ABSTRACT
Objective:
To investigate the association between the risk of tooth agenesis and single-nucleotide polymorphisms (SNPs) of MSX1 and PAX9 genes in nonsyndromic cleft patients.
Materials and Methods:
The subjects were 126 Korean nonsyndromic cleft patients. Tooth agenesis type (TAT) was classified as none (0); cleft area (1); cleft area + other area (2); and other area (3) based on agenesis of the maxillary lateral incisor (MXLI) and another tooth within or outside the cleft area. TAT was further grouped into two subcategories (0 and 1) and four subcategories (0, 1, 2, and 3). Three SNPs of MSX1 and 10 SNPs of PAX9 were investigated using Fisher's exact test and logistic regression analysis.
Results:
Although the association between genotype distribution of PAX9-rs7142363 and TAT was significant (P < .05 in four subcategories), genotypic odds ratios (GORs) of SNPs in each TAT were not meaningful. However, for MSX1-rs12532 and PAX9-rs2073247, associations between genotypic distribution and TAT were significant (P < .01 in four subcategories and P < .05 in two subcategories; P < .01 in two subcategories, respectively). In cleft area, GORs of MXLI agenesis in genotypes GA of MSX1-rs12532 and CT of PAX9-rs2073247 were increased by 3.14-fold and 4.15-fold compared with genotype GG of MSX1-rs12532 and CC of PAX9-rs2073247, respectively (P <. 01; P < .05). In cleft area + other area, the GOR of agenesis of MXLI and another tooth in genotype AA of MSX1-rs12532 was increased by fivefold compared with genotype GG (P < .05).
Conclusion:
Genetic disturbances of MSX1 and PAX9 genes are associated with tooth agenesis within and outside the cleft area.
Keywords: SNP, MSX1, PAX9, Tooth agenesis, Nonsyndromic cleft, Association analysis
INTRODUCTION
Because development of the oral cleft and formation of the tooth germ are closely related in terms of timing and anatomic position, dental anomalies including tooth agenesis both within and outside the cleft area have been reported to be more frequent in persons with nonsyndromic cleft lip with or without cleft palate (NS-CL ± P) than in individuals with nonsyndromic cleft lip with or without cleft palate (NS-CL ±P) than in the non-cleft individuals.1–6 Tooth agenesis in cleft patients affects esthetics, function, and periodontal health; causes collapse of the dental arch; and creates psychosocial problems. Therefore, an interdisciplinary approach is required to allow patients to receive more effective and efficient treatment.
The MSX1 and PAX9 genes are known to contribute to tooth agenesis of the posterior teeth and the maxillary lateral incisor.7–14 The MSX1 genes with a homeodomain and the PAX9 genes with a paired domain encode transcription factors that are essential for craniofacial and dental development of the mesenchyme.15–19 Generally, mutations in MSX1 and PAX9 cause loss of function because of haploinsufficiency and reduce the amount of functional protein available to maintain tooth development; this results in abnormalities in odontogenesis, such as arrest of the tooth bud.15–19
To date, only a few studies have addressed the genetic basis of oral cleft with or without tooth agenesis in humans.20–22 Van den Boogaard et al.20 and Liang et al.21 suggested that tooth agenesis and oral cleft were associated with nonsense mutations of MSX1, such as Ser104stop in exon 1 in a Dutch family and Q189X in exon 2 in a Chinese family, respectively. However, Liang et al.21 also reported that sequence analysis of PAX9 did not reveal mutation in any of the affected individuals studied. Modesto et al.22 investigated single-nucleotide polymorphisms (SNPs) in MSX1 of CL±P with or without tooth agenesis compared with non-cleft individuals and reported that the 101C>G variant occurred more frequently in patients with both NS-CL±P and tooth agenesis, whereas the *6C>T variant was found more often in those with NS-CL±P. However, these studies have several limitations, such as small sample size, inclusion of non-cleft individuals, or no classification of tooth agenesis within and outside the cleft area.
Given that not all patients with NS-CL±P have tooth agenesis, it is necessary to investigate the genetic basis of NS-CL±P with tooth agenesis compared with NS-CL±P without tooth agenesis. The MSX1 and PAX9 genes are known to contribute to both cleft and tooth agenesis.16,18,20–23 However, no study has investigated whether tooth agenesis within the cleft area is associated with isolated genetic disturbance or local tissue defect as part of clefting.
Therefore, the purpose of this study was to investigate the association between the risk of tooth agenesis and the SNPs of MSX1 and PAX9 genes in Korean nonsyndromic cleft patients. The null hypothesis was that there was no significant association between genotypic distribution of SNPs in the MSX1 and PAX9 genes and tooth agenesis type (TAT) in patients with cleft lip and alveolus (CLA) and cleft lip and palate (CLP).
MATERIALS AND METHODS
The study samples consisted of 126 Korean nonsyndromic cleft patients (82 males and 44 females; 28 patients with CLA and 98 patients with CLP). Subjects with longitudinal serial records and panoramic radiographs were selected from the Department of Orthodontics, Seoul National University Dental Hospital (SNUDH). The study protocol was approved by the Institutional Review Board at SNUDH (IRB CRI-G07002).
Subjects were classified according to cleft type and the status and location of missing teeth. Diagnosis of nonsyndromic CLA and CLP was made through clinical inspections by highly trained orthodontists. Tooth agenesis was identified from serial panoramic radiographs based on the age of the subject and considering the fact that the mean delay in tooth formation of the cleft children was approximately 4 to 6 months relative to that of non-cleft children.1,24 Regardless of size and morphology, any permanent tooth on either side of the alveolar cleft between the maxillary central incisor and canine was considered existence of the maxillary lateral incisor (MXLI).25,26
TAT was divided into none; cleft area only (missing of the MXLI within the cleft area only), cleft area + other area (missing of the MXLI within the cleft area and another maxillary tooth outside the cleft area), and other area only (missing of another maxillary tooth outside the cleft area only) (Figure 1). The status of those who were missing another maxillary tooth outside the cleft area did not include agenesis of the maxillary third molar. TAT was further grouped into two subcategories (none and cleft area only) and four subcategories (none, cleft area only, cleft area + other area, and other area only).
Figure 1.
A. Tooth agenesis type (TAT)-0 ‘None (no missing tooth)’ with bilateral cleft lip and palate (CLP) B. TAT-1 ‘Cleft area only [missing of the maxillary lateral incisor (MXLI) within the cleft area only]’ with unilateral CLP. C. TAT-2 ‘Cleft area + Other area (missing of MXLI within the cleft area and another maxillary tooth outside the cleft area)’ with unilateral CLP. D. TAT-3 ‘Other area only (missing of another maxillary tooth outside the cleft area only)’ with unilateral CLP. An asterisk represents a missing tooth.
Peripheral venous blood samples of patients were collected at SNUDH after obtaining written informed consent. Genomic DNA samples were extracted from peripheral venous blood lymphocytes by the protein precipitation method using a commercial DNA extraction kit (Quiagen Inc, Valencia, Calif) and were genotyped using the VeraCode Technology program (Illumina Inc, San Diego, Calif) at SNP Genetics Inc (Seoul, Korea).
SNP markers located from 2kb ∼ 5′ to 2kb ∼ 3′ of the MSX1 and PAX9 genes were obtained from literature review and the National Center for Biotechnology Information dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/). Three SNP markers of the MSX1 gene (rs3821949, rs12532, and rs4464513) and 10 SNP markers of the PAX9 gene (rs2295221, rs7142363, rs2073247, rs17104928, rs17176643, rs11156925, rs17104939, rs17104944, rs17104965, and rs1884213) with minor allele frequency (MAF) greater than 1% in the Japanese population were selected using the Web-based program, TAG SNP selection (TagSNP; http://snpinfo.niehs.nih.gov/guide.htm#snptag).27–29
Fisher's exact test was used to investigate the correlation between TAT and genotypic distribution of SNPs in MSX1 and PAX9 genes. Logistic regression analysis was performed to calculate the genotypic odds ratio of SNPs in MSX1 and PAX9 genes according to the genotypes in each TAT.
RESULTS
Demographic Information for Cleft Type, TAT, and Gender
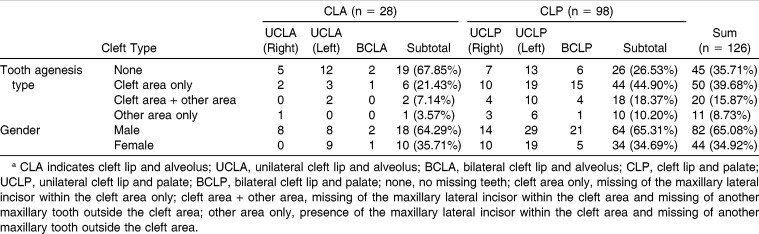
Among 126 Korean patients with NS-CL±P (82 males and 44 females; 28 patients with CLA and 98 patients CLP), TAT distribution was cleft area only (39.7%), cleft area + other area (15.9%), other area only (8.7%), and none (35.7%) (Table 1).
Table 1.
Demographic Information Regarding Cleft Type, Tooth Agenesis Type, and Gendera
Distribution of Tooth Agenesis
The most frequently missing teeth were the MXLI (73.6%) and the maxillary second premolar (16.5%) (Table 2).
Table 2.
Distribution of the Tooth Agenesis of the Maxillary Dentitiona
Association Between TAT and SNPs of MSX1 and PAX9 Genes
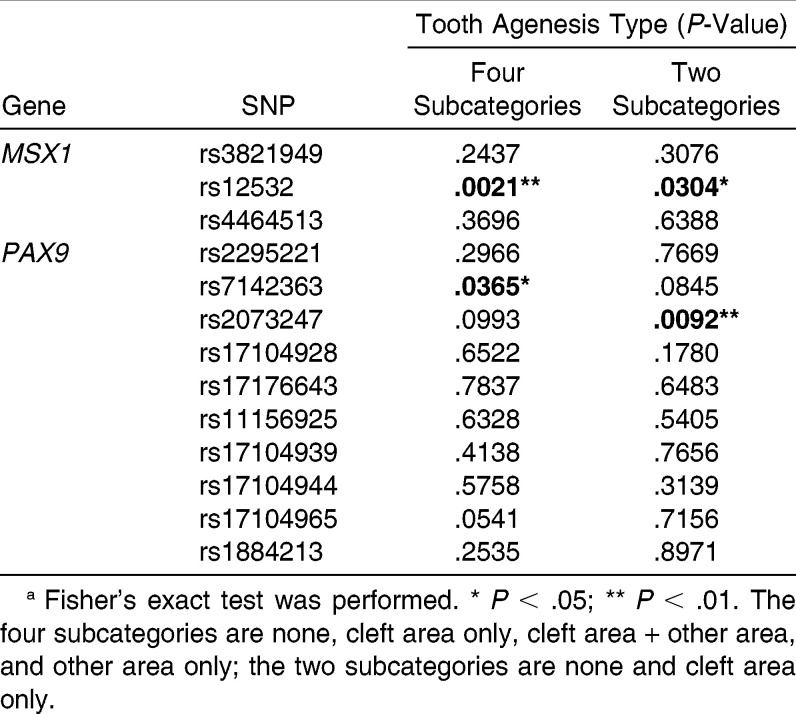
In the four subcategories of none, cleft area only, cleft area + other area, and other area only, MSX1-rs12532 and PAX9-rs7142363 showed a significant association with tooth agenesis (P < .01 and P < .05, respectively, Table 3). In the two subcategories of none and cleft area only, MSX1-rs12532 and PAX9-rs2073247 were significantly associated with tooth agenesis (P < .05 and P < .01, respectively, Table 3). These three candidate SNPs (MSX1-rs12532, PAX9-rs7142363, and PAX9-rs2073247) were further examined to assess genotypic distribution and genotypic odds ratios (GORs).
Table 3.
Association Between Tooth Agenesis Type and Single-Nucleotide Polymorphisms (SNPs) of the MSX1 and PAX9 Genesa
Genotypic Distribution and GORs of MSX1-rs12532 According to TAT
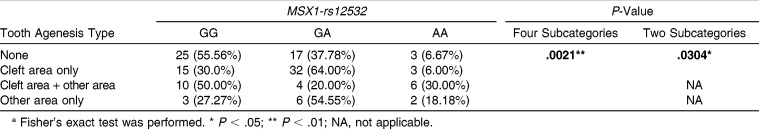
For MSX1-rs12532, there was a significant association between genotypic distribution and TAT in both the four subcategories and the two subcategories (P < .01 and P < .05, respectively, Table 4). The GOR of MXLI agenesis in genotype GA of MSX1-rs12532 was significantly increased 3.14-fold in cleft area only compared with genotype GG (95% confidence interval [CI] = 1.32–7.48, P < .01;Table 5). The GOR of MXLI and another maxillary tooth agenesis in genotype AA of MSX1-rs12532 was also significantly increased fivefold in cleft area + other area compared with genotype GG (95% CI = 1.04–23.98, P < .05, Table 5).
Table 4.
Genotypic Distributions of MSX1-rs12532 According to Tooth Agenesis Type in Four Subcategories and Two Subcategoriesa
Table 5.
Genotypic Odds Ratios (GORs) of Genotypes GA and AA Compared with Genotype GG of MSX1-rs12532 in Each Tooth Agenesis Typea
Genotypic Distribution and GORs of PAX9-rs7142363 and rs2073247 According to TAT
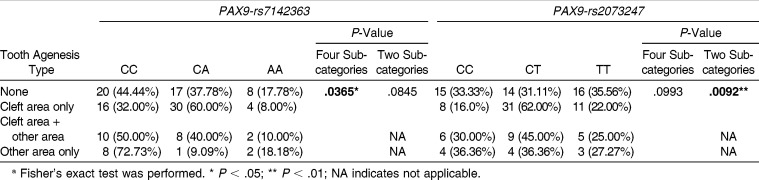
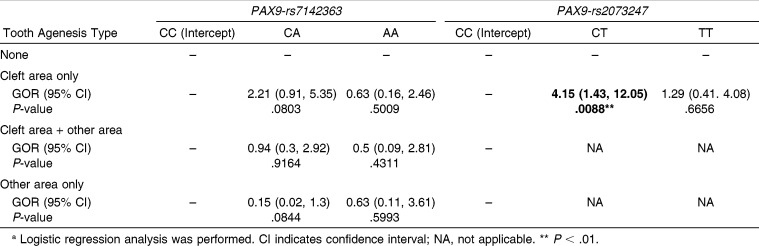
Although the association between genotype distribution of PAX9-rs7142363 and TAT was not significant in the two subcategories (P >.05, Table 6), this association was significant in the four subcategories (P < .05, Table 6). However, PAX9-rs2073247 showed the opposite tendency: the genotypic distribution of PAX9-rs2073247 exhibited a significant association with TAT in the two subcategories (P < .01, Table 6), but not in the four subcategories (P > .05, Table 6).
Table 6.
Genotypic Distributions of PAX9-rs7142363 and rs2073247 According to Tooth Agenesis Type in Four Subcategories and Two Subcategoriesa
Although the GORs of genotypes of PAX9-rs7142363 in each TAT were not meaningful (Table 7), the GOR of tooth agenesis of MXLI in genotype CT of PAX9-rs2073247 was significantly increased 4.15-fold in cleft area only compared with genotype CC (95% CI = 1.43–12.05, P < .01, Table 7).
Table 7.
Genotypic Odds Ratios (GORs) of Genotypes CA and AA Compared with Genotype CC of PAX9-rs7142363 and of Genotypes CT and TT Compared with Genotype CC of PAX9-rs2073247 in Each Tooth Agenesis Typea
DISCUSSION
Because mutations in PAX9 or MSX1 are known to cause agenesis of the posterior teeth7–10,17 and MXLI,11–14 we categorized NS-CL±P samples into four types of tooth agenesis—none (0), cleft area only (1), cleft area + other area (2), and other area only (3)—and grouped them into two subcategories (0 and 1) and four subcategories (0, 1, 2, and 3).
In this study, MSX1-rs12532 showed a significant association with tooth agenesis in both the four subcategories and the two subcategories of TAT (P < .01 and P < .05, respectively, Table 4). MXLI agenesis was significantly higher in patients with genotype GA compared with those with genotype GG (3.14-fold, P < .01, Table 5). In addition, agenesis of MXLI and another maxillary tooth was significantly higher in patients with genotype AA compared with those with genotype GG (fivefold, P < .05, Table 5). These findings suggest that this SNP might be associated with agenesis of MXLI and another maxillary tooth.
However, the results of this study were somewhat different from those of previous studies. In three patients with sporadic nonsyndromic oligodontia, Pawlowska et al.11 observed SNPs at MSX1-rs12532 in a patient with presence of MXLI and SNP at MSX1-rs8670 in a patient with absence of MXLI. Because these SNPs may be relatively common, they concluded that these polymorphisms would not be expected to have any pronounced phenotypic effect.11 In addition, although Paixão-Côrtes et al.13 found three polymorphic sites in the untranslated region of MSX1 exon 2 (rs8670, rs1095, and rs12532), there was no statistical difference in allele and genotype distributions between patients and control subjects. The polymorphism at rs8670 was not included in the present study, but it has previously been observed in individuals with MXLI agenesis.11,14 However, because this polymorphism at rs8670 is also relatively common, additional genes might be involved in this phenotype.14 The discordance between previous studies and this study seem to originate from differences between oligodontia patients and cleft patients.
For PAX9-rs7142363, a significant association was found in the four subcategories of TAT (P < .05, Table 6) but not in the two subcategories (Table 6). However, a meaningful GOR was not demonstrated in either category (Table 7), suggesting that this SNP might not be related to MXLI agenesis. However, the low percentage of genotype CA in the other area only compared with that of the cleft area only (9.1% vs 60.0%, respectively, Table 6) suggests a possible association between this SNP and TAT in cleft patients (GOR = 2.21, P = .0803 in cleft area only; GOR = 0.15, P = .0844 in other area only, compared with none, Table 7). These results indicate that statistical power might be increased as the sample size increases. Therefore, further studies are needed to investigate the relationship between PAX9-rs7142363 and MXLI agenesis using a larger sample size.
In contrast, PAX9-rs2073247 showed a significant association in the two subcategories of TAT (P < .01, Table 6), but not in the four subcategories (Table 6). In particular, the frequency of genotype CT was significantly increased in cleft area only (4.15-fold, P < .01, Table 7). These results indicate that PAX9-rs2073247 might be a potential candidate marker for MXLI agenesis susceptibility in Korean cleft patients. However, PAX9-rs2073247 and other polymorphisms located adjacent to this locus in the promoter region are also known to be associated with third molar agenesis. Saito et al.30 and Bianchi et al.31 reported that the CC genotype of the PAX9-rs2073247 showed a positive association with third molar agenesis. Peres et al.32 also suggested that the GT haplotype of PAX9-rs 2073244 and rs2076246 polymorphisms was associated with hypodontia, in most cases including third molar agenesis. Therefore, further studies are needed to investigate the relationship between PAX9-rs2073247 and MXLI agenesis.
In previous studies on the association between NS-CL±P and SNPs of the MSX1 and PAX9 genes, the A allele at MSX1-rs3821949 was associated with a significantly increased risk of NS-CL±P (GOR = 1.64, 95% CI = 1.03–2.63, P < .05, additive model)28 and the G/A heterozygote at PAX9-rs17104928 showed a significant association with NS-CL±P (GOR = 2.88, 95% CI = 1.42 into 5.84, P < .01).29 However, in the present study, these two SNPs of MSX1 and PXA9 did not show any significant association with tooth agenesis (Tables 3 through 7), whereas MSX1-rs12532 and PAX9-rs7142363 and rs2073247 did. These findings suggest that occurrence of oral cleft or tooth agenesis might depend on the locus of the SNP within the MSX1 gene or PAX9 gene.
The results of this study might be helpful for improving our understanding of the effects of genetic variation of MSX1 and PAX9 genes on tooth agenesis within and outside the cleft area. However, there are several factors to consider when interpreting the results obtained in this study. First, it would be better to increase the sample size to increase the statistical power and to avoid unnecessary statistical errors. Second, it is necessary to examine the genotypic distribution in non-cleft individuals with and without tooth agenesis to verify the exact role of SNPs in tooth agenesis. Third, functional consequences of these SNPs should be verified. Fourth, further studies are needed to investigate the influence of interactions of these genes with other genes, environmental factors, or ethnic differences.
CONCLUSIONS
Because an association between the risk of tooth agenesis and SNPs of MSX1 and PAX9 genes was found in nonsyndromic cleft patients, the null hypothesis was rejected.
Genetic disturbances of MSX1 and PAX9 genes are associated with tooth agenesis within and outside the cleft area in addition to the local tissue defect of clefting.
ACKNOWLEDGMENT
This research was supported by the Basic Science Research Program, the National Research Foundation of Korea (NRF 2009-0069859) funded by the Ministry of Education, Science and Technology, Republic of Korea. The authors thank all participants who donated samples and acknowledge Jung Sun Cho (Hallym University College of Medicine) for assistance during this work, as well as Duk Hwan Kim, Yong Ick Ji, Eunhyun Jung, and Se Young Cho for their contributions to DNA preparation for genotyping (Center for Genome Research, Samsung Biomedical Research Institute, Seoul, Korea).
REFERENCES
- 1.Ranta R. A review of tooth formation in children with cleft lip/palate. Am J Orthod Dentofacial Orthop. 1986;90:11–18. doi: 10.1016/0889-5406(86)90022-3. [DOI] [PubMed] [Google Scholar]
- 2.Shapira Y, Lubit E, Kuftinec MM. Hypodontia in children with various types of clefts. Angle Orthod. 2000;70:16–21. doi: 10.1043/0003-3219(2000)070<0016:HICWVT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Larmour CJ, Mossey PA, Thind BS, Forgie AH, Stirrups DR. Hypodontia—a retrospective review of prevalence and etiology. Part I. Quintessence Int. 2005;36:263–270. [PubMed] [Google Scholar]
- 4.Baek SH, Kim NY. Congenital missing permanent teeth in Korean unilateral cleft lip and alveolus and unilateral cleft lip and palate patients. Angle Orthod. 2007;77:88–93. doi: 10.2319/113005-419R.1. [DOI] [PubMed] [Google Scholar]
- 5.Al Jamal GA, Hazza'a AM, Rawashdeh MA. Prevalence of dental anomalies in a population of cleft lip and palate patients. Cleft Palate Craniofac J. 2010;47:413–420. doi: 10.1597/08-275.1. [DOI] [PubMed] [Google Scholar]
- 6.Camporesi M, Baccetti T, Marinelli A, Defraia E, Franchi L. Maxillary dental anomalies in children with cleft lip and palate: a controlled study. Int J Paediatr Dent. 2010;20:442–450. doi: 10.1111/j.1365-263X.2010.01063.x. [DOI] [PubMed] [Google Scholar]
- 7.Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 8.Nieminen P, Arte S, Tanner D, et al. Identification of a nonsense mutation in the PAX9 gene in molar oligodontia. Eur J Hum Genet. 2001;9:743–746. doi: 10.1038/sj.ejhg.5200715. [DOI] [PubMed] [Google Scholar]
- 9.Frazier-Bowers SA, Guo DC, Cavender A, et al. A novel mutation in human PAX9 causes molar oligodontia. J Dent Res. 2002;81:129–133. [PubMed] [Google Scholar]
- 10.Hansen L, Kreiborg S, Jarlov H, Niebuhr E, Eiberg H. A novel nonsense mutation in PAX9 is associated with marked variability in number of missing teeth. Eur J Oral Sci. 2007;115:330–333. doi: 10.1111/j.1600-0722.2007.00457.x. [DOI] [PubMed] [Google Scholar]
- 11.Pawlowska E, Janik-Papis K, Wisniewska-Jarosinska M, Szczepanska J, Blasiak J. Mutations in the human homeobox MSX1 gene in the congenital lack of permanent teeth. Tohoku J Exp Med. 2009;217:307–312. doi: 10.1620/tjem.217.307. [DOI] [PubMed] [Google Scholar]
- 12.Pinho T, Silva-Fernandes A, Bousbaa H, Maciel P. Mutational analysis of MSX1 and PAX9 genes in Portuguese families with maxillary lateral incisor agenesis. Eur J Orthod. 2010;32:582–588. doi: 10.1093/ejo/cjp155. [DOI] [PubMed] [Google Scholar]
- 13.Paixão-Côrtes VR, Braga T, Salzano FM, et al. PAX9 and MSX1 transcription factor genes in non-syndromic dental agenesis. Arch Oral Biol. 2011;56:337–344. doi: 10.1016/j.archoralbio.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Boeira BR, Jr, Echeverrigaray S. Polymorphism in the MSX1 gene in a family with upper lateral incisor agenesis. Arch Oral Biol. 2012;57:1423–1428. doi: 10.1016/j.archoralbio.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 16.Peters H, Neubüser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumlongras D, Lin JY, Chapra A, et al. A novel missense mutation in the paired domain of PAX9 causes non-syndromic oligodontia. Hum Genet. 2004;114:242–249. doi: 10.1007/s00439-003-1066-6. [DOI] [PubMed] [Google Scholar]
- 18.Nakatomi M, Wang XP, Key D, et al. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 19.Boeira BR, Jr, Echeverrigaray S. Dentistry and molecular biology: a promising field for tooth agenesis management. Tohoku J Exp Med. 2012;226:243–249. doi: 10.1620/tjem.226.243. [DOI] [PubMed] [Google Scholar]
- 20.van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 21.Liang J, Zhu L, Meng L, Chen D, Bian Z. Novel nonsense mutation in MSX1 causes tooth agenesis with cleft lip in a Chinese family. Eur J Oral Sci. 2012;120:278–282. doi: 10.1111/j.1600-0722.2012.00965.x. [DOI] [PubMed] [Google Scholar]
- 22.Modesto A, Moreno LM, Krahn K, King S, Lidral AC. MSX1 and orofacial clefting with and without tooth agenesis. J Dent Res. 2006;85:542–546. doi: 10.1177/154405910608500612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slayton RL, Williams L, Murray JC, et al. Genetic association studies of cleft lip and/or palate with hypodontia outside the cleft region. Cleft Palate Craniofac J. 2003;40:274–279. doi: 10.1597/1545-1569(2003)040<0274:GASOCL>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai MC, King NM, Wong HM. Dental development of Chinese children with cleft lip and palate. Cleft Palate Craniofac J. 2008;45:289–296. doi: 10.1597/07-019. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki A, Watanabe M, Nakano M, Takahama Y. Maxillary lateral incisors of subjects with cleft lip and/or palate: part 2. Cleft Palate Craniofac J. 1992;29:380–384. doi: 10.1597/1545-1569_1992_029_0380_mliosw_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro LL, das Neves LT, Costa B, Gomide MR. Dental development of permanent lateral incisor in complete unilateral cleft lip and palate. Cleft Palate Craniofac J. 2002;39:193–196. doi: 10.1597/1545-1569_2002_039_0193_ddopli_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 27.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim NY, Kim YH, Park JW, Baek SH. Association between MSX1 SNPs and Nonsyndromic Cleft Lip with or without Cleft Palate in the Korean Population. J Korean Med Sci. 2013;28:522–526. doi: 10.3346/jkms.2013.28.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JK, Park JW, Kim YH, Baek SH. Association between PAX9 single-nucleotide polymorphisms and nonsyndromic cleft lip with or without cleft palate. J Craniofac Surg. 2012;23:1262–1266. doi: 10.1097/SCS.0b013e31824e27c7. [DOI] [PubMed] [Google Scholar]
- 30.Saito CPB, Bianchi FJ, Peres RCR, Line SRP. Suggestive associations between polymorphisms in PAX9, MSX1 genes and third molar agenesis in humans. Curr Genomics. 2006;7:191–196. [Google Scholar]
- 31.Bianchi FJ, de Oliveira TF, Saito CB, Peres RC, Line SR. Association between polymorphism in the promoter region (G/C-915) of PAX9 gene and third molar agenesis. J Appl Oral Sci. 2007;15:382–386. doi: 10.1590/S1678-77572007000500002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peres RC, Scarel-Caminaga RM, do Espírito Santo AR, Line SR. Association between PAX-9 promoter polymorphisms and hypodontia in humans. Arch Oral Biol. 2005;50:861–871. doi: 10.1016/j.archoralbio.2005.02.003. [DOI] [PubMed] [Google Scholar]



![Figure 1. A. Tooth agenesis type (TAT)-0 ‘None (no missing tooth)’ with bilateral cleft lip and palate (CLP) B. TAT-1 ‘Cleft area only [missing of the maxillary lateral incisor (MXLI) within the cleft area only]’ with unilateral CLP. C. TAT-2 ‘Cleft area + Other area (missing of MXLI within the cleft area and another maxillary tooth outside the cleft area)’ with unilateral CLP. D. TAT-3 ‘Other area only (missing of another maxillary tooth outside the cleft area only)’ with unilateral CLP. An asterisk represents a missing tooth.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8636/8722836/d5c1144686ad/i0003-3219-83-6-1036-f01.jpg)