Abstract
Motivation
Antimicrobial resistance (AMR) is one of the biggest global problems threatening human and animal health. Rapid and accurate AMR diagnostic methods are thus very urgently needed. However, traditional antimicrobial susceptibility testing (AST) is time-consuming, low throughput and viable only for cultivable bacteria. Machine learning methods may pave the way for automated AMR prediction based on genomic data of the bacteria. However, comparing different machine learning methods for the prediction of AMR based on different encodings and whole-genome sequencing data without previously known knowledge remains to be done.
Results
In this study, we evaluated logistic regression (LR), support vector machine (SVM), random forest (RF) and convolutional neural network (CNN) for the prediction of AMR for the antibiotics ciprofloxacin, cefotaxime, ceftazidime and gentamicin. We could demonstrate that these models can effectively predict AMR with label encoding, one-hot encoding and frequency matrix chaos game representation (FCGR encoding) on whole-genome sequencing data. We trained these models on a large AMR dataset and evaluated them on an independent public dataset. Generally, RFs and CNNs perform better than LR and SVM with AUCs up to 0.96. Furthermore, we were able to identify mutations that are associated with AMR for each antibiotic.
Availability and implementation
Source code in data preparation and model training are provided at GitHub website (https://github.com/YunxiaoRen/ML-iAMR).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The rise of antimicrobial resistance (AMR) is one of the greatest threats to global health, food security and societal development. Estimates indicate that the number of yearly deaths will be at 10 million worldwide with a cost of $100 trillion if no steps to tackle AMR are taken by 2050 (Naylor et al., 2018). Traditional antimicrobial susceptibility testing (AST) is widely used for AMR analysis in clinical practice. However, this approach requires professional facilities and technicians for implementation and is viable only for cultivable bacteria (Boolchandani et al., 2019). Recently, many studies highlight the potential of machine learning methods in predicting AMR combining sequencing methods and well-known databases with phenotypic information for AMR (Boolchandani et al., 2019; Liu et al., 2020; Lv et al., 2021). For instance, Yang et al. (2018) and Kouchaki et al. (2018) analyzed AMR using different machine learning algorithms [e.g. support vector machine (SVM), logistic regression (LR) and random forest (RF)] trained on whole-genome sequencing and achieved high accuracy on AMR prediction. Deep learning algorithms also showed significant potential for predicting new antibiotic drugs, AMR genes and AMR peptides (Arango-Argoty et al., 2018; Stokes et al., 2020; Veltri et al., 2018). However, these studies focused on genome variants (such as single-nucleotide polymorphisms, SNPs) or other features only related to resistant genes identified in previous studies or resistant databases. The potential of machine learning models for predicting AMR without using known resistance mutation databases or annotated genes remains to be clarified.
To use machine learning methods for the classification of AMR, the input sequences (here: genomic sequences) need to be encoded into numerical values. A practical and informative encoding method for the whole-genome sequence is, thus, crucial for downstream analysis. There are various encoding methods for sequences (Spänig and Heider, 2019), e.g. one-hot encoding or label encoding. One-hot encoding, also referred to as sparse encoding, encodes the DNA sequence into a binary matrix, which is then vectorized and used as input for the machine learning models. Label encoding is another simple and straightforward encoding method, where each label is assigned a unique integer.
Thus, in this study, we use label encoding, one-hot encoding and Chaos Game Representation (CGR) to encode the genomic data. CGR is a recurrent iterative function system, which can be used to visualize sequences by building fractals from sequences of symbols, i.e. from an alphabet . Jeffrey (1990) was the first who applied the CGR algorithm to DNA sequences, i.e. n = 4 and , thus the resulting fractals are constructed from squares. Since the development of the CGR and its application in life science, it has been used for the analysis and alignment-free comparison of whole-genome sequences (Joseph and Sasikumar, 2006; Kania and Sarapata, 2021; Lichtblau, 2019). It has been shown that CGR is an excellent representation for genomes and that CGR-driven phylogeny leads to reliable predictions (Deschavanne et al., 1999). In particular, the comparison between genomes using CGR is straightforward and fast (Hoang et al., 2016). CGR has been used, for instance, for a fast comparison of SARS-CoV2 strains (Sengupta et al., 2020). Extensions of CGR include color grids (Deschavanne et al., 1999) and frequency matrix chaos game representation (FCGR) (Almeida et al., 2001). Wang et al. (2005) used FCGR to calculate the image distance between genomes to generate phylogenetic trees. Rizzo et al. (2016) showed that deep neural networks (DNNs) trained on genomes encoded with FCGR yielded very accurate predictions. They used a convolutional neural network (CNN) to divide bacteria into three different phyla, order, family and genus, and showed very high accuracy for the method.
While most existing studies on CGR encoding focused on CGR for DNA, there also exist a smaller number of studies dealing with other alphabets, e.g. the encoding of protein sequences. Yu et al. (2004) used the CGR algorithm for protein classification by separating the amino acids into four groups based on their properties and used multifractal and correlation analysis to construct a phylogenetic tree of Archaea and Eubacteria. In other approaches, the amino acids were retranslated into DNA for CGR (Yang et al., 2009). Sun et al. (2020) used a three-dimensional CGR representation for protein classification, and Löchel et al. (2020) used FCGR for resistance prediction in HIV-1 with CNNs.
Thus, in this study, we analyzed the potential of different statistical and machine learning methods, including LR, SVM, RF and CNN with label encoding, one-hot encoding and FCGR encoding for predicting AMR based on whole-genome sequencing of Escherichia coli (E.coli).
2 Materials and methods
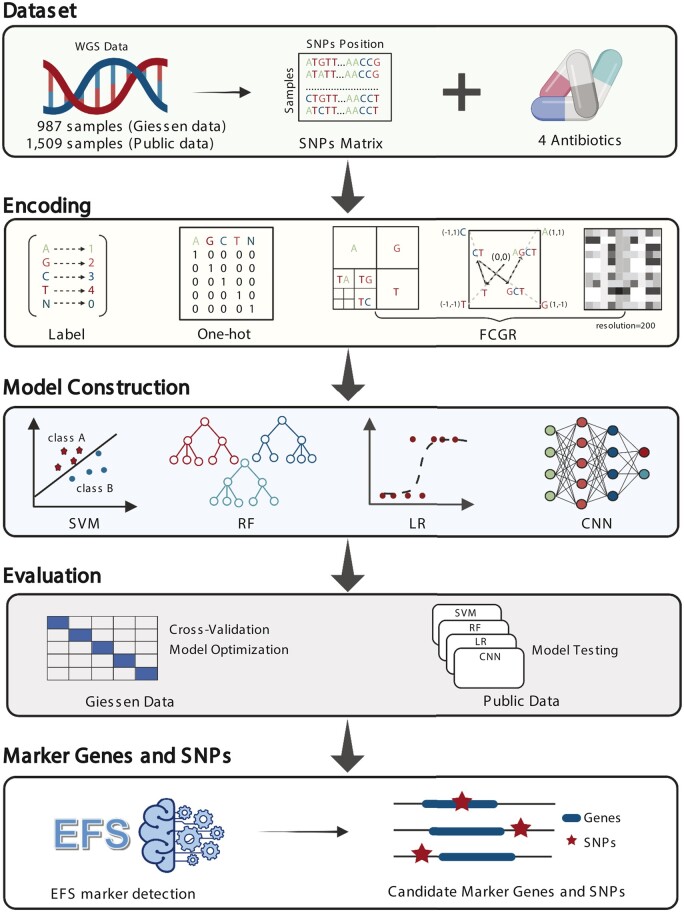
The workflow of the study is shown in Figure 1.
Fig. 1.
Workflow of the study. WGS data from Giessen and the public data from Moradigaravand et al. (2018) were processed, and single nucleotide polymorphisms (SNPs) were called. The SNP data were encoded by label encoding, one-hot encoding and FCGR encoding for subsequent machine learning. The Giessen dataset was used to train and validate the four machine learning algorithms using cross-validation. The public data were used for the final evaluation of the models. Finally, we analyzed the association of SNPs and SNPs-adjacent genes with AMR using EFS. Created with BioRender.com
2.1 Data collection and sample phenotype
Escherichia coli is an important model organism that can cause severe infections in humans and animals, it also represents a significant resistance gene pool that may be responsible for treatment failure in humans and veterinary medicine (Poirel et al., 2018).
In our study, we used two datasets, referred to as the Giessen data and the public data. The first dataset (Giessen) was collected as part of our study and contains whole-genome sequencing data (WGS) and corresponding phenotypic information for several antibiotics for, in total, 987 E.coli strains. These isolates were obtained from human and animal clinical samples. Antimicrobial susceptibility testing was performed using the VITEK® 2 system (bioMérieux, Nürtingen, Germany) and interpreted following EUCAST guidelines. DNA isolation and whole-genome sequencing were performed, as described by Falgenhauer et al. (2020).
The latter dataset (public) consists of WGS of 1509 E.coli strains and corresponding phenotypic information (Moradigaravand et al., 2018). In our study, we focused on the four antibiotics ciprofloxacin (CIP), cefotaxime (CTX), ceftazidime (CTZ) and gentamicin (GEN).
CIP belongs to the class of fluoroquinolones and is widely used to treat various infections, including gastroenteritis, respiratory tract infections or urinary tract infections (Heeb et al., 2011). CIP is particularly effective against Gram-negative bacteria, such as E.coli. However, due to overuse, resistances evolve rapidly. CTX and CTZ belong to the class of cephalosporins and are also widely used to treat various infections, such as meningitis, pneumonia, urinary tract infections, sepsis and gonorrhea. They are broad-spectrum antibiotics with activity against numerous Gram-positive and Gram-negative bacteria, including E.coli. Nevertheless, resistance is also increasing noticeably (Gums et al., 2008; Sharma, 2013).
GEN belongs to the aminoglycoside class and is widely used to treat various infections, including meningitis, pneumonia, urinary tract infections and sepsis. It is active against a wide range of bacterial infections, mostly Gram-negative bacteria including E.coli. It binds to the 30S subunit of the bacterial ribosome and negatively affects protein synthesis (Garneau-Tsodikova and Labby, 2016).
We used data of 900 isolates with resistance information for CIP (418 resistant, 482 susceptible), 930 isolates with resistance information for CTX (455 resistant, 475 susceptible), 841 isolated for CTZ (291 resistant, 550 susceptible) and 926 isolates for GEN (216 resistant, 710 susceptible).
While the CIP and CTX data are balanced, the Giessen datasets are imbalanced on the CTZ and GEN data (34% and 23% resistant isolates, respectively). The public dataset is imbalanced for all antibiotics. For CIP, CTX, CTZ and GEN, there are only 267, 115, 73 and 101 resistant samples, representing 18%, 8%, 5% and 7% of all isolates in the public dataset, respectively.
The summary of the datasets is shown in Table 1.
Table 1.
Overview of the datasets
| Drug | CIP | CTX | CTZ | GEN | ||||
|---|---|---|---|---|---|---|---|---|
| Source | Giessen | Public | Giessen | Public | Giessen | Public | Giessen | Public |
| Resistant | 418 | 267 | 455 | 115 | 291 | 73 | 216 | 101 |
| Susceptible | 482 | 1229 | 475 | 1313 | 550 | 1398 | 710 | 1398 |
| Total | 900 | 1496 | 930 | 1428 | 841 | 1471 | 926 | 1489 |
2.2 Variants calling of whole-genome sequencing data
The raw whole-genome sequencing reads were first quality checked and filtered by fastp (Chen et al., 2018). The filtered reads were then aligned to the E.coli reference genome (E.coli K-12 strain. MG1655) using BWA-mem (Li et al., 2009). Bcftools (Danecek et al., 2021) was used for calling variants. Samtools (Li and Durbin, 2009) was used to sort the aligned reads, and vcftools (Danecek et al., 2011) was used to filter the raw variants. We used default parameters for all tools.
2.3 SNPs pre-processing and encoding
We first extracted reference alleles, variant alleles and their positions, and merged all isolates based on the position of reference alleles. We filtered out the loci without variation (N replaces a locus without variation), and we built the final SNP matrix, where the rows represent the samples and columns are the variant alleles.
To encode the SNPs for subsequent machine learning, we used label encoding, one-hot encoding and FCGR encoding. For the label encoding, the A, G, C, T and N in the SNP matrix were converted to 1, 2, 3, 4 and 0. In one-hot encoding, the DNA sequence is encoded into a binary matrix, which is subsequently vectorized. For the FCGR encoding, we used the R package kaos to transform the sequences into an image-like matrix with a resolution of 200 (Löchel et al., 2020).
2.4 Machine learning and model evaluation
We used four machine learning methods, including LR, SVM, RF and CNN. For LR, RF and SVM, we used the Scikit-learn python package (Pedregosa et al., 2011). LR was used with default parameters, except that we used 1000 iterations. RF was used with default parameters and 200 trees. For SVM, we used a linear kernel and default parameters.
We implemented CNNs using the Keras (https://keras.io/) package and TensorFlow (https://tensorflow.org). The CNN architecture is based on eleven hidden layers, including four convolutional layers, two batch normalization layers, two pooling layers, one flattening layer, one fully connected layer and one dropout layer. The structure of the networks for label encoding and one-hot encoding are the same, which differ from FCGR encoding-based CNNs only in the convolutional layers and pooling layers (see Supplementary Fig. S1). For FCGR, we used the Conv2D and MaxPooling2D function, while the CNN for the label encoding used the 1D versions instead.
We used eight filters in the first two convolution layers with a kernel size of three, rectified linear unit activation function and same padding. The last two convolution layers used 16 filters instead. The pool size of all pooling layers is two. We used the softmax activation function in the final fully connected layer and compiled the model with Adam optimization and cross-entropy loss.
2.5 Statistical evaluation
We optimized the machine learning models on the Giessen data using five times 5-fold stratified cross-validation. We applied an up-sampling strategy to balance the samples in the training set. For the final evaluation on the public data, we analyzed the performance on the raw public dataset and on a balanced set using a down-sample strategy.
We evaluated the models using the receiver operating characteristics curve (ROC) and the area under the curve (AUC). We also calculated precision and recall for all models. Statistical comparisons were made by the DeLong test (Demler et al., 2012).
2.6 Marker genes identification located around SNPs
To identify the SNPs that are associated with resistance, we performed a marker gene identification using the EFS R package (Neumann et al., 2017). The EFS package aggregates eight feature selection methods for binary classification tasks (Neumann et al., 2016). We used EFS with default parameters. We then annotated the corresponding genes of SNPs using SnpEff software (Cingolani et al., 2012).
3 Results
3.1 Performance of different machine learning methods for predicting AMR on Giessen data
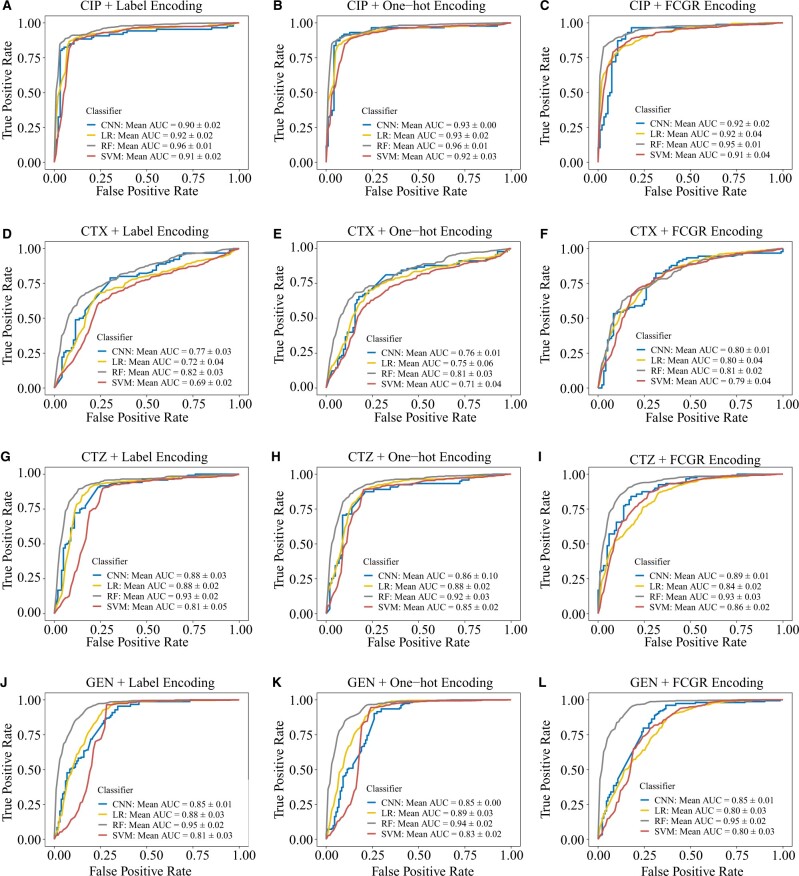
We used the filtered SNPs matrix encoded by label encoding, one-hot encoding and FCGR encoding from the Giessen dataset to train the four machine learning methods LR, RF, SVM and CNN. The performance of the four machine learning models was evaluated using five times 5-fold cross-validation. The ROC curves and AUC values of the different machine learning models range from 0.69 to 0.96, demonstrating that all models can effectively predict AMR compared with random null models (Fig. 2). We observed that the mean AUC of the RFs was higher than for LR, SVM and CNN classifiers for all antibiotics with both encoding methods (Fig. 2). In particular, RFs were significantly better than LR (P = 0.03), SVMs (P = 0.01) and CNNs (P = 0.02) for CIP with label encoding (Supplementary Fig. S2). RFs were also better than the other three classifiers for GEN with label encoding and FCGR encoding (P < 0.05). For CTZ, RFs significantly outperformed SVMs with all encoding methods (P < 0.05) (Supplementary Fig. S2). For CTX, RFs are significantly better than LR and SVM with label encoding and one-hot encoding (P < 0.05), while there are no significant differences if the FCGR encoding is used (Supplementary Fig. S2).
Fig. 2.
ROC curves for the models with label encoding, one-hot encoding and FCGR encoding on the Giessen data. First row: ROC curves for CIP with label encoding (A), one-hot encoding (B) and FCGR encoding (C), respectively. Second row: ROC curves for CTX with label encoding (D), one-hot encoding (E) and FCGR encoding (F), respectively. Third row: ROC curves for CTZ with label encoding (G), one-hot encoding (H) and FCGR encoding (I), respectively. Fourth row: ROC curves for GEN with label encoding (J), one-hot encoding (K) and FCGR encoding (L), respectively
Moreover, all models show high precision and recall using label (Table 2), one-hot (Table 3) and FCGR encoding (Table 4) for CIP. For CTZ and GEN, the models show high recall but lower precision, which may be related to the imbalanced resistant and susceptible isolates. In sum, RF, CNN, LR and SVM can predict AMR for CIP, CTZ, GEN and CTX with three encoding methods in E.coli.
Table 2.
Results of the four machine learning models with label encoding on the Giessen data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.88 ± 0.04 | 0.75 ± 0.04 | 0.81 ± 0.02 | 0.76 ± 0.03 | 0.87 ± 0.01 | 0.65 ± 0.10 | 0.89 ± 0.03 | 0.91 ± 0.02 |
| LR | 0.88 ± 0.05 | 0.71 ± 0.04 | 0.81 ± 0.03 | 0.77 ± 0.02 | 0.90 ± 0.03 | 0.69 ± 0.08 | 0.92 ± 0.05 | 0.96 ± 0.03 |
| RF | 0.92 ± 0.04 | 0.75 ± 0.03 | 0.84 ± 0.03 | 0.79 ± 0.02 | 0.89 ± 0.03 | 0.73 ± 0.07 | 0.90 ± 0.06 | 0.97 ± 0.03 |
| SVM | 0.85 ± 0.03 | 0.69 ± 0.02 | 0.78 ± 0.03 | 0.75 ± 0.02 | 0.89 ± 0.04 | 0.73 ± 0.03 | 0.89 ± 0.03 | 0.96 ± 0.03 |
Table 3.
Results of the four machine learning models with one-hot encoding on the Giessen data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.87 ± 0.05 | 0.75 ± 0.00 | 0.84 ± 0.01 | 0.80 ± 0.00 | 0.90 ± 0.01 | 0.71 ± 0.03 | 0.84 ± 0.03 | 0.87 ± 0.05 |
| LR | 0.89 ± 0.05 | 0.71 ± 0.04 | 0.80 ± 0.03 | 0.78 ± 0.02 | 0.89 ± 0.03 | 0.73 ± 0.08 | 0.89 ± 0.05 | 0.95 ± 0.02 |
| RF | 0.92 ± 0.05 | 0.75 ± 0.01 | 0.82 ± 0.02 | 0.80 ± 0.03 | 0.90 ± 0.02 | 0.73 ± 0.07 | 0.90 ± 0.07 | 0.97 ± 0.03 |
| SVM | 0.86 ± 0.05 | 0.68 ± 0.03 | 0.77 ± 0.03 | 0.76 ± 0.03 | 0.89 ± 0.03 | 0.69 ± 0.06 | 0.89 ± 0.06 | 0.95 ± 0.04 |
Table 4.
Results of the four machine learning models with FCGR encoding on the Giessen data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.87 ± 0.04 | 0.74 ± 0.04 | 0.81 ± 0.03 | 0.75 ± 0.02 | 0.91 ± 0.03 | 0.84 ± 0.04 | 0.87 ± 0.06 | 0.96 ± 0.01 |
| LR | 0.79 ± 0.08 | 0.70 ± 0.04 | 0.73 ± 0.05 | 0.69 ± 0.04 | 0.85 ± 0.04 | 0.79 ± 0.05 | 0.85 ± 0.04 | 0.86 ± 0.02 |
| RF | 0.91 ± 0.03 | 0.74 ± 0.01 | 0.82 ± 0.02 | 0.80 ± 0.02 | 0.87 ± 0.03 | 0.72 ± 0.07 | 0.90 ± 0.07 | 0.98 ± 0.01 |
| SVM | 0.81 ± 0.03 | 0.72 ± 0.03 | 0.73 ± 0.01 | 0.69 ± 0.02 | 0.88 ± 0.03 | 0.81 ± 0.05 | 0.87 ± 0.03 | 0.92 ± 0.03 |
3.2 Evaluation of the models on public data
We performed a further evaluation of our models using the public data of E.coli of Moradigaravand et al. (2018). The public data are highly imbalanced and thus performance metrics are difficult to interpret. Thus, to evaluate the performance of the models, we performed a down-sampling to balance the public data. For completeness, results for the imbalanced set are shown in Supplementary Tables S1–S3.
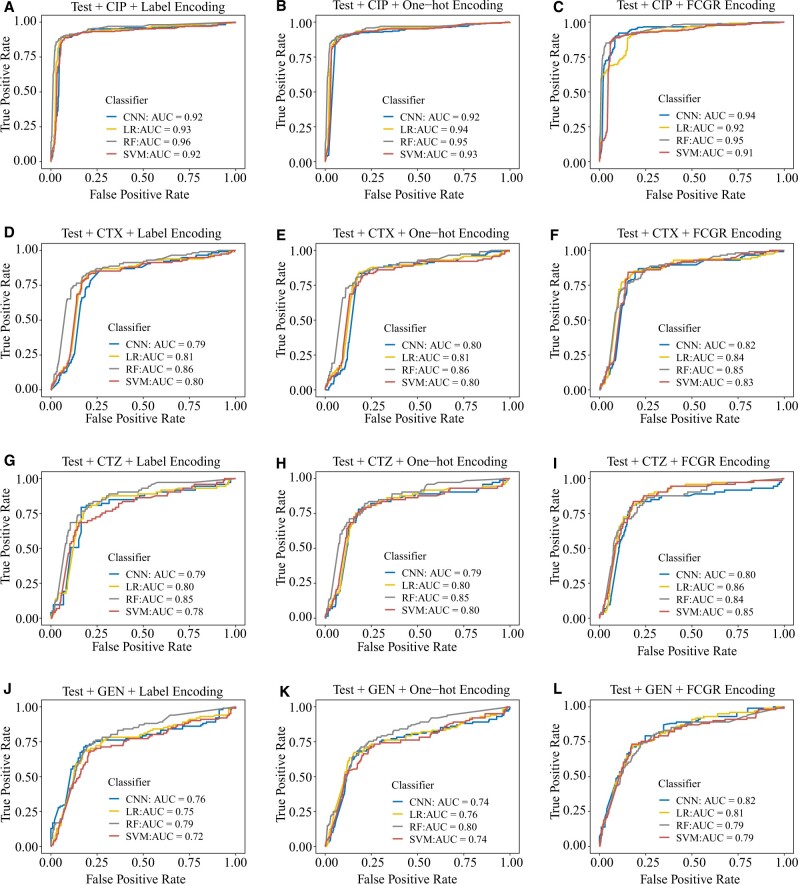
The resulting ROC curves clearly show that the machine learning models generalize well and can predict AMR (Fig. 3). The AUCs of RFs are higher compared with those from LR, SVM and CNN with three encoding approaches, except for CTZ and GEN with FCGR encoding. Consistent with the results from the Giessen data, all classifiers have high precision and recall for three encoding methods (Tables 5–7).
Fig. 3.
ROC curves for the models with label, one-hot and FCGR encoding on the public data. First row: ROC curves for CIP with label encoding (A), one-hot encoding (B) and FCGR encoding (C), respectively. Second row: ROC curves for CTX with label encoding (D), one-hot encoding (E) and FCGR encoding (F), respectively. Third row: ROC curves for CTZ with label encoding (G), one-hot encoding (H) and FCGR encoding (I), respectively. Fourth row: ROC curves for GEN with label encoding (J), one-hot encoding (K) and FCGR encoding (L), respectively
Table 5.
Evaluation of the machine learning models with label encoding on the public data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.94 | 0.71 | 0.79 | 0.84 | 0.88 | 0.88 | 0.81 | 0.70 |
| LR | 0.93 | 0.76 | 0.80 | 0.82 | 0.90 | 0.84 | 0.75 | 0.62 |
| RF | 0.95 | 0.75 | 0.81 | 0.83 | 0.90 | 0.85 | 0.77 | 0.61 |
| SVM | 0.94 | 0.71 | 0.75 | 0.77 | 0.87 | 0.84 | 0.74 | 0.60 |
Note: Precision and recall are calculated based on balanced data using down-sampling.
Table 7.
Evaluation of the machine learning models with FCGR encoding on the public data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.84 | 0.71 | 0.72 | 0.74 | 0.93 | 0.89 | 0.86 | 0.71 |
| LR | 0.85 | 0.77 | 0.79 | 0.80 | 0.89 | 0.87 | 0.86 | 0.74 |
| RF | 0.92 | 0.77 | 0.83 | 0.83 | 0.88 | 0.89 | 0.78 | 0.59 |
| SVM | 0.88 | 0.78 | 0.77 | 0.75 | 0.90 | 0.86 | 0.86 | 0.74 |
Note: Precision and recall are calculated based on balanced data using down-sampling.
Table 6.
Evaluation of the machine learning models with one-hot encoding on the public data
| Classifiers/drug | Precision | Precision | Precision | Precision | Recall | Recall | Recall | Recall |
|---|---|---|---|---|---|---|---|---|
| CIP | CTX | CTZ | GEN | CIP | CTX | CTZ | GEN | |
| CNN | 0.95 | 0.83 | 0.84 | 0.80 | 0.90 | 0.83 | 0.78 | 0.62 |
| LR | 0.90 | 0.80 | 0.76 | 0.81 | 0.90 | 0.85 | 0.78 | 0.63 |
| RF | 0.90 | 0.78 | 0.73 | 0.81 | 0.90 | 0.86 | 0.78 | 0.63 |
| SVM | 0.89 | 0.78 | 0.75 | 0.73 | 0.88 | 0.83 | 0.77 | 0.55 |
Note: Precision and recall are calculated based on balanced data using down-sampling.
3.3 Marker genes associated with antibiotic resistance
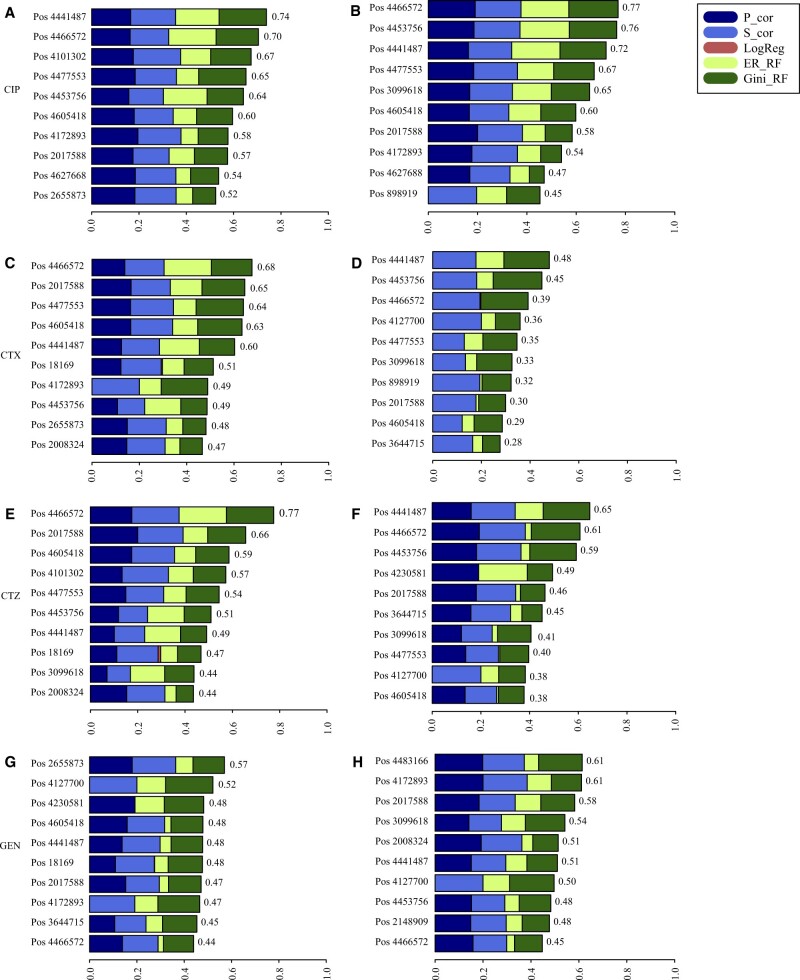
We performed an SNP association study on the Giessen and public data using the EFS R package with default parameters. In this analysis, we did not include the known resistance genes. Thus, we aimed at identifying secondary mutations that contribute to the resistance directly or indirectly, e.g. compensatory mutations. This data-driven approach does not need AMR expert knowledge and can also be used and predict resistance even without knowing the resistance genes but by identification of the secondary mutations. EFS provided a ranking of the SNPs for each antibiotic. The ten most important SNPs for each antibiotic are shown in Figure 4. These SNPs are part of 19 different genomic regions. We then annotated and analyzed the corresponding genes of these regions (Table 8).
Fig. 4.
EFS analysis for each antibiotic for both datasets. The left four figures are the identified ten most important SNPs for CIP (A), CTX (C), CTZ (E) and GEN (G) from the Giessen dataset. The right figures are the corresponding SNPs from the public dataset
Table 8.
SNPs and corresponding genes associated with AMR
| SNP Position | Gene location | SNP annotation | Gene | Gene biotype | Drug |
|---|---|---|---|---|---|
| 18169 | 17489 → 18655 | Synonymous | nhaA | CDS | CTX, CTZ, GEN |
| 898919 | 898518 → 899645 | Synonymous | rlmC | CDS | CIP, CTX |
| 2008324 | 2008277 → 2009482 | Synonymous | yedE | CDS | CTX, CTZ, GEN |
| 2017588 | 2016554 → 2017927 | synonymous | fliI | CDS | CIP, CTX, CTZ, GEN |
| 2148909 | 2147674 → 2149026 | Synonymous | yegD | CDS | GEN |
| 2655873 | 2655075 → 2656358 | Synonymous | pepB | CDS | CIP, CTX, GEN |
| 3099618 | 3098558 → 3099565 | Upstream gene | yggM | CDS | CIP, CTX, CTZ, GEN |
| 3644715 | 3643140 → 3645182 | Synonymous | prlC | CDS | CTX, CTZ, GEN |
| 4101302 | 4100810 → 4101430 | Missense | sodA | CDS | CIP, CTZ |
| 4127700 | 4127286 → 4127894 | Synonymous | yiiX | CDS | CTX, CTZ, GEN |
| 4172893 | 4172057 → 4173085 | Missense | murB | CDS | CIP, CTX, GEN |
| 4230581 | 4230354 → 4231226 | Synonymous | rluF | CDS | CTZ, GEN |
| 4441487 | 4439872 → 4441215 | Upstream gene | ytfL | CDS | CIP, CTX, CTZ, GEN |
| 4453756 | 4453583 → 4454578 | Synonymous | yjfF | CDS | CIP, CTX, CTZ, GEN |
| 4466572 | 4466299 → 4467246 | Synonymous | treR | CDS | CIP, CTX, CTZ, GEN |
| 4477553 | 4477307 → 4478311 | Missense | argI | CDS | CIP, CTX, CTZ |
| 4483166 | 4480982 → 4483837 | Synonymous | valS | CDS | GEN |
| 4605418 | 4604875 → 4605663 | Synonymous | fhuF | CDS | CIP, CTX, CTZ, GEN |
| 4627668 | 4627315 → 4628547 | Synonymous | nadR | CDS | CIP |
Note: The first column shows the positions of the identified SNPs for the four antibiotics. The second column and third column show the gene location and SNP annotation. The fourth column and fifth column show the genes annotated from SNPs and gene biotype. The final column is the antibiotics that are associated with the SNPs.
Some of these genes are well-known genes conferring antibiotic resistance, such as marA. marA is a gene related to multiple drug resistance (Abdolmaleki et al., 2019). In comparison, the other genes have not been well studied so far. For instance, the gene nhaA (associated with CTX, CTZ and GEN resistance) displays a Na+/H+ antiport activity in E.coli that can regulate the permeability, which may further affect drug resistance (Padan et al., 2004). The gene rlmC encodes a 23S RNA methyltransferase that methylates the 23S rRNA, of antibiotic binding sites and is related to antibiotic resistance (Pletnev et al., 2020; Stojković et al., 2016). It has been reported that the gene fliI encodes a virulence factor, and some studies focused on the correlation between antimicrobial resistance and bacterial virulence (Beceiro et al., 2013; Deng et al., 2019). The gene pepB encodes the peptidase B, which is related to the production of bacteriocins, narrow-spectrum antimicrobial peptides produced by bacteria (Suzuki et al., 2001; Telhig et al., 2020). MurB is the key biosynthetic enzyme involved in the synthesis of peptidoglycan, the key component of the cell wall (Nasiri et al., 2017; Walsh and Wencewicz, 2014). In sum, the marker genes and SNPs identified by EFS can be used as a reference for further AMR studies.
4 Discussion
This study analyzed four different machine learning methods (RFs, LR, SVMs and CNNs) for predicting four antibiotic resistances in E.coli based on whole-genome sequence data with three different encoding schemes, namely, label encoding, one-hot encoding and FCGR encoding. Moreover, our goal was to identify mutations (secondary mutations) contributing to resistance beyond known resistance genes. Thus, we used a reference genome for E.coli without known resistance genes. Our study confirmed that label encoding, one-hot encoding and FCGR encoding could encode genomic data for preparing the input data for subsequent machine learning and deep learning methods. Our results show that the four machine learning methods can effectively predict AMR without the need for a database of known resistance genes or SNPs, which is an essential prerequisite for AMR prediction in less well-studied pathogens and drugs. Furthermore, we provide potential genes and SNPs associated with AMR based that can be used as a reference for the subsequent experiments.
Previous studies reported different SNPs in the bacterial genome associated with multiple drug resistance (Brimacombe et al., 2007; Figueroa et al., 2019; Shi et al., 2019; Su et al., 2019; Yang et al., 2018). However, these studies mainly focused on partial SNPs based on available AMR databases (Yang et al., 2018). Machine learning based on the complete set of SNPs from whole-genome sequencing gives further insights and can be used to identify novel biological mechanisms of resistance.
Encoding the genomic features into a readable format for ma-chine learning and deep learning is an essential step. Label encoding, one-hot encoding and CGR encoding can convert SNPs into machine-recognizable formats very efficiently. Our study used the three approaches to encode SNPs and yield excellent predictions for both encoding methods. Many studies indicated that CNNs outperform other machine learning algorithms in image classification, which was the rationale for incorporating FCGR as an encoding scheme.
We compared four machine learning methods, including RFs, LR, SVMs and CNNs. Overall, the four machine learning methods showed good performance in predicting the four antibiotic resistances of E.coli. We also demonstrated that our models generalize well on unseen data, as proven by validating the results based on an independent public dataset. We were also able to identify SNPs associated with resistance. However, the marker genes located around the SNPs associated with AMR need experimental validation.
Although we only focused on four antibiotics in this study, our method can easily be applied to other antibiotics and can also be extended to other resistance-related SNPs of other pathogens, also from species other than bacteria. Furthermore, our approach can also be applied to other biomedical areas, e.g. for cancer resistance prediction. More importantly, our method may have huge potential in systems medicine, to improve the diagnosis, targeted therapy and disease prevention.
There are also some limitations in our study. For example, we only used SNP data in our models that have been called based on a single reference genome. This, however, spares many genomic regions that might be important resistance factors. This is especially true for diverse species like E.coli. One approach to mitigate this issue would be the selection of more suitable or multiple reference genomes. Another option potentially leading to a more holistic set of potential SNPs would be to use an artificial pseudo-pan-genome incorporating many genomes of a particular species as a reference within the SNP detection workflow. However, other features, e.g. transcriptomics or proteomics data, might be important for AMR as well (Moradigaravand et al., 2018). Moreover, several other important drugs have not been taken into account yet. However, they may be analyzed with the same methodology when enough data are available.
5 Conclusion
We investigated four machine learning methods for predicting AMR to four different drugs in E.coli from whole-genome sequence data with label encoding, one-hot encoding and FCGR encoding. Our results demonstrated that all methods perform very well also for unseen data. Overall, our study provides a new machine learning-driven approach for resistance prediction and thus, may improve treatment of patients in the future.
We evaluated the performance based on cross-validation on our own data and tested the model performance on public data. Moreover, we identified potential SNPs and corresponding genes that are associated with AMR.
We could demonstrate that label encoding, one-hot encoding and FCGR encoding can be used for whole-genome sequence analyses. Moreover, we provide a comprehensive evaluation of different machine learning algorithms for AMR prediction in E.coli. The results of the study give a rich reference resource for further research on both experimental and computational aspects of AMR.
Supplementary Material
Acknowledgements
The authors thank Moradigaravand et al. for making their data publicly available.
Funding
This work was financially supported by the German Federal Ministry of Education and Research (BMBF) [031L0209B] (Deep-iAMR).
Data availability
The public data is publicly available (see material and methods). The Giessen data is available upon request.
Conflict of Interest: none declared.
Contributor Information
Yunxiao Ren, Department of Data Science in Biomedicine, Faculty of Mathematics and Computer Science, Philipps-University of Marburg, Marburg 35032, Germany.
Trinad Chakraborty, Institute of Medical Microbiology, Justus Liebig University Giessen, Giessen 35392, Germany; German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany.
Swapnil Doijad, Institute of Medical Microbiology, Justus Liebig University Giessen, Giessen 35392, Germany; German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany.
Linda Falgenhauer, German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany; Institute of Hygiene and Environmental Medicine, Justus Liebig University Giessen, Giessen 35392, Germany; Hessisches universitäres Kompetenzzentrum Krankenhaushygiene, Giessen 35392, Germany.
Jane Falgenhauer, Institute of Medical Microbiology, Justus Liebig University Giessen, Giessen 35392, Germany; German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany.
Alexander Goesmann, German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany; Department of Bioinformatics and Systems Biology, Justus Liebig University Giessen, Giessen 35392, Germany.
Anne-Christin Hauschild, Department of Data Science in Biomedicine, Faculty of Mathematics and Computer Science, Philipps-University of Marburg, Marburg 35032, Germany.
Oliver Schwengers, German Center for Infection Research, Partner site Giessen-Marburg-Langen, Giessen 35392, Germany; Department of Bioinformatics and Systems Biology, Justus Liebig University Giessen, Giessen 35392, Germany.
Dominik Heider, Department of Data Science in Biomedicine, Faculty of Mathematics and Computer Science, Philipps-University of Marburg, Marburg 35032, Germany.
References
- Abdolmaleki Z. et al. (2019) Phenotypic and genotypic characterization of antibiotic resistance in the methicillin-resistant Staphylococcus aureus strains isolated from hospital cockroaches. Antimicrob. Resist. Infect. Control, 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.S. et al. (2001) Analysis of genomic sequences by chaos game representation. Bioinformatics, 17, 429–437. [DOI] [PubMed] [Google Scholar]
- Arango-Argoty G. et al. (2018) DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome, 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A. et al. (2013) Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev., 26, 185–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boolchandani M. et al. (2019) Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet., 20, 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe M. et al. (2007) Antibiotic resistance and single-nucleotide polymorphism cluster grouping type in a multinational sample of resistant mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother., 51, 4157–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. ; 1000 Genomes Project Analysis Group. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P. et al. (2021) Twelve years of SAMtools and BCFtools. GigaScience, 10, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demler O.V. et al. (2012) Misuse of DeLong test to compare AUCs for nested models. Stat. Med., 31, 2577–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. et al. (2019) Horizontal gene transfer contributes to virulence and antibiotic resistance of vibrio harveyi 345 based on complete genome sequence analysis. BMC Genomics, 20, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschavanne P.J. et al. (1999) Genomic signature: characterization and classification of species assessed by chaos game representation of sequences. Mol. Biol. Evol., 16, 1391–1399. [DOI] [PubMed] [Google Scholar]
- Falgenhauer L. et al. (2020) Cross-border emergence of clonal lineages of ST38 Escherichia coli producing the OXA-48-like carbapenemase OXA-244 in Germany and Switzerland. Int. J. Antimicrob. Agents, 56, 106157. [DOI] [PubMed] [Google Scholar]
- Figueroa J. et al. (2019) Analysis of single nucleotide polymorphisms (SNPs) associated with antibiotic resistance genes in Chilean Piscirickettsia salmonis strains. J. Fish Dis., 42, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Garneau-Tsodikova S., Labby K.J. (2016) Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. MedChemComm, 7, 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gums J.G. et al. (2008) Differences between ceftriaxone and cefotaxime: microbiological inconsistencies. Ann. Pharmacother., 42, 71–79. [DOI] [PubMed] [Google Scholar]
- Heeb S. et al. (2011) Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev., 35, 247–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. et al. (2016) Numerical encoding of DNA sequences by chaos game representation with application in similarity comparison. Genomics, 108, 134–142. [DOI] [PubMed] [Google Scholar]
- Jeffrey H. (1990) Chaos game representation of gene structure. Nucleic Acids Res., 18, 2163–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J., Sasikumar R. (2006) Chaos game representation for comparison of whole genomes. BMC Bioinformatics, 7, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A., Sarapata K. (2021) The robustness of the chaos game representation to mutations and its application in free-alignment methods. Genomics, 113, 1428–1437. [DOI] [PubMed] [Google Scholar]
- Kouchaki S. et al. ; CRyPTIC Consortium. (2019) Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinformatics, 35, 2276–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtblau D. (2019) Alignment-free genomic sequence comparison using FCGR and signal processing. BMC Bioinformatics, 20, 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. (2020) Evaluation of machine learning models for predicting antimicrobial resistance of Actinobacillus pleuropneumoniae from whole genome sequences. Front. Microbiol., 11, doi: 10.3389/fmicb.2020.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchel H.F. et al. (2020) Deep learning on chaos game representation for proteins. Bioinformatics, 36, 272–279. [DOI] [PubMed] [Google Scholar]
- Lv J. et al. (2021) A review of artificial intelligence applications for antimicrobial resistance. Biosafety Health, 3, 22–31. [Google Scholar]
- Moradigaravand D. et al. (2018) Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol., 14, e1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiri M.J. et al. (2017) New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front. Microbiol., 8, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor N.R. et al. (2018) Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob. Resist. Infect. Control, 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann U. et al. (2016) Compensation of feature selection biases accompanied with improved predictive performance for binary classification by using a novel ensemble feature selection approach. BioData Mining, 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann U. et al. (2017) EFS: an ensemble feature selection tool implemented as r-package and web-application. BioData Min., 10, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan E. et al. (2004) NhaA of escherichia coli, as a model of a pH-regulated na+h+antiporter. Biochim. Biophys. Acta (BBA) Bioenerget., 1658, 2–13. [DOI] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Pletnev P. et al. (2020) Comprehensive functional analysis of Escherichia coli ribosomal RNA methyltransferases. Front. Genet., 11, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L. et al. (2018) Antimicrobial resistance in escherichia coli. Microbiol. Spectrum, 6, doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo R. et al. (2016) Classification experiments of DNA sequences by using a deep neural network and chaos game representation. In: Proceedings of the 17th International Conference on Computer Systems and Technologies. ACM, Palermo, Italy, pp. 222–228.
- Sengupta D.C. et al. (2020) Similarity studies of corona viruses through chaos game representation. Comput. Mol. Biosci., 10, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. (2013) Prevalence and antibiogram of extended spectrum beta-lactamase (ESBL) producing gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J. Clin. Diagn. Res., 7, 2173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. et al. (2019) Antimicrobial resistance genetic factor identification from whole-genome sequence data using deep feature selection. BMC Bioinformatics, 20, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spänig S., Heider D. (2019) Encodings and models for antimicrobial peptide classification for multi-resistant pathogens. BioData Mining, 12, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojković V. et al. (2016) Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating enzyme. Nucleic Acids Res., 44, 8897–8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J.M. et al. (2020) A deep learning approach to antibiotic discovery. Cell, 180, 688–702.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M. et al. (2019) Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol., 57, e01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. et al. (2020) A novel numerical representation for proteins: three-dimensional chaos game representation and its extended natural vector. Comput. Struct. Biotechnol. J., 18, 1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H. et al. (2001) Purification and characterization of aminopeptidase b from Escherichia coli k-12. Biosci. Biotechnol. Biochem., 65, 1549–1558. [DOI] [PubMed] [Google Scholar]
- Telhig S. et al. (2020) Bacteriocins to thwart bacterial resistance in gram negative bacteria. Front. Microbiol., 11, 586433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri D. et al. (2018) Deep learning improves antimicrobial peptide recognition. Bioinformatics, 34, 2740–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C.T., Wencewicz T.A. (2014) Prospects for new antibiotics: a molecule-centered perspective. J. Antibiot., 67, 7–22. [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. (2005) The spectrum of genomic signatures: from dinucleotides to chaos game representation. Gene, 346, 173–185. [DOI] [PubMed] [Google Scholar]
- Yang J.-Y. et al. (2009) Prediction of protein structural classes by recurrence quantification analysis based on chaos game representation. J. Theor. Biol., 257, 618–626. [DOI] [PubMed] [Google Scholar]
- Yang Y. et al. (2018) Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics, 34, 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.-G. et al. (2004) Chaos game representation of protein sequences based on the detailed HP model and their multifractal and correlation analyses. J. Theor. Biol., 226, 341–348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public data is publicly available (see material and methods). The Giessen data is available upon request.
Conflict of Interest: none declared.