Early detection of allograft rejection is critical to the successful management of transplant recipients. Tissue biopsy has been the “gold standard” for diagnosis of active rejection (AR), but is invasive and has poor reproducibility.1 Conventional noninvasive biomarkers, such as changes in serum creatinine, are available for detecting AR, but are limited due to low sensitivity and specificity.2 Thus, there is a need for new noninvasive markers that have high accuracy for detecting AR.
Donor-derived cell-free DNA fraction (dd-cfDNA[%]) is a promising noninvasive biomarker for detecting allograft rejection. However, dd-cfDNA(%) can be artificially depressed by high levels of circulating cfDNA, which can occur in patients who are obese, have had recent surgery, have medical complications, or received certain medications.3 This can potentially lead to false negative results.
Recently, two studies provided preliminary evidence indicating that the absolute quantity of dd-cfDNA may show better performance for detecting AR than dd-cfDNA(%).4,5 In this study we present our results from an assay that utilizes a new two-threshold algorithm that combines both dd-cfDNA(%) and absolute quantity of dd-cfDNA (copies/ml), with the goal of increasing test sensitivity, particularly through improved detection in patients where cfDNA levels are high.
The study included 41 patients undergoing allograft management at the University of California at Los Angeles Medical Center, who received dd-cfDNA testing as part of routine clinical care between November 6, 2019 and September 18, 2020. Patients who were under 18, pregnant, had an organ transplant other than kidney, or a blood transfusion within 2 weeks of enrollment were excluded. Ethics approval was provided by the University of California at Los Angeles Institutional Review Board (IRB12–000991). The study was performed in full adherence to the Declaration of Helsinki, and clinical and research activities reported are consistent with the Principles of the Declaration of Istanbul as outlined in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
Laboratory testing was performed at Natera Inc.’s Clinical Laboratory Improvement Amendments–certified and College of American Pathologists–accredited laboratory (San Carlos, CA), as previously described, amplifying cfDNA using massively multiplexed-PCR, targeting 13,926 single nucleotide polymorphisms.6 dd-cfDNA(%) was measured according to a previously published methodology;6 the absolute concentration of dd-cfDNA was calibrated to give the quantity of dd-cfDNA (cp/ml). The new two-threshold algorithm combined the previously validated dd-cfDNA(%) cutoff (≥1%) and a previously established dd-cfDNA quantity cutoff of ≥78 cp/ml. Samples exceeding either threshold were considered at high risk for AR.
Matched biopsy results (for cause biopsies occurring within 4 weeks of dd-cfDNA testing) were available for 16 patients; 14 out of 16 occurred within 2 weeks of the dd-cfDNA test. Biopsy samples were analyzed and graded by pathologists according to standard practice using Banff 2017 classification.7 Samples without biopsy were classified as not having AR, on the basis of clinical assessment (stable according to serum creatinine and other clinical indicators). AR was found in nine of 16 (56%) biopsies performed, with five classified as T cell–mediated rejection, one as antibody-mediated rejection, and three as mixed type (antibody-mediated rejection/T cell–mediated rejection).
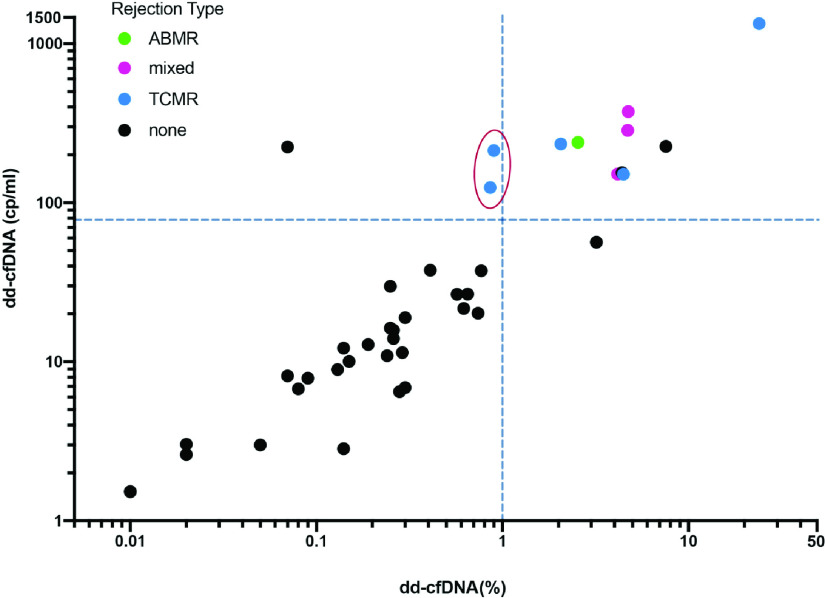
We calculated sensitivity and specificity for each algorithm for the 41 patients in the sample. The original method, on the basis of the ≥1% dd-cfDNA cutoff, had a sensitivity of seven out of nine (77.8%; 95% confidence interval [95% CI], 40.0% to 97.2%) and a specificity of 29 out of 32 (90.6%; 95% CI, 75.0% to 98.0%). Applying the two-threshold algorithm to the dataset, yielded a sensitivity of nine out of nine (100%; 95% CI, 66.4% to 100%), and a specificity of 28 out of 32 (87.5%; 95% CI, 71.0% to 96.5%) (Figure 1).
Figure 1.
Plot of dd-cfDNA quantity (cp/ml) by dd-cfDNA(%). Green, pink, and blue data points show patients with biopsy proven antibody-mediated rejection, mixed, and T cell–mediated rejection, respectively. Black data points show patients with no evidence of AR at biopsy. The vertical dashed line shows the cutoff point for dd-cfDNA(%) (≥1%); the horizontal dashed line shows the cutoff for the new quantity of dd-cfDNA threshold (≥78 cp/ml). The samples to the right of the vertical dashed line, with dd-cfDNA(%) values ≥1%, are considered high risk according to the original single-threshold algorithm. The samples in the upper left quadrant, with dd-cfDNA <1% and dd-cfDNA ≥78 cp/ml, are considered low risk for rejection by the single threshold algorithm, and high risk for rejection by the two-threshold algorithm. The two data points in the red oval show the patients with AR identified by the new algorithm. The samples in the lower left quadrant are considered at low risk for rejection by both algorithms.
Our results suggest that using both dd-cfDNA quantity and dd-cfDNA(%) to assess the rejection status of allografts can improve performance over using dd-cfDNA(%) alone. Consistent with expectations, the three patients whose risk determination changed with the introduction of the new threshold had high total cfDNA levels (≥88th percentile), and thus depressed donor fractions that led, in two patients, to false negative results when using the ≥1% donor fraction cutoff alone (Figure 1, red oval).
Importantly, although the incorporation of the quantity of dd-cfDNA threshold improved the sensitivity of the assay, specificity showed a minimal decline. Maintaining high specificity is critical to minimizing unnecessary biopsies and is considered a key benefit of adding dd-cfDNA testing to current standard of care biomarkers.4
This study has several limitations. One is the small sample size, and another is that a number of the patients considered to be nonrejecting were adjudicated by clinical evidence alone, without biopsy pathology information. Further research with a larger, biopsy-matched cohort could improve performance estimates of this methodology.
In conclusion, this study suggests the combination of the quantity of dd-cfDNA threshold and the previously validated dd-cfDNA(%) threshold holds promise for improved sensitivity in the detection of AR in patients receiving a renal allograft, while maintaining high specificity.
Disclosures
E. Ahmed, G. Fehringer, P.R. Billings, and H. Tabriziani are employees and equity holders at Natera, Inc. G. Fehringer reports consultancy agreements with Natera, Inc. S. Bunnapradist reports consultancy agreements with CareDx; Research Funding from Alexion, Astellas, CareDx, Merck, and Angion; honoraria from BMS, Veloxis, CareDX, and Sanofi; and speakers bureau from BMS, Veloxis, CareDX, Natera, and Vitaeris. H. Tabriziani reports consultancy agreements, ownership interest, scientific advisor or membership, and speakers bureau with Natera; and other interests/relationships with HossMed, Inc. P. Billings reports consultancy agreements with OmniSeq Inc, Alveo Technologies, Target BioPharma Inc., and Baebies Inc; ownership interest in Natera Inc., OmniSeq Inc, MissionBio Inc, 23andMe, LabCorp Inc, ThermoFisher Inc, LungLifeAI Inc., Fabric Genomics Inc., Precision Biosciences Inc, DNArx Inc., and Iliad Bioscience Inc; and scientific advisor or membership with OmniSeq Inc, MissionBio Inc, Fabric Genomics Inc., LungLifeAI Inc., Alveo Technologies, TargetBioPharma, Inc, and Baebies Inc. The remaining author has nothing to disclose.
Funding
This study was sponsored by Natera, Inc.
Acknowledgments
P. Billings, S. Bunnapradist, and H. Tabriziani conceptualized the study; S. Bunnapradist and P. Homkrailas were responsible for data curation; S. Bunnapradist was responsible for funding acquisition and project administration; E. Ahmed was responsible for methodology and formal analysis; S. Bunnapradist and P. Homkrailas wrote the original draft; and P. Billings, G. Fehringer, and H. Tabriziani reviewed and edited the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Veronese FV, Manfro RC, Roman FR, Edelweiss MI, Rush DN, Dancea S, et al. : Reproducibility of the Banff classification in subclinical kidney transplant rejection. Clin Transplant 19: 518–521, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Sigdel TK, Sarwal MM: The proteogenomic path towards biomarker discovery. Pediatr Transplant 12: 737–747, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gauthier P, Aleshin A, Shchegrova S, Mckenna T, Kalashnikova E, Sharma S, et al.: Factors influencing background cell-free DNA levels: Implications for donor-derived cell-free DNA assessment in transplant patients. ATC Virtual Conference, 2020
- 4.Oellerich M, Shipkova M, Asendorf T, Walson PD, Schauerte V, Mettenmeyer N, et al. : Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant 19: 3087–3099, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitlam JB, Ling L, Skene A, Kanellis J, Ierino FL, Slater HR, et al. : Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant 19: 1037–1049, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Altuğ Y, Liang N, Ram R, Ravi H, Ahmed E, Brevnov M, et al. : Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation 103: 2657–2665, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. : The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 18: 293–307, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]