Abstract
Motivation
Single-cell transcriptomics profiling technologies enable genome-wide gene expression measurements in individual cells but can currently only provide a static snapshot of cellular transcriptional states. RNA velocity analysis can help infer cell state changes using such single-cell transcriptomics data. To interpret these cell state changes inferred from RNA velocity analysis as part of underlying cellular trajectories, current approaches rely on visualization with principal components, t-distributed stochastic neighbor embedding and other 2D embeddings derived from the observed single-cell transcriptional states. However, these 2D embeddings can yield different representations of the underlying cellular trajectories, hindering the interpretation of cell state changes.
Results
We developed VeloViz to create RNA velocity-informed 2D and 3D embeddings from single-cell transcriptomics data. Using both real and simulated data, we demonstrate that VeloViz embeddings are able to capture underlying cellular trajectories across diverse trajectory topologies, even when intermediate cell states may be missing. By considering the predicted future transcriptional states from RNA velocity analysis, VeloViz can help visualize a more reliable representation of underlying cellular trajectories.
Availability and implementation
Source code is available on GitHub (https://github.com/JEFworks-Lab/veloviz) and Bioconductor (https://bioconductor.org/packages/veloviz) with additional tutorials at https://JEF.works/veloviz/. Datasets used can be found on Zenodo (https://doi.org/10.5281/zenodo.4632471).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Current technologies for high-throughput single-cell transcriptomics profiling provide a static snapshot of the transcriptional states of individual cells. Still, the continuum of transcriptional states for cells along dynamic processes such as organ development or tumorigenesis can be used to infer how cell states may change over time (Saelens et al., 2019; Tritschler et al., 2019). Notably, RNA velocity analysis can be applied to infer dynamics of gene expression and predict the future transcriptional state of a cell from single-cell RNA-sequencing and imaging data (La Manno et al., 2018; Xia et al., 2019).
To interpret such cell state changes from RNA velocity analysis, current approaches project the observed current and predicted future transcriptional states onto 2-dimensional (2D) embeddings to visualize the putative directed cellular trajectory (Bastidas-Ponce et al., 2019; La Manno et al., 2018; Zhang et al., 2019; Zywitza et al., 2018). Previously used 2D embeddings include those derived from principal components (PC), t-distributed Stochastic Neighbor Embeddings (t-SNE), Uniform Manifold Approximation and Projection (UMAP) and diffusion maps (Coifman et al., 2005; van der Maaten et al., 2008; McInnes et al., 2018) established using the observed single-cell transcriptional states. However, these approaches can yield different representations of the underlying cellular trajectory. Furthermore, in dynamic processes where intermediate cell states are not well represented (e.g. rare or missing) due to their transient nature or due to technical limitations in sample collection and processing, current 2D embeddings may be unable to capture global relationships between cell subpopulations thereby hindering downstream interpretation of cell state changes (Kester et al., 2018; Weinreb et al., 2018). Although alternative non-visual methods such as identifying dynamic driver-genes have been developed to help interpret information from RNA velocity analysis (Bergen et al., 2020), visual representation of cellular trajectories remains an important approach to understanding the overall relationships between cell states.
Here, we developed VeloViz to visualize cellular trajectories by incorporating information about each cell’s predicted future transcriptional state inferred from RNA velocity analysis. Using both real and simulated data representing cellular trajectories, we demonstrate that VeloViz embeddings are better able to consistently capture underlying cellular trajectories across diverse trajectory topologies compared with other evaluated methods. Likewise, given simulated and sparsely sampled cellular trajectories with missing intermediate cell states, we find that VeloViz embeddings are able to more robustly retain the overall cell state relationships in the underlying trajectories compared with other evaluated methods.
2 Materials and methods
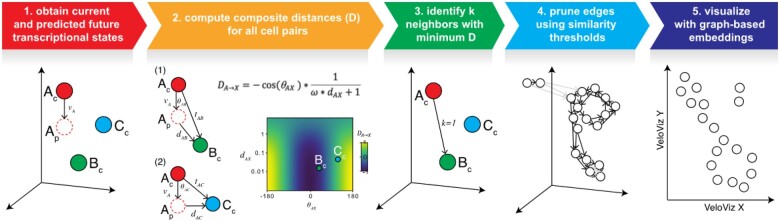
To create an RNA velocity-informed embedding, VeloViz uses each cell’s current observed and predicted future transcriptional states inferred from RNA velocity analysis to represent cells in the population as a graph (Fig. 1, Supplementary Information S1). Briefly, starting with spliced and unspliced RNA counts from single-cell RNA-sequencing (scRNA-seq) data or cytoplasmic and nuclear RNA counts from single-cell molecular imaging data, the predicted future transcriptional state of cells are inferred using RNA velocity analysis pipelines such as velocyto (La Manno et al., 2018) or scVelo (Bergen et al., 2020). We then optionally restrict to over-dispersed genes (Fan et al., 2016) and unit scale each gene’s variance, as well as mean center each gene’s expression for the observed current and predicted future transcriptional states, followed by dimensionality reduction by projecting these observed current and predicted future transcriptional states into a common PC space. Using this reduced dimensional representation of the observed current and predicted future transcriptional states, VeloViz then computes a composite distance between all cell pairs in the population. The composite distance between two cells, Cell A and Cell B, takes into account: (i) their transcriptional dissimilarity, defined as the Euclidean distance in the common PC space between Cell A’s predicted future state and Cell B’s observed current state () and (ii) their velocity similarity, defined as the cosine correlation between Cell A’s velocity vector and the change vector representing the transition from Cell A to Cell B (). An additional tuning parameter (ω) weighs the relative importance of the transcriptional similarity and the velocity similarity components.
| (1) |
Fig. 1.
Overview of VeloViz. RNA velocity-informed embeddings are created by VeloViz in five steps: (1) The observed current (Xc) and predicted future (Xp) transcriptional cell states are inferred from RNA velocity analysis and reduced into a common PC space; (2) composite distances (D) between all cell pairs are computed. The composite distance from Cell A to Cell X takes into account the similarity in transcriptional profiles (dAX) between Cell X’s observed current (Xc) and Cell A’s predicted future transcriptional state (Ap), and the cosine correlation between Cell A’s RNA-velocity (vA) and the change vector (tAX) representing a transition from Cell A’s current state (Ac) to Cell X’s current state (Xc). A distance weight (ω) is used to adjust the relative importance of transcriptional similarity and cosine correlation in the composite distance; (3) each cell is represented as a node in a graph, and for each cell, graph edges are assigned to the k cells with the minimum composite distances. Edge weights are computed based on composite distances as weightAB = max(D) – DAB; (4) edges assigned in (3) are pruned (in grey) using transcriptional and velocity similarity thresholds. Edge shade corresponds to edge weight computed based on composite distance, with darker arrows representing edges with larger weights; (5) the resulting graph can be visualized as a 2D or 3D embedding using graph-based embedding approaches
In this manner, the composite distance will be minimized when Cell A’s predicted future transcriptional state is similar to Cell B’s observed current transcriptional state and when the direction of Cell A’s RNA velocity is similar to the direction of the transition from Cell A to Cell B. Based on these composite distances, VeloViz creates a k-nearest neighbor graph by assigning k directed, weighted edges from each cell to the k neighboring cells with smallest composite distances. Edges are further pruned based on parameters that specify the minimum transcriptional and velocity similarity to remove spurious cell state relationships. Finally, the pruned graph can be visualized in 2D or 3D using graph layout or graph-embedding approaches such as force-directed layout algorithms (Fruchterman and Reingold, 1991) or UMAP (McInnes et al., 2018).
3 Results
3.1 Comparing VeloViz to other embeddings
To evaluate the performance of VeloViz, we first assessed VeloViz’s ability to capture cellular trajectories in simulated data representing cycling or branching trajectories (Supplementary Information S2). We compared the VeloViz embeddings to more conventional PC, t-SNE, UMAP and diffusion map embeddings. To evaluate how accurately each embedding captured the ground truth trajectory, we calculated a trajectory consistency (TC) score [Supplementary Information S3 (Boggust et al., 2019)] where high TC scores indicate more accurate representations of the ground truth trajectory. For the simulated cycling trajectory, all evaluated embeddings were able to capture the cycling structure of the trajectory except for the PC embedding (Supplementary Fig. S1A). The TC score for the VeloViz embedding was further higher than that of the PC, t-SNE and UMAP embeddings. For the simulated branching trajectory, the TC score for the VeloViz embedding was higher than TC scores for the t-SNE, UMAP and diffusion map embeddings (Supplementary Fig. S1B and C). Likewise, we evaluated VeloViz’s ability to capture simultaneously cellular trajectories in conjunction with terminally differentiated cell-types using simulated data representing both cycling or branching trajectories with a stable cell population. For the simulated cycling trajectory with a stable cell population, all evaluated embeddings were able to correctly distinguish the cycling and stable populations except for the PC embedding (Supplementary Fig. S1D). Likewise, the VeloViz, t-SNE, UMAP and diffusion map embeddings preserved the cycling trajectory, while the PC embedding did not. The TC score for the VeloViz embedding was higher than that of the other embeddings. For the simulated branching trajectories with a stable cell population, all embeddings were able to separate the dynamic and stable populations, but only the VeloViz and PC embeddings were able to capture the underlying branching trajectory of the dynamic population (Supplementary Fig. S1E and F). This is again reflected in the TC scores, which are consistently higher for the VeloViz and PC embeddings compared to the TC scores for the t-SNE, UMAP and diffusion map embeddings. These simulation results demonstrate that VeloViz is able to capture trajectories of various topologies compared to other embeddings, which may be better suited for specific topologies.
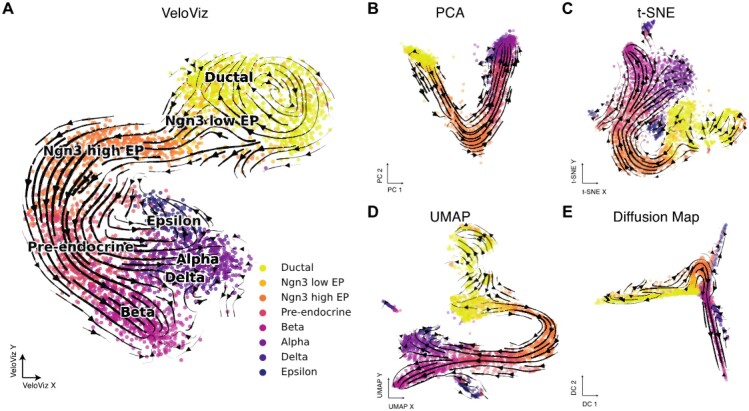
Next, we assessed VeloViz’s ability to capture cellular trajectories in scRNA-seq data. We applied VeloViz to scRNA-seq data of mouse spermatogenic cells (Supplementary Information S4), where we expect a developmental progression from spermatogonial stem cells to more differentiated spermatids (Hermann et al., 2018). For this simple, linear cellular trajectory, VeloViz was able to capture the overall expected trajectory from secondary spermatocytes to early, mid, then late round spermatids (Supplementary Fig. S2). Generally, PCA, t-SNE, UMAP and diffusion map were also able to capture this expected trajectory. To assess VeloViz’s ability to capture more complex trajectory structures, we applied VeloViz to scRNA-seq data of the developing mouse pancreas (Supplementary Information S5), where we expect to see both cycling and branching topologies at different stages of the trajectory. Briefly, we expect cycling ductal cells to give rise to endocrine progenitor-precursor (EP) cells, which become pre-endocrine cells that then differentiate into four hormone producing endocrine cell-types (Alpha, Beta, Delta and Epsilon cells) (Bastidas-Ponce et al., 2019). We observed that while all evaluated embeddings captured the progression of EP cells toward pre-endocrine cells, VeloViz, UMAP and t-SNE embeddings also captured the terminal branching differentiation into the different endocrine cell-types, which is not clear in the PC or diffusion map embeddings (Fig. 2). In addition, VeloViz was better able to capture the cycling structure of ductal cells. Overall, these results indicate that VeloViz embeddings are able to recapitulate expected trends from real scRNA-seq data.
Fig. 2.
VeloViz reconstructs trajectories from pancreatic endocrinogenesis scRNA-seq data. 2D embeddings visualizing pancreatic endocrinogenesis using VeloViz (A), PCA (B), t-SNE (C), UMAP (D) and diffusion mapping (E). Cells are colored by cell state annotations provided in Bergen et al. (2020). Arrows show the projection of velocities derived from dynamical velocity modeling (Bergen et al., 2020) onto the embeddings
To explore the potential of using VeloViz with velocity estimated from other data types, we further applied VeloViz to multiplexed error-robust fluorescent in situ hybridization (MERFISH) data (Xia et al., 2019) of cycling cultured U-2 OS cells (Supplementary Information S6). Again, we compared the VeloViz embedding to embeddings constructed using PCA, t-SNE and UMAP and found that all evaluated embeddings, including VeloViz, were able to capture the expected cycling trajectory (Supplementary Fig. S3). In this manner, we find that VeloViz is able to capture cellular trajectories of diverse topologies using both simulated and real data from multiple single-cell transcriptomics technologies.
3.2 Performance with missing intermediate cell states
To visualize how cell states may change over time, current visualization approaches rely on a reasonably uniform sampling of the continuum of transcriptional states for cells along dynamic processes in the collected data. However, in dynamic processes where certain intermediate cell states may be rare or short-lived or differentially impacted by cell isolation protocols, such intermediate cell states may be lost during data collection (Fan et al., 2020; Krishnaswami et al., 2016; MacLean et al., 2018; Moffitt et al., 2018; Slyper et al., 2020; Villani et al., 2017). Such a loss of intermediate cell states in the collected data could lead to differentiation gaps in the observed cellular trajectory. Because current visualization approaches such as t-SNE and UMAP preferentially preserve local cell-cell relationships, given such data with missing intermediate cell states, we hypothesized that the resulting embeddings could split the visualized trajectories into distinct components before and after the differentiation gap (Heiser and Lau, 2020; Kobak and Berens, 2019), thereby no longer accurately reflecting the underlying trajectories and potentially hindering downstream interpretation of cell state relationships. In contrast, we hypothesized that by incorporating information about each cell’s predicted future transcriptional state, VeloViz could more robustly construct representative cellular trajectories even when the collected data of sampled cell states contain missing intermediate cell states or differentiation gaps in the underlying trajectory.
To evaluate these hypotheses, we first used simulated and real single-cell transcriptomics data where some intermediate cells were removed, creating a trajectory gap. Again, because t-SNE and UMAP preferentially preserve local cell-cell relationships, we hypothesized that these embeddings would result in two distinct clusters of cells before and after the simulated gap (Heiser and Lau, 2020; Kobak and Berens, 2019). Therefore, in addition to TC scores, we calculated a gap distance (Supplementary Information S3), which measures the distance in the 2D embedding space between cells before and after the simulated gap in the trajectory. Embeddings that preserve the underlying trajectory despite this simulated gap will have a smaller gap distance. A small gap distance between cells that are part of the same trajectory will facilitate a clearer depiction of the underlying cell transitions compared to a large gap distance which may erroneously suggest that the cells are unrelated.
Indeed, for the simulated cycling trajectory where cells corresponding to a segment of the cycle were removed (Supplementary Information S2), VeloViz was the only evaluated embedding able to clearly represent the cycling structure of the trajectory (Supplementary Fig. S4A). The gap distance in the VeloViz embedding was also smaller than in t-SNE, UMAP and diffusion map embeddings. Likewise, for the simulated branching trajectories where cells corresponding to a segment of an intermediate branch were removed (Supplementary Information S2), only VeloViz and PCA were able to preserve the underlying topology (Supplementary Fig. S4B and C). The gap distance in the VeloViz embedding was smaller than that in the t-SNE, UMAP and diffusion map embeddings. In contrast, t-SNE and UMAP split cells before and after the simulated gap into distinct clusters as expected. TC scores were also consistently higher for VeloViz than with t-SNE, UMAP and diffusion map embeddings. Similar trends were observed with simulated data that included both dynamic cycling and branching populations with missing intermediate cell states along with a stable cell population (Supplementary Fig. S4D–F).
Likewise, for the U-2 OS MERFISH data, to simulate missing intermediate cell states, we removed cells in the G2/M cell cycle phase. Briefly, we identified cells in the G2/M cell cycle phase by computing for each cell a G2/M score based on the aggregated expression of canonical G2/M phase genes (Supplementary Information S6). As before, we compared the VeloViz embedding to those constructed with PCA, t-SNE and UMAP. We found that VeloViz was better able to retain the cycling trajectory despite the missing G2/M cells compared to the other evaluated embeddings (Supplementary Fig. S5).
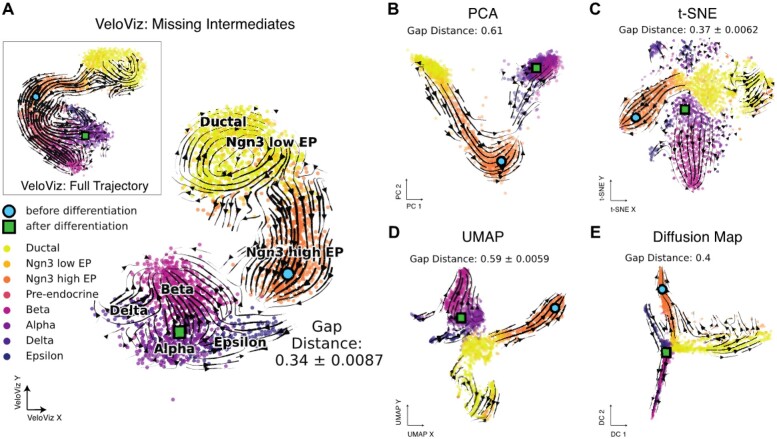
Similarly, for the developing mouse pancreas scRNA-seq data, to simulate missing intermediate cell states, we removed pre-endocrine cells and used cell latent time (Bergen et al., 2020) to identify cells before and after pre-endocrine cells in the developmental trajectory and to calculate differentiation gap distances in the recalculated embeddings (Supplementary Information S5). Notably, while all embeddings depicted the transition from ductal cells to endocrine progenitors, the subsequent transition from endocrine progenitors into terminal endocrine cell-types was best captured by VeloViz. As expected, t-SNE and UMAP split ductal and endocrine progenitor cells from terminal endocrine cell-types, which is reflected in the differentiation gap distances (Fig. 3). In particular, the position of endocrine progenitors and terminal endocrine cells and the resulting velocity streams may lead to the interpretation that these two cell populations are differentiating in two separate trajectories.
Fig. 3.
VeloViz reconstructs trajectories from pancreatic endocrinogenesis scRNA-seq data with missing intermediates. (A) VeloViz 2D embedding visualizing pancreatic endocrinogenesis with pre-endocrine intermediates removed creating a differentiation gap in the developmental trajectory. Inset shows the VeloViz embedding of the full dataset. Cells are colored by cell state annotations provided in Bergen et al. (2020). Arrows show the projection of velocities derived from dynamical velocity modeling onto the VeloViz embeddings. Differentiation gap distances measure the median distance in the 2D embedding between the 300 cells before and after pre-endocrine cells in the developmental trajectory (Supplementary Information S3iii). Blue circle and green square indicate the median coordinates of cells before and after pre-endocrine cells in the developmental trajectory, respectively. (B–E) 2D embeddings visualizing pancreatic endocrinogenesis with removed pre-endocrine intermediates using PCA, t-SNE, UMAP and diffusion mapping, respectively, with arrows showing the projection of velocities derived from dynamical RNA velocity modeling
Still, because low dimensional representations can vary depending on parameter choices, we explored the effect of changing these parameters on t-SNE and UMAP visualizations to see if certain parameter choices would yield visualizations more representative of the underlying cellular trajectory. For t-SNE embeddings, the perplexity parameter affects the extent to which the embedding reflects global versus local structure, with higher values resulting in embeddings that better preserve global structure (Kobak and Berens, 2019). However, with a differentiation gap in the trajectory, the t-SNE embeddings result in two distinct clusters of cells before and after the trajectory gap, even at large perplexity values (Supplementary Fig. S6). Likewise, for UMAP, we varied the values of two parameters: minimum distance, which controls how densely packed points are in the embedding with small values resulting in more dense clusters, and the number of neighbors, which functions similarly to perplexity in t-SNE (McInnes et al., 2018). As with t-SNE, when embedding data with a simulated gap, UMAP is unable to capture the expected trajectory even at large values of number of neighbors (Supplementary Fig. S7). This indicates that when intermediate cell states are missing, t-SNE and UMAP embeddings may be unable to recapitulate the expected underlying trajectory structure regardless of parameter choices.
While the simulation of missing intermediate states by explicitly removing cells represents an extreme case, we note that missing intermediate states may also occur with sparsely sampled datasets. To illustrate how this may occur, we subsampled the full pancreas dataset by factors of 4 and 10, resulting in datasets with 924 and 369 cells, respectively (Supplementary Information S5). We then used only the 924 or 369 cells to construct 2D embeddings using t-SNE, UMAP and VeloViz (Supplementary Fig. S8). Again, given such sparsely sampled datasets, both t-SNE and UMAP represented cells before the transition from ductal to endocrine progenitors as a distinct cluster from cells after this transition. In contrast, when constructing an embedding using VeloViz, the expected transitions from ductal cells to endocrine progenitors, pre-endocrine, then alpha, beta, delta and epsilon cells, was well represented. This suggests that the predicted future transcriptional states inferred from RNA velocity analysis can provide an additional source of information to help ensure that low dimensional embeddings accurately capture underlying cellular trajectories, even when sparse cell sampling limits the number of observed intermediate cell states. Overall, we find that VeloViz is able to visualize a more reliable representation of underlying trajectories even when intermediate cell states may be missing.
3.3 Scalability
Given the increasing availability of large single-cell transcriptomics datasets (Lähnemann et al., 2020), we sought to evaluate the scalability of VeloViz with increasing cell numbers. Briefly, we down-sampled a dataset of approximately 10,000 cells (10X Genomics, 2020) to create datasets ranging from 100 to 9,295 cells. For each dataset, we calculated RNA velocity using velocyto.R and constructed an embedding using VeloViz while evaluating runtime and memory usage (Supplementary Information S7). We find that both runtime and memory usage of VeloViz scales linearly with the number of cells and is comparable to that of RNA velocity calculations by velocyto.R (Supplementary Fig. S8).
4 Discussion
To facilitate better visual representation of relationships between cell states in single-cell transcriptomic data, we developed VeloViz to create low dimensional embeddings that incorporate dynamic information inferred from RNA velocity analysis. We find that VeloViz is able to visualize cellular trajectories of diverse topologies and capture global cell state relationships, even when intermediate cell states may be missing. Particularly when intermediate cell states are missing, we find that visualization with t-SNE and UMAP may result in distinct clusters containing cells before and after differentiation gaps, potentially leading to the erroneous interpretation that these cells are part of biologically distinct subpopulations rather than the same biological trajectory. In contrast, VeloViz is able to retain the underlying cellular trajectory and connect cells before and after differentiation gaps even when intermediate cell states are missing. As demonstrated, such missing intermediate cell states may result from insufficient sampling, though other biological and technical limitations of data collection may also lead to their preferential loss (Fan et al., 2020; Krishnaswami et al., 2016; MacLean et al., 2018; Moffitt et al., 2018; Slyper et al., 2020; Villani et al., 2017). Though it may be difficult to determine a priori whether a dynamic process or collected dataset contains such missing intermediate cell states, VeloViz is robust to such potential missing intermediate cell states and thus provides an additional approach for visualizing cellular trajectories.
However, several limitations of VeloViz should be considered when using VeloViz embeddings to interpret putative cellular trajectories. Embeddings constructed using VeloViz incorporate multiple user inputted hyperparameters (Supplementary Information S1). We explored the effects of changing these parameters on the visualization of simulated cycling trajectories with missing intermediates and the resulting TC scores (Supplementary Fig. S10). We found that the VeloViz embedding was most robust to changes in cosine similarity threshold (tt) and most sensitive to changes in k. However, without a priori knowledge of expected relationships between cell subpopulations, it may be challenging to find the optimal parameter set that yields the most representative embedding. Furthermore, different components of the trajectory being visualized, such as gaps versus branching structures, may have different optimal parameters. Thus, a range of hyperparameters may need to be explored to evaluate the stability of visualized cellular trajectories. Further limitations of VeloViz extend from the limitations of RNA velocity analysis in general. Notably, RNA velocity analysis can only infer cell state changes that are determined by changes in gene expression. Other molecular features such as alternative splicing, chromatin state, post-translational modifications, differential localization and cell microenvironment that contribute to cell state changes are not considered in RNA velocity analysis, and therefore these cell state changes will not be represented in the VeloViz embedding (Tritschler et al., 2019; Weinreb et al., 2018). In addition, it remains unknown the degree to which cell state changes are stochastic i.e. the probability that two cells with similar transcriptional states will develop differently. This stochasticity may limit the accuracy of predicting future cell state based on current gene expression dynamics. Ultimately, insights gained from RNA velocity analysis should be considered within the context of other available data, such as differential gene expression, mutational analysis and targeted experimental validation.
Overall, by taking into account the predicted future transcriptional states of cells from RNA velocity analysis, VeloViz provides an additional approach for visualizing putative cellular trajectories to aid in the interpretation of cellular dynamics from single-cell transcriptomics data.
Supplementary Material
Acknowledgements
The authors thank Brendan Miller, Feiyang Huang and Tobi Adelaja for their feedback in the preparation of this manuscript.
Funding
This work was supported by the National Science Foundation [2047611]. L.A. was also supported by the National Institutes of Health [T32-GM136577].
Conflict of Interest: none declared.
Contributor Information
Lyla Atta, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Center for Computational Biology, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD 21211, USA; Medical Scientist Training Program, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Arpan Sahoo, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Computer Science, Johns Hopkins University, Baltimore, MD 21218, USA.
Jean Fan, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21218, USA; Center for Computational Biology, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD 21211, USA; Department of Computer Science, Johns Hopkins University, Baltimore, MD 21218, USA.
References
- 10X Genomics. (2020) 10k mouse E18 combined cortex, hippocampus and subventricular zone cells, single indexed.
- Bastidas-Ponce A. et al. (2019) Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development, 146, dev.173849. [DOI] [PubMed] [Google Scholar]
- Bergen V. et al. (2020) Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol., 38, 1408–1414. [DOI] [PubMed] [Google Scholar]
- Boggust A. et al. (2019) Embedding comparator: visualizing differences in global structure and local neighborhoods via small multiples. ArXiv, 191204853 Cs.
- Coifman R.R. et al. (2005) Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl. Acad. Sci. USA, 102, 7426–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. et al. (2016) Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods, 13, 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. et al. (2020) Single-cell transcriptomics in cancer: computational challenges and opportunities. Exp. Mol. Med., 52, 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchterman T.M.J., Reingold E.M. (1991) Graph drawing by force-directed placement. Softw. Pract. Exp., 21, 1129–1164. [Google Scholar]
- Heiser C.N., Lau K.S. (2020) A quantitative framework for evaluating single-cell data structure preservation by dimensionality reduction techniques. Cell Rep., 31, 107576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P. et al. (2018) The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep., 25, 1650–1667.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester L. et al. (2018) Single-cell transcriptomics meets lineage tracing. Cell Stem Cell, 23, 166–179. [DOI] [PubMed] [Google Scholar]
- Kobak D., Berens P. (2019) The art of using t-SNE for single-cell transcriptomics. Nat. Commun., 10, 5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswami S.R. et al. (2016) Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc., 11, 499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G. et al. (2018) RNA velocity of single cells. Nature, 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lähnemann D. et al. (2020) Eleven grand challenges in single-cell data science. Genome Biol., 21, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L. et al. (2008) Visualizing data using t-SNE. J. Mach. Learn. Res., 9, 2579–2605. [Google Scholar]
- MacLean A.L. et al. (2018) Exploring intermediate cell states through the lens of single cells. Curr. Opin. Syst. Biol., 9, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L. et al. (2018) UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw., 3, 861. [Google Scholar]
- Moffitt J.R. et al. (2018) Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science, 362, 5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W. et al. (2019) A comparison of single-cell trajectory inference methods. Nat. Biotechnol., 37, 547–554. [DOI] [PubMed] [Google Scholar]
- Slyper M. et al. (2020) A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med., 26, 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler S. et al. (2019) Concepts and limitations for learning developmental trajectories from single cell genomics. Development, 146, dev170506. [DOI] [PubMed] [Google Scholar]
- Villani A.-C. et al. (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science, 356, eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C. et al. (2018) Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl. Acad. Sci. USA, 115, E2467–E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C. et al. (2019) Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA, 116, 19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. (2019) Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell, 179, 829–845.e20. [DOI] [PubMed] [Google Scholar]
- Zywitza V. et al. (2018) Single-cell transcriptomics characterizes cell-types in the subventricular zone and uncovers molecular defects impairing adult neurogenesis. Cell Rep., 25, 2457–2469.e8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.