Abstract
As the main component of the Gram-negative bacterial cell wall, lipopolysaccharide (LPS) is well documented as an inducer of inflammation in bovine mammary cells. Lycium barbarum (goji) polysaccharides (LBP) have been used in nonruminants as prebiotics to improve growth performance, immune ability, and antioxidant capacity. We aimed to investigate the underlying effects of LBPs on proinflammatory responses in LPS-stimulated primary bovine mammary epithelial cells (bMECs). Cells were isolated from mammary tissue of three lactating Holstein cows without clinical disease (30.26 ± 3.1 kg/d of milk yield; 175 ± 6 DIM). For the pre-experimental treatment, bMECs were precultured with serum-free medium for 12 h. Treatments were as follows: pretreatment with culture medium devoid of LPS or LBP for 30 h (CON); CON for 24 h followed by challenge with 2 μg/mL LPS for 6 h (LPS); pretreatment with 100 or 300 μg/mL LBP for 24 h followed by LPS challenge (2 μg/mL) for 6 h (LBP(100)+LPS; LBP(300)+LPS). To further determine if the effect of LBP on immuneregulation is peroxisome proliferator-activated receptor-γ (PPARγ) activation dependent, an inhibitor of PPARγ, GW9662, at a concentration of 1 μM was used. Cells treated with LBP at 100, 300, and 500 μg/mL had upregulated protein abundance of PPARγ, while PGC1α had a higher expression only at 300 μg/mL of LBP treatment. Compared with CON, cells pretreated with LBP at 100 and 300 μg/mL had greater protein abundance of SCD1 and SREBP1. 5-Ethynyl-2′-deoxyuridine (EdU) staining and cell wound healing assays showed that the negative effect of LPS alone on cell proliferation was reversed by pretreatment with LBP at both 100 and 300 μg/mL. Upregulation of gene and protein abundance of proinflammatory factors and cytokines (COX-2, NLRP3, TNF-α, IL-1β, and IL-6) induced by LPS stimulation were alleviated by LBP pretreatment at 300 μg/mL (more than 2-fold decrease). Compared with LPS challenge alone, phosphorylation of proteins involved in NF-κB (IκBα and p65) and MAPK (p38, JNK, and ERK) pathways was downregulated following LBP treatment. Additionally, inhibition of PPARγ by GW9662 weakened the protective effect of LBP on LPS-induced protein abundance of phosphorylated p65, COX-2, IL-1β, and TNF-α. These results indicated that the protective effect of LBP on LPS-induced bMECs inflammatory responses is PPARγ activation-dependent. As such, this knowledge might help design strategies for intervening against the detrimental effects of bovine mastitis.
Interpretive summary
Current research examined Lycium barbarum polysaccharides (LBP) for combating LPS-induced inflammatory responses in primary bovine mammary epithelial cells. We uncovered a preventive role of LBP in reducing detrimental effects induced by LPS including inhibition of NF-κB and MAPK along with peroxisome proliferator-activated receptor-γ (PPARγ) activation. The decrease in cell proliferation due to LPS was curtailed by pretreatment with LBP. Moreover, the effect of LBP on regulation of inflammatory responses in bovine mammary epithelial cell was PPARγ dependent. Collectively, data suggest that LBP reverses LPS-induced inflammatory response via MAPK/NF-κB signaling in a PPARγ-activation-dependent manner. Thus, the study provides new insights into therapeutic strategies for combating mastitis using LBP and highlighted the link between PPARγ and regulation of mammary cell inflammation.
Keywords: Lycium barbarum polysaccharides, bovine mammary cells, PPARγ, inflammatory responses, NF-κB signaling pathway
Lycium barbarum polysaccharides offsets LPS-induced inflammation through a MAPK/NFKB/PPARɣ pathway.
Introduction
Infectious bovine mastitis is characterized by bacterial invasion of the mammary gland and threatens milk production, which results in economic losses (De Vliegher et al., 2012). Primary bovine mammary epithelial cells (bMECs) are a useful resource for studies of mechanisms associated with bacterial infection of the mammary gland (Thomas et al., 2016; Chen et al., 2020b). For example, invasion of Gram-negative bacteria such as Escherichia coli can be mimicked by addition of lipopolysaccharide (LPS) in vitro or in vivo (Giovannini et al., 2017; Carl-Fredrik et al., 2018).
Challenge of mammary cells with LPS stimulates the innate immunity response, including toll-like receptor 4, NF-κB, and mitogen-activated protein kinase (MAPK) signaling (Takeda et al., 2003; Shizuo et al., 2006). During an LPS challenge, NF-κB and MAPK signaling are triggered by the activation of myeloid differentiation factor 88 (MyD88) and c-Jun N-terminal kinase (JNK), respectively (Li et al., 2013; Fu et al., 2014; Jiang et al., 2020). Challenge with LPS also increases abundance of TNF-α, IL-1β, and IL-6 in bovine mammary tissue (Wellnitz and Kerr, 2004; Wang et al., 2016b), which has led to the suggestion that targeting NF-κB would be an effective therapeutic option against mastitis when aiming to suppress the proinflammatory cytokine response of mammary cells (Loor et al., 2011).
Recent studies have revealed that dietary supplementation with selenium or methionine hastens LPS-induced inflammatory responses through the inhibition of NF-κB translocation and MAPK signaling in bMEC (Wang et al., 2018; Dai et al., 2020a). Thymol and stevioside have also been shown to possess anti-inflammatory properties in LPS-stimulated mouse mammary epithelial cells through the regulation of NF-κB and MAPK signaling pathways (Liang et al., 2014; Wang et al., 2014). A negative effect of bovine mammary inflammation due to mastitis on peroxisome proliferator-activated receptor-γ (PPARγ) signaling and inhibition of lipogenesis was first reported more than 10 yr ago (Moyes et al., 2009). More recent work has also underscored the relevance of PPARγ in the regulation of inflammation induced by H2O2 through MAPK/NF-κB pathway in skeletal muscle cells (Kim et al., 2017). Because the PPAR pathway in ruminants can be manipulated by nutrients (Bionaz et al., 2013), it is a logical target for interventions designed to counteract the negative effects of localized inflammation within mammary cells.
Lycium barbarum polysaccharides (LBPs) extracted from Chinese wolfberry (goji) are known as a prebiotic dietary supplement in China (Liu et al., 2015). Along with its multiple positive biological functions against cancer development, viral infections, aging, cardiovascular problems, and immune function in humans, the benefits of LBP also extend to livestock including broilers and piglets (Yu et al., 2005; Cheng et al., 2015; Chen et al., 2018, 2020a). Dietary LBP supplementation at 2,000 mg/kg improved growth performance, antioxidant capacity, and immune function in broilers (Long et al., 2020). In piglets, LBP led to downregulation of E. coli and Firmicutes numbers in the ileum and cecum and enhanced antioxidant defenses (Chen et al., 2020a). Although LBP has shown promise as an alternative to antibiotic additives (Hao et al., 2021), whether LBP could suppress or hasten the mammary cell inflammatory response when exposed to LPS remains unknown. Thus, the present study aimed to investigate the effect of LBP on LPS-stimulated bMEC on the MAPK/PPARγ/NF-κB signaling pathway.
Materials and Methods
Chemicals
LBP with a purity of more than 80% was purchased from Solarbio (P7850, Solarbio Life Sciences, Beijing, China). Polysaccharide fractions were identified by high-performance anion-exchange chromatography as methods indicated in Supplementary Figure S1. The LPS used in all experiments was from E. coli O111:B4 lyophilized powder (L2630, Sigma, St. Louis, MO).
Ethics
All experimental procedures were approved by the Animal Experiment Committee of Yangzhou University (YZU202003-321) in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals published by the Ministry of Science and Technology, China in 2004. All experimental protocols were performed in accordance with approved guidelines and regulations.
Cell culture conditions
Culture of primary bMEC was performed as described previously (Chen et al., 2019; Ma et al., 2019). Briefly, mammary tissue obtained from three mid-lactating Holstein cows (30.26 ± 3.1 kg/d of milk yield, 2.67 parity, and 175 ± 6 DIM) without incidence of clinical disease was used for cell isolation and purification. Cells from each cow were pooled into a single sample. All experiments were performed with cells at the fourth to sixth passage. Cells (2 × 105) were seeded in six-well plates with overnight incubation in DMEM with 10% heat inactivated fetal bovine serum (DMEM/F-12, 31330095 Gibco, CA) and antibiotics (penicillin 100 IU/mL; streptomycin 100 μg/mL). All media supplements were purchased from Gibco (Thermo Fisher Scientific, CA). Cells were maintained at 37 °C in a humidified 5% CO2 incubator.
Experimental design
The inflammation model was tested and optimized with respect to LPS concentration as described previously (Xu et al., 2021b). For the pre-experimental treatment, the bMECs were precultured with serum-fasting medium (DMEM/F-12) for 12 h. Treatments were as follows: pretreatment with culture media (DMEM/F-12 with 10% heat inactivated FBS) devoid of LPS or LBP for 30 h (CON), CON for 24 h followed by challenge with 2 μg/mL LPS for 6 h (LPS), pretreatment with 100 or 300 μg/mL LBP for 24 h followed by LPS challenge (2 μg/mL) for 6 h (LBP(100)+LPS; LBP(300)+LPS). Additionally, to verify that the effect of LBP on immune-modulation is PPARγ activation dependent, the inhibitor of PPARγ, GW9662, at a concentration of 1 μM was applied.
Flow cytometry
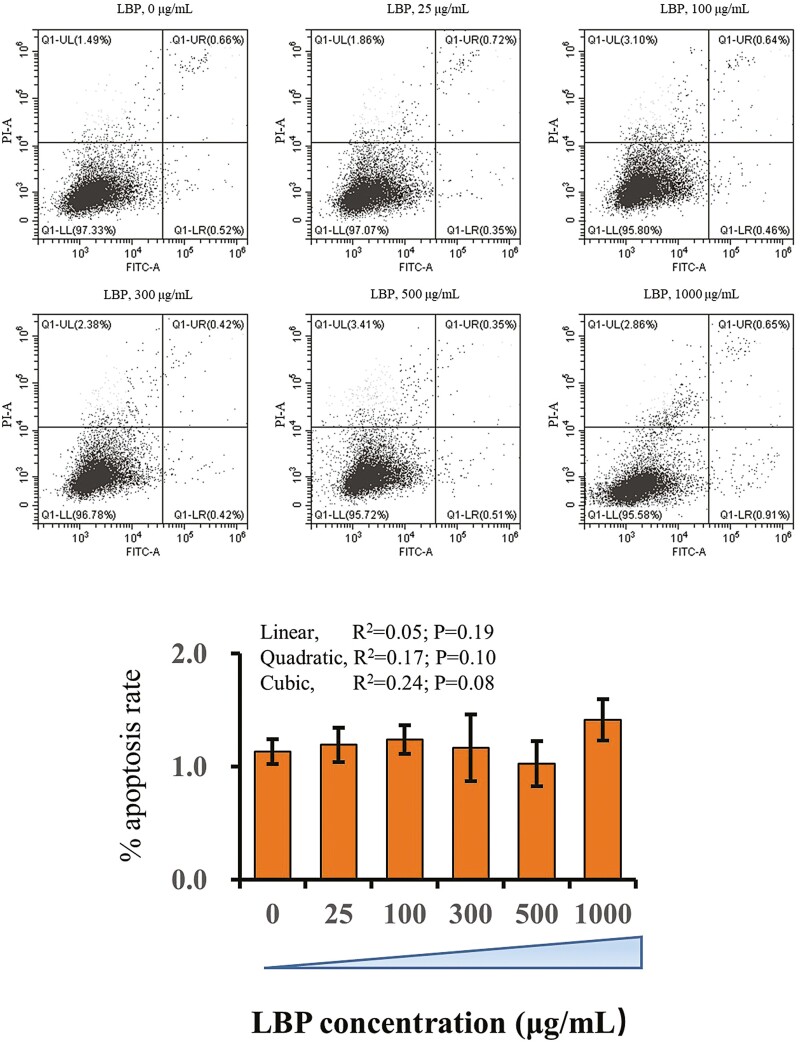
The bMECs were seeded into six-well plates (2 × 105 cells/well). Cells were maintained in medium with 10% (v/v) fetal bovine serum and the various treatments selected for this study. Cells were treated with LBP at concentrations of 0, 25, 100, 300, 500, and 1,000 μg/mL. The apoptotic effect of LBP on differentiating bMECs was evaluated using an Annexin V staining kit (#11858777001, Sigma-Aldrich). Cells were collected via trypsin, stained, and analyzed by flow cytometry using the FACSCalibur (BD Biosciences, Franklin Lakes, NJ).
Wound healing assay
Cells were seeded in six-well plates (2 × 105 cells/well) and wounded by scraping with a micropipette tip after treatments had been applied. The spreading of wound closure was observed after 12 h. Images were captured using a phase-contrast microscope (Nikon, Tokyo, Japan) either immediately or 12 h after wounding. Wound closure rate was determined using image J software (LOCI).
RNA extraction and quantitative real-time PCR analysis
Total RNA was isolated with RNA isolater (No.: R401-01, Vazyme, Nanjing, China) according to the manufacturer’s instructions (https://v1.cecdn.yun300.cn/100001_20052050 05%2FR401-%20%E8%AF%B4%E6%98%8E% E4%B9%A6.pdf). cDNA was synthesized using HiScript III RT SuperMix (R323-01, Vazyme) and then purified with a purification kit (Axygen, Tewksbury, MA). qRT-PCR was performed using HiScript II One Step qRT-PCR SYBR Green Kit (Vazyme) on an Applied Biosystems QuantStudio 5 Real-Time PCR System (Applied Biosystems, Foster City, CA) according to a previous study (Xu et al., 2018). Primers were designed with Premier 6.0 software (Premier Biosoft International, Palo Alto, CA) as reported in previous publications(Xu et al., 2015, 2017). The genes GAPDH, RPS9, and UXT were used as internal controls, and their geometric mean used to normalize target gene expression data. The validity of the selected internal control genes as references for normalizing gene expression in mammary samples has been previously reported (Zhou et al., 2018). The 2−ΔΔCt method was used for relative quantification (Pfaffl, 2001).
EdU detection
Following the manufacturer’s protocols, the BeyoClick 5-ethynyl-2′-deoxyuridine (EdU) cell proliferation kit with Alexa Fluor 555 (Beyotime, Shanghai, China) was used to measure the ability of bMEC to proliferate under different treatments. Cells were incubated with 10 μM EdU solution for 2 h, subsequently fixed with 4% paraformaldehyde for 20 min at room temperature (RT), and permeabilized with 0.3% Triton X-100 in PBS for 15 min at RT. For nuclear staining, cells were incubated with Hoechst for 10 min at RT protected from light. Finally, cells were imaged with a fluorescence microscope DMi8 Microsystems GmbH (Leica, Wetzlar, Germany) at 200× magnification.
Western blotting
Western blot was performed using protocols described previously (Chen et al., 2019). Briefly, equal amounts of protein isolated from bMEC by RIPA lysis buffer (Beyotime, Shanghai, China) were separated on 4% to 20% SDS polyacrylamide gels. Proteins were transferred onto nitrocellulose membranes (Millipore, Billerica, MA), which were incubated with primary antibodies overnight at 4 °C. After washing six times, blots were incubated with horseradish peroxidase-coupled secondary antibodies. Differences in protein transfer efficiency between blots were normalized with GAPDH quantification. The gray values of the bands of each target protein were quantified with Bio-Rad image system analysis software (Bio-Rad, Hercules, CA). Primary antibodies for p-P65, p65, p-IκBα, IκBα, p-p38 (Thr180/Tyr182), p38, p-JNK (Thr183/Tyr185), JNK, p-ERK (Thr202/Tyr204), ERK, COX-2, IL-1β, TNF-α, NLRP3, PPARγ, PGC1α, and SCD1 were purchased from Cell Signaling Technology (Danvers, MA; #3033, #8242, #2859, #4812, #4511, #8690, #4668, #9252, #4370, #9102, #12282, #12703, #6945, #15101, #2435, #2178, #2794), and were diluted 1:1,000 for incubation. Primary antibodies for SREBP1c and GAPDH were purchased from Abcam Corporation (ab28481 and ab8245) and were diluted 1:5,000 for incubation.
Immunofluorescence
Immunofluorescence was performed using protocols described previously (Xu et al., 2018). Mammary cells (2 × 104 cells/well) were plated onto 12-well plates, fixed with 4% paraformaldehyde for 15 min, then washed three times with PBS and incubated with 0.3% or 0.5% Triton X-100 for 15 min at RT to increase the permeability. Cells were washed three times with PBS, incubated for 1 h with 5% BSA at 37 °C, and then incubated at 4 °C overnight with primary antibody (the same as that used in the Western blot analysis) in PBS containing 1% BSA and 0.3 Triton X-100 (T9284, Sigma-Aldrich). After PBS washes, cells were stained for 1 h with FITC labeled goat anti-rabbit FITC secondary antibody in a dark 37 °C room and then washed three times with PBS. DAPI (1 μg/mL; D8417, Sigma-Aldrich) was used for nuclear counterstaining for 5 min, and then cells were washed three times. Cells were imaged using a DMi8 Microsystems GmbH (Leica).
Statistics
Statistical analyses were performed in a similar fashion as described in a previous publication(Xu et al., 2021a). Briefly, abundance data were log-2 transformed prior to analysis to fit normal distribution of residuals. The resulting LS means were log-2 back-transformed for ease of interpretation and reported in figures. In the dose–response experiments, contrast analysis was performed to assess the linear, quadratic, and cubic effects of LBP on all response criteria. All data were analyzed using one-way ANOVA with Dunnett’s post-test by SAS Statistics (v 9.2, SAS Institute Inc., Cary, NC). Data are expressed as the means ± standard error of the means (mean ± SEM). Differences with P-values <0.05 were considered statistically significant. Experiments were performed in triplicate, with three replicates in each experiment.
Results
Cytotoxicity of LBP in bMEC
There was no significant effect of LBP dose on cell apoptosis. The doses of LBP used in the present study did not change apoptotic rate of bMEC during the experiments (Figure 1).
Figure 1.
Flow cytometry analysis of bMEC treated with doses ranging from 0 to 1,000 μg/mL LBP. Cells were treated with LBP for 24 h at doses of 0, 25, 100, 300, 500, and 1,000 μg/mL. (A) X-axis indicates the absorbance of FITC-A; Y-axis indicates the absorbance of fluorescein isothiocyanate−Annexin V. (B) Apoptosis percentages in cultures exposed to doses ranging from 0 to 10 mM of metformin. Results are expressed as means ± SEM. *P < 0.05 vs. the NC group. These data are representative of three independent experiments.
LBP activates PPARγ in a dose-dependent manner
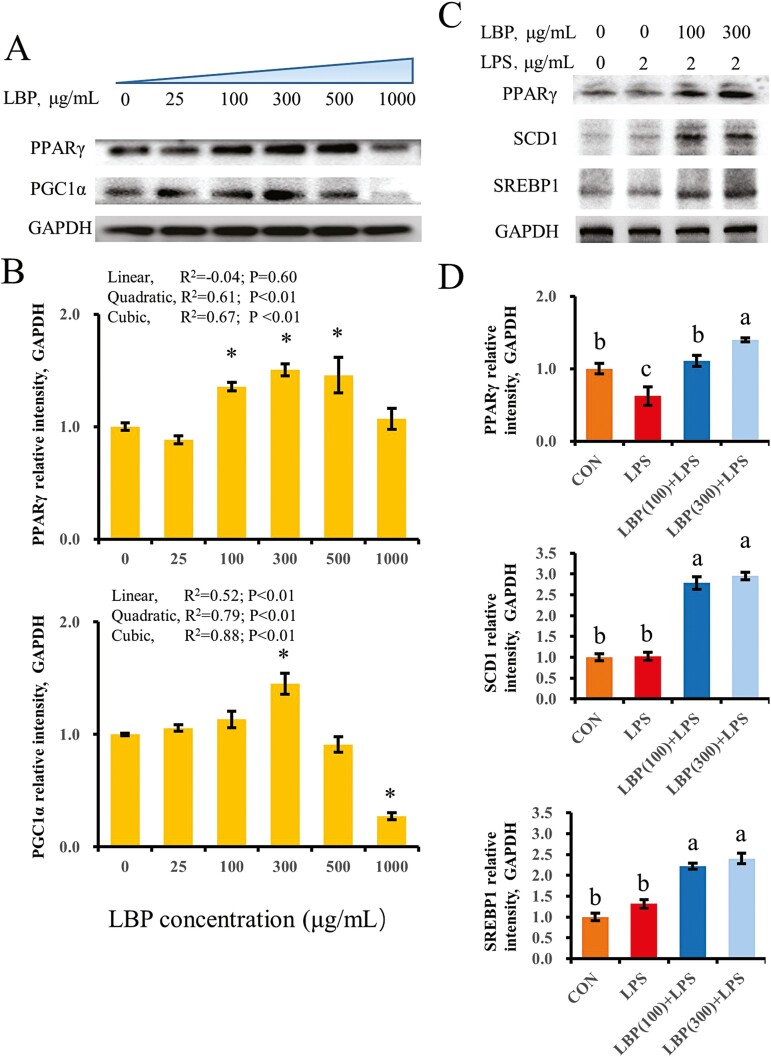
To indicate a trend across doses and a plateau, less frequently a nonmonotonic relationship or a steep change in slope, data were analyzed using linear, quadratic, and cubic contrasts to assess “linear”, “quadratic”, or “cubic” effects of dose in this study. Compared with cells treated with 0 μg/mL of LBP, concentrations of 100, 300, and 500 μg/mL upregulated protein abundance of PPARγ (quadratic, P < 0.05; cubic, P < 0.05; Figure 2A and B). However, among cells treated with LBP from 0 to 1,000 μg/mL, protein abundance of PGC1α in cells supplemented with LBP at 300 μg/mL was the greatest (P < 0.05). Furthermore, compared with the control group, supplementation of LBP at 1,000 μg/mL downregulated PGC1α protein abundance (P < 0.05). Stimulation with LPS downregulated protein abundance of PPARγ as compared with control cells (P < 0.05). However, protein abundance of SCD1 and SREBP1 was not affected by LPS challenge alone (P > 0.05). In contrast, supplementation with LBP at 100 and 300 μg/mL reversed the downregulation of PPARγ induced by LPS. It is noteworthy that, compared with the LPS group, SCD1 and SREBP1 protein abundance were also upregulated in cells stimulated with LPS and supplemented with LBP at 100 and 300 μg/mL (P < 0.05) (Figure 2C and D).
Figure 2.
Activation of PPARγ in cells supplemented with LBP in a dose-dependent way. Results are expressed as means ± SEM. (A, B) Immunoblotting and intensity from the respective blots. Cells were treated with LBP for 24 h at concentrations of 0, 25, 100, 300, 500, and 1,000 μg/mL. *P < 0.05 vs. the cell without treatment with LBP at 0 μg/mL. (C, D) Immunoblotting and intensity from the respective blots. Cells were pretreated with or without LBP at doses of 100 or 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. Letters in superscript indicate that the difference between groups was significant (P < 0.05). Protein abundance was normalized by the respective abundance of GAPDH.
Effect of LBP on proliferation and wound healing of LPS-challenged bMEC
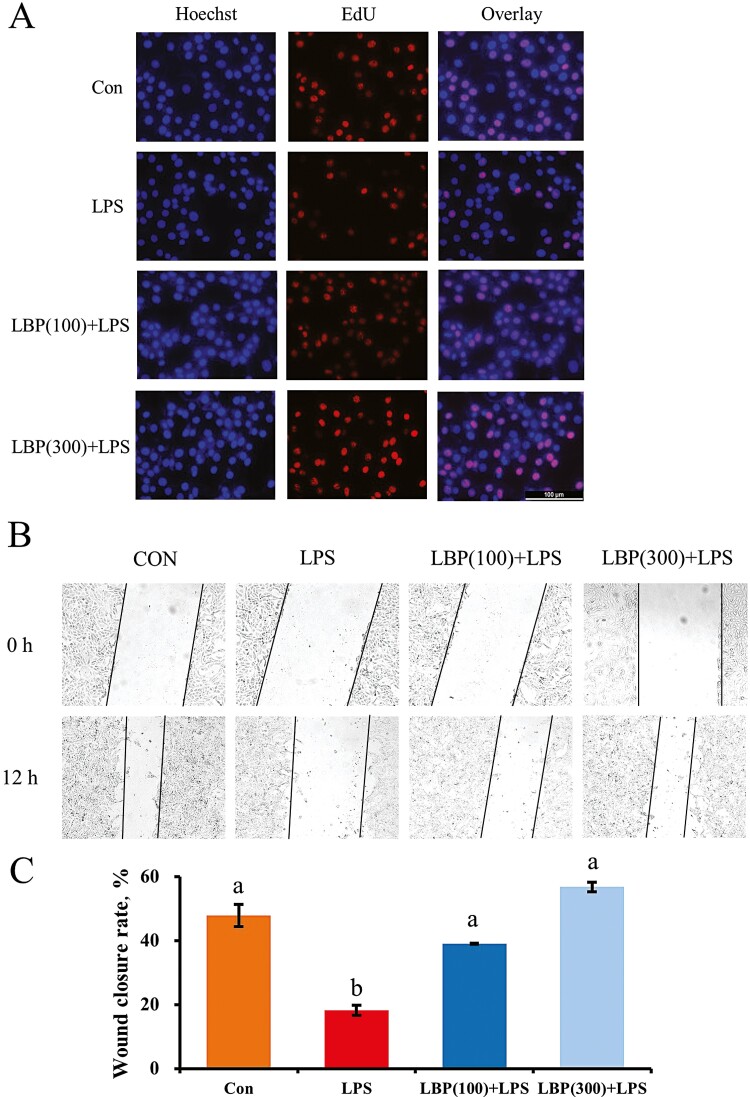
Measurements of proliferation and wound healing of bMECs were conducted to assess the cells activity in LPS stimulation and pretreatment of LBP. Compared with the control, EdU staining of LPS-stimulated cells decreased markedly (Figure 3). However, supplementation with LBP at 100 and 300 μg/mL blocked the inhibitory effect on cell proliferation in LPS-primed cells. As shown in results of the wound healing experiment, compared with the control group, LPS had an inhibitory effect on cell migration with respect to the enlarged wound gap after 12 h. In contrast, supplementation of LBP closed the wound at both 100 and 300 μg/mL.
Figure 3.
Proliferation of cells pretreated with LBP followed by LPS stimulation using 5-ethynyl-2′-deoxyuridine (EdU) and wound healing assay. (A) EdU assay for determination of cell proliferation. Cells were pretreated with or without LBP at doses of 100 or 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. (B) Wound healing assay was performed for the migration ability of cells pretreated with LBP at dosses of 100 or 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. Results are expressed as means ± SEM. Letters in superscript indicate that the difference between groups was significant (P < 0.05).
LBP suppresses LPS-induced pro-inflammatory gene and protein abundance
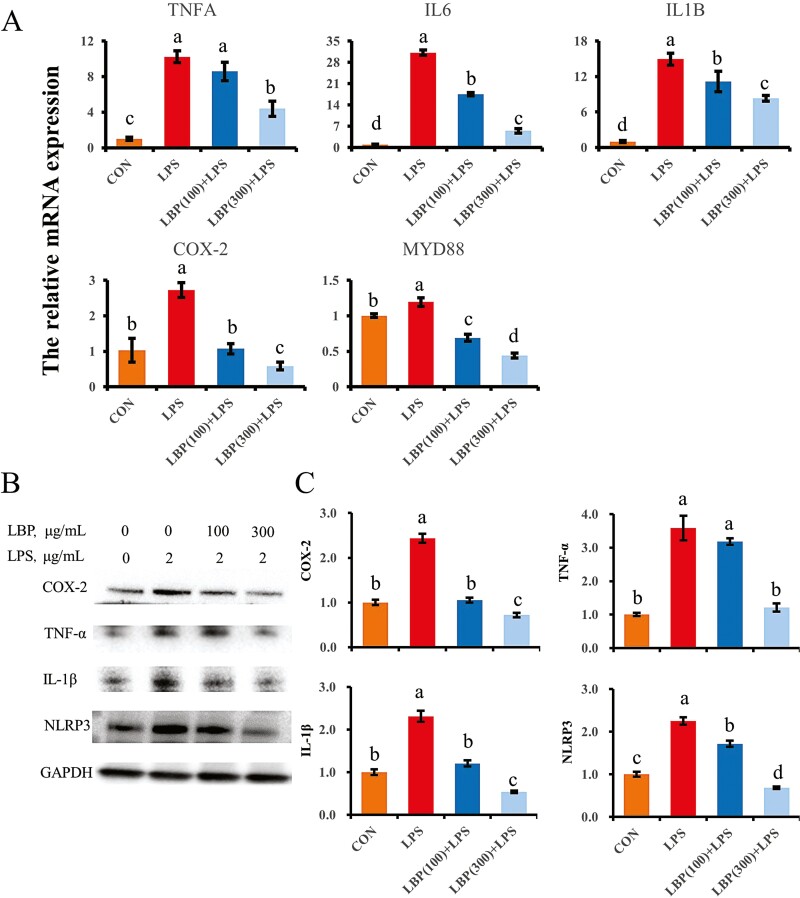
Upregulation of genes related to inflammation (TNFA, IL6, and IL1B) was detected after challenge with LPS (Figure 4). Compared with LPS alone, supplementation with LBP at 100 and 300 μg/mL downregulated IL6 and IL1B mRNAs (P < 0.05). In addition, compared with LPS alone, LBP at 300 μg/mL downregulated abundance of TNFA (P < 0.05), whereas 100 μg/mL of LBP did not change TNFA abundance (P > 0.05). LPS also upregulated COX-2 mRNA abundance, while pretreatment with LBP reversed this effect (P < 0.05). Similarly, pretreatment with LBP inhibited the LPS upregulation of MYD88 mRNA abundance especially at 300 μg/mL (P < 0.05).
Figure 4.
LBP reversed the genes and proteins expression of proinflammatory factors induced by LPS challenge. (A) Abundance of genes associated with pro-inflammation, normalized by the geometric mean of the internal control genes (GAPDH, UXT, and RPS9). Abundance of genes in CON group was set as 1.0. (B, C) Protein levels of proinflammatory factors in each group of bMEC. Results are expressed as means ± SEM. Letters in superscript indicate that the difference between groups was significant (P < 0.05).
There was a consistent upregulation of pro-inflammatory protein abundance in response to LPS stimulation, whereas pretreatment with LBP following LPS challenge downregulated protein abundance of TNF-α and IL-1β compared with LPS alone (P < 0.05). Furthermore, LBP supplementation at concentrations of 100 and 300 μg/mL reversed upregulation of COX-2 and NLRP3 protein abundance induced by LPS stimulation.
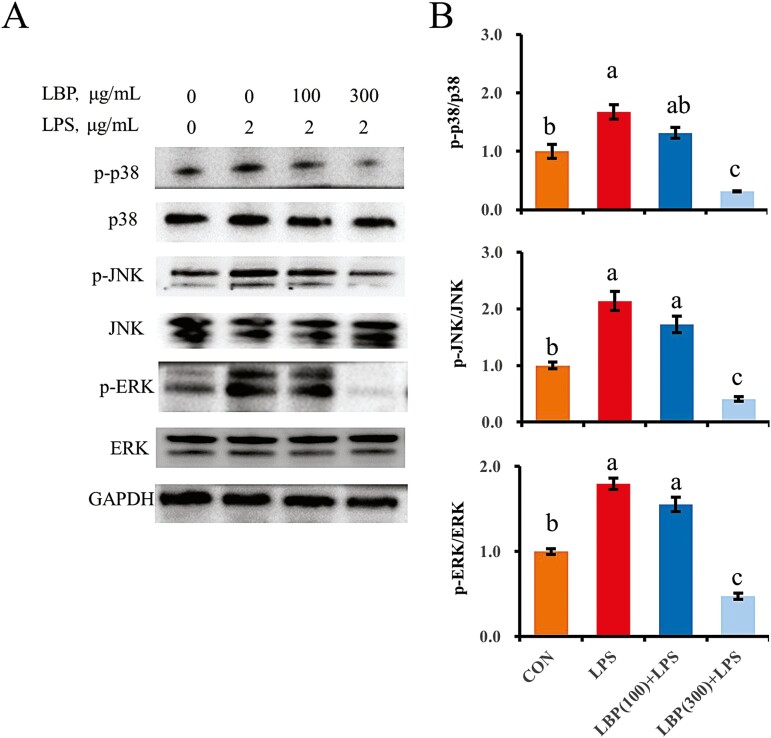
LBP inhibits the LPS-induced activation of the MAPK pathway
Challenge with LPS activated the MAPK pathway in bMEC, which was manifested in the increased ratio of phosphorylated p38 (Thr180/Tyr182) to total p38, phosphorylated JNK (Thr183/Tyr185) to total JNK and phosphorylated ERK (Thr202/Tyr204) to total ERK (P < 0.05; Figure 5). Although LBP at a concentration of 100 μg/mL had no effects, LBP at 300 μg/mL downregulated the ratio of phosphorylated p38 to total p38, phosphorylated JNK to total JNK and phosphorylated ERK to total ERK compared to the LPS group (P < 0.05).
Figure 5.
The suppressive effect of LBP on the LPS-induced activation of MAPK signaling pathway. Immunoblotting and intensity from the respective blots represent for the phosphorylated p38, phosphorylated JNK, and phosphorylated ERK. Cells were pretreated with or without LBP at doses of 100 or 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. Results are expressed as means ± SEM. Letters in superscript indicate that the difference between groups was significant (P < 0.05).
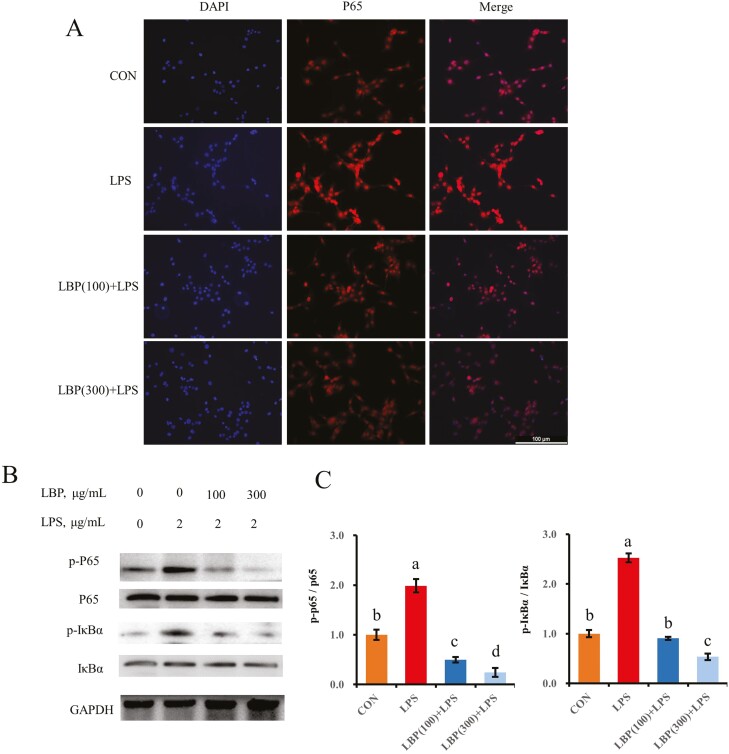
Effect of LBP on LPS-induced activation of the NF-κB pathway
Compared with the control, protein abundance of phosphorylated IκBα and NF-κB subunit p65 was upregulated by LPS challenge (P < 0.05; Figure 6). However, compared with the LPS group, a decrease of phosphorylated IκBα and p65 was observed in cells pretreated with LBP at 100 and 300 μg/mL (P < 0.05). In addition, there was translocation of p65 into the nucleus in cells treated with LPS (Figure 6A). However, pretreatment with LBP inhibited this LPS-induced NF-κB nuclear translocation.
Figure 6.
The inhibitory effect of LBP pretreatment on the NF-κB signaling pathway. (A) Immunofluorescence for detecting p65 activity. After fixation and permealization steps, cells were stained with antibody against subunit p65 of NF-κB (Cy3-labeled secondary antibody) and counterstained with DAPI (blue) for nuclear staining. (B, C) Immunoblotting and intensity from the respective blots represent for the phosphorylated p65 and phosphorylated IκBα. Cells were pretreated with or without LBP at doses of 100 or 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. Results are expressed as means ± SEM. Letters in superscript indicate that the difference between groups was significant (P < 0.05).
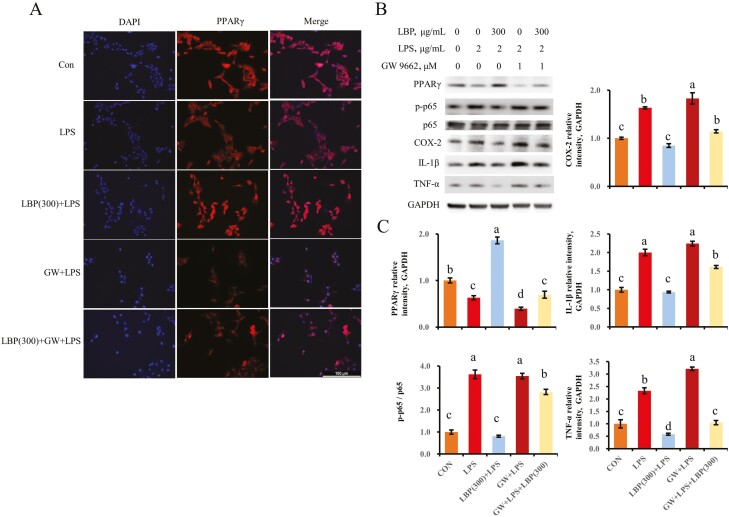
PPARγ activation is involved in the LBP-mediated anti-inflammatory response
Challenge with LPS led to lower staining of bMEC when compared with the control group (Figure 7A). A strong staining of PPARγ was observed in cells pretreated with LBP at 300 μg/mL following LPS stimulation. However, the antagonist of PPARγ GW 9664 inhibited staining of the cells pretreated with LBP at 300 μg/mL.
Figure 7.
PPARγ is required for the LBP-mediated inhibition of proinflammatory proteins expression. (A) Immunofluorescence for detecting PPARγ activity. After fixation and permealization steps, cells were stained with antibody against PPARγ (Cy3-labeled secondary antibody) and counterstained with DAPI (blue) for nuclear staining. GW9662 was represented for the inhibitory effect of PPARγ activity. (B, C) Immunoblotting and intensity from the respective blots. Cells were pretreated with or without LBP at dose of 300 μg/mL for 24 h, followed by the induction of 2 μg/mL LPS for 6 h. GW9662 was represented for the inhibitory effect of PPARγ activity. Results are expressed as means ± SEM. Letters in superscript indicate that the difference between groups was significant (P < 0.05).
As indicated by upregulation (P < 0.05) of TNF-α, IL-1β, and COX-2 in bMEC pretreated with LBP and the PPARγ inhibitor following LPS challenge, the supplementation with LBP failed to reduce the pro-inflammatory response when PPARγ was blocked. In addition to the interdiction of suppressive effect of LBP on the expression of proinflammatory factors, the phosphorylation of p65 was also not decreased due to pretreatment with the PPARγ inhibitor (Figure 7B and C).
Discussion
Alternatives to antibiotics have been proposed as one of the most-effective ways to reduce drug resistance and counteract detrimental effects of inflammation during bacterial infection(Liu et al., 2020). Work with many cell types has underscored that PPARγ signaling serves an important anti-inflammatory function (Are et al., 2008; Zenhom et al., 2011; Kundu et al., 2014; Hasan et al., 2019). However, the mechanisms driving the protective effects of LBP on the pro-inflammatory responses in bMEC remain unknown. The present study indicated that PPARγ is involved in the anti-inflammatory function of LBP through the inhibition of the MAPK/NF-κB pathway in bMEC.
Regulatory effects of PPARs have been reported at both cellular and whole body levels and especially in the context of lipid metabolism (Lamichane et al., 2018; Hong et al., 2019). The fact that protein abundance of PPARγ and PGC1α was upregulated in response to LBP at 100 and 300 μg/mL suggested that this compound might be able to trigger biological effects in vivo. PPARγ targets the transcription of lipogenic genes including SCD1 and SREBP1c in bMEC (Kadegowda et al., 2009). Thus, the upregulation of SCD1 and SREBP1 protein abundance in this study was consistent with activation of PPARγ driven by LBP supplementation, and also indicated this compound can activate fatty acid synthesis in bovine mammary cells. In addition to these metabolic effects, results from EdU assay and scratch test both appeared to indicate that LBP might have a positive effect on mammary cell proliferation as reported in nonruminant cells (Zhang et al., 2019). Clearly, these metabolic and cellular effects associated with the supply of LBP merit further investigation in vivo.
The anti-inflammatory effect of LBP has been reported previously in rodent models(Xiao et al., 2012). Cytokines such as TNF-α, IL-6, and IL-1β are released during stimulation with endotoxins and mastitis pathogens (Nakajima et al., 1997; Blum et al., 2017). Thus, the downregulation of mRNA and protein abundance of TNFA, IL6, and IL1B in response to LBP after LPS stimulation provides evidence of an anti-inflammatory effect of LBP in bovine cells. In monocytes and macrophages, the enzyme COX-2 responds to inflammation through the synthesis of prostaglandins (Hinson et al., 1996). The fact that LBP attenuated the LPS-induced upregulation of COX-2 in bMEC as well as the inflammasome NLRP3 is further evidence of its anti-inflammatory effect. Clearly, LBP can help dampen inflammatory responses in bovine cells, but the underlying mechanisms for its effects could involve multiple molecules and encompass different forms of regulation.
From a mechanism standpoint, our data suggested that MAPK and NF-κB signaling pathways are closely involved in the LBP-mediated inhibition of inflammatory responses. The suppression of phosphorylated subtypes (p38, JNK, and ERK) of MAPK in response to LBP suggested an important immune-modulatory effect. Although the interaction of activated MAPKs and transcription factors such as NF-κB are unknown in bovine mammary epithelial cells, available lines of evidence indicate that the LPS-induced upregulation of cytokines is under control of both signaling pathways(Kang et al., 2004; Wang et al., 2016a; Dai et al., 2020b). The NF-κB complex of transcription factors acts as a master switch in regulating a number of proinflammatory genes, and its activation relies on disengaging from IκB and translocating from the cytoplasm to the nucleus(Hayden and Ghosh, 2011). Thus, the fact that LBP inhibited the degradation of IκBα and phosphorylation of NF-κB subunit p65 induced by LPS along with the reduction in translocation of NF-κB into the nucleus suggested that this compound effectively suppressed the LPS stimulated proinflammatory responses.
In addition to the role in regulating lipid metabolism, accumulating evidence indicates that PPARγ is also involved in the modulation of E. coli- or LPS-induced inflammatory responses in bMEC (He et al., 2017; Ma et al., 2019). Those data led us to examine the relevance of this nuclear receptor on the anti-inflammatory effects of LBP. The fact that the PPARγ antagonist GW9662 blocked the positive effects of LBP on inflammation and PPARγ protein abundance during LPS challenge suggested that this compound is able to trigger PPAR signaling. This idea is also supported by the reversal of the PPARγ-mediated suppression of COX-2, IL-1β, and TNF-α abundance when cells were incubated with GW9662. Although potential interactions between PPARγ with regulators of the NF-κB complex were not determined in the current study, the direct impact of PPARγ on NF-κB has been linked with its enzymatic properties in an epigenetic fashion (Chen et al., 2003; Hou et al., 2012). Hence, the mechanisms whereby PPARγ elicits epigenetic regulation of immune molecules in bMEC remains to be determined. In the current study, we clarified that upregulation of PPARγ led to inactivation of NF-κB, while the contribution of epigenetic regulation needs to be further studied in bMEC.
In conclusion, LPS-induced inflammatory reactions in bovine mammary epithelial cells are closely regulated by immune molecules under the control of NF-κB and MAPK complexes. The LBP molecule can activate PPARγ and inhibit core elements of the inflammatory reaction under the control of NF-κB and MAPK complexes. These findings allow for the development of therapeutic approaches for the treatment of bovine mastitis using LBP.
Supplementary Material
Glossary
Abbreviations
- COX
cyclooxygenase
- EdU
5-ethynyl-2′-deoxyuridine
- ERK
extracellular regulated protein kinases
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IL
interleukin
- IκB
inhibitor of NF-κB
- JNK
c-Jun N-terminal kinase
- LBP
Lycium barbarum (goji) polysaccharides
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor kappa B
- PGC1α
peroxisome proliferator-activated receptor-γ coactlvator-1α
- PPAR
peroxisome proliferators-activated receptors
- RPS9
ribosomal protein S9
- RT-qPCR
real-time quantitative polymerase chain reaction
- TNF
tumor necrosis factor
- UXT
ubiquitously expressed transcript
Conflict of Interest Statement
The authors declare no real or perceived conflicts of interest.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 32102731, 31872324).
Authors’ Contributions
X.T., L.J., and M.Y. analyzed and interpreted the data. L.R., L.X., and W.X. performed the experiment, and X.T., H.P., and Y.Z. were major contributor in writing the manuscript. Y.Z. supports to the financial cost throughout the experiment and generate the ideas. All authors read and approved the final manuscript.
Literature Cited
- Are, A., Aronsson L., Wang S., Greicius G., Lee Y. K., Gustafsson J. A., Pettersson S., and Arulampalam V.. . 2008. Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL-10 levels in colonic epithelial cells. Proc. Natl. Acad. Sci. USA. 105:1943–1948. doi: 10.1073/pnas.0711734105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bionaz, M., Chen S., Khan M. J., and Loor J. J.. . 2013. Functional role of PPARs in ruminants: potential targets for fine-tuning metabolism during growth and lactation. PPAR Res. 2013:684159. doi: 10.1155/2013/684159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, S. E., Heller E. D., Jacoby S., Krifucks O., and Leitner G.. . 2017. Comparison of the immune responses associated with experimental bovine mastitis caused by different strains of Escherichia coli. J. Dairy Res. 84:190–197. doi: 10.1017/S0022029917000206. [DOI] [PubMed] [Google Scholar]
- Carl-Fredrik, J., Josef D., Ann-Marie G., Ida W., Moazzami A. A., Karin s., and Gunnar P.. . 2018. The effect of lipopolysaccharide-induced experimental bovine mastitis on clinical parameters, inflammatory markers, and the metabolome: a kinetic approach. Front. Immunol. 9:1487. doi: 10.3389/fimmu.2018.01487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Chu S., Wang X., Sun Y., Xu T., Mao Y., Loor J. J., and Yang Z.. . 2019. MiR-16a regulates milk fat metabolism by targeting large tumor suppressor kinase 1 (LATS1) in bovine mammary epithelial cells. J. Agric. Food Chem. 67:11167–11178. doi: 10.1021/acs.jafc.9b04883. [DOI] [PubMed] [Google Scholar]
- Chen, L., Li W., Qi D., and Wang D.. . 2018. Lycium barbarum polysaccharide protects against LPS-induced ARDS by inhibiting apoptosis, oxidative stress, and inflammation in pulmonary endothelial cells. Free Radic. Res. 52:480–490. doi: 10.1080/10715762.2018.1447105. [DOI] [PubMed] [Google Scholar]
- Chen, J., Long L., Jiang Q., Kang B., Li Y., and Yin J.. . 2020a. Effects of dietary supplementation of Lycium barbarum polysaccharides on growth performance, immune status, antioxidant capacity and selected microbial populations of weaned piglets. J. Anim. Physiol. Anim. Nutr. (Berl.). 104:1106–1115. doi: 10.1111/jpn.13247. [DOI] [PubMed] [Google Scholar]
- Chen, F., Wang M., O’Connor J. P., He M., Tripathi T., and Harrison L. E.. . 2003. Phosphorylation of PPARgamma via active ERK1/2 leads to its physical association with p65 and inhibition of NF-kappabeta. J. Cell. Biochem. 90:732–744. doi: 10.1002/jcb.10668. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Zhang Y., Zhou J., Lu L., Wang X., Liang Y., Loor J. J., Gou D., Xu H., and Yang Z.. . 2020b. Tea tree oil prevents mastitis-associated inflammation in lipopolysaccharide-stimulated bovine mammary epithelial cells. Front. Vet. Sci. 7:496. doi: 10.3389/fvets.2020.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Zhou Z. W., Sheng H. P., He L. J., Fan X. W., He Z. X., Sun T., Zhang X., Zhao R. J., Gu L., . et al. 2015. An evidence-based update on the pharmacological activities and possible molecular targets of Lycium barbarum polysaccharides. Drug Des. Dev. Ther. 9:33–78. doi: 10.2147/DDDT.S72892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, H., Coleman D. N., Hu L., Martinez-Cortés I., Wang M., Parys C., Shen X., and Loor J. J.. . 2020a. Methionine and arginine supplementation alter inflammatory and oxidative stress responses during lipopolysaccharide challenge in bovine mammary epithelial cells in vitro. J. Dairy Sci. 103:676–689. doi: 10.3168/jds.2019-16631. [DOI] [PubMed] [Google Scholar]
- Dai, H., Wei G., Wang Y., Ma N., Chang G., and Shen X.. . 2020b. Sodium butyrate promotes lipopolysaccharide-induced innate immune responses by enhancing mitogen-activated protein kinase activation and histone acetylation in bovine mammary epithelial cells. J. Dairy Sci. 103:11636–11652. doi: 10.3168/jds.2020-18198. [DOI] [PubMed] [Google Scholar]
- De Vliegher, S., Fox L. K., Piepers S., McDougall S., and Barkema H. W.. . 2012. Invited review: Mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J. Dairy Sci. 95: 1025–1040. doi: 10.3168/jds.2010-4074. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Gao R., Cao Y., Guo M., Wei Z., Zhou E., Li Y., Yao M., Yang Z., and Zhang N.. . 2014. Curcumin attenuates inflammatory responses by suppressing TLR4-mediated NF-κB signaling pathway in lipopolysaccharide-induced mastitis in mice. Int. Immunopharmacol. 20:54–58. doi: 10.1016/j.intimp.2014.01.024. [DOI] [PubMed] [Google Scholar]
- Giovannini, A. E. J., van den Borne B. H. P., Wall S. K., Wellnitz O., Bruckmaier R. M., and Spadavecchia C.. . 2017. Experimentally induced subclinical mastitis: are lipopolysaccharide and lipoteichoic acid eliciting similar pain responses? Acta Vet. Scand. 59:40. doi: 10.1186/s13028-017-0306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Z., Li Z., Huo J., Li J., Liu F., and Yin P.. . 2021. Effects of Chinese wolfberry and Astragalus extract on the antioxidant capacity of Tibetan pig liver. PLoS One 16:e0245749. doi: 10.1371/journal.pone.0245749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, A. U., Rahman A., and Kobori H.. . 2019. Interactions between host PPARs and gut microbiota in health and disease. Int. J. Mol. Sci. 20:387. doi: 10.3390/ijms20020387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden, M. S., and Ghosh S.. . 2011. NF-κB in immunobiology. Cell Res. 21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Liu W., Shi M., Yang Z., Zhang X., and Gong P.. . 2017. Docosahexaenoic acid attenuates LPS-stimulated inflammatory response by regulating the PPARγ/NF-κB pathways in primary bovine mammary epithelial cells. Res. Vet. Sci. 112:7–12. doi: 10.1016/j.rvsc.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Hinson, R. M., Williams J. A., and Shacter E.. . 1996. Elevated interleukin 6 is induced by prostaglandin E2 in a murine model of inflammation: possible role of cyclooxygenase-2. Proc. Natl. Acad. Sci. USA. 93:4885–4890. doi: 10.1073/pnas.93.10.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, F., Pan S., Guo Y., Xu P., and Zhai Y.. . 2019. PPARs as nuclear receptors for nutrient and energy metabolism. Molecules 24:2545. doi: 10.3390/molecules24142545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Moreau F., and Chadee K.. . 2012. PPARγ is an E3 ligase that induces the degradation of NFκB/p65. Nat. Commun. 3:1300. doi: 10.1038/ncomms2270. [DOI] [PubMed] [Google Scholar]
- Jiang, A., Zhang Y., Zhang X., Wu D., Liu Z., Li S., Liu X., Han Z., Wang C., Wang J., . et al. 2020. Morin alleviates LPS-induced mastitis by inhibiting the PI3K/AKT, MAPK, NF-κB and NLRP3 signaling pathway and protecting the integrity of blood-milk barrier. Int. Immunopharmacol. 78:105972. doi: 10.1016/j.intimp.2019.105972. [DOI] [PubMed] [Google Scholar]
- Kadegowda, A. K., Bionaz M., Piperova L. S., Erdman R. A., and Loor J. J.. . 2009. Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J. Dairy Sci. 92:4276–4289. doi: 10.3168/jds.2008-1932. [DOI] [PubMed] [Google Scholar]
- Kang, J. S., Yoon Y. D., Lee K. H., Park S. K., and Kim H. M.. . 2004. Costunolide inhibits interleukin-1beta expression by down-regulation of AP-1 and MAPK activity in LPS-stimulated RAW 264.7 cells. Biochem. Biophys. Res. Commun. 313:171–177. doi: 10.1016/j.bbrc.2003.11.109. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., Lee Y. H., Chang Y. U., and Yi H. K.. . 2017. PPARγ regulates inflammatory reaction by inhibiting the MAPK/NF-κB pathway in C2C12 skeletal muscle cells. J. Physiol. Biochem. 73:49–57. doi: 10.1007/s13105-016-0523-3. [DOI] [PubMed] [Google Scholar]
- Kundu, P., Ling T. W., Korecka A., Li Y., D’Arienzo R., Bunte R. M., Berger T., Arulampalam V., Chambon P., Mak T. W., . et al. 2014. Absence of intestinal PPARγ aggravates acute infectious colitis in mice through a lipocalin-2-dependent pathway. PLoS Pathog. 10:e1003887. doi: 10.1371/journal.ppat.1003887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichane, S., Dahal Lamichane B., and Kwon S. M.. . 2018. Pivotal roles of peroxisome proliferator-activated receptors (PPARs) and their signal cascade for cellular and whole-body energy homeostasis. Int. J. Mol. Sci. 19:949. doi: 10.3390/ijms19040949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Fu Y., Zhang W., Su G., Liu B., Guo M., Li F., Liang D., Liu Z., Zhang X., . et al. 2013. Salidroside attenuates inflammatory responses by suppressing nuclear factor-κB and mitogen activated protein kinases activation in lipopolysaccharide-induced mastitis in mice. Inflamm. Res. 62:9–15. doi: 10.1007/s00011-012-0545-4. [DOI] [PubMed] [Google Scholar]
- Liang, D., Li F., Fu Y., Cao Y., Song X., Wang T., Wang W., Guo M., Zhou E., Li D., . et al. 2014. Thymol inhibits LPS-stimulated inflammatory response via down-regulation of NF-κB and MAPK signaling pathways in mouse mammary epithelial cells. Inflammation 37:214–222. doi: 10.1007/s10753-013-9732-x. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Jia Y., Yang K., Li R., Xiao X., Zhu K., and Wang Z.. . 2020. Metformin restores tetracyclines susceptibility against multidrug resistant bacteria. Adv. Sci. (Weinh). 7:1902227. doi: 10.1002/advs.201902227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Xu J., Zhu R., Zhu Y., Zhao Y., Chen P., Pan C., Yao W., and Gao X.. . 2015. Fingerprinting profile of polysaccharides from Lycium barbarum using multiplex approaches and chemometrics. Int. J. Biol. Macromol. 78:230–237. doi: 10.1016/j.ijbiomac.2015.03.062. [DOI] [PubMed] [Google Scholar]
- Long, L. N., Kang B. J., Jiang Q., and Chen J. S.. . 2020. Effects of dietary Lycium barbarum polysaccharides on growth performance, digestive enzyme activities, antioxidant status, and immunity of broiler chickens. Poult. Sci. 99:744–751. doi: 10.1016/j.psj.2019.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loor, J. J., Moyes K. M., and Bionaz M.. . 2011. Functional adaptations of the transcriptome to mastitis-causing pathogens: the mammary gland and beyond. J. Mammary Gland Biol. Neoplasia 16:305–322. doi: 10.1007/s10911-011-9232-2. [DOI] [PubMed] [Google Scholar]
- Ma, N., Chang G., Huang J., Wang Y., Gao Q., Cheng X., Liu J., and Shen X.. . 2019. cis-9, trans-11-Conjugated linoleic acid exerts an anti-inflammatory effect in bovine mammary epithelial cells after Escherichia coli stimulation through NF-κB signaling pathway. J. Agric. Food Chem. 67:193–200. doi: 10.1021/acs.jafc.8b05500. [DOI] [PubMed] [Google Scholar]
- Moyes, K. M., Drackley J. K., Morin D. E., Bionaz M., Rodriguez-Zas S. L., Everts R. E., Lewin H. A., and Loor J. J.. . 2009. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARgamma signaling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics 10:542. doi: 10.1186/1471-2164-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, Y., Mikami O., Yoshioka M., Motoi Y., Ito T., Ishikawa Y., Fuse M., Nakano K., and Yasukawa K.. . 1997. Elevated levels of tumor necrosis factor-alpha (TNF-alpha) and interleukin-6 (IL-6) activities in the sera and milk of cows with naturally occurring coliform mastitis. Res. Vet. Sci. 62:297–298. doi: 10.1016/s0034-5288(97)90209-5. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuo, A., Satoshi U., and Osamu T.. . 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Takeda, K., Kaisho T., and Akira S.. . 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Thomas, F. C., Mullen W., Tassi R., Ramírez-Torres A., Mudaliar M., McNeilly T. N., Zadoks R. N., Burchmore R., and David Eckersall P.. . 2016. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 1. High abundance proteins, acute phase proteins and peptidomics. Mol. Biosyst. 12:2735–2747. doi: 10.1039/c6mb00239k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Bi C., Wang Y., Sun J., Meng X., and Li J.. . 2018. Selenium ameliorates Staphylococcus aureus-induced inflammation in bovine mammary epithelial cells by inhibiting activation of TLR2, NF-κB and MAPK signaling pathways. BMC Vet. Res. 14:197. doi: 10.1186/s12917-018-1508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Guo M., Song X., Zhang Z., Jiang H., Wang W., Fu Y., Cao Y., Zhu L., and Zhang N.. . 2014. Stevioside plays an anti-inflammatory role by regulating the NF-κB and MAPK pathways in S. aureus-infected mouse mammary glands. Inflammation 37:1837–1846. doi: 10.1007/s10753-014-9915-0. [DOI] [PubMed] [Google Scholar]
- Wang, J., Guo C., Wei Z., He X., Kou J., Zhou E., Yang Z., and Fu Y.. . 2016a. Morin suppresses inflammatory cytokine expression by downregulation of nuclear factor-κB and mitogen-activated protein kinase (MAPK) signaling pathways in lipopolysaccharide-stimulated primary bovine mammary epithelial cells. J. Dairy Sci. 99:3016–3022. doi: 10.3168/jds.2015-10330. [DOI] [PubMed] [Google Scholar]
- Wang, X. G., Ju Z. H., Hou M. H., Jiang Q., Yang C. H., Zhang Y., Sun Y., Li R. L., Wang C. F., Zhong J. F., . et al. 2016b. Correction: deciphering transcriptome and complex alternative splicing transcripts in mammary gland tissues from cows naturally infected with Staphylococcus aureus mastitis. PLoS One 11:e0167666. doi: 10.1371/journal.pone.0167666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellnitz, O., and Kerr D. E.. . 2004. Cryopreserved bovine mammary cells to model epithelial response to infection. Vet. Immunol. Immunopathol. 101:191–202. doi: 10.1016/j.vetimm.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Xiao, J., Liong E. C., Ching Y. P., Chang R. C., So K. F., Fung M. L., and Tipoe G. L.. . 2012. Lycium barbarum polysaccharides protect mice liver from carbon tetrachloride-induced oxidative stress and necroinflammation. J. Ethnopharmacol. 139:462–470. doi: 10.1016/j.jep.2011.11.033. [DOI] [PubMed] [Google Scholar]
- Xu, T., Lu X., Arbab A. A. I., Wu X., Mao Y., Loor J. J., and Yang Z.. . 2021a. Metformin acts to suppress β-hydroxybutyric acid-mediated inflammatory responses through activation of AMPK signaling in bovine hepatocytes. J. Anim. Sci. 99. doi: 10.1093/jas/skab153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Ma N., Wang Y., Shi X., Chang G., Loor J. J., and Shen X.. . 2018. Sodium butyrate supplementation alleviates the adaptive response to inflammation and modulates fatty acid metabolism in lipopolysaccharide-stimulated bovine hepatocytes. J. Agric. Food Chem. 66:6281–6290. doi: 10.1021/acs.jafc.8b01439. [DOI] [PubMed] [Google Scholar]
- Xu, T. L., Seyfert H. M., and Shen X. Z.. . 2017. Epigenetic mechanisms contribute to decrease stearoyl-CoA desaturase 1 expression in the liver of dairy cows after prolonged feeding of high-concentrate diet. J. Dairy Sci. 101:2506–2518. doi: 10.3168/jds.2017-12878. [DOI] [PubMed] [Google Scholar]
- Xu, T., Tao H., Chang G., Zhang K., Xu L., and Shen X.. . 2015. Lipopolysaccharide derived from the rumen down-regulates stearoyl-CoA desaturase 1 expression and alters fatty acid composition in the liver of dairy cows fed a high-concentrate diet. BMC Vet. Res. 11:52. doi: 10.1186/s12917-015-0360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Wu X., Lu X., Liang Y., Mao Y., Loor J. J., and Yang Z.. . 2021b. Metformin activated AMPK signaling contributes to the alleviation of LPS-induced inflammatory responses in bovine mammary epithelial cells. BMC Vet. Res. 17:97. doi: 10.1186/s12917-021-02797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. S., Leung S. K., Lai S. W., Che C. M., Zee S. Y., So K. F., Yuen W. H., and Chang R. C.. . 2005. Neuroprotective effects of anti-aging oriental medicine Lycium barbarum against beta-amyloid peptide neurotoxicity. Exp. Gerontol. 40:716–727. doi: 10.1016/j.exger.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Zenhom, M., Hyder A., de Vrese M., Heller K. J., Roeder T., and Schrezenmeir J.. . 2011. Prebiotic oligosaccharides reduce proinflammatory cytokines in intestinal Caco-2 cells via activation of PPARγ and peptidoglycan recognition protein 3. J. Nutr. 141:971–977. doi: 10.3945/jn.110.136176. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Zheng L., and Yuan Z.. . 2019. Lycium barbarum polysaccharides promoted proliferation and differentiation in osteoblasts. J. Cell. Biochem. 120:5018–5023. doi: 10.1002/jcb.27777. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Zhou Z., Peng J., and Loor J. J.. . 2018. Methionine and valine activate the mammalian target of rapamycin complex 1 pathway through heterodimeric amino acid taste receptor (TAS1R1/TAS1R3) and intracellular Ca2+ in bovine mammary epithelial cells. J. Dairy Sci. 101:11354–11363. doi: 10.3168/jds.2018-14461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.