Abstract
Animal metagenomic studies, in which host-associated microbiomes are profiled, are an increasingly important contribution to our understanding of the physiological functions, health and susceptibility to diseases of livestock. One of the major challenges in these studies is host DNA contamination, which limits the sequencing capacity for metagenomic content and reduces the accuracy of metagenomic profiling. This is the first study comparing the effectiveness of different sequencing methods for profiling bovine vaginal metagenomic samples. We compared the new method of Oxford Nanopore Technologies (ONT) adaptive sequencing, which can be used to target or eliminate defined genetic sequences, to standard ONT sequencing, Illumina 16S rDNA amplicon sequencing, and Illumina shotgun sequencing. The efficiency of each method in recovering the metagenomic data and recalling the metagenomic profiles was assessed. ONT adaptive sequencing yielded a higher amount of metagenomic data than the other methods per 1 Gb of sequence data. The increased sequencing efficiency of ONT adaptive sequencing consequently reduced the amount of raw data needed to provide sufficient coverage for the metagenomic samples with high host-to-microbe DNA ratio. Additionally, the long reads generated by ONT adaptive sequencing retained the continuity of read information, which benefited the in-depth annotations for both taxonomical and functional profiles of the metagenome. The different methods resulted in the identification of different taxa. Genera Clostridium, which was identified at low abundances and categorized under Order “Unclassified Clostridiales” when using the 16S rDNA amplicon sequencing method, was identified to be the dominant genera in the sample when sequenced with the three other methods. Additionally, higher numbers of annotated genes were identified with ONT adaptive sequencing, which also produced high coverage on most of the commonly annotated genes. This study illustrates the advantages of ONT adaptive sequencing in improving the amount of metagenomic data derived from microbiome samples with high host-to-microbe DNA ratio and the advantage of long reads in preserving intact information for accurate annotations.
Keywords: adaptive sequencing, host contamination, long read, metagenome, Oxford Nanopore
Introduction
Rapid development of metagenomic studies, including the sequencing techniques and metagenomic profiling analyses, is enabling increased application of metagenomic studies in various environments (Thompson et al. 2017). The common sites for animal-associated metagenomic studies are gastrointestinal tracts, reproductive tracts, respiratory tracts, skin, oral and amniotic fluids. The application of metagenomic studies in animals has focused on the contributions of the metagenomes to host health (Oba et al., 2021), susceptibility to diseases (Rodrigues Hoffmann, 2017; Borsanelli et al., 2018; Mach et al., 2021), physiological purposes (Ault et al., 2019; Quereda et al., 2020), and anatomical differences (Glendinning et al., 2017).
Sequencing methods, which have been adopted for metagenomic studies, are targeted short-read sequencing, untargeted short-read sequencing, and untargeted long-read sequencing. Targeted shotgun sequencing, also known as amplicon sequencing, utilizes polymerase chain reaction (PCR) to selectively sequence regions of interest (Thomas et al. 2006; Heil et al., 2019). Different PCR primers have different binding affinities to different species; therefore, the sequencing targets are not conserved across all microbes (Pirolo et al., 2021). Hence, the choice of sequencing target introduces biases, for example under- or over-representation, to the metagenomic profiles. Additionally, amplicon sequencing does not provide information regarding the actual abundances of the identified species and the functional aspects of the metagenomic profile (Gupta et al., 2019).
Untargeted short-read sequencing overcomes the shortcomings of amplicon sequencing by yielding metagenomic profiles of greater resolution and higher accuracy due to their unbiased nature. As a result, shotgun sequencing can allow identification of more genera, including undiscovered genera and provides insights into the functional potential of the metagenomes, which are beneficial to investigate animal-associated metagenomes (Brumfield et al., 2020). The pitfalls of using shotgun sequencing for profiling animal-associated metagenomes include the high amount of host contamination and low microbial content at certain sites, including saliva and reproductive tracts (Marotz et al., 2018). Additionally, short reads can be difficult to assign accurately to taxa due to sequence homology, and profiles can be distorted due sequencing bias.
Long-read sequencing, including PacBio and Oxford Nanopore Technologies (ONT), have been applied to metagenomic studies shortly after they were introduced as genomic sequencing methods (Haro-Moreno et al., 2020). Long-read sequencing addresses the major limitation of short-read sequencing on the assembly contiguity, which is often hampered by repeat elements in genomes (Haro-Moreno et al., 2020). Long-read sequencing enables sequencing of longer fragments and retrieves complete genes, operons and repetitive elements before assembly, which are beneficial for accurate metagenomic annotations and profile recovery. Additionally, multiple genes and non-coding regions have often been deciphered in one single long read, thus revealing the actual genomic structures which consequently contributed to assembling the high-quality metagenome-assembled genomes (MAGs) from metagenomic samples (Overholt et al., 2020; Xie et al., 2020; Ciuffreda et al., 2021). Complete and circular genomes were reported from long-read metagenomic studies, with and without coupling with short-read data for error-corrections (Moss et al., 2020; Cuscó et al., 2021). The MAGs generated with long-read data were much longer, if not complete, hence recovering more genomic elements as well as the gene arrangements which are useful for accurate strain tracking and providing more meaningful insights to the functions of the metagenomes (Quick, 2019; Haro-Moreno et al., 2020; Maguire et al., 2021).
Adaptive sequencing is the real-time selection of sequencing targets on the ONT platform. This method allows the enrichment or depletion of user specified sequences (Loose et al., 2016; Payne et al., 2020). ONT adaptive sampling was demonstrated to be beneficial in enriching low abundant species in a synthetic mock community consisting of 7 bacteria with known proportions (Martin et al., 2021). ONT adaptive sampling can allow deeper sequencing of a metagenome by specifying that the contaminating host DNA be depleted from the sequencing run.
Sequencing depth is a critical variable factor to obtain meaningful and representative metagenomic data from untargeted animal-associated metagenomes with high host-to-microbe DNA ratio (Zaheer et al., 2018; Pirolo et al., 2021). Studies have demonstrated that deep coverage is essential for the sensitivity and accuracy of untargeted sequencing for metagenomic profiling, both for taxonomical and functional aspects (Gweon et al. 2019; Pereira-Marques et al., 2019). To avoid inefficient and uneconomical sequencing, investigations into depleting the host DNA prior to sequencing have improved overall resolution without expensive deep-sequencing (Heravi et al., 2020; Yap et al., 2020). Nonetheless, questions arise if the depletion and selective extraction methods alter the actual profiles of the animal-associated metagenomes.
In this study, we investigate the effect of the depletion mode of ONT adaptive sequencing on the bovine vaginal microbiome. We test the hypothesis that by applying ONT adaptive sequencing to enrich for sequences of microbial origin, without distorting the observed community.
Methods and Materials
Sample collection and extraction
Vaginal samples were collected from 24 nonpregnant heifers and cows from cattle properties in Northern Queensland under UQ Animal Ethics Approval AE30009 by an experienced veterinarian, who conducted health assessments on the animals prior to sample collection. The Tricamper (DAF Queensland, Australia) sampling tool was utilized to collect the bovine vaginal swab following the manufacturer’s protocol. Upon removing the Tricamper from the vagina, the other end of the Tricamper was blocked to prevent spillage. The swab sample was immediately preserved in a 10-mL tube preloaded with 5 mL phosphate-buffered saline (PBS) by excising the head of the Tricamper device.
The samples were kept on ice during delivery and were processed within 6 h upon arrival to the laboratory. Each sample was first vortexed for 15 s and followed by an additional 15 s vortexing after the Tricamper head was removed from the tube. The vaginal mucus samples from the tubes were then transferred into a sterile 15 mL tube. The vaginal samples were pelleted by centrifugation and the supernatant was removed. The samples were extracted using Qiagen DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) according to manufacturer’s instruction for Gram-positive bacteria.
Two separate approaches were taken to investigate the effect of adaptive sequencing. The first was sequencing a pooled sample with multiple sequencing approaches, and the second was to use biological replication to determine if the adaptive sampling approach consistently improved the target data yield. For the pooled sample, the extracted DNA from five Droughtmaster heifers were pooled together into a single sample that was split into four technical replicates, labeled as 5DH, to allow testing of the four different sequencing methods. Separately, DNA from the vaginal swabs of 19 animals was sequenced using the untargeted sequencing methods, Illumina shotgun (n = 3), regular ONT (n = 2) and ONT adaptive (n = 14).
Metagenomics sequencing
The purity and quantity of the DNA was examined using NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). The purity of the DNA was assessed by measuring the absorbance ratio at A260/280. From the pooled 5DH sample, two technical replicates of 20 ng each were sent for 16S rDNA amplicon (27F-519R) and Illumina shotgun sequencing at the Australian Centre of Ecogenomics (University of Queensland, Brisbane, Australia). Sequencing was performed on the NextSeq500 platform using NextSeq 500/550 High Output v2 2 × 150 bp paired-end chemistry to obtain 1 Gbp for each sample. In addition, the 3 Illumina shotgun samples (biological replicates) were sequenced using the same conditions.
Libraries for ONT regular and adaptive sequencing were prepared using the ONT SQK-LSK109 kit (ONT, Cambridge, UK) according to the manufacturer’s instructions. The library was loaded onto an individual MinION flowcells FLO-MIN106D and sequenced with an ONT GridION Mk1 sequencer loaded with MinKNOW version 21.05.8 (ONT). For adaptive sequencing, the adaptive sampling mode was turned on and the genomes of both ARS-UCD1.2 Bos taurus genome (GCA_002263795.2) and Brahman genome (Ross, 2019) were supplied for depletion. The sequencing was stopped when 1 Gbp of data was obtained. Base-calling was conducted using Guppy version 5.0.11 (ONT).
Sequence trimming and host-contamination examination
The quality of the paired-end short reads was evaluated using FastQC 0.11.4 (Andrew, 2010) before the paired-end sequences were merged using PEAR 0.9.11 (Zhang et al., 2014). The merged short reads were trimmed with Trimmomatic 0.39.1 (Bolger et al., 2014) using the single-end mode.
The quality of the long reads generated using ONT was evaluated using NanoPlot 1.3.0 (De Coster et al., 2018). Adapters on the long reads were removed using Porechop 0.2.4 (Wick et al., 2017) while NanoFilt 2.7.0 (De Coster et al., 2018) was applied to filter out reads which were lesser than 5 in quality score.
All the short reads and long reads were mapped against the ARS-UCD1.2 Bos taurus genome (GCA_002263795.2) and Brahman genome (Ross, 2019) using Minimap2 2.17 (r941) (Li, 2018) to examine the percentage of bovine DNA in the sample sequenced with different technologies. Three pairwise t-tests were used to determine if the observed differences in metagenome proportion were significantly different with a P-value threshold of 0.05.
Contig construction
The processed short reads generated from Illumina shotgun sequencing were assembled into contigs using Megahit v1.2.9 (Li et al., 2016) with minimum contig length of 200bp while the filtered long reads generated with ONT were assembled into contigs using Flye 2.8.3-b1725 (Kolmogorov et al. 2020) with the meta mode for the uneven coverage in metagenomic samples. Additionally, a co-assembly was constructed using all the reads with Flye 2.8.3-b1725 (Kolmogorov et al. 2020). Each set of reads generated with different sequencing technologies was mapped to the co-assembly using Minimap2 2.17 (r941) (Li, 2018) to obtain the alignment file for coverage depth calculation using the jgi_summarize_bam_contig_depths script from Metabat2 v2.15 (Kang et al., 2019).
Metagenomics classification
Read-based taxonomic classification of the reads was performed using Kraken v2.1.2 (Wood et al., 2019). To ensure a more targeted and efficient search for the metagenomic samples in this study, instead of the standard database suggested by Kraken2, a customized database was constructed with the build script provided by Kraken2. The customized database used in this study was built with the complete genomes of archaea, bacteria, and fungi, which were downloaded from NCBI RefSeq (Pruitt et al., 2005) with their low complexity sequences masked. The abundances of the organisms in the metagenomic samples sequenced with different methods were estimated using the Bracken v2.6.2 (Lu et al., 2017). The downstream analysis and visualization of the results were conducted using the snake pipeline wrapper, Kraken2-classification v1.0.0 (Siranosian and Bhatt, 2021), with modifications to better describe the results in this study.
Functional annotations were called from the assembled contigs using the annotation pipeline MetaErg 1.2.0 (Dong and Strous, 2019). Briefly, the predicted ORFs were subjected to HMMs profile similarity searches against several databases, including Pfam-A (Finn et al. 2016), TIGRFAM (Haft et al., 2013), FOAM (Prestat et al. 2014), metabolic-hmms (Anantharaman et al. 2016), and casgenes.hmm (Burstein et al., 2017). MetaErg also performed DIAMOND (double index alignment of next-generation sequencing data) searches against SwissProt (Bairoch and Apweiler, 2000) and the MetaErg in-built database GenomeDB. Mapping files generated from searches against SwissProt, FOAM and TIGRFAMs databases were incorporated in MinPath (Ye and Doak, 2009) to infer to KEGG (Kanehisa and Goto, 2000) and MetaCyc (Karp et al., 2002) metabolic pathways.
Results
After quality filtering, Illumina amplicon and shotgun short-read sequencing resulted in 1.62 and 1.12 Gbp of sequencing data, respectively, for the 5DH sample (Table S1). ONT regular and adaptive sequencing yielded 0.68 and 0.78 Gbp of sequencing data, respectively, for the 5DH sample after removal of low-quality reads. There was a higher number of reads in samples sequenced with Illumina amplicon and shotgun sequencing as higher numbers of short reads were required to achieve 1 Gbp of raw data.
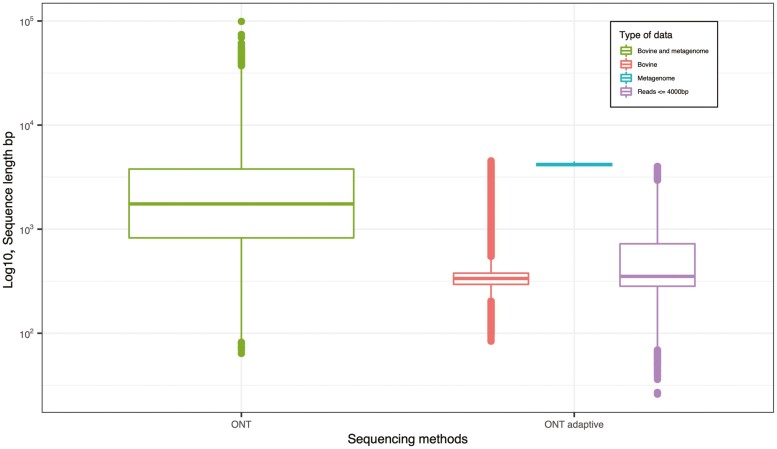
For the ONT adaptive sequencing depletion mode (Figure 1), there were 3 possible decision for the sequence read: (i) “stop_receiving” were the reads that did not map to the provided bovine genomes and were sequenced completely – these are presumed to be metagenomic reads; (ii) “unblock” reads were reads that mapped to the bovine genome are were rejected from the pore for further sequencing, and (iii) “no_decision” where reads were too short (≤4,000 bp) for the ONT adaptive sampling to determine if the read should be accepted or rejected for sequencing. In our ONT adaptive sampling, only 1,634 reads sequenced from the 5DH sample were identified as metagenome sample (“unblock”) and 1,350,892 reads were bovine sample (“stop_receiving”) (Figure 1). A total of 1,599,257 reads were too short (“no_decision”) for the adaptive sequencing algorithm to determine if they were of bovine origin or not.
Figure 1.
Type of sequence data in the pooled heifer vaginal sample (5DH) sequenced by Oxford Nanopore Technologies (ONT) and ONT adaptive sequencing. The data sequenced by ONT sequencing was non-differentiative and hence labelled as “Bovine and metagenome”. The data sequenced by ONT adaptive sequencing was categorized into “Bovine”, “Metagenome”, and “No decision”, the later one was reads which were too short (≤4,000 bp) for ONT adaptive sequencing to determine their origins.
Each non-amplicon read set was mapped against the bovine genome to remove potential host contamination. Illumina shotgun sequencing resulted in 11.79 Mbp (1.02%) of microbiome sequence data, while ONT and ONT adaptive sequencing resulted in 2.45 Mbp (0.23%) and 21.29 Mbp (2.75%) of microbiome sequence data, respectively, from the 5DH sample after host contamination was removed (Table S1).
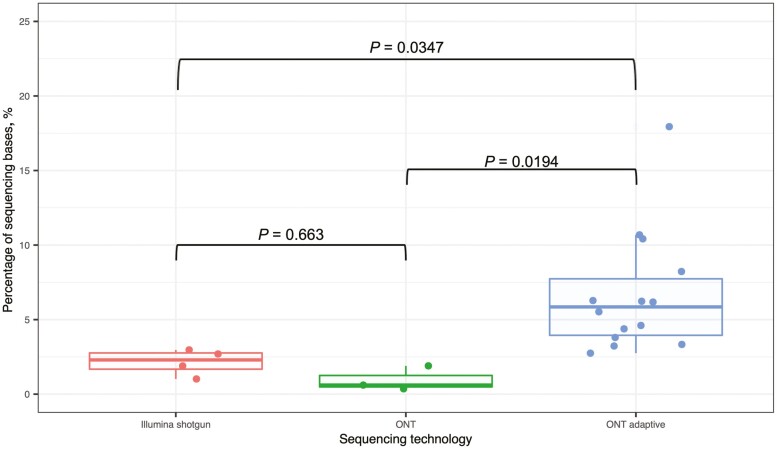
Even though there were only 2.75% of metagenomic data recovered from the 5DH sample sequenced with ONT adaptive sequencing, it provided the equivalent of 10.65X coverage to a 2 MB microbial genome. While Illumina shotgun and regular ONT sequencing provided 5.9X and 1.23X of coverage, respectively, to a 2-MB microbial genome. Our results demonstrated a 10-fold enrichment for microbiome sequence using adaptive sequencing on the ONT platform. Provided there was no other limitation to the sequencing capacity, a sample sequenced with ONT adaptive sequencing requires 7.28 Gbp of data to provide 100X coverage to a 2-MB microbial genome. Illumina shotgun and regular ONT sequencing require 19.6 and 88.6 Gbp of raw data respectively. Therefore, ONT adaptive sequencing significantly decreased the amount of raw data needed to achieve high coverage of the microbiome DNA, which in turn reduces the cost and computational effort for samples with high host-to-microbe DNA ratio. Further analysis including the biological replicates sequenced with different methods reinforced that ONT adaptive sequencing significantly improved the cattle-to-metagenome ratio in the samples when compared with Illumina shotgun (P = 0.0347) and regular ONT sequencing technologies (P = 0.0194) (Figure 2).
Figure 2.
The proportions of metagenome in the bovine vaginal samples sequenced by Illumina shotgun (n = 4), Oxford Nanopore Technologies (ONT) sequencing (n = 3) and ONT adaptive sequencing (n = 15). The significance of differences between the untargeted sequencing technologies was calculated using pairwise t-test.
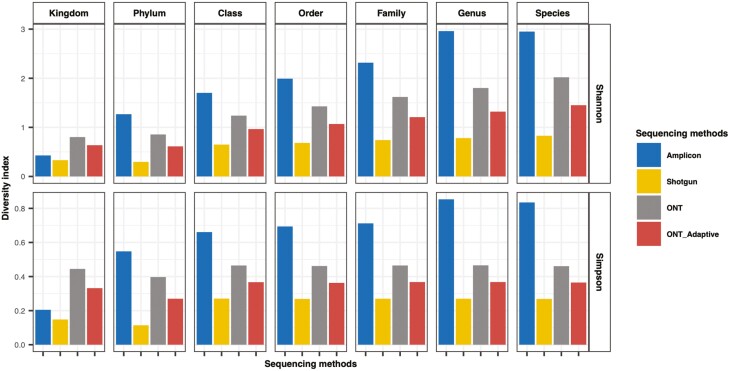
To examine the impact of sequencing technologies on the metagenomic profile, the downstream analyses focused on the profiles generated from the 5DH sample sequenced with different technologies. In the rarefaction analysis none of the sample curves reached a plateau (Supplementary Figure S1), indicating that the sequencing method did not capture the maximum metagenomic diversity in 1 Gbp of sequencing data. The rarefaction analysis showed that ONT methods increased the number of observed species more quickly than Illumina methods. ONT adaptive sequencing recovered a higher number of species in 1 Gbp of raw data compared with normal ONT sequencing, which contained a higher proportion of host DNA (Table S1). Consistent with this, Shannon and Simpson alpha diversity analyses demonstrated that the sample sequenced with ONT consistently resulted in a higher diversity than sample sequenced with ONT adaptive sequencing and Illumina shotgun sequencing (Figure 3).
Figure 3.
Alpha diversity indexes at every taxonomy level in the pooled heifer vaginal sample (5DH) sequenced with different sequencing methods including 16S rDNA amplicon, Illumina shotgun, Oxford Nanopore Technologies (ONT) and ONT adaptive sequencing.
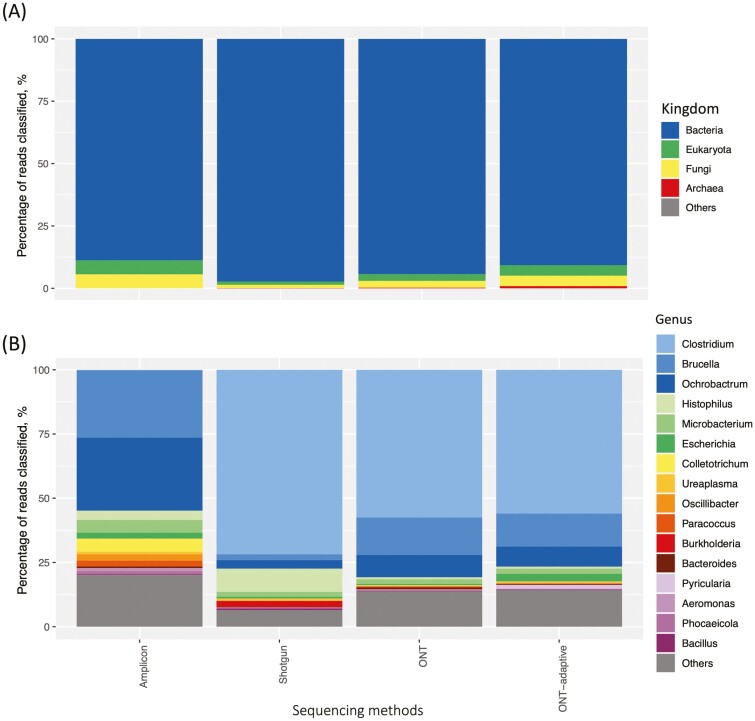
Taxonomical profiles retrieved from the 5DH sample were mostly from the Kingdom of Bacteria, followed by Eukaryota, Fungi, and Archaea (Figure 4A). Samples sequenced with Illumina methods recovered higher number of organisms from the Kingdom Bacteria, 88.83% and 92.16%, respectively, for Illumina amplicon and shotgun sequencing. Although the taxonomical classification was also dominated by Kingdom Bacteria, higher number of organisms from Kingdom Eukaryota and Fungi was recovered in samples sequenced with long-read sequencing than those with short-read sequencing.
Figure 4.
Percentages of classified reads at (A) Kingdom level and (B) Genus level in the pooled heifer vaginal sample (5DH) sequenced with different sequencing methods including 16S rDNA amplicon, Illumina shotgun, Oxford Nanopore Technologies (ONT), and ONT adaptive sequencing.
The taxonomical profile at Genus level disclosed that the sample sequenced with amplicon sequencing showed different profiles than the sample processed with other sequencing methods (Figure 4B). Interestingly, bacteria from genera Clostridium were detected at a low abundance (0.12%) in the sample sequenced with amplicon sequencing, instead genera Ochrobactrum (13.83%) and Brucella (12.71%) were the 2 most abundant genera. Additionally, genera which were identified at lower abundances in samples sequenced with other methods, including Colletotrichum (2.46%) and Microbacterium (2.40%), were more abundant in the sample sequenced with amplicon sequencing.
Functional annotation was conducted on the co-assembly generated with the sequence data obtained from Illumina shotgun, ONT and ONT adaptive sequencing and the relative coverages were calculated. There were 10 genes annotated in the sample sequenced with ONT sequencing (Supplementary Table S2). On the other hand, there were 706 genes and pathways in the sample sequenced with Illumina shotgun and ONT adaptive sequencing methods. However, the sample sequenced with ONT adaptive sequencing showed higher coverages on most of the common annotated genes (Supplementary Figure S2 and Figure S3).
Discussion
The inefficiency of host-associated metagenomic study was depicted in this study. Only 1.02%, 0.23%, and 2.75% of the metagenomic data were recovered from the 5DH sample sequenced with Illumina shotgun, ONT and ONT adaptive sequencing, respectively. Adaptive sequencing outperformed other sequencing methods by recovering higher percentage of metagenomic data.
With the assistance of the adaptive sampling mechanism, ONT adaptive sequencing first sequences each detected DNA strand for up to 5 s before rejecting it for further sequencing (Loose et al., 2016; Payne et al., 2020). In this study, the depletion mode of ONT adaptive sequencing was applied, in which the DNA strands were rejected if they mapped the provided bovine genomes during the first five seconds of sequencing. Our ONT adaptive sequencing result indicated that there was a high number of reads which were rejected. The high rate of read rejection was consistent with the results portrayed by the samples sequenced with Illumina shotgun and regular ONT sequencing, which were detrimentally affected by the high host-to-microbe DNA ratio in bovine vaginal metagenomic samples. ONT adaptive sequencing was relatively less affected because the adaptive technique frees up the sequencing capacity after rejecting the bovine DNA reads.
The rarefaction curves indicate that all sequencing methods tested in this study did not capture the maximum sample diversity with 1 Gbp of raw data. The rarefaction curves demonstrated the high efficiencies of ONT and ONT adaptive sequencing in taxonomical profiling as high numbers of species were recovered when sample sizes were small. The longer reads produced by ONT and ONT adaptive sequencing supplied longer and likely intact information for taxonomical classification, resulting in higher species identification as similar species were not collapsed together. Since clinical samples were used here instead of a mocked community with known proportion of each species, the accuracy of diversity indices reported in the sample sequenced with different methods remain unknown. Still, the different diversity indices in the same sample sequenced by different sequencing methods indicated that sequencing methods introduced biases to the sample diversity.
The different taxonomical profile depicted in the sample sequenced by amplicon sequencing was attributed to the selective amplification nature of the chosen primer pairs (Schmalenberger et al., 2001). The amplicon sequencing in this study targeted the V1–V3 hypervariable regions of the 16S rDNA gene, which has differential binding affinity to the 16S rDNA gene in microbes (Soergel et al., 2012). The choice of amplicon target resulted in a distorted resolution and taxonomic prospect of the entire metagenome in comparison to the other methods described in this study.
Low detection of genera Clostridium was previously reported in healthy bovine vaginal samples sequenced with amplicon sequencing (Rodrigues et al., 2015; Clemmons et al., 2017). In several other 16S rDNA amplicon studies, the genus Clostridium was described as Order “unclassified Clostridiales”, indicating the limitation of amplicon sequencing for accurate and deep taxonomic classification (Shpigel et al., 2017; Ault et al., 2019). Additionally, the loss of Order Clostridiales as a commensal was thought to be an indicator of bovine mild vulvovaginitis (Shpigel et al., 2017). The unbiasedness of Illumina shotgun and ONT sequencing was hypothesized to contribute to the increased annotation depth for genera Clostridium.
The dominance of genera Histophilus in both healthy cows (Schlafer et al., 2007; Quereda et al., 2020) and cows with reproductive diseases (Bano et al., 2011; Rodrigues et al., 2015) was also addressed in previous studies. Histophilus was described as an obligate commensal member in bovine genital mucosal surfaces as well as an opportunistic pathogen under favorable conditions (Bisgaard, 1995; Sandal and Inzana, 2010). On the other hand, genus Brucella and Ochrobactrum were not reported as the top abundant genera in other bovine vaginal metagenomic studies (Jeon and Galvão, 2018; Ong et al., 2021). Genus Burkholderia in bovine vaginal metagenome was identified as the indicator of exposure to toxic fescue, which is associated with reduced reproductive success (Ratton et al., 2018). Nonetheless, since the taxonomical profiles were generated from clinical samples instead of a mocked community, the actual abundances of the detected species remain unknown in this study.
The ONT adaptive mechanism increased the number of reads belonging to the bovine vaginal metagenome, which was retained following host contamination removal. Large amounts of data were filtered from the sample sequenced by regular ONT sequencing, hence the number of functional genes detected was detrimentally affected. All the functional genes detected in the sample sequenced with ONT adaptive sequencing were also recalled in sample sequencing with Illumina shotgun sequencing, however at low coverages. The limited information encoded with the short and highly fragmented reads hindered the detection of functional genes (Quick, 2019).
Conclusion
Our results demonstrated that ONT adaptive sequencing is a more effective sequencing method for metagenomic samples with high host contamination because it overcame the impacts of high host-to-microbe DNA ratio, which affected both Illumina shotgun and regular ONT sequencing. Moreover, a significantly higher number of functional genes were recalled in sample sequenced with ONT adaptive sequencing as the long reads were more intact and thus provided more information for accurate metagenomic profiling. Nonetheless, the tested sample was pooled from actual bovine vaginal swabs hence the accuracy of the annotations could not be investigated. Future research should investigate the accuracy of metagenomic profiles recovered by ONT adaptive sequencing using mocked samples with known concentration of several different microbial species. This method will increase the accessibility of metagenomic data from samples with high host-to-microbe DNA ratios, including vaginal and salivary microbiomes.
Supplementary Material
Acknowledgement
The authors thank Meat and Livestock Australia Donor Company project P.PSH.0799 for the funding of this research. We also thank Dr Geoffry Fordyce for collecting the bovine vaginal samples used in this study. We would also like to thank Dr Paul Marshall for his kind assistance in running Oxford Nanopore Technologies adaptive sampling on the GridION Mk1 sequencer.
Glossary
Abbreviations
- MAG
metagenome-assembled genomes
- PCR
polymerase chain reaction
Conflict of Interest Statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Anantharaman, K., Brown C. T., Hug L. A., Sharon I., Castelle C. J., Probst A. J., Thomas B. C., Singh A., Wilkins M. J., Karaoz U., . et al. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. doi: 10.1038/ncomms13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew S. 2010. FastQC: a quality control tool for high throughput sequence data., Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Ault, T. B., Clemmons B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., Edwards J. L., Myer P. R., and Pohler K. G.. . 2019. Bacterial taxonomic composition of the postpartum cow uterus and vagina prior to artificial insemination1. J. Anim. Sci. 97:4305–4313. doi: 10.1093/jas/skz212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A., and Apweiler R.. . 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45–48. doi: 10.1093/nar/28.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano L, Bonci M, Drigo I, Tonon E, Mazzolini E, Carminato A, Granato A, Ceglie L, Natale A, Zanette G, . et al. 2011. Recurrent detection of Histophilus somni in the genital tract of dairy cattle with reproductive failures in Italy. Large Anim. Rev. 17:171–176. https://www.cabdirect.org/cabdirect/FullTextPDF/2011/20113374203.pdf [Google Scholar]
- Bisgaard, M. 1995. Taxonomy of the family Pasteurellaceae Pohl 1981. In: Donachie W., Lainson F. A., and Hodgson J. C., editors, Haemophilus, Actinobacillus, and Pasteurella. Boston, MA: Springer. p. 1–7. [Google Scholar]
- Bolger, A. M., Lohse M., and Usadel B.. . 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsanelli, A. C., Lappin D. F., Viora L., Bennett D., Dutra I. S., Brandt B. W., and Riggio M. P.. . 2018. Microbiomes associated with bovine periodontitis and oral health. Vet. Microbiol. 218:1–6. doi: 10.1016/j.vetmic.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Brumfield, K. D., Huq A., Colwell R. R., Olds J. L., and Leddy M. B.. . 2020. Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data. PLoS One. 15:e0228899. doi: 10.1371/journal.pone.0228899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein, D., Harrington L. B., Strutt S. C., Probst A. J., Anantharaman K., Thomas B. C., Doudna J. A., and Banfield J. F.. . 2017. New CRISPR-Cas systems from uncultivated microbes. Nature 542:237–241. doi: 10.1038/nature21059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda, L., Rodríguez-Pérez H., and Flores C.. . 2021. Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 19:1497–1511. doi: 10.1016/j.csbj.2021.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons, B. A., Reese S. T., Dantas F. G., Franco G. A., Smith T. P. L., Adeyosoye O. I., Pohler K. G., and Myer P. R.. . 2017. Vaginal and uterine bacterial communities in postpartum lactating cows. Front. Microbiol. 8:1047. doi: 10.3389/fmicb.2017.01047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuscó, A., Pérez D., Viñes J., Fàbregas N., and Francino O.. . 2021. Long-read metagenomics retrieves complete single-contig bacterial genomes from canine feces. BMC Genomics. 22:330. doi: 10.1186/s12864-021-07607-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster, W., D’Hert S., Schultz D. T., Cruts M., and Van Broeckhoven C.. . 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 34:2666–2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X., and Strous M.. . 2019. An integrated pipeline for annotation and visualization of metagenomic contigs. Front. Genet. 10:999. doi: 10.3389/fgene.2019.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., Potter S. C., Punta M., Qureshi M., Sangrador-Vegas A., . et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44:D279–D285. doi: 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning, L., Collie D., Wright S., Rutherford K. M. D., and McLachlan G.. . 2017. Comparing microbiotas in the upper aerodigestive and lower respiratory tracts of lambs. Microbiome. 5:145. doi: 10.1186/s40168-017-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Mortensen M. S., Schjørring S., Trivedi U., Vestergaard G., Stokholm J., Bisgaard H., Krogfelt K. A., and Sørensen S. J.. . 2019. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2:291. doi: 10.1038/s42003-019-0540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gweon, H. S., Shaw L. P., Swann J., De Maio N., AbuOun M., Niehus R., Hubbard A. T. M., Bowes M. J., Bailey M. J., Peto T. E. A., . et al. ; REHAB consortium. 2019. The impact of sequencing depth on the inferred taxonomic composition and AMR gene content of metagenomic samples. Environ. Microbiome. 14:7. doi: 10.1186/s40793-019-0347-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft, D. H., Selengut J. D., Richter R. A., Harkins D., Basu M. K., and Beck E.. . 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41:D387–D395. doi: 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro-Moreno, J. M., López-Pérez M., and Rodríguez-Valera F.. . 2020. Long read metagenomics, the next step? bioRxiv. 2020.2011.2011.378109. doi: 10.1101/2020.11.11.378109 [DOI] [Google Scholar]
- Heil, B. A., Paccamonti D. L., and Sones J. L.. . 2019. Role for the mammalian female reproductive tract microbiome in pregnancy outcomes. Physiol. Genomics. 51:390–399. doi: 10.1152/physiolgenomics.00045.2019 [DOI] [PubMed] [Google Scholar]
- Heravi, F. S., Zakrzewski M., Vickery K., and Hu H.. . 2020. Host DNA depletion efficiency of microbiome DNA enrichment methods in infected tissue samples. J. Microbiol. Methods. 170:105856. doi: 10.1016/j.mimet.2020.105856 [DOI] [PubMed] [Google Scholar]
- Jeon, S. J., and Galvão K. N.. . 2018. An advanced understanding of uterine microbial ecology associated with metritis in dairy cows. Genomics Inform. 16:e21. doi: 10.5808/GI.2018.16.4.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M., and Goto S.. . 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D. D., Li F., Kirton E., Thomas A., Egan R., An H., and Wang Z.. . 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 7:e7359. doi: 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp, P. D., Riley M., Paley S. M., and Pellegrini-Toole A.. . 2002. The MetaCyc database. Nucleic Acids Res. 30:59–61. doi: 10.1093/nar/30.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov, M., Bickhart D. M., Behsaz B., Gurevich A., Rayko M., Shin S. B., Kuhn K., Yuan J., Polevikov E., Smith T. P. L., . et al. 2020. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods. 17:1103–1110. doi: 10.1038/s41592-020-00971-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Luo R., Liu C. M., Leung C. M., Ting H. F., Sadakane K., Yamashita H., and Lam T. W.. . 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102:3–11. doi: 10.1016/j.ymeth.2016.02.020 [DOI] [PubMed] [Google Scholar]
- Loose, M., Malla S., and Stout M.. . 2016. Real-time selective sequencing using nanopore technology. Nat. Methods. 13:751–754. doi: 10.1038/nmeth.3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser F. P., Thielen P, and Salzberg S. L.. . 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3:e104. [Google Scholar]
- Mach, N., Baranowski E., Nouvel L. X., and Citti C.. . 2021. The Airway Pathobiome in complex respiratory diseases: a perspective in domestic animals. Front. Cell. Infect. Microbiol. 11:583600. doi: 10.3389/fcimb.2021.583600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, M., Kase J. A., Roberson D., Muruvanda T., Brown E. W., Allard M., Musser S. M., and González-Escalona N.. . 2021. Precision long-read metagenomics sequencing for food safety by detection and assembly of Shiga toxin-producing Escherichia coli in irrigation water. PLoS One. 16:e0245172. doi: 10.1371/journal.pone.0245172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotz, C. A., Sanders J. G., Zuniga C., Zaramela L. S., Knight R., and Zengler K.. . 2018. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 6:42. doi: 10.1186/s40168-018-0426-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S., Heavens D., Lan Y., Horsfield S., Clark M. D., and Leggett R. M.. . 2021. Nanopore adaptive sampling: a tool for enrichment of low abundance species in metagenomic samples. bioRxiv. 2021.2005.2007.443191. doi: 10.1101/2021.05.07.443191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, E. L., Maghini D. G., and Bhatt A. S.. . 2020. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 38:701–707. doi: 10.1038/s41587-020-0422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba, P. M., Carroll M. Q., Alexander C., Somrak A. J., Keating S. C. J., Sage A. M., and Swanson K. S.. . 2021. Dental chews positively shift the oral microbiota of adult dogs. J. Anim. Sci. 99:skab100. doi: 10.1093/jas/skab100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, C. T., Turni C., Blackall P. J., Boe-Hansen G., Hayes B. J., and Tabor A. E.. . 2021. Interrogating the bovine reproductive tract metagenomes using culture-independent approaches: a systematic review. Anim. Microbiome. 3:41. doi: 10.1186/s42523-021-00106-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholt, W. A., Hölzer M., Geesink P., Diezel C., Marz M., and Küsel K.. . 2020. Inclusion of Oxford Nanopore long reads improves all microbial and viral metagenome-assembled genomes from a complex aquifer system. Environ. Microbiol. 22:4000–4013. doi: 10.1111/1462-2920.15186 [DOI] [PubMed] [Google Scholar]
- Payne, A., Holmes N., Clarke T., Munro R., Debebe B., and Loose M.. . 2020. Nanopore adaptive sequencing for mixed samples, whole exome capture and targeted panels. bioRxiv. 2020.2002.2003.926956. doi: 10.1101/2020.02.03.926956 [DOI] [Google Scholar]
- Pereira-Marques, J., Hout A., Ferreira R. M., Weber M., Pinto-Ribeiro I., van Doorn L. J., Knetsch C. W., and Figueiredo C.. . 2019. Impact of Host DNA and sequencing depth on the taxonomic resolution of whole metagenome sequencing for microbiome analysis. Front. Microbiol. 10:1277. doi: 10.3389/fmicb.2019.01277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirolo, M., Espinosa-Gongora C., Bogaert D., and Guardabassi L.. . 2021. The porcine respiratory microbiome: recent insights and future challenges. Anim. Microbiome. 3:9. doi: 10.1186/s42523-020-00070-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestat, E., David M. M., Hultman J., Taş N., Lamendella R., Dvornik J., Mackelprang R., Myrold D. D., Jumpponen A., Tringe S. G., . et al. 2014. FOAM (functional ontology assignments for metagenomes): a hidden Markov model (HMM) database with environmental focus. Nucleic Acids Res. 42:e145. doi: 10.1093/nar/gku702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K. D., Tatusova T., and Maglott D. R.. . 2005. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501–D504. doi: 10.1093/nar/gki025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quereda, J. J., Barba M., Mocé M. L., Gomis J., Jiménez-Trigos E., García-Muñoz Á., Gómez-Martín Á., González-Torres P., Carbonetto B., and García-Roselló E.. . 2020. Vaginal Microbiota changes during estrous cycle in dairy heifers. Front. Vet. Sci. 7:371. doi: 10.3389/fvets.2020.00371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J. 2019. Ultra-long read nanopore sequencing methods for metagenomics. J Biomol Tech. 30:S63–S63. [Google Scholar]
- Ratton, A. E., Chewning S., Meyer L. R., Atchley J. A., Powell J. G., Tucker J. D., Hubbell D. S. III, Zhao J., and Koltes J. E.. . 2018. 505 Toxic fescue exposure alters vaginal microbial communities of Crossbred beef cows. J. Anim. Sci. 96(suppl_2):269–270. doi: 10.1093/jas/sky073.502 [DOI] [Google Scholar]
- Rodrigues, N. F., Kästle J., Coutinho T. J., Amorim A. T., Campos G. B., Santos V. M., Marques L. M., Timenetsky J., and de Farias S. T.. . 2015. Qualitative analysis of the vaginal microbiota of healthy cattle and cattle with genital-tract disease. Genet. Mol. Res. 14:6518–6528. doi: 10.4238/2015.June.12.4 [DOI] [PubMed] [Google Scholar]
- Rodrigues Hoffmann, A. 2017. The cutaneous ecosystem: the roles of the skin microbiome in health and its association with inflammatory skin conditions in humans and animals. Vet. Dermatol. 28:60–e15. doi: 10.1111/vde.12408 [DOI] [PubMed] [Google Scholar]
- Ross E. M. 2019. Characterisation of the Brahman genome. P.PSH.0868 Final Report., Locked Bag 1961, North Sydney NSW 2059, Australia, Limited MaLA, Meat and Livestock Australia Limited. [Google Scholar]
- Sandal, I., and Inzana T. J.. . 2010. A genomic window into the virulence of Histophilus somni. Trends Microbiol. 18:90–99. doi: 10.1016/j.tim.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Schlafer D, Miller R, Maxie M. . 2007. Jubb, Kennedy, and Palmer’s pathology of domestic animals. St. Louis Missouri: Elsevier. [Google Scholar]
- Schmalenberger, A., Schwieger F., and Tebbe C. C.. . 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557–3563. doi: 10.1128/AEM.67.8.3557-3563.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigel, N. Y., Adler-Ashkenazy L., Scheinin S., Goshen T., Arazi A., Pasternak Z., and Gottlieb Y.. . 2017. Characterization and identification of microbial communities in bovine necrotic vulvovaginitis. Vet. J. 219:34–39. doi: 10.1016/j.tvjl.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Siranosian B, and Bhatt A.. . 2021. bhattlab/Kraken2_classification, vVersion 1.0.0.
- Soergel, D. A., Dey N., Knight R., and Brenner S. E.. . 2012. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 6:1440–1444. doi: 10.1038/ismej.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, R. K., Nickerson E., Simons J. F., Jänne P. A., Tengs T., Yuza Y., Garraway L. A., LaFramboise T., Lee J. C., Shah K., . et al. 2006. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat. Med. 12:852–855. doi: 10.1038/nm1437 [DOI] [PubMed] [Google Scholar]
- Thompson, L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., Prill R. J., Tripathi A., Gibbons S. M., Ackermann G., . et al. ; Earth Microbiome Project Consortium. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 551:457–463. doi: 10.1038/nature24621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, R. R., Judd L. M., Gorrie C. L., and Holt K. E.. . 2017. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3:e000132. doi: 10.1099/mgen.0.000132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, D. E., Lu J., and Langmead B.. . 2019. Improved metagenomic analysis with Kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, H., Yang C., Sun Y., Igarashi Y., Jin T., and Luo F.. . 2020. PacBio long reads improve metagenomic assemblies, gene catalogs, and genome binning. Front. Genet. 11:516269. doi: 10.3389/fgene.2020.516269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, M., Feehily C., Walsh C. J., Fenelon M., Murphy E. F., McAuliffe F. M., van Sinderen D., O’Toole P. W., O’Sullivan O., and Cotter P. D.. . 2020. Evaluation of methods for the reduction of contaminating host reads when performing shotgun metagenomic sequencing of the milk microbiome. Sci. Rep. 10:21665. doi: 10.1038/s41598-020-78773-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., and Doak T. G.. . 2009. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 5:e1000465. doi: 10.1371/journal.pcbi.1000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaheer, R., Noyes N., Ortega Polo R., Cook S. R., Marinier E., Van Domselaar G., Belk K. E., Morley P. S., and McAllister T. A.. . 2018. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 8:5890. doi: 10.1038/s41598-018-24280-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Kobert K., Flouri T., and Stamatakis A.. . 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 30:614–620. doi: 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.