signatures of colorectal cancer in urine: a systematic review and meta-analysis. Cancers,13, 2534. [DOI] [PMC free article] [PubMed]
- Marot G. et al. (2009) Moderated effect size and P-value combinations for microarray meta-analyses. Bioinformatics, 25, 2692–2699. [DOI] [PubMed] [Google Scholar]
- Mutter S. et al. (2019) Statistical reporting of metabolomics data: experience from a high-throughput NMR platform and epidemiological applications. Metabolomics, 16, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z. et al. (2021) Comprehensive meta-analysis of COVID-19. Global Metab. Datasets Metab., 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzke M. et al. ; On behalf of the METTEN Study Group. (2019) Stratification of cancer and diabetes based on circulating levels of formate and glucose. Cancer Metab., 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proenza A.M. et al. (2003) Breast and lung cancer are associated with a decrease in blood cell amino acid content. J. Nutr. Biochem., 14, 133–138. [DOI] [PubMed] [Google Scholar]
- Rattray N.J.W. et al. (2018) Beyond genomics: understanding exposotypes through metabolomics. Hum. Genomics, 12, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair K., Dudley E. (2019) Metabolomics and biomarker discovery. Adv. Exp. Med. Biol., 1140, 613–633. [DOI] [PubMed] [Google Scholar]
- Sumner L.W. et al. (2007) Proposed minimum reporting standards for chemical analysis: chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofte N. et al. (2020) Plasma metabolomics identifies markers of impaired renal function: a meta-analysis of 3089 persons with type 2 diabetes. J. Clin. Endocrinol. Metab., 105, 1–13. [DOI] [PubMed] [Google Scholar]
- Tolstikov V. et al. (2020) Current status of metabolomic biomarker discovery: impact of study design and demographic characteristics. Metabolites, 10, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant M.R. et al. (2019) Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat. Commun, 10, 3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. et al. (2021) Powerful p-value combination methods to detect incomplete association. Sci. Rep., 11, 6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X. et al. (2018) Biotransformation-based metabolomics profiling method for determining and quantitating cancer-related metabolites. J. Chromatogr. A, 1580, 80–89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


