Abstract
Summary
Recent efforts to identify novel bacterial structured noncoding RNA (ncRNA) motifs through searching long, GC-rich intergenic regions (IGRs) have revealed several new classes, including the recently validated HMP-PP riboswitch. The DIMPL (Discovery of Intergenic Motifs PipeLine) discovery pipeline described herein enables rapid extraction and selection of bacterial IGRs that are enriched for structured ncRNAs. Moreover, DIMPL automates the subsequent computational steps necessary for their functional identification.
Availability and implementation
The DIMPL pipeline is freely available as a Docker image with an accompanying set of Jupyter notebooks. Full instructions for download and use are available at https://github.com/breakerlab/dimpl.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Discovery and validation of the over 45 known classes of metabolite- or elemental ion-binding riboswitches (McCown et al., 2017) have relied extensively on large-scale computational approaches based on comparative sequence analysis (Weinberg et al., 2007, 2010, 2017). However, these large-scale approaches may struggle to identify new classes of riboswitches, which are predicted to exist by the thousands but are likely much rarer than known classes (Breaker, 2011; McCown et al., 2017). Genome-level filtering of bacterial intergenic regions (IGRs) by nucleic acid composition and length (Brewer et al., 2021; Meyer et al., 2009; Stav et al., 2019) was developed to address the challenges of discovering these rarer riboswitch classes. This approach has already enabled the discovery and validation of the SAM-V (Meyer et al., 2009; Poiata et al., 2009), HMP-PP (Atilho et al., 2019) and NAD-II (Panchapakesan et al. 2021) riboswitch classes and the discovery of dozens of new intergenic motif candidates in the first genomes analyzed. However, until now this approach has required time-consuming manual analysis using several bioinformatic tools and lacked well-defined techniques to define genomic regions for further analysis that are enriched for noncoding RNAs (ncRNAs).
In this article, we introduce DIMPL (Discovery of Intergenic Motifs PipeLine), a bioinformatics pipeline which automates the process of total genome analysis by extracting IGRs, filtering them by length and nucleic acid composition, and collecting the data necessary to identify candidate motifs and assign their possible functions. DIMPL also provides reproducible techniques for identifying genomic regions enriched for ncRNA through support vector machine (SVM) classifiers. Although our primary objective in creating DIMPL was to accelerate the discovery of novel riboswitch classes, it can also be used to identify a wide-range of other intergenic nucleic acid and protein motifs such as upstream open reading frames, short open reading frames, ribosomal protein leader sequences, selfish genetic elements and other structured RNA motifs of unknown function.
2 Results
2.1 Pipeline overview
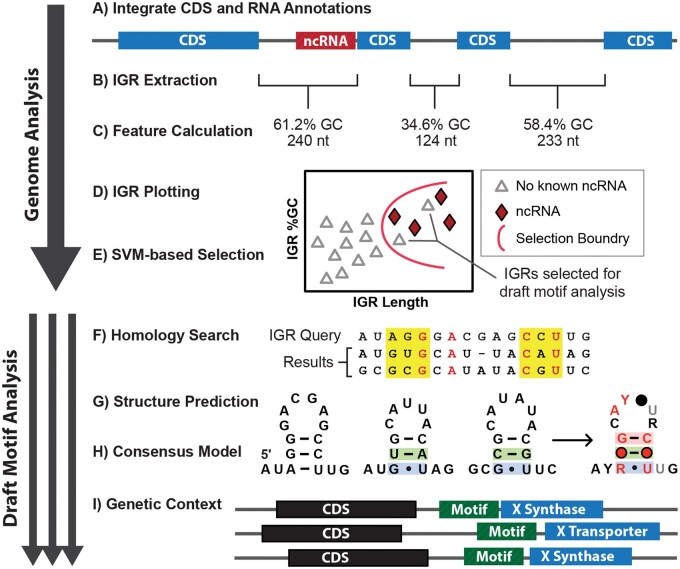
The DIMPL computational pipeline consists of two primary stages: (1) genome analysis and (2) draft motif analysis. For the genome analysis stage of DIMPL, the user begins by entering the Uniprot ID for a microbial genome for which there are Rfam annotations. DIMPL proceeds to automatically request the latest genomic sequence and protein annotations (Fig. 1A) accessible via NCBI Entrez (Agarwala et al., 2016) and the corresponding RNA family annotations provided by the Rfam MySQL Database (Kalvari et al., 2018). All IGRs located between protein-coding open reading frames are then extracted and labeled (Fig. 1B and C) with their percentage of G and C nucleotides relative to the total nucleotides in the IGR (%GC content), length and the presence of any known ncRNA motifs. DIMPL then generates an interactive graph (Fig. 1D) showing the IGRs plotted by their %GC content and length with labels for IGRs with known RNA families. This genome plot can help evaluate the suitability of the selected genome for analysis using the GC-IGR search approach. Ideal genomes will have strong separation between the cluster of IGRs containing known ncRNAs and those the bulk of IGRs with no known annotation.
Fig. 1.
Overview of DIMPL. Process are divided into the two stages: genome analysis (A–E) and draft motif analysis (F–I). Annotations for sections D (Stav et al., 2019) and H (Rivas et al., 2017; Weinberg et al., 2011) have been reported previously
In the next step, the tool uses a SVM classifier (Fig. 1E) to identify IGRs with no annotated ncRNAs that have similar GC content and length to other IGRs with known structured ncRNAs. DIMPL then performs a BLASTX search (Camacho et al., 2009) on the selected IGRs to ensure they do not contain unannotated protein coding regions. Any unannotated protein coding regions discovered in the search are removed from the selected IGRs, which are discarded in their entirety if the remaining IGR no longer meets the length and %GC content requirements for the selection.
The draft motif analysis portion of DIMPL is performed in parallel on all IGRs that have met the selection criteria. The process begins by using Infernal 1.1.3 (Nawrocki and Eddy, 2013) to search each selected IGR’s sequence (Fig. 1F) against a database of all microbial IGRs derived from NCBI’s RefSeq (O’Leary et al., 2016). The collection of homologous sequences from a single IGR search forms the ‘draft motif’ that is further analyzed in several steps. First, representatives with identical nucleotide sequences are removed. Next, the draft motifs are analyzed via CMfinder 0.4.18 (Yao et al., 2006) to look for possible RNA secondary structure features (Fig. 1G). All realigned motifs generated by CMfinder are evaluated for evidence of statistical significance for predicted nucleotide covariations. Subsequently, the consensus sequence and structural model for each motif is generated (Fig. 1H) using R-scape 1.4.0 (Rivas et al., 2017), which integrates the RNA drawing algorithm R2R (Weinberg et al., 2011). Draft motifs are also checked for the presence of coding regions using RNAcode (Washietl et al., 2011). Finally, for each draft motif, DIMPL uses GenomeView (Spies et al., 2018) to visualize the genetic contexts (Fig. 1I) of the motif’s representatives to aid in determining a possible function for the candidate RNA motif. A draft motif’s most strongly supported alignment can then be analyzed by one or more additional cycles of Infernal homology searches, which take advantage of the proposed secondary structure to expand the number of representatives found.
2.2 Details on SVM enrichment
The SVM enrichment of IGRs in DIMPL uses a radial basis-function (RBF) kernel and is implemented with scikit-learn (Pedregosa et al., 2011). The SVM classifier is trained de novo for each genome analyzed using the IGR %GC content and nucleotide length as the features, the presence/absence of a structured RNA as the class labels and a set of hyperparameters that have been weighted to select a contiguous region of a genome’s %GC versus length plot. The primary purpose of the SVM classifier is to perform an enrichment of IGRs that reduces the number subjected to the more computationally intensive steps in the pipeline. Applying the SVM-RBF algorithm allows DIMPL to accomplish this goal in a systematic and reproducible manner.
2.3 Usage
The DIMPL pipeline is built primarily in Python and is distributed as a Docker image (Merkel, 2014) with all the necessary tools already installed. Along with the Docker image, DIMPL includes a set of detailed Jupyter notebooks that walk users through the steps of the pipeline, display interactive graphs and assemble results from analysis tools. For computationally intensive steps such as BLAST, Infernal and CMfinder that are typically performed on a high-performance computing cluster, DIMPL exports compressed tar files containing the necessary bash scripts and data files that can be configured for a custom compute environment. Detailed instructions are included in the Supplementary Information of this article. Sample datasets, preprocessed search database files, and the source code are available at www.github.com/breakerlab/dimpl.
3 Conclusion
DIMPL provides an integrated collection of tools to streamline the process of identifying novel structured ncRNA motifs, including new riboswitch candidates, on a genome-wide scale. It relies on established methods of enriching bacterial IGRs for ncRNA motif discovery (Stav et al., 2019) and quickly assembles the combination of structural and genetic context information that are key to identifying the function of the newly discovered motifs. This pipeline should permit the rapid analysis of each new bacterial genome for novel and rare ncRNA classes, which will aid in the discovery of novel classes of riboswitches and ribozymes.
Supplementary Material
Acknowledgements
We thank Aya Narunsky, Gadareth Higgs, Diane Yu and other members of the Breaker laboratory for helpful discussions.
Funding
This work was supported by grants from the National Institutes of Health [GM022778, AI136794] and by the Howard Hughes Medical Institute [Investigator grant to R.R.B.]. Some computational efforts were assisted by infrastructure and support staff from the High Performance Computing facilities at the Yale Center for Research Computing, which is funded by Yale University and by the National Institutes of Health [S10RR029676].
Conflict of Interest: none declared.
Contributor Information
Kenneth I Brewer, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8103, USA.
Glenn J Gaffield, Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8103, USA.
Malavika Puri, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520-8103, USA.
Ronald R Breaker, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8103, USA; Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8103, USA; Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520-8103, USA.
References
- Agarwala R. et al. (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 44, D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilho R.M. et al. (2019) A bacterial riboswitch class for the thiamin precursor HMP-PP employs a terminator-embedded aptamer. eLife, 8, e45210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R.R. (2011) Prospects for riboswitch discovery and analysis. Mol. Cell, 43, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer K.I. et al. (2021) Comprehensive discovery of novel structured noncoding RNAs in 26 bacterial genomes. RNA Biol. Doi:10.1080/15476286.2021.1917891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I. et al. (2018) Non-coding RNA analysis using the Rfam database. Curr. Protoc. Bioinformatics, 62, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown P.J. et al. (2017) Riboswitch diversity and distribution. RNA, 23, 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M.M. et al. (2009) Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique’. BMC Genomics, 10, 268–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel D. (2014) Docker: lightweight Linux containers for consistent development and deployment. Linux J., 2. [Google Scholar]
- Nawrocki E.P., Eddy S.R. (2013) Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics, 29, 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary N.A. et al. (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res., 44, D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchapakesan S.S.S. et al. (2021) A second riboswitch class for the enzyme cofactor NAD+. RNA, 27, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Poiata E. et al. (2009) A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA, 15, 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E. et al. (2017) A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods, 14, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N. et al. (2018) GenomeView—an extensible python-based genomics visualization engine. BioRxiv, 355636. [Google Scholar]
- Stav S. et al. (2019) Genome-wide discovery of structured noncoding RNAs in bacteria. BMC Microbiol., 19, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S. et al. (2011) RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data. RNA, 17, 578–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. et al. (2007) Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res., 35, 4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. et al. (2010) Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol., 11, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. et al. (2011) R2R-software to speed the depiction of asthentic consensus RNA secondary structures. BMC Bioinformatics, 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. et al. (2017) Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res., 45, 10811–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z. et al. (2006) CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics, 22, 445–452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.