Abstract
Summary
UCSC Xena platform provides huge amounts of processed cancer omics data from large cancer research projects (e.g. TCGA, CCLE and PCAWG) or individual research groups and enables unprecedented research opportunities. However, a graphical user interface-based tool for interactively analyzing UCSC Xena data and generating elegant plots is still lacking, especially for cancer researchers and clinicians with limited programming experience. Here, we present UCSCXenaShiny, an R Shiny package for quickly searching, downloading, exploring, analyzing and visualizing data from UCSC Xena data hubs. This tool could effectively promote the practical use of public data, and can serve as an important complement to the current Xena genomics explorer.
Availability and implementation
UCSCXenaShiny is an open source R package under GPLv3 license and it is freely available at https://github.com/openbiox/UCSCXenaShiny or https://cran.r-project.org/package=UCSCXenaShiny. The docker image is available at https://hub.docker.com/r/shixiangwang/ucscxenashiny.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Over the past decade, large research programs including TCGA (The Cancer Genome Atlas) (Weinstein et al., 2013), ICGC (International Cancer Genome Consortium) (Zhang et al., 2011), PCAWG (Pan-cancer analysis of whole genomes) (Campbell et al., 2020), GTEx (Genotype-Tissue Expression) (Ardlie et al., 2015), CCLE (Cancer Cell Line Encyclopedia) (Barretina et al., 2012) have generated large amounts of molecular data characterizing the different omics landscapes (including genomics, transcriptomics, proteomics and epigenomics) of thousands of tumors. The data have been uniformly preprocessed, curated and stored in data hubs of UCSC Xena (https://xenabrowser.net/datapages/) along with many public cancer datasets from individual research groups, providing unprecedented opportunities for either simple or systematic exploration of cancer behaviors and mechanisms at multiple molecular layers in individual cancer type or across cancer types (Goldman et al., 2020).
Despite the fact that UCSC Xena provides a functional genomics explorer (https://xenabrowser.net/) to allow users to explore and analyze its multi-omics and clinical/phenotype data, it is still difficult for cancer researchers to rapidly explore all available UCSC Xena datasets, find what they need in their research, and download or analyze the corresponding data. Besides, the analysis features and visualization quality provided by UCSC Xena platform have room for improvement. Advanced functionalities for analyzing different molecular profiles from specified data hubs including TCGA, CCLE and PCAWG, and generating publication-ready result plots are still lacking.
In 2019, we developed UCSCXenaTools, an open-source R package for retrieving metadata and data from more than one thousand public UCSC Xena datasets (Wang and Liu, 2019). However, this package lacks analysis and visualization capabilities, and only provides a low-level application program interface (API) for accessing data. Thus, it is not suitable for cancer researchers with limited programming experience. Here, we are motivated to present UCSCXenaShiny, an R/CRAN package containing a web application based on the R Shiny framework (https://shiny.rstudio.com/) for quickly searching, retrieving, exploring, analyzing and visualizing data from UCSC Xena data hubs. This tool could effectively promote the practical use of UCSC Xena public data, and serve as an important complement to the functionality of current Xena functional genome explorer.
2 Tool description
UCSCXenaShiny uses both the R package interface (i.e. R functions) and the Shiny application interface to allow the user to efficiently retrieve and analyze data from UCSC Xena data hubs. The architecture of UCSCXenaShiny can be classified into three layers (Fig. 1). The first layer retrieves data from UCSC Xena data hubs and is built on the top of UCSCXenaTools (Wang and Liu, 2019). The second layer is implemented as an R package, it provides almost all core data and analysis features as built-in datasets and public functions (i.e. API) of the R package (Supplementary Table S1). The third layer is implemented as an R Shiny application and provides a graphical user interface for interactive exploration and analysis of UCSC Xena data. A demo of this Shiny is deployed at https://shiny.hiplot.com.cn/ucsc-xena-shiny/ for public use. UCSCXenaShiny has more functionalities compared with other UCSC Xena related tools (including UCSC Xena browser, UCSCXenaTools and xenaPython) (Supplementary Table S2).
Fig. 1.
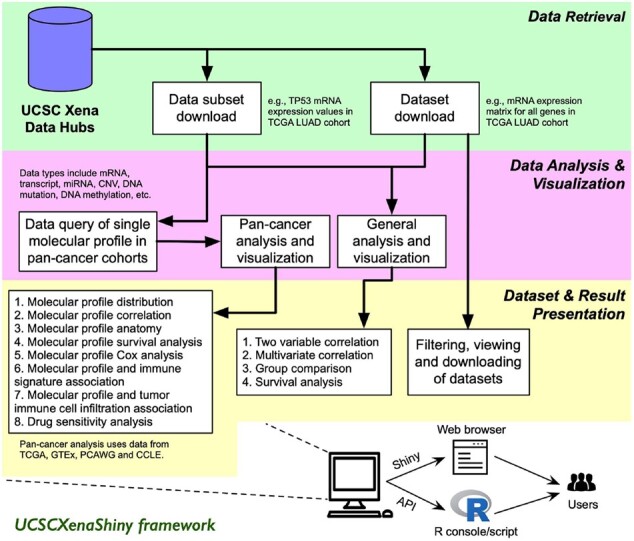
The architecture diagram of UCSCXenaShiny
2.1 R function interface
The public functions of UCSCXenaShiny can be divided into three categories based on their functionalities: (i) data retrieval; (ii) analysis and visualization of pan-cancer studies; (iii) advanced analysis and visualization (Supplementary Table S1).
2.2 Shiny interface
Its Shiny web application is the highlight of the UCSCXenaShiny software. It is a web-based software to provide interactive data retrieval, analysis and visualization for users. Similar to the R function interface, there are three core web pages: ‘Repository’, ‘General Analysis’ and ‘Quick PanCan Analysis’. The ‘Repository’ page contains a clickable table of UCSC Xena datasets, dataset filter widgets based on data hubs, cohorts, data types, keywords, etc., and corresponding action buttons (Supplementary Fig. S1). The current ‘General Analysis’ page contains four common analysis modules for exploring relationships between continuous variables, value difference between sample groups and survival curve difference between sample groups (Supplementary Fig. S2). The ‘Quick PanCan Analysis’ page contains several analysis modules for well-known pan-cancer data including TCGA, GTEx (Supplementary Fig. S3) and CCLE (Supplementary Fig. S4). For example: (i) comparisons of molecular profiles among samples, such as mRNA expression between TCGA tumor and normal control tissue (Supplementary Fig. S3A) or across different types of cancer (Maag, 2018) (Supplementary Fig. S3B) or CCLE cell lines (Supplementary Fig. S4A); (ii) association analysis between two molecular profiles with TCGA (Supplementary Fig. S3C) or CCLE (Supplementary Fig. S4B) data; (iii) association studies between a certain molecular profile and tumor/immune features, such as TMB (tumor mutational burden)/MSI (microsatellite instability)/stemness (Supplementary Fig. S3D) and immune gene signatures (Li et al., 2020; Thorsson et al., 2018) (Supplementary Fig. S3E); (iv) Kaplan–Meier survival analysis among samples with different levels of a molecular profile (Supplementary Fig. S3F); (v) association analysis between survival hazard ratio and a molecular profile with the Cox model across TCGA cancer types (Supplementary Fig. S3G); (vi) association analysis between mRNA expression of a gene (list) and cell line drug response (Supplementary Fig. S4C); (vii) exploring drug response differences between samples with different gene expression levels (Supplementary Fig. S4D).
3 Implementation
UCSCXenaShiny has been developed with R version ≥3.5 and Shiny following a modular and robust design of both R package and Shiny application. Continuous integration tests with CRAN R package is done automatically after each code commit to help test functionality and detect program bugs in a timely manner. Instructions on how to install, use UCSCXenaShiny and run the Shiny application are presented in the public GitHub repository (https://github.com/openbiox/UCSCXenaShiny). A detailed manual of built-in data and public R functions are organized and described in the package reference page (https://openbiox.github.io/UCSCXenaShiny/reference/index.html). Instructions including texts and videos on how to use functionalities of the Shiny application are documented in the help page of the Shiny application. Tooltips are adopted to help users to understand and customize the parameter setting. The Shiny application also shows data table behind each result plot and provides data download buttons to facilitate the archiving of data and result reproducibility.
4 Conclusion
In recent years, several bioinformatics platforms or tools, such as cBioPortal (Cerami et al., 2012), Genomic Data Commons (GDC) data portal (Grossman et al., 2016), ICGC Data Portal (Zhang et al., 2019), CVCDAP (Guan et al., 2020) and UCSC Xena (Goldman et al., 2020) have been constructed for the analysis and visualization of cancer genomics data (Supplementary Table S3). UCSCXenaShiny works as a UCSC Xena client, cBioportal, ICGC data portal, GDC data portal are independent data portals. Compared with these other data portals, UCSC Xena platform is featured with a comprehensive collection of public cancer genome datasets, and combined analysis between public and researchers’ own data (Goldman et al., 2020). However, UCSC Xena only provided limited number of analysis tools. For efficient cancer genome data download, integration, exploration and visualization, we built UCSCXenaShiny to allow a wide range of users to perform interactive analysis of UCSC Xena data by either programming or graphical interface operation. Since its release, UCSCXenaShiny has been downloaded for more than 10 000 times around the world (according to the API for CRAN package download counts, from the RStudio CRAN mirror, https://cranlogs.r-pkg.org/). We believe that UCSCXenaShiny could effectively promote the practical use of public cancer data and serve as an important complement to the functionality of current Xena functional genome explorer.
Supplementary Material
Acknowledgements
The authors thank UCSC Xena team for the helpful discussions about UCSC Xena related user questions. They thank Openbiox bioinformatics innovation collaboration group (https://github.com/openbiox/) for organizing the authors to launch and proceed this project. They thank Hiplot platform (https://hiplot.org/) to freely deploy the Shiny application of UCSCXenaShiny for common use. They thank FigureYa (Blogger, WeChat Official Account) for the technical support in figure drafting and revision. They thank ShanghaiTech University High Performance Computing Public Service Platform for computing services.
Funding
This work was supported in part by The National Natural Science Foundation of China [31771373, 81472594, 81770781], Shanghai Science and Technology Commission [21ZR1442400] and startup funding from ShanghaiTech University.
Conflict of Interest: none declared.
Contributor Information
Shixiang Wang, School of Life Science and Technology, ShanghaiTech University, 201203 Shanghai, China; Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 200031 Shanghai, China; University of Chinese Academy of Sciences, 100049 Beijing, China.
Yi Xiong, Department of Neurosurgery, Xiangya Hospital, Central South University, 410008 Changsha, China; Hunan International Scientific and Technological Cooperation Base of Brain Tumor Research, Xiangya Hospital, Central South University, 410008 Changsha, China; Xiangya School of Medicine, Central South University, 410013 Changsha, China.
Longfei Zhao, School of Pharmaceutical Sciences, Zhengzhou University, 450001 Zhengzhou, China.
Kai Gu, Roche Diagnostics (Shanghai) Limited, 201107 Shanghai, China.
Yin Li, Department of Thoracic Surgery, Zhongshan Hospital, Fudan University, 200032 Shanghai, China.
Fei Zhao, University of Chinese Academy of Sciences, 100049 Beijing, China; CAS Center for Excellence in Molecular Plant Sciences, 200032 Shanghai, China.
Jianfeng Li, State Key Laboratory of Medical Genomics, Shanghai Institute of Hematology, National Research Center for Translational Medicine, Rui-Jin Hospital, Shanghai Jiao Tong University, School of Medicine, 200025 Shanghai, China; School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, 200240 Shanghai, China.
Mingjie Wang, State Key Laboratory of Medical Genomics, Shanghai Institute of Hematology, National Research Center for Translational Medicine, Rui-Jin Hospital, Shanghai Jiao Tong University, School of Medicine, 200025 Shanghai, China.
Haitao Wang, Center for Precision Medicine Research and Training, Faculty of Health Sciences, University of Macau, 999087 Macau SAR, China.
Ziyu Tao, School of Life Science and Technology, ShanghaiTech University, 201203 Shanghai, China.
Tao Wu, School of Life Science and Technology, ShanghaiTech University, 201203 Shanghai, China.
Yichao Zheng, School of Pharmaceutical Sciences, Zhengzhou University, 450001 Zhengzhou, China.
Xuejun Li, Department of Neurosurgery, Xiangya Hospital, Central South University, 410008 Changsha, China; Hunan International Scientific and Technological Cooperation Base of Brain Tumor Research, Xiangya Hospital, Central South University, 410008 Changsha, China.
Xue-Song Liu, School of Life Science and Technology, ShanghaiTech University, 201203 Shanghai, China.
References
- Ardlie K.G. et al. ; GTEx Consortium. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P.J. et al. (2020) Pan-cancer analysis of whole genomes. Nature, 578, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.J. et al. (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol., 38, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman R.L. et al. (2016) Toward a shared vision for cancer genomic data. N. Engl. J. Med., 375, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.Q. et al. (2020) CVCDAP: an integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucleic Acids Res., 48, W463–W471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.W. et al. (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res., 48, W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag J.L.V. (2018) gganatogram: an R package for modular visualisation of anatograms and tissues based on ggplot2. F1000Res., 7, 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsson V. et al. ; Cancer Genome Atlas Research Network. (2018) The immune landscape of cancer. Immunity, 48, 812–830.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu X. (2019) The UCSCXenaTools R package: a toolkit for accessing genomics data from UCSC Xena platform, from cancer multi-omics to single-cell RNA-seq. J. Open Source Softw., 4, 1627. [Google Scholar]
- Weinstein J.N. et al. ; Cancer Genome Atlas Research Network. (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet., 45, 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J. et al. (2019) The International Cancer Genome Consortium Data Portal. Nat. Biotechnol., 37, 367–369. [DOI] [PubMed] [Google Scholar]
- Zhang J.J. et al. (2011) International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database Oxford, 2011, bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


