Abstract
Sp1-like proteins are defined by three highly homologous C2H2 zinc finger motifs that bind GC-rich sequences found in the promoters of a large number of genes essential for mammalian cell homeostasis. Here we report that TIEG2, a transforming growth factor β-inducible Sp1-like protein with antiproliferative functions, represses transcription through recruitment of the mSin3A-histone deacetylase complex. The interaction of TIEG2 with mSin3A is mediated by an alpha-helical repression motif (α-HRM) located within the repression domain (R1) of TIEG2. This α-HRM specifically associates with the second paired amphipathic helix (PAH2) domain of mSin3A. Mutations in the TIEG2 α-HRM domain that disrupt its helical structure abolish its ability to both bind mSin3A and repress transcription. Interestingly, the α-HRM is conserved in both the TIEG (TIEG1 and TIEG2) and BTEB (BTEB1, BTEB3, and BTEB4) subfamilies of Sp1-like proteins. The α-HRM from these proteins also mediates direct interaction with mSin3A and represses transcription. Surprisingly, we found that the α-HRM of the Sp1-like proteins characterized here exhibits structural and functional resemblance to the Sin3A-interacting domain previously described for the basic helix-loop-helix protein Mad1. Thus, our study defines a mechanism of transcriptional repression via the interactions of the α-HRM with the Sin3-histone deacetylase complex that is utilized by at least five Sp1-like transcriptional factors. More importantly, we demonstrate that a helical repression motif which mediates Sin3 interaction is not an exclusive structural and functional characteristic of the Mad1 subfamily but rather has a wider functional impact on transcriptional repression than previously demonstrated.
The Sp1-like family of transcription factors is characterized by the presence of three highly homologous C-terminal zinc finger motifs that are capable of binding GC-rich DNA sequences. These GC-rich motifs are present in the promoters of more than a thousand different gene products (10, 17, 21, 23). Currently, the Sp1-like proteins identified contain at least 16 members that can be classified into several subgroups, including Sp (Sp1, Sp2, Sp3, and Sp4), BTEB (BTEB1), KLF (BKLF, BKLF3, EKLF, GKLF, BTEB2/IKLF, and LKLF), CPBP (CPBP and UKLF), TIEG (TIEG1 and TIEG2), and Ap-2rep. The detailed nomenclature and classification of these proteins can be found in several recent reviews (7, 8, 26, 34). Several new members, including BTEB3 (J. Kaczynski et al., unpublished data), BTEB4 (A. Conley et al., unpublished data), Sp5 (14), and SP6/KLF14 (27), have recently been added to this growing family of proteins. Because many of the genes essential for the regulation of cell growth (6, 18, 28, 30), differentiation (2, 9), and apoptosis (21, 32) contain Sp1-like binding sites, it is not surprising that members of the Sp1 family are important regulators of mammalian cell homeostasis. Additionally, Sp1-like proteins are critical for normal development. Studies with animal models have shown that disruption of Sp1-like genes in mice is associated with abnormalities in early and late embryonic development, as well as decreased postnatal survival (3, 14, 18, 19, 22, 24, 25, 31, 36). Thus, elucidation of the molecular mechanisms by which the Sp1-like proteins regulate transcription will greatly advance our knowledge of cell growth control and morphogenesis.
We and others have previously identified two novel Sp1-like proteins, TIEG1 and TIEG2 (6, 29). The TIEG proteins function as transcriptional regulators capable of binding GC-rich sequences and repressing transcription (5, 6). In addition, the TIEG proteins are negative regulators of cell growth (6, 32). Deletion and site-directed mutagenesis analysis have defined three independent repressor domains (R1, R2, and R3) conserved within the amino terminus of TIEG proteins that are a defining feature of this subfamily of Sp1-like transcription factors (5). To gain insight into how these proteins function, our laboratory has been pursuing the characterization of the mechanisms used by these proteins to repress gene expression.
In this study, we have identified a 160-kDa TIEG2 R1-interacting protein as mSin3A and shown that mSin3A functions as a corepressor with TIEG2. Deletion mutagenesis demonstrates that R1 associates with mSin3A through interaction with the second paired amphipathic helix (PAH2) domain. Thus, R1 represents a functional Sin3 interaction domain for TIEG2. Furthermore, sequence analysis and circular dichroism (CD) data indicate that the primary and secondary structures of the TIEG2 Sin3-interacting domain (SID) is conserved in other members of the Sp1-like repressor protein family, including TIEG1 (5), BTEB1 (16), BTEB3 (Kaczynski et al., unpublished data), and BTEB4 (Conley et al., unpublished data). Each of these Sp1-like transcription factors contains a conserved sequence that may adopt an alpha-helical structure and is sufficient to mediate transcriptional repression and mSin3A interaction. Thus, we have named this structural motif the alpha-helical repression motif (α-HRM). Although we found that a core sequence within the α-HRM is also conserved within the Mad1 SID (1, 4, 12), sequences outside this region differ significantly between the α-HRM of Sp1-like repressors and the Mad1 SID. Our data suggest that this conserved α-HRM mediates interaction with the corepressor protein mSin3A and represents a transcriptional repression mechanism utilized by at least five different Sp1-like transcriptional repressors. The differences between the Sp1-like and Mad1 SID motifs and their role in transcriptional regulation are discussed.
MATERIALS AND METHODS
Vectors and plasmid construction.
The vectors used in the present study include glutathione S-transferase (GST) expression vectors pGEX-5x-1 and pGEX-6p-1 (Pharmacia, Piscataway, N.J.) for cloning, expression, and purification of GST fusion proteins in bacteria; DNA binding domain (DBD) effector pM GAL4, containing the yeast transcription factor GAL4 DBD (Clontech, Palo Alto, Calif.) fused with various repressor domains for transcriptional regulatory assays; vector pCMV-Tag2 (Stratagene, La Jolla, Calif.) for expression of FLAG epitope-tagged full-length TIEG2, mSin3A, and various mSin3A deletion constructs; and the pCMV-Myc/nuc vector (Invitrogen, Carlsbad, Calif.), carrying three carboxyl-terminal nuclear localization signals for nucleus-targeted expression. For GAL4 assays, a reporter vector carrying the firefly luciferase gene cloned downstream from five tandem GAL4 DNA binding sites and the thymidine kinase promoter was used. To control for transfection efficiency, a reporter plasmid carrying the Renilla luciferase gene under the control of the Rous sarcoma virus (RSV) promoter was used.
The GST fusion constructs were generated as follows. The repression domains of TIEG2 (R1, amino acids [aa] 1 to 79; R2, aa 151 to 162; R3, aa 273 to 351; Nterm, aa 1 to 371), previously described by Cook et al. (5), were cloned in frame into GST fusion vectors. The Mad1 SID, corresponding to aa 2 to 25 of the mouse Mad1 protein, and the putative SID-like domain of TIEG1 (aa 1 to 72) were amplified by PCR and cloned into GST fusion vectors. The putative SID-like repression domains of BTEB1 (aa 2 to 25), BTEB3 (aa 2 to 25), and BTEB4 (aa 2 to 25) were generated by primer annealing and extension and cloned into the GST fusion vectors. Standard PCR-based methods were used to generate mutations in TIEG2-R1 (ΔE29P and ΔA30P) and the mouse Mad1 SID (ΔA12P and ΔL16P) (12).
The plasmid containing the full-length mSin3A cDNA was kindly provided by R. N. Eisenman (Fred Huntington Cancer Center). Deletion constructs of the mSin3A-encoding gene were generated by using standard PCR techniques and were cloned in frame with the FLAG epitope of the pCMV-Tag2 vector. Fragments containing TIEG2-R1, the Mad1 SID, and their respective mutant forms were subcloned into the GAL4 DBD expression vector. The putative SID-like domains of BTEB1, BTEB3, and BTEB4 were released from the GST fusion vector and subcloned into the pM GAL4 DBD expression vector. Both wild-type and mutant TIEG2-R1 and the PAH2 of mSin3A were subcloned into the pCMV-Myc/nuc vector carrying three carboxyl-terminal nuclear localization signals for nucleus-targeted expression. All constructs were verified by direct sequencing (Mayo Molecular Biology Core Facility).
Transcriptional reporter assays.
The Chinese hamster ovary (CHO) cell line was obtained from the American Type Culture Collection (Manassas, Va.). CHO cells were cultured in Ham's F12 medium supplemented with 5% fetal bovine serum, 5% normal calf serum, 100 U of streptomycin per ml, and 100 U of penicillin per ml (Life Technologies, Rockville, Md.). The transfection conditions used with this cell line and the luciferase reporter assay were described previously (13). Briefly, 5 × 104 CHO cells were plated in 24-well tissue culture dishes and transfected 24 h later with 30 ng of the pGL3 firefly reporter plasmid carrying five tandem GAL4 DNA binding sites (GAL4-luc) and various amounts of GAL4 effector plasmid, as indicated, using Lipofectamine (Life Technologies). As a control for basal transcriptional activity, the reporter constructs were cotransfected with an effector plasmid carrying the GAL4 DBD alone. pcDNA3.1+ plasmid DNA (Invitrogen) was added to make the total quantity of DNA 300 ng per well. Following 4 h of Lipofectamine treatment, cells were washed with fresh medium and allowed to recover for 24 h. Following recovery, cells were lysed and luciferase assays were performed with a Turner 20/20 luminometer and the Dual-Luciferase Reporter Assay System in accordance with the manufacturer's (Promega, Madison, Wis.) suggestions. As a control for transfection efficiency, all conditions included cotransfection with 3 ng of Renilla luciferase control plasmid driven by the RSV promoter (RSV-pRL). In all experiments, firefly luciferase values were normalized to Renilla luciferase activity. For treatment with histone deacetylase (HDAC) inhibitors, CHO cells were cultured following Lipofectamine treatment in a medium containing either 75 nM trichostatin A (TSA) or 2.5 μM suberoylanilide hydroxamic acid (SAHA). Where indicated, cells were cotransfected with pCMV-Myc/nuc constructs expressing nucleus-targeted TIEG2 R1, TIEG2 R1m, and PAH2 of mSin3A or FLAG-tagged full-length mSin3A. Studies were performed in triplicate in at least three independent experiments with similar results.
GST fusion protein purification, in vitro translation, and pulldown assays.
GST and GST fusion protein expression was induced in BL21 cells (Stratagene) by the addition of 2 mM isopropyl-β-d-thiogalactopyranoside and incubation for 2 h. Cells were lysed and subsequently purified by using glutathione Sepharose 4B affinity chromatography in accordance with the manufacturer's (Pharmacia) suggestions. For GST pulldown assays using cell extracts, approximately 5 × 106 CHO cells were labeled with [35S]methionine for 4 h at 37°C and lysed at 4°C for 20 min in lysis buffer (150 mM NaCl, 0.5% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 20 mM MgCl2) supplemented with Complete Protease Inhibitor cocktail (Roche/Boehringer Mannheim, Indianapolis, Ind.). Cell extracts were precleared by incubation with GST and glutathione-conjugated Sepharose beads for 30 min at 4°C, followed by centrifugation at 500 × g for 5 min. The supernatant was transferred to fresh tubes and incubated with GST, GST-Nterm, GST-R1, GST-R2, or GST-R3 and additional glutathione-conjugated Sepharose beads for 2 h at 4°C. Complexes were pelleted by centrifugation at 500 × g for 5 min, washed five times with lysis buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was treated with AutoFluor (National Diagnostics, Atlanta, Ga.), dried, and exposed for autoradiography at −80°C. Where indicated, pulldown assays using unlabeled CHO cells were performed as described above. Complexes were pelleted, separated by SDS-PAGE, and transferred to a polyvinylidene difluoride membrane for Western blot analysis using rabbit polyclonal antibody against mSin3A or goat polyclonal antibody against HDAC1 (Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-rabbit or -goat peroxidase-conjugated secondary antibodies (Sigma, St. Louis, Mo.), and Lumilight (Roche/Boehringer Mannheim). The [35S]methionine-labeled, FLAG-tagged full-length mSin3A and mSin3A deletion constructs were produced by in vitro translation using the TNT coupled transcription-translation system under the conditions described by the manufacturer (Promega). GST pulldown assays using the in vitro-translated proteins was performed as described above for the cell extracts.
Immunoprecipitation and Western blot analysis.
To detect TIEG2 and mSin3A interaction, approximately 5 × 106 CHO cells were transiently transfected with 10 μg of the FLAG-tagged full-length TIEG2 plasmid using Lipofectamine. At 24 h posttransfection, cells were washed once with phosphate-buffered saline, harvested, and lysed in lysis buffer for 30 min at 4°C. Immunoprecipitations were performed by using anti-FLAG M2 agarose-conjugated antibody (Sigma). To detect the interaction between endogenous TIEG2 and mSin3A, 107 CHO cells were harvested and lysed and immunoprecipitations were carried out with anti-TIEG2 monoclonal antibody (Transduction Laboratories, Lexington, Ky.) and protein G-agarose beads (Santa Cruz Biotechnology). Whole-cell lysates and immunoprecipitated samples were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and analyzed by Western blot assay for mSin3A and HDAC1 coimmunoprecipitation as described above for GST pulldown assays. For detection of endogenous TIEG2 in immunoprecipitated samples, affinity-purified rabbit polyclonal anti-TIEG2 antibody was used (generated by Research Genetics, Huntsville, Ala.). To determine the expression levels of GAL4 constructs, 5 × 106 CHO cells were transfected for 24 h with 5 μg of GAL4 effector plasmids expressing various repressor domains. Cells were metabolically labeled with [35S]methionine prior to lysis, and immunoprecipitations were carried out by using anti-GAL4 DBD agarose-conjugated antibodies (Sigma). Immunocomplexes were separated by SDS-PAGE and subjected to autoradiography. For control of nucleus-targeted expression constructs, 5 × 106 CHO cells were transiently transfected with 10 μg of Myc-nuc constructs and immunoprecipitations were performed by using anti-Myc epitope polyclonal antibodies (Santa Cruz Biotechnology). Immunocomplexes were separated by SDS-PAGE and subjected to Western analysis by using mouse monoclonal anti-Myc epitope antibodies (Santa Cruz Biotechnology) as performed for GST pulldown complexes.
Peptide synthesis and far-UV CD spectroscopy.
All peptides for CD analysis were synthesized and purified by Bio-Synthesis Inc. (Lewisville, Tex.). The sequences for the wild-type mouse Mad1 SID, TIEG2 R1, and BTEB3 SID-like domains are GMNIQLLLEAADYLER, ILEQTDMEAVEALVCMSS, and AAAYVDHFAAECLVSM, respectively. Mutant peptides of these sequences contained prolines in place of the underlined residues. Peptide concentrations for far-UV CD measurements were determined by using the Edelhoch method (11). Briefly, aliquots of each peptide were added to 8 M guanidine hydrochloride (GdHCl) for a final concentration of 6 M GdHCl. The absorption spectrum of the resulting solutions was measured over a range 240 to 400 nm by using a CARY 2200 (Varian) spectrophotometer and corrected for turbidity when necessary. Subsequently, the A280 (tryptophan-containing peptides) and A272 (tyrosine-containing peptides) were used along with published values of the molar absorptivity of tryptophan and tyrosine in 6 M GdHCl to calculate peptide concentrations. The concentration of the TIEG2, BTEB3, and Mad1-SID mutant peptides was 200 μM, and that of the MAD1-SID wild-type peptide was 30 μM. Far-UV (180 to 250 nm) CD spectra were obtained at 0, 20, and 50% trifluoroethanol (TFE) by using a J-710 spectropolarimeter (JASCO) with U-type cells and a path length of 0.0148 cm (TIEG2 R1 and BTEB3) or 0.048 cm (Mad1 SID). Typically, three spectra were accumulated and subsequently averaged, except for the wild-type Mad1 SID, for which smoothing was also performed. Spectra were recorded by using a scan speed of 20 nm/min, a response time of 2 s, and a bandwidth of 2 nm.
RESULTS
Identification of a 160-kDa protein as a putative corepressor for TIEG2 R1.
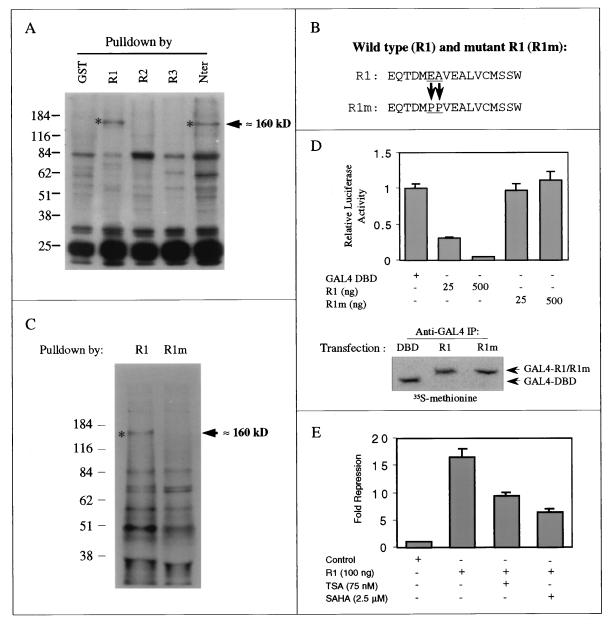
TIEG2 contains a potent transcriptional repression domain (R1) containing the minimal 17-aa sequence (aa 24 to 41) located within the N-terminal region. R1 is structurally and functionally conserved between TIEG1 and TIEG2 (5). In this study, we have investigated the molecular mechanisms underlying the function of R1 by initially focusing on the TIEG2 protein. To identify a corepressor(s) that may mediate the repression function of this domain, we performed GST pulldown assays with cells prelabeled with [35S]methionine using GST fusion proteins containing the repression domains of TIEG2. Figure 1A reveals that a 160-kDa protein specifically binds to R1 and the N terminus of TIEG2 but not to GST alone or to R2 and R3. This result suggests that this 160-kDa protein specifically interacts with R1.
FIG. 1.
Identification of a 160-kDa protein as a putative corepressor for TIEG2 R1. (A) Pulldown assays of [35S]methionine-labeled CHO cells were performed as described in Materials and Methods, by using purified GST fusion proteins expressing the N terminus, R1, R2, or R3 of TIEG2. Note that a 160-kDa protein is present in complexes pulled down by the N terminus and R1 of TIEG2 (asterisks) but not by R2 or R3. Protein molecular weight markers are shown on the left. (B) Wild-type TIEG2 R1 corresponds to aa 1 to 79, containing the minimum repressor sequence (aa 24 to 41). Mutant R1 (R1m) contains a double-proline substitution (arrowheads) at positions 29 and 30 (ΔE29P and ΔA30P). (C) Comparative pulldown assay of [35S]methionine labeled CHO cells using GST fusion proteins expressing R1 or R1m. The 160-kDa protein (asterisk) is present in the complex pulled down by wild-type R1 but not by mutant R1. Protein molecular size markers are shown on the left. (D) The GAL4 DBD alone or fused with wild-type R1 (GAL4-R1) or mutant R1 (GAL4-R1m) was cotransfected into CHO cells along with reporter plasmids as described in Materials and Methods. Relative luciferase activities determined by using the reporter construct with the GAL4 DBD alone and TIEG2 constructs are plotted. Note that potent repression activity is associated with GAL4-R1 but not with GAL4-R1m. To control for expression of the GAL4 constructs, we performed GAL4 immunoprecipitation assays of transfected, [35S]methionine-labeled CHO cells. Note that the levels of GAL4 R1 and GAL4 R1m expression are comparable. (E) The GAL4 reporter assay using GAL4-R1 in the presence of the HDAC inhibitors TSA and SAHA was performed as described in Materials and Methods. The repression activity of GAL4-R1 in the presence of TSA and SAHA is normalized to that of the GAL4 DBD alone under the same treatment. Note that TSA and SAHA significantly relieved the repression activity of GAL4-R1.
Based on a low-resolution secondary-structure prediction algorithm, we previously proposed that R1 adopts an α-helical conformation (5). Thus, we studied the effect of proline mutagenesis, which disrupts α-helix formation, on the binding of R1 to the 160-kDa protein. The minimal repressor sequences of wild-type and mutant R1 are shown in Fig. 1B. Mutant R1 has the residues glutamic acid (aa 29) and alanine (aa 30) mutated to prolines. By using GST pulldown analysis, we demonstrate that proline mutations in R1 abolish its ability to interact with the 160-kDa protein (Fig. 1C). In addition, we used the GAL4-based transcriptional reporter assay to determine what effect proline mutagenesis has on R1-mediated repression. For this purpose, we cotransfected CHO cells with plasmids carrying either wild-type or mutant R1 fused to the yeast GAL4 DBD (GAL4-R1 and GAL4-R1m, respectively) and a GAL4 reporter vector. Figure 1D shows that mutant R1, expressed at a level similar to that of wild type R1, completely lost its ability to repress transcription. This result suggests that R1 of TIEG2 requires a functional interaction with the 160-kDa protein to repress gene expression.
A common mechanism used to repress transcription is the posttranslational modification of histone proteins, which results in changes in chromatin structure. To determine if R1 of TIEG2 utilizes HDAC activity to repress transcription, we performed a GAL4-based transcriptional reporter assay in the presence of two known HDAC inhibitors. The data presented in Fig. 1E show that both TSA and SAHA relieve the R1-mediated repression by two- and threefold, respectively, in a manner similar to the well-characterized transcription factor Mad1 (data not shown). To control for the inhibitory effects of TSA and SAHA on the basal promoter, we normalized the relative luciferase activity of GAL4-R1 to that of the GAL4 DBD alone in order to determine the net effect of the HDAC inhibitors on R1-mediated repression. The results of this experiment suggest that HDAC activity is important for R1-mediated repression. We therefore hypothesized that the 160-kDa protein functions as a corepressor with R1 and, furthermore, that the repression mediated by R1 involves HDAC activity.
Identification of the 160-kDa protein as mSin3A.
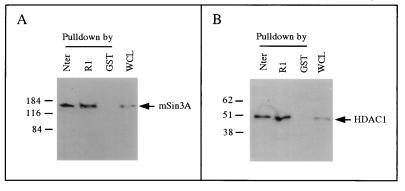
Among the previously described corepressor proteins, mSin3A has a molecular mass of approximately 160 kDa, which correlates well with the size of the R1 binding protein identified in Fig. 1A. In addition, mSin3A is present in a corepressor complex containing HDAC1 and HDAC2. To test directly whether the 160-kDa R1 binding protein is mSin3A, we performed a GST pulldown assay using CHO cell lysates and Western blot analysis. Figure 2A shows that antibodies against mSin3A specifically recognize a single band of 160 kDa on a Western blot of samples pulled down with GST-R1 and GST-Nter. In addition, we found that a deletion mutant expressing only R2 and R3 (aa 42 to 371) does not pull down mSin3A (data not shown). As a control, a band of the same size was also detected in whole-cell lysates prior to the pulldown assay but not in samples pulled down with GST. These results suggest that CHO cells constitutively express mSin3A and that this protein can specifically interact with TIEG2 through its R1 domain. In addition, Fig. 2B shows that HDAC1 also is associated with TIEG2 R1. Thus, HDAC1 appears to reside in the same complex with mSin3A and TIEG2. The R1-mSin3A interaction remained stable in lysis buffer containing up to 300 mM NaCl (data not shown). Taken together, these data suggest that recruitment of the mSin3A corepressor complex is the mechanism of TIEG2 R1-mediated repression.
FIG. 2.
Interaction of TIEG2 and mSin3A in vitro. (A) A GST pulldown assay using GST fusion proteins and unlabeled CHO cell extracts combined with Western blot analysis was used to analyze TIEG2-interacting proteins. The anti-mSin3A antibody recognizes a single band of approximately 160 kDa (arrow) in complexes pulled down by GST-R1 and GST-Nter but not by GST alone. As a control, a band of the same size is also observed in whole-cell lysate (WCL; 20 μg per lane) prior to pulldown. (B) The pulled-down complexes were also subjected to Western blot analysis using anti-HDAC1 antibody. The antibody recognizes a single band of approximately 50 kDa (arrow) in complexes pulled down by GST-R1 and GST-Nter, but not by GST alone, which is consistent with the size of HDAC1. As a control, a band of the same size is also observed in whole-cell lysate (20 μg per lane) prior to pulldown. The values on the left are molecular sizes in kilodaltons.
Endogenous mSinA is associated with TIEG2 in vivo
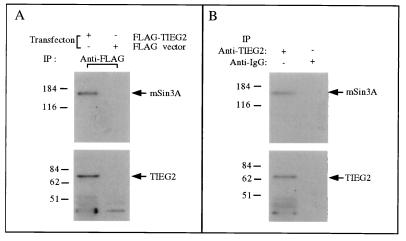
To address the question of whether full-length TIEG2 interacts with mSin3A in intact cells, we transfected a plasmid expressing full-length TIEG2 cloned in frame with the FLAG epitope (FLAG-TIEG2) into CHO cells and performed an immunoprecipitation assay. We detected the presence of TIEG2 and mSin3A in the immunocomplex by Western blot analysis using anti-TIEG2 and mSin3A antibodies, respectively. Figure 3A shows that TIEG2 and mSin3A are specifically precipitated by using the anti-FLAG antibody from cells transfected with the FLAG-TIEG2 plasmid but not in control cells transfected with the parental FLAG vector. This result indicates that overexpression of full-length TIEG2 can interact with endogenous mSin3A in mammalian cells. To further analyze the physiological relevance of this interaction, we performed immunoprecipitation and Western blot analysis of nontransfected CHO cells. The result, shown in Fig. 3B, reveals that endogenous TIEG2, immunoprecipitated by an anti-TIEG2 monoclonal antibody, and endogenous mSin3A are associated with each other. Taken together, these data show that TIEG2 interacts with Sin3A both in vitro and in vivo.
FIG. 3.
Endogenous mSin3A interaction with TIEG2. (A) CHO cells transfected with FLAG epitope-tagged full-length TIEG2 (FLAG-TIEG2) and the parental vector (FLAG vector) were subjected to immunoprecipitation (IP) with anti-FLAG antibody as described in Materials and Methods. The immunocomplexes were analyzed by Western blot analysis with antibodies against mSin3A and TIEG2 (arrows). Note that mSin3A is present in the immunocomplexes from cells transfected with FLAG-TIEG2 but not the FLAG vector. (B) Immunoprecipitation from untransfected CHO cell lysates using anti-TIEG2 antibody. The immunocomplexes were analyzed for endogenous mSin3A and TIEG2 by Western blot analysis as in panel A. Note that endogenous mSin3A is present in the complex immunoprecipitated with a TIEG2-specific antibody. The values on the left are molecular sizes in kilodaltons.
mSin3A is required for TIEG2 R1-mediated repression activity.
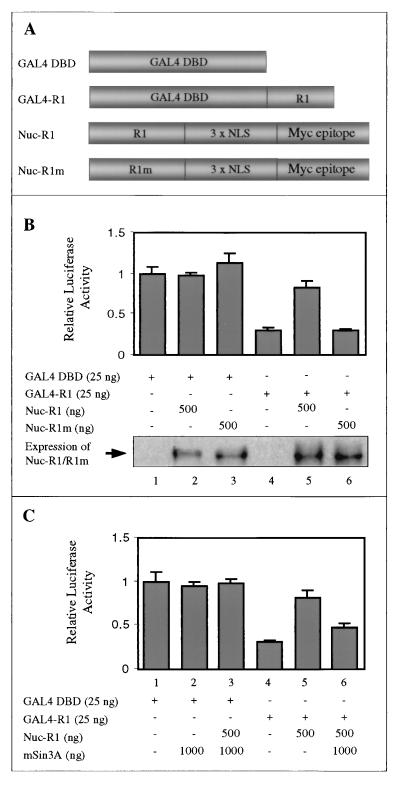
To further investigate the functional role of mSin3A in R1-mediated repression, we performed GAL4-based competition assays. We first examined whether nucleus-targeted overexpression of R1, which should compete with GAL4-R1 for any titratable factor involved in transcriptional repression (e.g., mSin3A), is sufficient to abolish GAL4-R1-mediated transcriptional repression. For this purpose, CHO cells were cotransfected with GAL4-R1 and increasing amounts of a plasmid containing wild-type or mutant R1 fused to the potent nuclear localization signals (Nuc-R1 and Nuc-R1m, respectively) of the pCMV/myc/nuc vector. A schematic representation of each construct used is shown in Fig. 4A. Figure 4B shows that GAL4-R1 exhibits strong repression activity (lane 4), as previously shown in Fig. 1D. Cotransfection of Nuc-R1 relieved the repression activity of GAL4-R1 (lane 5). However, cotransfection of the Nuc-R1m construct had no effect on repression activity (lane 6). As a control, we show that Nuc-R1 and Nuc-R1m are expressed at comparable levels and have no significant effect on the reporter activity of the control GAL4 vector alone (lanes 2 and 3, respectively).
FIG. 4.
mSin3A is required for R1-mediated repression. (A) Schematic diagram of expression vectors carrying the GAL4 DBD, GAL4-R1, and R1 and R1m fused with the Myc epitope and the three tandem nuclear localization signals (NLS) of the pCMV/Myc/nuc vector (Nuc-R1 and Nuc-R1m, respectively). (B) CHO cells were cotransfected as indicated with various effector plasmids along with reporter constructs. Basal transcriptional activity was determined by using the GAL4 reporter and the GAL4 DBD alone (lane 1). GAL4-R1 exhibits strong repression activity (lane 4), as shown in Fig. 1D. Cotransfection with Nuc-R1 led to a release of R1-mediated repression (lane 5). Cotransfection with Nuc-R1m had no effect on the repression activity of R1 (lane 6). Nuc-R1 and Nuc-R1m had no significant effect on reporter activity compared to that of GAL4 DBD alone (lanes 2 and 3, respectively). To control for expression of nucleus-targeted constructs, we performed immunoprecipitation and Western blot analysis by using anti-Myc antibodies. Note that the expression levels of Nuc-R1 (lanes 2 and 5) and Nuc-R1m (lanes 3 and 6) are comparable. (C) GAL4 assays were performed as in panel B, along with an expression vector carrying FLAG epitope-tagged full-length mSin3A. Note that addition of mSin3A partially restored the GAL4-R1-mediated repression activity that was relieved by Nuc-R1 (lane 6). As a control, we showed that mSin3A, alone or cotransfected with Nuc-R1, has no significant transcriptional activity (lanes 2 and 3, respectively).
A defining feature of corepressor proteins is their ability to stimulate the repression activity of their target transcription factors. We next examined the ability of full-length mSin3A to enhance R1-mediated transcriptional repression by using the GAL4 reporter assay. As shown in Fig. 4C, we first inhibited GAL4-R1-induced transcriptional repression partially by cotransfection with Nuc-R1 (lane 5), similar to the result shown in Fig. 4B. We then cotransfected full-length mSin3A to determine if this protein could restore GAL4-R1-mediated repression. Indeed, overexpression of full-length mSin3A significantly restored the repression activity of GAL4-R1 (lane 6). As a control, cotransfection of either mSin3A or mSin3A and Nuc-R1 with the GAL4 DBD vector alone had no significant effect on reporter activity (lanes 2 and 3, respectively). Therefore, these data suggest that mSin3A functions as a corepressor with TIEG2 and is required for R1-mediated transcriptional repression.
TIEG2 R1 interacts with PAH2 of mSin3A.
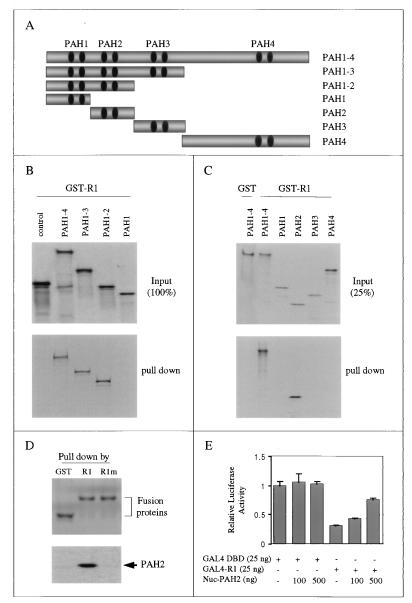
The mSin3A protein is highly modular and contains four PAH domains that mediate protein-protein interactions with distinct transcription factors and HDAC proteins to modify chromatin structure. To determine which region(s) of mSin3A is required for interaction with R1, we performed pulldown assays by using GST-R1 and in vitro-translated, [35S]methionine-labeled mSin3A protein fragments. A detailed map of each construct is shown in Fig. 5A. Figure 5B shows that R1 interacts with the full-length mSin3A protein (PAH1 to -4) and mSin3A deletion proteins containing PAH1 and-2 and PAH1 to-3, but not PAH1 and the luciferase control, suggesting that R1 interacts with the PAH2 domain of mSin3A. To determine whether the PAH2 region alone is sufficient to bind R1, we performed pulldown assays by using mSin3A constructs containing the individual PAH domains. The results in Fig. 5C show that GST-R1 directly binds only PAH2 and full-length mSin3A. Furthermore, Fig. 5D shows that mutations in R1 abolish its ability to bind PAH2, which is consistent with the data shown in Fig. 1C for full-length mSin3A binding. Therefore, we conclude that the PAH2 domain of mSin3A is sufficient to mediate specific interaction with TIEG2 R1 in vitro.
FIG. 5.
Mapping of the TIEG2 R1-interaction domain of mSin3A. (A) Schematic representation of vectors expressing either full-length or various deletion mutant forms of mSin3A. (B) We performed pulldown assays by using GST-R1 with in vitro-translated, [35S]methionine-labeled proteins expressing various deletion constructs of mSin3A, as indicated. The input (100%) of the in vitro-translated proteins is shown at the top, and the pulldown complexes are shown at the bottom. Note that GST-R1 interacts with the mSin3A constructs PAH1–4, PAH1–3, and PAH1–2. (C) A pulldown assay was performed by using GST-R1 with in vitro-translated individual PAH domains of mSin3A. The input (25%) of in vitro-translated proteins is shown at the top, and the pulldown complexes are shown at the bottom. Note that GST-R1 interacts only with full-length mSin3A and PAH2 and not with the other PAH domains. GST alone, used as a control, does not interact with full-length mSin3A. (D) A pulldown assay using GST-R1, GST-R1m, and GST alone with in vitro-translated, [35S]methionine-labeled PAH2 was performed. Note that PAH2 is pulled down only by R1 and not by mutant R1 or GST alone. (E) GAL4 reporter assay showing the squelching effect of nucleus-targeted PAH2 on R1-mediated repression. The relative luciferase activity of GAL4-R1 cotransfected with increasing concentrations of Nuc-PAH2 is plotted. Nuclear overexpression of PAH2 does not affect significantly the basal promoter activity of the reporter mediated by the GAL4 DBD alone but significantly reduces GAL4-R1-mediated repression activity.
To further study the in vivo participation of the PAH2 domain of mSin3A in R1-mediated repression, we performed GAL4-based competition assays by using a nucleus-targeted PAH2 domain (Nuc-PAH2). The HDAC1 or -2 binding region of mSin3 is located between PAH3 and PAH4 (20) and is required for mSin3A-mediated repression. We hypothesized that nuclear overexpression of the PAH2 domain alone should compete with binding of endogenous mSin3A to R1 and relieve R1-mediated repression. Indeed, Fig. 5E shows that overexpression of Nuc-PAH2 results in a dose-dependent release of GAL4-R1-mediated repression but exhibits no significant effect on the reporter activity of the GAL4 vector alone. This suggests that although PAH2 does not possess active repression activity, it is sufficient to mediate the interaction between mSin3A and R1. Therefore, based on these data, together with the results shown in Fig. 4C, we conclude that the interaction between TIEG2 R1 and PAH2 is necessary for R1-mediated repression through mSin3A-HDACs.
The α-helical structure is conserved in the TIEG and BTEB subfamily of Sp1-like repressor proteins.
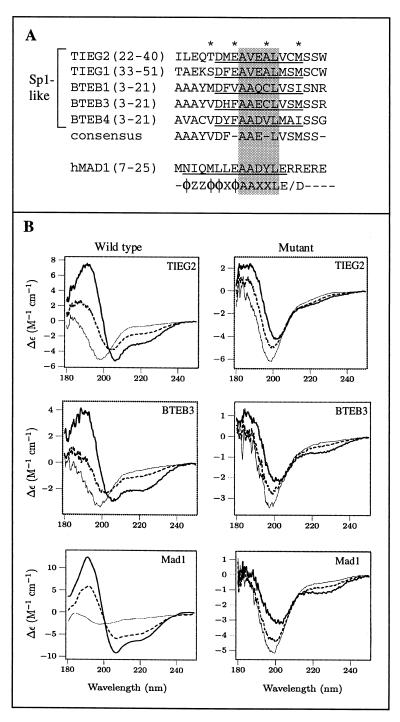
We have previously shown that the TIEG proteins function as potent transcriptional repressors (5). To understand the mechanisms used by Sp1-like proteins to regulate cell growth and differentiation, we have identified two novel Sp1-like proteins that belong to the BTEB subfamily, BTEB3 (Kaczynski et al., unpublished data) and BTEB4 (Conley et al., unpublished data). Interestingly, we have found that the primary sequence is conserved not only in R1 of TIEG1 and TIEG2 but also in the N terminus of the BTEB proteins. In Fig. 6A, we compare these conserved sequences with the Mad1 SID and the PAH2 interaction consensus sequence as defined by Brubaker et al. Note that the Sp1-like proteins analyzed display amino acid identity at four residues (asterisks) and that a core sequence is conserved between these Sp1-like proteins and the Mad1 SID and PAH2 interaction consensus sequence (shaded residues). As mentioned earlier, a low-resolution secondary-structure prediction algorithm had previously allowed us to propose that R1 of TIEG2 adopts an α-helical conformation (5). We have analyzed and compared the secondary structure of the conserved domains within the TIEG and BTEB proteins by using the PSIpred V2.0 Server and found that the putative α-helical nature is also conserved (Fig. 6A, underlined residues). To determine whether these domains could adopt an α-helical conformation, we measured the CD spectra of synthesized peptides from representative members of the Sp1-like subfamilies (TIEG2 and BTEB3). Each wild-type peptide was analyzed along with a peptide containing proline substitutions (as indicated in Materials and Methods). In the case of the TIEG2 R1 peptide, the proline mutations are identical to the R1 mutant used in GST pulldown assays. Each peptide was measured in 20 mM phosphate buffer (pH 7.2) containing 0, 20, or 50% TFE. In the absence of TFE, all of the peptides studied lacked secondary structure (Fig. 6B). At TFE concentrations of 20 and 50%, the TIEG2 R1, BTEB3, and Mad1 SID peptides adopt a helical conformation, as indicated by the strong negative peaks at 208 and 222 nm in the CD spectra. In contrast, the peptides containing proline substitutions display little or no induced helical structure at the same TFE concentrations. It should be noted that the wild-type Mad1 SID aggregates at concentrations of greater than 30 μM (data not shown), which is consistent with the finding that the Mad1 SID aggregates in solution at concentrations required for nuclear magnetic resonance (NMR) analysis (4). The CD data suggest that the TIEG2 R1 and BTEB3 peptides have a tendency similar to that of the Mad1 SID to adopt an α-helical conformation (10). Taken together, these results suggest not only that the primary sequence is homologous between the TIEG and BTEB proteins but also that the secondary helical structure is conserved.
FIG. 6.
A conserved α-helical motif is present in TIEG and BTEB proteins. (A) Sequence alignment of regions in the TIEG and BTEB proteins homologous to TIEG2 R1. Identical residues within this region of these Sp1-like proteins are indicated by asterisks. The amino acid residues corresponding to the conserved α-helical region are underlined. The consensus sequence is shown below. For comparison, the mouse Mad1 SID, along with the PAH2 interaction motif, as defined by Brubaker et al. (4), is included. Note that the sequences outside the AAXXL core (shaded residues) differ significantly between the Sp1-like proteins and the Mad1 SID. (B) CD analysis of peptides containing consensus sequences from TIEG2 R1, BTEB3, the Mad1 SID, and their respective mutant forms was performed as described in Materials and Methods. Note that the wild-type peptides are random coils in the absence of TFE (thin line) but become helical in the presence of 20% (dashed line) and 50% (thick line) TFE, as evidenced by the strong negative peaks at 208 and 222 nm. This trend is not observed in the mutant peptides, which remain largely unstructured under the same conditions. The spectrum of each peptide represents the average of three scans. Smoothing was performed for the wild-type Mad1 SID only.
The conserved α-HRM motif is sufficient to mediate mSin3A interaction and repression.
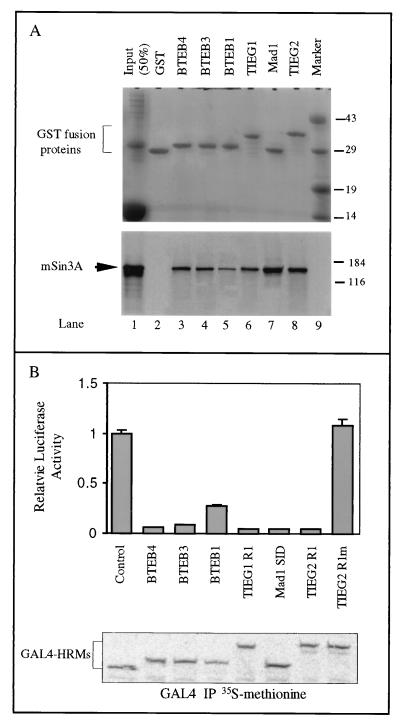
The sequence homology and CD data shown above suggest that the conserved α-helical domain is a general mechanism by which Sp1-like proteins interact with the corepressor mSin3A. To test this hypothesis, we performed GST pulldown assays by using GST fusion proteins containing the conserved domains of the Sp1-like proteins and in vitro-translated, [35S]methionine-labeled full-length mSin3A. The results shown in Fig. 7A demonstrate that the conserved α-helical domains from TIEG1, BTEB1, BTEB3, and BTEB4 pulled down mSin3A, similar to TIEG2 R1 and the Mad1 SID. This result demonstrates that these conserved α-helical motifs are sufficient to mediate interaction with mSin3A. In addition, we determined whether these conserved domains are sufficient to mediate transcriptional repression. For this purpose, we performed the GAL4 reporter assay by using GAL4 constructs expressing the conserved α-helical motifs from TIEG1, BTEB1, BTEB3, and BTEB4. Figure 7B shows that these motifs are potent repressors of transcription similar to TIEG2 R1 and the Mad1 SID when expressed at comparable levels. Thus, our data suggest the presence of a conserved α-helical repression motif (α-HRM) in the TIEG and BTEB subfamilies of Sp1-like proteins that mediates transcriptional repression activity through interaction with the corepressor mSin3A.
FIG. 7.
The α-HRM is sufficient to mediate interaction with mSin3A and repress transcription. (A) Pulldown assay using GST fusion proteins expressing the α-HRM from TIEG proteins, BTEB proteins, the Mad1 SID, and in vitro-translated, [35S]methionine-labeled full-length mSin3A was performed. Following separation by SDS-PAGE, pulled down complexes were Coomassie stained to show equal input of fusion proteins (top) and exposed for autoradiography to reveal the presence of [35S]methionine-labeled mSin3A (bottom). Lane 1 shows 50% input of in vitro-translated mSin3A. Note that the α-HRM from TIEG1 and BTEBs (lanes 3 to 6) interacts with mSin3A, similar to TIEG2 R1 (lane 8) and the Mad1 SID (lane 7). Protein molecular size markers (lane 9) are shown on the right (sizes are in kilodaltons). (B) GAL4 reporter assays with the α-HRMs from TIEG proteins, BTEB proteins, and the Mad1 SID were performed as described in Materials and Methods. The relative luciferase activities of the GAL4 DBD alone and that of the GAL4 DBD fused with different α-HRMs are plotted as a histogram. Note that the α-HRMs from TIEG1 and BTEB proteins strongly repress transcription, similar to TIEG2 R1 and the Mad1 SID. As shown previously, TIEG2 R1m does not exhibit any repression activity. To control for expression of the GAL4 constructs, we performed GAL4 immunoprecipitation (IP) assays with transfected, [35S]methionine-labeled CHO cells. Note that the expression levels of all of the GAL4 constructs are comparable.
DISCUSSION
The present report describes the identification and functional characterization of the α-HRM, a novel mSin3A-interacting domain that is conserved in several Sp1-like transcriptional repressors. This study originated from the observation that R1 of TIEG2 binds mSin3A with high affinity. Indeed, detailed biochemical and functional analyses have demonstrated that the TIEG2 α-HRM domain interacts specifically with the PAH2 domain of mSin3A to repress transcription. Interestingly, we had previously proposed that R1 (a peptide containing the α-HRM domain) had the potential to adopt an α-helical conformation based on secondary-structure prediction algorithms (5). In this study, by using CD analysis, we confirmed that, indeed, this domain has the propensity to form an α-helix. In addition, we showed that an α-helical SID similar to the TIEG α-HRM domains is conserved in the N termini of BTEB1, BTEB3, and BTEB4. Thus, our data demonstrate that this α-HRM domain is a defining structural and functional feature of these Sp1-like transcription factors, linking the function of these proteins to HDAC-mediated transcription repression via mSin3A binding (15).
A striking finding of this study is that the α-HRM domain of the Sp1-like proteins shows some structural and functional resemblance to the SID previously described in the basic helix-loop-helix protein Mad1 (12). Members of the Mad family of repressor proteins interact with the PAH2 domain of mSin3A via the SID located at the amino terminus (1). CD and mutational analyses demonstrated that the Mad1 SID adopts an amphipathic α-helical conformation in solution and that this α-helical structure is necessary for interaction with PAH2 (12). More recently, the prediction of the helical nature of the Mad1 SID was directly confirmed through the use of NMR by solving the solution structure of the Mad1 SID bound to the PAH2 domain (4). Both domains undergo mutual folding transitions upon complex formation, thus generating an unusual left-handed, four-helix bundle structure in the PAH2 domain and an amphipathic α-helix in the Mad1 SID. Based on the high-resolution NMR structure of the PAH2-SID complex, in conjunction with mutational sequence analysis, a PAH2 domain-interacting sequence motif was proposed (4), providing, for the first time, a unique example of a short structural motif that is involved in the interaction of Mad-like proteins with mSin3A. Thus, these studies demonstrated that the SID is a distinct characteristic of this helix-loop-helix subfamily of proteins.
The fact that the TIEG2 α-HRM and the Mad1 SID interact with the same PAH2 domain prompted us to investigate whether these peptides share structural similarities that may explain these interactions. Although BLAST (http://www.ncbi.nlm.nih.gov/BLAST) searches failed to find similarities between the α-HRM and the Mad1 SID, a forced sequence comparison done by using the BoxShade program revealed a low level of homology between the TIEG2 α-HRM and the Mad1 SID. This homology extends to both the presence of a core consensus sequence, AA/VXXL (Fig. 6A, shaded residues), and similar helical propensities (Fig. 6B). However, Fig. 6A shows that there are distinct sequence differences among these proteins outside of the AA/VXXL core (Fig. 6A). Note that overall, the α-HRM domains of the TIEG and BTEB proteins are more similar to each other than to the Mad1 SID. This poor sequence homology outside of the conserved core can explain, at least in part, why database searches done with the Mad1 SID were unable to identify the α-HRM of the Sp1-like proteins. Nevertheless, in spite of this difference, the data presented in this report demonstrate that both types of sequences function as a binding site for the PAH2 domain of Sin3A and underscore the importance of detailed biochemical characterizations in guiding functional domain annotation. We do not know whether the PAH2 domain adopts a similar conformation upon binding to the α-HRM of TIEG2 compared to the Mad1 SID. Ongoing structural studies in our laboratory are aimed at defining this interaction. The results of these studies should be helpful in defining the structural bases for the interactions of different repressors with mSin3A and may eventually offer the possibility of predicting such interactions.
It is also important to discuss the potential impact of our results on the understanding of the functional properties of Sp1-like proteins. The activation function of several Sp1-like proteins has been extensively characterized. The picture emerging from these studies suggests that activation domains within these proteins interact with specific coactivator proteins to regulate either the basal transcriptional machinery or chromatin structure. However, the mechanism(s) of Sp1-like repressor proteins is much less well understood. Of the known Sp1-like repressors, only the molecular mechanisms of BKLF and the closely related protein BKLF3/KLF8, both of which associate with the mCtBP2 corepressors, have been reported (33, 35). A short consensus motif (PVDLS/T) in BKLF and BKLF3 appears to be required for the interaction of BKLF proteins with mCTBP2 (33, 35). However, it has not been shown if these short motifs are sufficient to mediate the interaction of these proteins with the corepressor and thus mediate transcriptional repression. It is possible that this domain recruits HDAC via mCTBP2 to silence the expression of target genes, although more studies are needed to test the validity of this hypothesis. Thus, the present study also expands our understanding of the molecular mechanism underlying the function of Sp1-like transcription repressors by defining a novel mechanism that is utilized by at least two subfamilies of Sp1-like proteins (TIEG and BTEB).
In conclusion, we have shown that, similar to Mad1, five Sp1-like transcriptional repressors, TIEG1, TIEG2, BTEB1, BTEB3, and BTEB4, contain an α-HRM that functions as a SID. In addition, we have demonstrated that this motif acts as a transcriptional repressor domain in vivo. It is important to emphasize that the TIEG and BTEB proteins are the first Sp1-like transcription factors shown to repress gene expression via the mSin3A-HDAC corepressor complex. More significantly, our study demonstrates that the α-HRM of Sp1-like repressors, in addition to the SID of the Mad subfamily of basic helix-loop-helix proteins, represents a broader mechanism for transcriptional repression. Together, these results extend our knowledge of the repertoire of molecular motifs used by mammalian cells to mediate repressor-corepressor interactions.
ACKNOWLEDGMENTS
We thank Brian Gebelein, Karen Hedin, and Vijay Shah for critically reviewing the manuscript. We kindly thank R. N. Eisenman for providing the mSin3A full-length cDNA. We also thank the members of the Mayo Molecular Biology Core Facility for oligonucleotide synthesis and DNA sequencing.
This work was supported by the Mayo Cancer Center and National Institutes of Health grant DK52913 to R.U.
REFERENCES
- 1.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 2.Billon N, Carlisi D, Datto M B, van Grunsven L A, Watt A, Wang X F, Rudkin B B. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene. 1999;18:2872–2882. doi: 10.1038/sj.onc.1202712. [DOI] [PubMed] [Google Scholar]
- 3.Bouwman P, Gollner H, Elsasser H P, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 2000;19:655–661. doi: 10.1093/emboj/19.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brubaker K, Cowley M S, Huang K, Loo L, Yochum G S, Ayer D E, Eisenman R N, Radhakrishnan I. Solution structure of the interacting domains of the mad-sin3 complex. Implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–665. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 5.Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- 6.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 7.Cook T, Gebelein B, Urrutia R. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann N Y Acad Sci. 1999;880:94–102. doi: 10.1111/j.1749-6632.1999.tb09513.x. [DOI] [PubMed] [Google Scholar]
- 8.Dang D T, Pevsner J, Yang V W. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denver R J, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem. 1999;274:23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- 10.Dynan W S, Saffer J D, Lee W S, Tjian R. Transcription factor Sp1 recognizes promoter sequences from the monkey genome that are simian virus 40 promoters. Proc Natl Acad Sci USA. 1985;82:4915–4919. doi: 10.1073/pnas.82.15.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 12.Eilers A L, Billin A N, Liu J, Ayer D E. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J Biol Chem. 1999;274:32750–32756. doi: 10.1074/jbc.274.46.32750. [DOI] [PubMed] [Google Scholar]
- 13.Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger–Krüppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison S M, Houzelstein D, Dunwoodie S L, Beddington R S. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with brachyury. Dev Biol. 2000;227:358–372. doi: 10.1006/dbio.2000.9878. [DOI] [PubMed] [Google Scholar]
- 15.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 16.Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 18.Kuo C T, Veselits M L, Leiden J M. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1890. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 19.Kuo C T, Veselits M L, Barton K P, Lu M M, Clendenin C, Leiden J M. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylase associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncog. 1996;7:101–125. [PubMed] [Google Scholar]
- 22.Marin M, Karism A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 24.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 25.Perkins A. Erythroid Krüppel like factor: from fishing expedition to gourmet meal. Int J Biochem Cell Biol. 1999;31:1175–1192. doi: 10.1016/s1357-2725(99)00083-7. [DOI] [PubMed] [Google Scholar]
- 26.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scohy S, Gabant P, Van Reeth T, Hertveldt V, Dreze P L, Van Vooren P, Riviere M, Szpirer J, Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 28.Shields J M, Christy R J, Yang V W. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramaniam M, Harris S A, Oursler M J, Rasmussen K, Riggs B L, Spelsberg T C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Chen X, Yang V W. Intestinal-enriched Krüppel like factor (Krüppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supp D M, Witte D P, Branford W W, Smith E P, Potter S S. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 32.Tachibana I, Imoto M, Adjei P, Gores G J, Subramaniam M, Spelsberg T C, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Investig. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Krüppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 35.van Vliet J, Turner J, Crossley M. Human Krüppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wani M A, Wert S E, Lingrel J B. Lung Krüppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J Biol Chem. 1999;274:21180–21185. doi: 10.1074/jbc.274.30.21180. [DOI] [PubMed] [Google Scholar]