Abstract
Periodic accumulation and destruction of mitotic cyclins are important for the initiation and termination of M phase. It is known that both APCCdc20 and APCHct1 collaborate to destroy mitotic cyclins during M phase. Here we show that this relationship between anaphase-promoting complex (APC) and Clb proteins is reversed in S phase such that the early Clb kinases (Clb3, Clb4, and Clb5 kinases) inactivate APCHct1 to allow Clb2 accumulation. This alternating antagonism between APC and Clb proteins during S and M phases constitutes an oscillatory system that generates undulations in the levels of mitotic cyclins.
Oscillation in the abundance of mitotic cyclins is a major driving force for the progression through mitosis. While mitotic entry requires the accumulation of these cyclins, the exit is dependent on their destruction.
Generally, a combination of transcriptional and posttranslational controls determines the appropriate levels of cyclins at particular stages of the cell cycle. In the budding yeast Saccharomyces cerevisiae, the transcription of mitotic cyclins Clb1 and Clb2 is cell cycle regulated such that their mRNA levels rise in late S phase, peak in G2/M, and finally decline at the end of M phase (4, 5, 16). Crucial to the posttranslational regulation of cyclin abundance is the ubiquitin-dependent proteolytic machinery. A ubiquitin ligase called the anaphase-promoting complex (APC) is essential for the mitotic cyclin degradation necessary for the exit from mitosis (26). In yeast, the APC is a multimeric complex consisting of at least 12 subunits (24) which requires the activator protein Cdc20 or its homolog Hct1 (also known as Cdh1) (9, 12, 18). While the APC activated by Cdc20 (APCCdc20) promotes chromosome segregation by causing destruction of the anaphase inhibitor Pds1, APCHct1 plays a role in the final exit from mitosis by mediating proteolytic degradation of the mitotic cyclin Clb2 (12, 18). Hct1 phosphorylation by mitotic kinases prevents it from activating APC (6, 23). The removal of this inhibitory phosphorylation by Cdc14 phosphatase (19) is a critical step which allows Hct1 to bind to and activate APC (7, 23). The activity of Cdc14 itself is under the control of the mitotic exit network pathway that includes regulatory proteins such as Tem1, Cdc15, and Net1 (25). Interestingly, Cdc20 function is also essential for the exit from mitosis (9, 21). It has been suggested elsewhere that Cdc20 promotes exit from mitosis solely by catalyzing the destruction of S-phase cyclin Clb5 (14). However, according to a less deterministic scheme, APCCdc20 facilitates activation of APCHct1 by causing progressive lowering of the inhibitory Clb-Cdc28 kinase activities by partial destruction of Clb proteins (3, 22). A reduction in the collective Clb kinase activities would allow a net increase in the dephosphorylation of Hct1 by Cdc14 phosphatase. A fully active APCHct1 can then target the remaining Clb2 for degradation during telophase, thus easing the cells out of mitosis. It is widely believed that, once activated in mitosis, this proteolytic machinery is turned off by the Cln kinases at the onset of the next cycle (2).
Although Clb5 along with other Clbs may prevent untimely exit from mitosis, we show here that the early Clbs collectively also have an essential role during S phase. We demonstrate that it is the early Clb kinase complexes (Clb3, Clb4, and Clb5) that inactivate APCHct1 in S phase. Their ability to suppress the APCHct1 activity early in the cycle is necessary for the effective expression and accumulation of late mitotic cyclins (such as Clb2) critical for the onset of mitosis. This regulation is a reversal of the scenario in M phase, where APC ensures the inactivation of Clb proteins. We suggest that such a periodic reversal of antagonism between APC and Clb proteins during S and M phases is critical in generating basic oscillations in the levels of major mitotic cyclins.
MATERIALS AND METHODS
Yeast media and reagents.
All strains used in this study are congenic to the wild-type strain W303. Cells were grown in yeast extract-peptone (YEP) or selective medium supplemented with 2% glucose (+Glu) or 2% raffinose plus galactose (+Raff +Gal). Kanamycin-resistant colonies were selected on plates containing G418 (200 mg/liter).
Strains and plasmids.
The strains (Table 1) were constructed by a combination of standard molecular genetic techniques such as gene transplacement, gene disruption, and tetrad dissection. Southern blot analysis was performed to confirm gene disruptions and transplacements.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| US164 | MATaclb2Δ:LEU2 bar1Δ:URA3 pADH-CLB2/TRP1/CEN his3 | U. Surana lab |
| US356 | MATabar1Δ his3 ura3 trp1 leu2 | U. Surana lab |
| US2337 | MATaclb5Δ:URA3 bar1Δ:URA3 CDC14-GFP-LEU2 HA3-HCT1-TRP1 his3 | This study |
| US2340 | MATabar1Δ:URA3 CDC14-GFP-LEU2 HA3-HCT1-TRP1 his3 | This study |
| US2352 | MATabar1Δ his3 ura3 leu2 pGAL-HA3-HCT1/TRP1/CEN | This study |
| US2354 | MATabar1Δ clb5Δ:URA3 his3 leu2 pGAL-HA3-HCT1/TRP1/CEN | This study |
| US2404 | MATabar1Δ his3 trp1 pMET3-CLB2-HA3/LEU2/CEN | This study |
| US2417 | MATaclb3Δ:TRP1 clb4Δ:HIS3 clb5Δ:URA3 GAL-CLB5::TRP1 | This study |
| US2435 | MATaclb3Δ:TRP1 clb4Δ:HIS3 clb5Δ:URA3 GAL-CLB5::TRP1 pMET3-CLB2-HA3/LEU2/CEN | This study |
| US2438 | MATaclb3Δ:TRP1 clb4Δ:HIS3 clb5Δ:URA3 GAL-CLB5::TRP1 pMET3-HA3-HCT1/LEU2/2μm | This study |
| US2439 | MATaclb3Δ:TRP1 clb4Δ:HIS3 clb5Δ:URA3 GAL-CLB5::TRP1 pMET3-CLN2-HA3/Kan/2μm | This study |
| US2440 | MATaclb3Δ:TRP1 clb4Δ:HIS3 clb5Δ:URA3 GAL-CLB5::TRP1 hct1Δ:KAN leu2 | This study |
CDC14-GFP was constructed by subcloning a 0.9-kb XhoI-KpnI green fluorescent protein (GFP) fragment (from Pam Silver's lab) into XhoI-KpnI sites introduced at the 3′ end of a 3.1-kb CDC14 fragment. GAL-HA3-HCT1 was made by ligating a SpeI-SalI (1.8-kb) fragment containing HA3-HCT1 from pWS 216 (W. Seufert) to XbaI-SalI-digested Ycplac22 carrying a GAL1-10 promoter. To construct MET3-CLB2-HA3, a MET3 promoter (0.6 kb, EcoRI-KpnI) was subcloned into EcoRI-KpnI-digested Ycplac111. This was then digested with EcoRI and ligated to a 2.1-kb EcoRI CLB2-HA3 fragment.
For MET3 HA3-HCT1, Ycplac111 carrying the MET3 promoter was cut with EcoRI, filled in, and ligated to a blunted HA3-HCT1 (1.8-kb SpeI-SalI) fragment from pWS 216 (W. Seufert). The construct was then transferred to YEplac181 using SalI-XbaI sites. An HCT1::KAN disruption cassette was made by replacing the NheI-BlpI fragment of HCT1 with the KAN MX2 resistance marker. The 3.3-kb EcoRI-XbaI fragment was used for gene disruption.
An MET3-CLN2-HA3 cassette was made by ligating a SalI-SpeI fragment containing the MET3 promoter and the 5′ end of CLN2 to a SpeI-EcoRI fragment containing the 3′ end of CLN2-HA3 with the 3′ untranslated region and part of a LEU2 marker. This cassette was subcloned onto a 2μm plasmid carrying a KAN MX2 marker. The plasmid backbone was constructed from pFL38 where the BglII-BglII fragment (URA3 sequences) had been replaced by a SalI-SpeI fragment (KAN MX2 marker). A ClaI fragment containing 2μm sequences was inserted at the NarI site of the multiple cloning site.
Cell synchronization, cell extracts, kinase assays, and Western blot analysis.
To obtain synchronous cultures, exponential-phase cells were grown in medium at 24°C containing either α factor (5 μg/ml for BAR1 cells and 0.8 μg/ml for bar1 cells) or 30 mg of hydroxyurea (HU) or nocodazole (15 μg/ml) per ml for 3 to 4 h. Lysates for whole-cell extracts were prepared, and kinase assays were performed according to the method of Surana et al. (17). Western blot analyses were performed as described in the work of Yeong et al. (22).
Northern blot hybridization, flow cytometry, and immunofluorescence staining.
The RNA extraction procedure, Northern blot analysis, flow cytometry analysis, and immunofluorescence staining were carried out as described by Lim et al. (8). Immunofluorescence signals were detected using the Leica DM microscope, and images were acquired using an attached Hamamatsu charge-coupled device camera driven by the MetaMorph (Universal Imaging Corporation) software.
RESULTS
Absence of Clb5 limits Clb2 accumulation in late S phase.
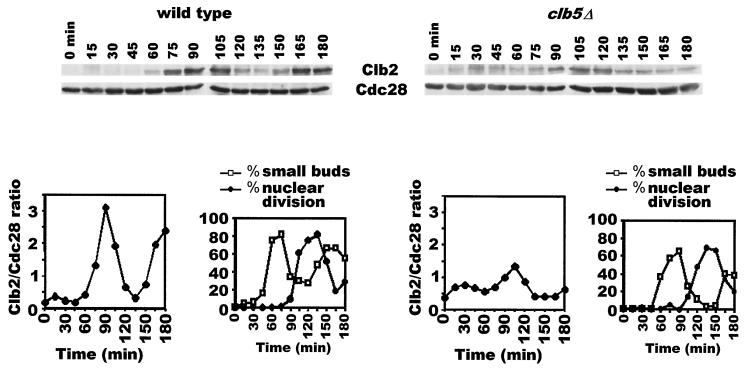
We have previously reported that Cdc20 function is essential for the final exit from mitosis (9). We had also shown previously that Cdc20's role in mitotic exit is to catalyze Clb protein destruction to varying degrees during the cell's approach to telophase, thus lowering the collective Clb kinase activities to facilitate Hct1 activation by Cdc14 phosphatase (22). Interestingly, a mutation in the S-phase cyclin CLB5 gene alone was found to be sufficient to allow cells to exit mitosis in the absence of the CDC20 gene (14), leading to the proposal that Cdc20 allows mitotic exit solely by causing destruction of Clb5 at metaphase. However, it seems puzzling that an S-phase cyclin, which is normally destroyed by the time that cells are in metaphase (11), could be responsible for inhibiting Hct1 just prior to exit from mitosis. To address this, we asked whether the absence of the CLB5 gene causes an overall change in the levels of mitotic cyclin. The wild-type and clb5Δ cells carrying CDC14-GFP (at its native locus) were synchronized in G1 by α-factor treatment and then released into α-factor-free medium at 24°C. In the wild-type strain, Clb2 protein rose approximately 15-fold relative to its level in α-factor arrest before beginning its decline at 120 min (Fig. 1, left panels). In clb5Δ cells, on the other hand, Clb2 levels increased only three- to fourfold compared to the level in G1 (Fig. 1, right panels). This is clearly not due to a difference in the extent of synchrony, because the two cultures showed comparable degrees of synchrony (see graphs). These results suggest that a lack of Clb5 function compromises significantly the cells' ability to build up Clb2 protein (and most likely Clb1), though the reduced amount is still sufficient to allow onset of mitosis. They also imply that the reason why deletion of the CLB5 gene allows exit from mitosis in the Cdc20-deficient cells is that, in the absence of Clb5, the collective Clb kinase activities do not build up to the wild-type level; consequently, Cdc20 function is no longer required to reduce the overall Clb kinase activities any further for the effective activation of Hct1 by Cdc14 phosphatase.
FIG. 1.
Reduced levels of Clb2 protein in the clb5Δ mutant. Wild-type cells (US2340) and clb5Δ cells carrying CDC14-GFP integrated at the CDC14 locus (US2337) were arrested in G1 with α factor and then released into YEP+Glu medium at a permissive temperature. Samples were analyzed for the budding index, nuclear division, and Clb2 and Cdc28 protein levels.
Early transcribed CLB3, CLB4, and CLB5 genes are collectively essential for accumulation of late mitotic cyclin Clb2.
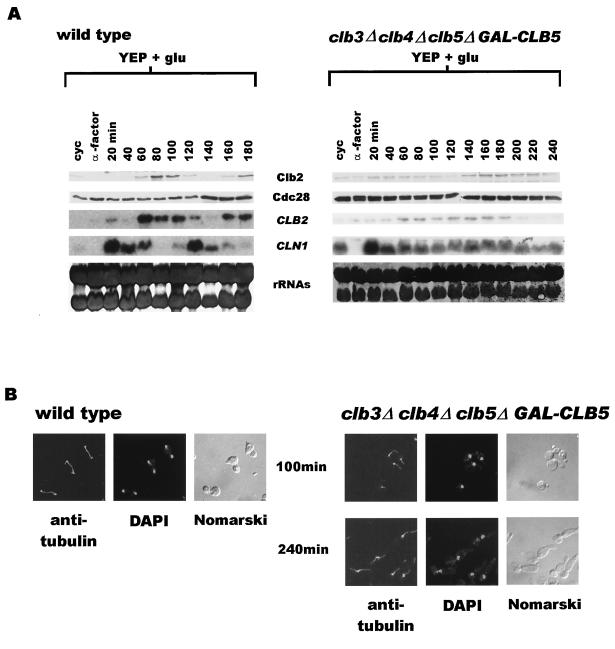
It has been reported previously that, while both clb3Δ clb4Δ and clb5Δ strains are viable (5, 13), clb3Δ clb4Δ clb5Δ triple mutant cells fail to survive (13); they arrest prior to mitosis with 2N DNA content and a single nucleus but completely lack a mitotic spindle. The explanation put forward for the inviability is that, in the absence of Clb3 and Clb4, Clb5 becomes essential for the biogenesis of mitotic spindle (13); thus, the clb3Δ clb4Δ clb5Δ mutant fails to form a spindle and eventually loses viability. Prompted by our finding that clb5Δ cells are compromised in their ability to accumulate Clb2, we determined the extent to which the collective deficiency in CLB3, CLB4, and CLB5 might affect the level of Clb2 protein. Wild-type and clb3Δ clb4Δ clb5Δ strains kept alive by GAL-CLB5 integrated at the TRP1 locus were synchronized in G1 by α-factor treatment and then allowed to resume cell cycle progression in glucose medium at 24°C. As expected, the abundance of both CLB2 transcript and the protein showed oscillatory behavior in the wild-type strain (Fig. 2A, left panel). CLN1 transcript appears slightly earlier and also shows characteristic fluctuation as cells continue to course through the cycle. The clb3Δ clb4Δ clb5Δ cells were released normally from their G1 arrest but soon accumulated at G2/M with an elongated bud, 2N DNA content (data not shown), and an undivided nucleus lacking a mitotic spindle (Fig. 2B, right panel). Surprisingly, the levels of both CLB2 transcript and the Clb2 protein remained low in these cells throughout the experiment (Fig. 2A). The CLN1 transcript appeared within 20 min of release from G1 arrest but, unlike that in wild-type cells, persisted throughout the time course. This is consistent with an earlier report that, in the absence of Clb kinases, cells are unable to turn off CLN1 transcription effectively (1). These results suggest that cells lacking CLB3, CLB4, and CLB5 can neither effectively transcribe the CLB2 gene nor accumulate Clb2 protein. Most likely, this is also true of the CLB1 gene since CLB1 and CLB2 are similarly regulated (4, 5).
FIG. 2.
(A) clb3Δ clb4Δ clb5Δ cells arrest in G2/M with low levels of CLB2 transcript and Clb2 protein. Wild-type cells (US356) and clb3Δ clb4Δ clb5Δ GAL-CLB5::TRP1 cells (US2417) were synchronized in G1 by α-factor treatment and then released in YEP+Glu medium at 24°C. Samples were collected at 20-min intervals and analyzed for the state of nuclear division, CLB2 and CLN1 RNA transcripts, and the levels of Clb2 and Cdc28 proteins. (B) clb3Δ clb4Δ clb5Δ cells arrest in G2/M without a mitotic spindle. Left panels, wild-type cells with long mitotic spindles in the 100-min sample from the experiment described for panel A; right panels, clb3Δ clb4Δ clb5Δ cells in 100- and 240-min samples, clearly lacking mitotic spindles. DAPI, 4′,6′-diamidino-2-phenylindole. (C) clb3Δ clb4Δ clb5Δ cells survive in the presence of MET3-CLB2-HA3. The clb3Δ clb4Δ clb5Δ GAL-CLB5::TRP1 control cells (US2417) were plated on either YEP+Gal (top left sector) or YEP+Glu (top right sector) plates. clb3Δ clb4Δ clb5Δ GAL-CLB5::TRP1 cells carrying MET3-CLB2-HA3 on a CEN plasmid (US2435) were plated on a −Met+Glu plate (bottom right sector). The plates were incubated at 24°C and photographed after 48 h. (D) Clb2 is unstable in S phase in clb3Δ clb4Δ clb5Δ cells. Wild-type cells (US2404) and clb3Δ clb4Δ clb5Δ cells carrying MET3-CLB2-HA3 on a CEN plasmid (US2435) were arrested in early S phase by growth in YEP+Glu+Met medium containing HU (30 mg/ml). After 3 h, the cultures were filtered and cells were resuspended in −Met+Glu+HU for 40 min. Methionine was added to the cultures to repress transcription, and samples, collected for a further 90 min, were analyzed for Clb2-HA3 and Cdc28 protein levels.
Taken together, these observations imply that the inviability of clb3Δ clb4Δ clb5Δ cells may be due to a severe deficiency of Clb1 and Clb2 proteins. Although the native promoter-driven CLB2 gene on a 2μm plasmid cannot rescue clb3Δ clb4Δ clb5Δ cells (13), strikingly, CLB2 expressed from the MET3 promoter on a CEN vector allows them to form healthy colonies (Fig. 2C). This observation is consistent with the notion that the clb3Δ clb4Δ clb5Δ cells die due to a lack of Clb2 protein (and perhaps Clb1) and suggests that, in the absence of Clb3, Clb4, and Clb5, the native promoter of CLB2 is not fully active.
Clb2 is unstable in clb3Δ clb4Δ clb5Δ cells.
It is known that the native CLB2 promoter is under positive feedback control; i.e., unlike the MET3 promoter, efficient transcription from the native promoter requires Clb2-Cdc28 activity (1). One reason why the clb3Δ clb4Δ clb5Δ mutant is unable to effectively transcribe CLB2 and accumulate Clb2 may be because Clb2 is unstable in these cells. To test this notion, wild-type and clb3Δ clb4Δ clb5Δ cells carrying MET3-CLB2-HA3 on a CEN vector were synchronized in early S phase by HU treatment in the presence of methionine (+Met) and then transferred to −Met medium to induce Clb2-HA3 protein. The induction was terminated after 40 min by the addition of methionine, and the stability of the protein was monitored by Western blot analysis. The cell cycle arrest was maintained throughout the experiment by the presence of HU in the medium at all times. The Clb2-HA3 protein, ectopically expressed during the short pulse period, remained stable in wild-type cells (Fig. 2D, left panel), indicated by its unchanged abundance at various time points. In clb3Δ clb4Δ clb5Δ cells, however, Clb2-HA3 begins to dissipate at 40 min and is barely detectable by 60 min, suggesting that the absence of Clb3, Clb4, and Clb5 makes Clb2 highly unstable (Fig. 2D, right panel).
clb3Δ clb4Δ clb5Δ cells are unable to phosphorylate Hct1 efficiently.
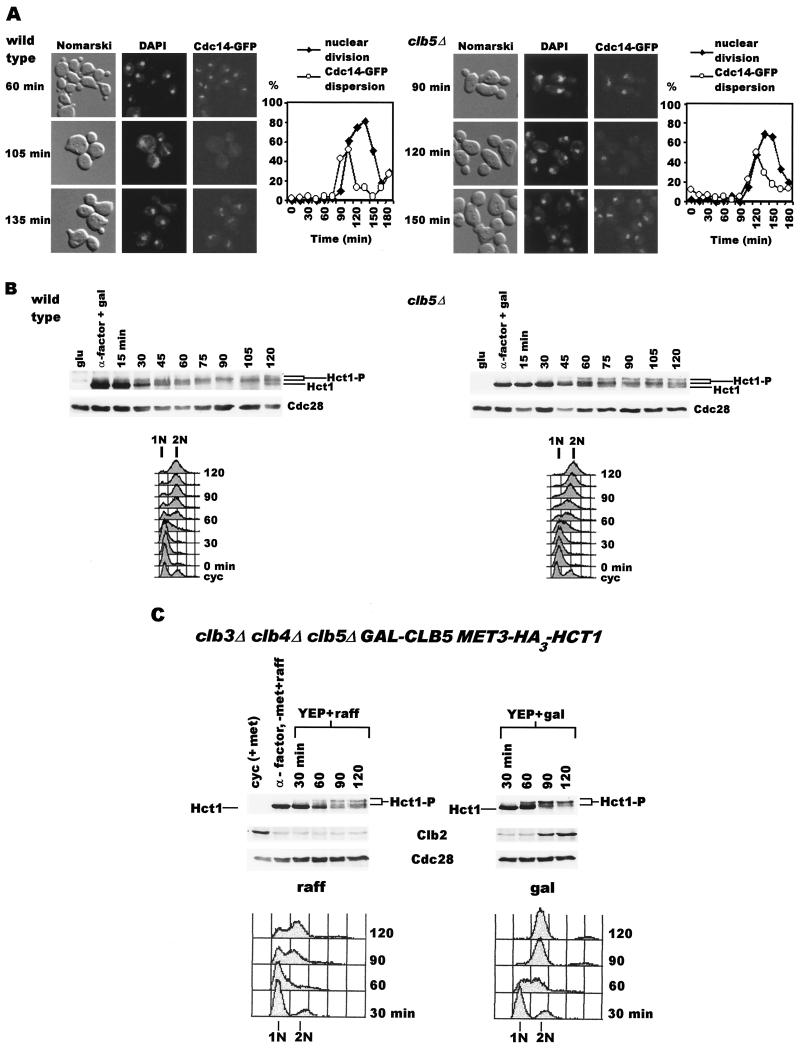
Since APCHct1 targets Clb2 for proteolytic degradation in telophase and in G1 of the subsequent division cycle, we suspected that the instability of Clb2 in clb3Δ clb4Δ clb5Δ cells might be due to hyperactive Hct1 during S and G2 phase. This may result either from premature release of Cdc14 from the nucleolus or from incomplete inactivation of APCHct1 (due to underphosphorylation of Hct1) during S phase. Therefore, we first examined the localization of Cdc14 in clb5Δ cells described for the experiment whose results are shown in Fig. 1. In both wild-type and clb5Δ cells, Cdc14-GFP remained sequestered in the nucleolus until the onset of sister chromatid separation, after which it was dispersed for a short period (Fig. 3A). The dispersion of Cdc14 from nucleolus during the metaphase-anaphase transition has been well documented elsewhere (15, 20). Hence, the deficiency of CLB5 does not lead to premature dispersion of Cdc14.
FIG. 3.
(A) Cdc14-GFP is not prematurely dispersed in clb5Δ cells. Wild-type (left panels) and clb5Δ (right panels) samples at various times are shown. The graphs depict the extents of nuclear division and Cdc14-GFP dispersion. DAPI, 4′,6′-diamidino-2-phenylindole. (B) Insufficient phosphorylation of Hct1 in a clb5Δ strain. Wild-type (US2352) and clb5Δ (US2354) cells carrying GAL-HA3-HCT1 were arrested in G1 by α-factor treatment in YEP+Raff+Gal at 24°C. The cultures were filtered, washed, and resuspended in YEP+Glu at 24°C. Samples were analyzed for DNA content, nuclear division index, Cdc28 protein levels, and Hct1 phosphorylation. (C) Inefficient phosphorylation of Hct1 in clb3Δ clb4Δ clb5Δ cells. clb3Δ clb4Δ clb5Δ cells carrying MET3-HA3-HCT1 on a 2μm plasmid (US2438) were synchronized in G1 by α-factor treatment for 3 h (the first 1.5 h in YEP+Raff+Met and the second 1.5 h in −Met+Raff medium). The culture was divided into two halves; one-half of the cells were resuspended in YEP+Raff+Met, and the other half were resuspended in YEP+Gal+Met. Samples were analyzed for DNA content, Hct1 phosphorylation, and Clb2 and Cdc28 protein levels.
The effect of Clb5 deficiency on the phosphorylation of Hct1 was determined in cells expressing Hct1 under the control of the GAL1 promoter since the wild-type level of Hct1 is not easily detectable in our Western blot analysis. Wild-type and clb5Δ cells carrying GAL-HA3-HCT1 on a CEN plasmid were subjected to the experimental conditions described for Fig. 1, and the extent of Hct1 phosphorylation was monitored at various times. During α-factor arrest, Hct1 is predominantly in nonphosphorylated form in both wild-type and clb5Δ cells (Fig. 3B). In wild-type cells, Hct1 appears as a single band at a higher position on the gel within 90 min after the release, indicating modification. However, clb5Δ cells fail to fully convert Hct1 to a single, low-mobility band (Fig. 3B); instead, multiple bands are seen throughout the course of the experiment. Since Hct1 contains at least nine potential phosphorylation sites (23), these multiple bands most likely represent different phosphorylated forms. These results suggest that clb5Δ cells are compromised in their ability to modify Hct1 completely. It is possible that a significant proportion of Hct1, which remains underphosphorylated in clb5Δ cells, is sufficient for the continuous destruction of Clb2 throughout the cell cycle, thereby preventing Clb2 from rising to the wild-type level (Fig. 1).
To determine if Hct1 is also underphosphorylated in the absence of CLB3, CLB4, and CLB5 genes, clb3Δ clb4Δ clb5Δ cells carrying one copy of GAL-CLB5 (integrated at the TRP1 locus) and MET3-HA3-HCT1 on a 2μm vector were synchronized in G1 by α-factor treatment for 3 h at 24°C. The last 1.5 h of α-factor treatment was carried out in −Met+Raff medium to induce the synthesis of HA3-Hct1. Cells were then released in either YEP+Raff+Met (no Clb5) or YEP+Raff+Gal+Met medium (Clb5 induction). Hct1 first appears as a nonphosphorylated, single band (Fig. 3C, α-factor lane). In the culture where Clb5 was induced (Fig. 3C, right panel), Hct1 phosphorylated to different extents can be seen within 60 min, as indicated by the appearance of low-mobility bands (Fig. 3C, right panel). By 90 min, most of Hct1 shifts upward. This is accompanied by the appearance of Clb2 protein (90- and 120-min lanes). In cells devoid of Clb5, a discernible amount of unphosphorylated Hct1 is still present even at 120 min (Fig. 3C, left panel). In addition, no Clb2 protein is detected in these cells. These findings suggest that cells deficient in Clb3, Clb4, and Clb5 are inefficient in phosphorylating Hct1 and are therefore unable to inactivate it completely. This results in the inability of these cells to accumulate Clb2. It is noteworthy that, in the absence of Clb3, Clb4, and Clb5, Hct1 is still phosphorylated to some extent though not sufficiently to allow growth. We suspect that this could be due to fully functional Clb6-Cdc28 kinase and Cln kinases present in these cells. However, the severe consequences elicited by the absence of Clb3, Clb4, and Clb5 suggest that phosphorylation by Clb6 and Cln kinases is clearly not sufficient to inactivate Hct1. These results imply that early Clb kinases (Clb3-Cdc28, Clb4-Cdc28, and Clb5-Cdc28) play a critical role, by inhibiting Hct1 through phosphorylation, in the accumulation of Clb2 to an appropriate level.
Deletion of HCT1 relieves G2/M arrest in clb3Δ clb4Δ clb5Δ cells.
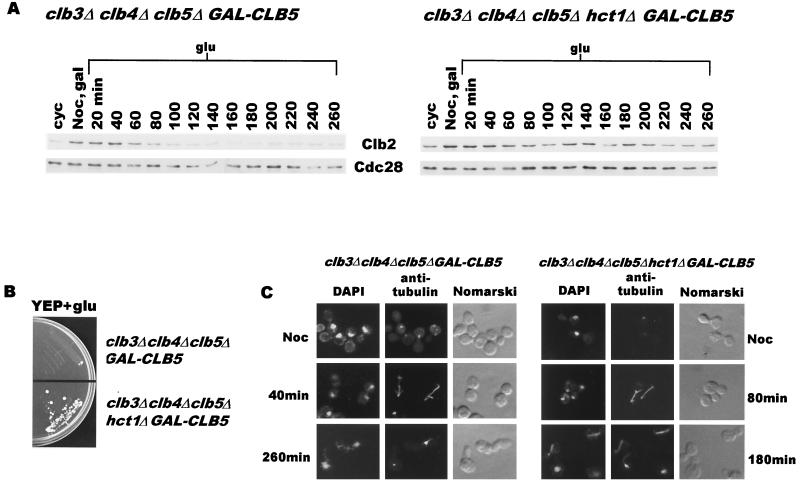
The data presented in the preceding sections argued that clb3Δ clb4Δ clb5Δ cells fail to enter mitosis because of their inability to inactivate Hct1. An important prediction of this claim is that a deletion of the HCT1 gene should allow clb3Δ clb4Δ clb5Δ cells to undergo nuclear division. We therefore constructed an hct1Δ clb3Δ clb4Δ clb5Δ quadruple-deletion mutant carrying one copy of GAL-CLB5 integrated at the TRP1 locus. While the clb3Δ clb4Δ clb5Δ triple mutant completely fails to grow on glucose plates, the hct1Δ clb3Δ clb4Δ clb5Δ mutant forms viable colonies (Fig. 4B). Microscopic examination revealed that not all quadruple mutant cells survived; however, they did appear to have undergone a few divisions before losing viability. We deemed it necessary to establish more clearly the ability of the quadruple mutant to undergo nuclear division. Since cells deficient in Hct1 are somewhat resistant to pheromone treatment, clb3Δ clb4Δ clb5Δ and hct1Δ clb3Δ clb4Δ clb5Δ cells carrying GAL-CLB5 were synchronized in a pre-nuclear-division state by treatment with the microtubule-disrupting agent nocodazole in galactose medium. Cells were then released into glucose medium to allow them to progress through the cycle in the absence of Clb5. As expected, clb3Δ clb4Δ clb5Δ cells reassembled mitotic spindles, underwent nuclear division within 40 min, and then exited mitosis as indicated by the disappearance of Clb2 protein (Fig. 4A, left panel). Within the next 60 min, they entered the next cycle (fresh buds) but eventually arrested with an elongated bud and an undivided nucleus devoid of mitotic spindle (Fig. 4C, left panels). As described in a previous section (Fig. 3), the level of Clb2 protein remained barely detectable after 100 min in these cells (Fig. 4A, left panel). The clb3Δ clb4Δ clb5Δ cells lacking HCT1 also reassembled the mitotic spindle, divided the nucleus, and exited mitosis. They entered the next cycle, reassembled the mitotic spindle, and continued through the cycle as indicated by the presence of divided nuclei with an elongated spindle (Fig. 4C). Moreover, the Clb2 protein also accumulates in these cells (Fig. 4A, left panel). However, the reaccumulation of Clb2 is not dramatic, perhaps due to a low degree of synchrony in cells released from nocodazole arrest. Nevertheless, these results suggest that HCT1 deletion indeed allows mitotic entry in clb3Δ clb4Δ clb5Δ cells. Thus, the collective essential function of these early transcribed CLB genes is to inactivate Hct1, making way for the accumulation of Clb2 (and Clb1) necessary for the onset of mitosis.
FIG. 4.
(A) Deletion of HCT1 relieves G2/M arrest in clb3Δ clb4Δ clb5Δ cells. clb3Δ clb4Δ clb5Δ GAL-CLB5::TRP1 (US2417) and clb3Δ clb4Δ clb5Δ hct1Δ GAL-CLB5::TRP1 (US2440) cells were treated with nocodazole (15 μg/ml) for 3.5 h in YEP+Gal at 24°C. The cells were washed and resuspended in YEP+Glu. Clb2 and Cdc28 protein levels were monitored in samples collected at 20-min intervals. (B) Deletion of HCT1 allows clb3Δ clb4Δ clb5Δ cells to grow on YEP-Glu medium. (C) Deletion of HCT1 relieves G2/M arrest in clb3Δ clb4Δ clb5Δ cells. The left panels show cells from the experiment described for panel A. clb3Δ clb4Δ clb5Δ cells form a mitotic spindle within 40 min after the release from nocodazole arrest but eventually are blocked in G2/M of the subsequent cycle without mitotic spindles (260-min sample). The right panels show that clb3Δ clb4Δ clb5Δ hct1Δ cells have reassembled spindles (80-min sample) after the release and continue to progress through the next cycle as indicated by the presence of mitotic spindles of various sizes (180-min sample). Noc, nocodazole; DAPI, 4′,6′-diamidino-2-phenylindole.
Clb-Cdc28 complexes are critical for switching off proteolysis in early S phase.
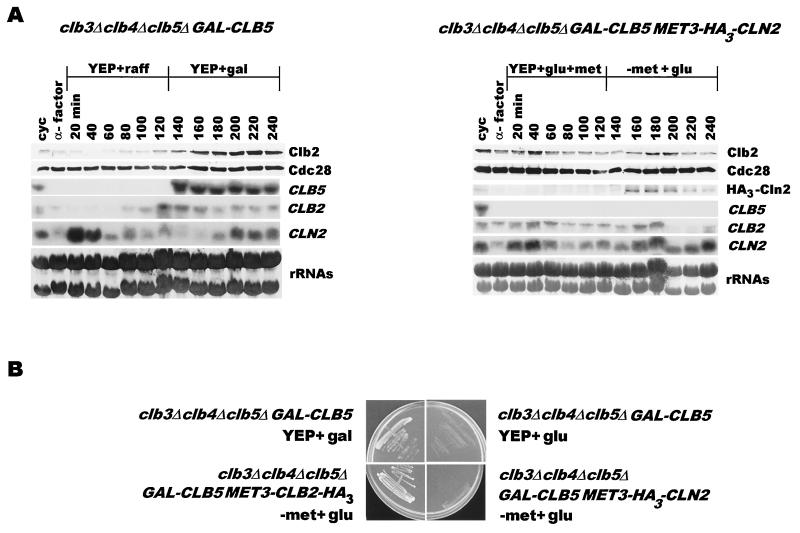
It is generally believed that the proteolytic machinery responsible for mitotic cyclin destruction is turned off early in the subsequent cycle by Cln-Cdc28 kinase (1). However, in direct contrast with this, our data suggest that APCHct1 activated during late mitosis is inactivated by Clb (Clb3, Clb4, and Clb5) kinases in the subsequent cycle. Consequently, clb3Δ clb4Δ clb5Δ cells are unable to inactivate APCHct1 and die due to a severe deficiency of Clb2 (and most likely of Clb1). If so, then the cyclin kinase that inactivates the proteolytic machinery would be expected to restore the accumulation of Clb2 protein in clb3Δ clb4Δ clb5Δ cells. We therefore compared the abilities of GAL-CLB5 (single copy) and MET3-CLN2 (on a 2μm vector) to reinstate the expression of CLB2. clb3Δ clb4Δ clb5Δ GAL-CLB5 cells were synchronized by α-factor treatment in YEP+Raff at 24°C and allowed to resume the cell cycle in YEP+Raff, while clb3Δ clb4Δ clb5Δ GAL-CLB5 cells carrying MET3-HA3-CLN2 on a 2μm plasmid were treated with α factor in YEP+Glu medium and were released in YEP+Glu+Met medium. After 2 h, galactose was added to cells without MET3-HA3-CLN2 to induce synthesis of Clb5, whereas those with MET3-HA3-CLN2 were shifted to −Met+Glu medium to induce Cln2. While both Clb2 protein and CLB2 transcript began to accumulate within 40 min of Clb5 induction, expression of Cln2 did not alter Clb2 expression, which remained at a basal level throughout the experiment (Fig. 5A). These results strengthen the notion that it is the early Clb kinases which make Clb2 accumulation possible. This is further supported by the fact that, while both GAL-CLB5 and MET3-CLB2 can efficiently rescue clb3Δ clb4Δ clb5Δ cells, a chronic expression of Cln2 from the MET3-CLN2 construct on a 2μm vector is unable to do so (Fig. 5B). However, it must be noted that our results do not completely rule out the possibility that Cln kinases may contribute toward APCHct1 inactivation to some extent, perhaps early in the cell cycle prior to S phase.
FIG. 5.
(A) Cln2 expression does not result in Clb2 accumulation. clb3Δ clb4Δ clb5Δ GAL-CLB5 (US2417) cells were arrested in G1 with α factor in YEP+Raff. The culture was filtered, and cells were resuspended in YEP+Raff for 2 h. Galactose (2%) was then added to induce the synthesis of Clb5 for 2 h. In parallel, clb3Δ clb4Δ clb5Δ GAL-CLB5 MET3-HA3-CLN2 (US2439) cells, synchronized in G1 by α-factor treatment in YEP+Glu+Met, were filtered and resuspended in YEP+Glu+Met for 2 h. Cells were then transferred to −Met+Glu medium to induce the synthesis of HA3-Cln2 for 2 h. Samples were analyzed for Clb2 and Cdc28 protein levels and CLB5, CLB2, and CLN2 RNA levels. (B) Overexpression of Cln2 is unable to restore viability in clb3Δ clb4Δ clb5Δ cells. clb3Δ clb4Δ clb5Δ GAL-CLB5 cells were plated on either glucose (upper right) or galactose (upper left) plates, whereas clb3Δ clb4Δ clb5Δ GAL-CLB5 MET3-HA3-CLN2 cells were plated on −Met+Glu medium. Plates were photographed after 3 days at 24°C.
Contribution of transcriptional regulation to oscillations in Clb2 abundance.
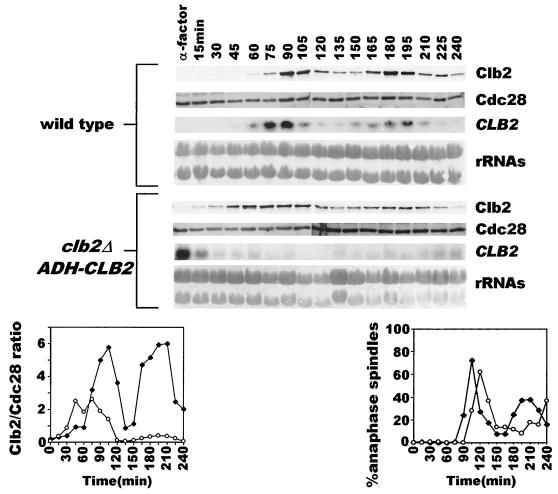
The results described above suggest that, while APCs act to inactivate Clb kinase complexes in M phase, some of the Clb kinases cooperate to inactivate APCHct1 during S phase to allow accumulation of mitotic cyclins. This also implies that such an alternating antagonism between APC and Clb kinases in S and M phases should be able to generate oscillations in Clb2 levels. If so, then to what extent does the fluctuating transcription of CLB2 contribute to these oscillations? We addressed this in a strain where CLB2 is transcribed, not periodically from its native promoter, but continuously from the ADH promoter. The wild-type strain and a strain in which the endogenous CLB2 gene had been replaced by ADH-CLB2 were synchronized in G1 by pheromone treatment at 24°C. Cells were allowed to resume cell cycle progression in pheromone-free medium. As expected, Clb2 protein showed oscillatory behavior for two synchronous cycles in wild-type cells (Fig. 6; lower panels, solid diamonds). Cells carrying ADH-CLB2 also exhibited the oscillatory pattern in the abundance of Clb2 (Fig. 6, lower panels, open circles). However, compared to the wild type, the amplitude of the oscillation in these cells was lower in the first cycle; the reduction in the amplitude was even more pronounced in the second division cycle. These observations imply that while the mutually repressive behavior of APC and Clb proteins is sufficient for producing basic oscillations in Clb2 protein, transcriptional regulation modulates the amplitude of these oscillations.
FIG. 6.
The oscillatory pattern in Clb2 protein levels is maintained even when CLB2 transcription is constitutive. Wild-type (US356) (♦) and clb2Δ ADH-CLB2 (US164) (○) cells, synchronized in G1 by α-factor treatment in YEP+Glu at 24°C, were released into YEP+Glu medium. The state of mitotic spindles and the levels of Clb2 and Cdc28 proteins and CLB2 transcript are shown.
DISCUSSION
A quick rise and a rapid fall in mitotic cyclin levels within a division cycle are important for the orderly progression through M phase. The cellular logistics which give rise to this oscillatory behavior are also of great interest because they promise a glimpse into the regulatory dynamics that generate such a pattern. The cyclic fashion in which the major mitotic cyclins appear and disappear has been attributed to the onset of cyclin transcription in late S phase and destruction in late telophase, respectively. As a coarse approximation, this indeed appears to be true. However, such a simplistic view overlooks entirely the dynamic realities of cellular events that generate such a periodic pattern.
It has been well documented elsewhere that the high mitotic kinase (such as Clb2 kinase) activity keeps Hct1 in an inactive state (6) and thus prevents cells from exiting mitosis. In vitro assays have shown that Hct1 can also be phosphorylated by Clb5 kinase (23). Consistent with these findings, it has been reported elsewhere that Clb5 destruction at the onset of anaphase is required to pave the way for Hct1 activation and hence mitotic exit (14). However, these studies do not in any way address the role that Clb5 (and other early Clb proteins) may play in inactivating proteolytic machinery during early S phase. Although APCHct1 activated in telophase is responsible for keeping mitotic cyclin levels to a minimum during G1, it becomes a detriment to cells as they attempt to reaccumulate mitotic cyclins (Clb1 and Clb2) at the onset of mitosis. In this report, we show that the inactivation of APCHct1 in early S phase is accomplished by the kinase activities associated with early transcribed Clb3, Clb4, and Clb5, which begin to accumulate soon after cells' passage through start. This role of early Clbs is essential such that, in the absence of Clb3, Clb4, and Clb5, cells fail to enter mitosis due to a lack of Clb2 (and Clb1) proteins.
The inability of the clb5Δ mutant to build up Clb2 protein to wild-type levels and to phosphorylate Hct1 efficiently provided the initial clue (Fig. 1 and 3). The severity of this defect becomes starkly apparent in cells deficient in Clb3, Clb4, and Clb5. The clb3Δ clb4Δ clb5Δ cells fail to accumulate Clb2 (and most likely Clb1) and therefore arrest in G2/M without mitotic spindles (Fig. 2). The fact that the deletion of the HCT1 gene restores viability and Clb2 accumulation in these cells strongly suggests that the low level of Clb2 in clb3Δ clb4Δ clb5Δ cells is due to their inability to inactivate Hct1 (Fig. 4). The instability of ectopically expressed Clb2 and insufficient phosphorylation of Hct1 in clb3Δ clb4Δ clb5Δ cells are both consistent with this hypothesis (Fig. 2D and 3). It should be noted that a sizable proportion of clb3Δ clb4Δ clb5Δ hct1Δ cells divide several times but die before forming a colony. This may be due to the buildup of a physiological imbalance caused by growth in the presence of multiple deletions to which some cells are unable to develop tolerance. Nevertheless, these results underscore the acute importance of shutting down proteolytic machinery in early S phase to allow the initial accumulation of Clb2 necessary for the initiation of the positive feedback loop for CLB2 transcription (1). They also strongly argue that the early Clb kinases collectively play a critical role of allowing accumulation of late mitotic cyclin for the onset of mitosis. The involvement of cyclin A in the accumulation of B-type cyclin (late cyclins) in mammalian cells has been reported earlier (10).
Our findings have a number of important implications. First, it has been suggested that Cdc20 facilitates mitotic exit by mediating Clb5 destruction in metaphase. This was based on the observation that a mutation in CLB5 allows a cdc20Δ pds1Δ double mutant to exit mitosis (14). We suspect that the reason why a mutation in CLB5 allows cdc20Δ pds1Δ cells to exit mitosis is that, in the absence of Clb5 function, the collective Clb kinase activities do not rise to the wild-type level (Fig. 1); consequently, Hct1 is not completely inactivated and cells no longer require Cdc20 function to reduce the kinase activities any further in order to effectively activate Hct1. We have also found that, in the absence of Pds1, cells' ability to build up Clb2-Cdc28 kinase is also compromised (data not shown). Second, our results also suggest that clb3Δ clb4Δ clb5Δ cells are inviable not because Clb5 is required for mitotic spindle formation in the absence of Clb3 and Clb4, as previously suggested (13); instead, a collectively essential function of the early expressed Clb3, Clb4, and Clb5 cyclins is to inactivate APCHct1 to make way for the accumulation of late mitotic cyclins. Third, the early Clb-Cdc28 kinases play a critical role in the inactivation of the mitotic proteolytic machinery in early S phase (Fig. 5); however, the possibility that Cln-Cdc28 kinases may contribute toward this inactivation during the early part of the division cycle cannot be completely ruled out. Last, the accumulation of Clb2 (and Clb1) is not a simple consequence of turning on CLB2 transcription; the inactivation of the cyclin destruction machinery plays a critical role in bringing the transcriptional positive feedback loop into effect.
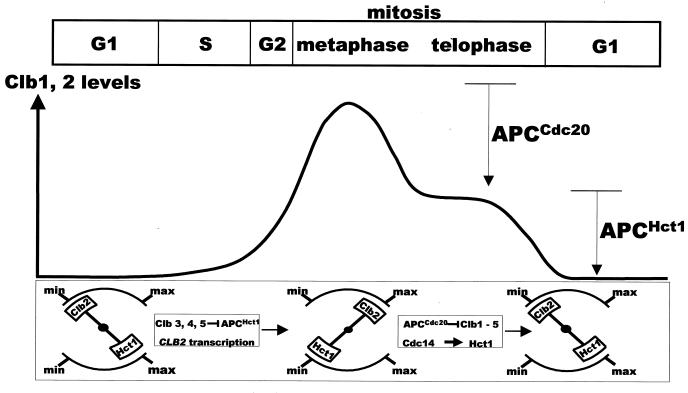
Complex processes such as mitosis are quite finely tuned to allow cells to adapt to changing intra- and extracellular contexts. For a process to be well honed, its on-off controls have to be overlaid with mechanisms that regulate the thresholds of activities of various effectors. Very often, the critical threshold of an effector is a result of interactions between multiple events. Our investigations suggest that the oscillatory pattern in cyclin abundance results from the interwoven functional relationships between various players. Central to these interactions is the antagonistic relationship between the APC and the Clb proteins. In our previous work, we had established that APCCdc20 and APCHct1 together inactivate the Clb kinases during mitosis (22). Here we have shown that APCHct1 is inactivated in early S phase of the subsequent cycle by early transcribed Clb3, Clb4, and Clb5 kinases to allow accumulation of late mitotic cyclin Clb2. In combination, these two sets of regulation can be seen to constitute a two-stroke engine. The forward stroke occurs in S phase when Clb2 accumulates as a result of APCHct1 inhibition by early Clb kinases. During the second (reverse) stroke, APCHct1 (activated by APCCdc20 and Cdc14) destroys Clb2 during mitosis. The occurrence of these two steps in tandem would therefore generate oscillations in Clb2 abundance. In a two-stroke process, it is important that, at the end of one stroke, the reverse stroke is initiated efficiently. In G2/M, the reverse stroke is set in motion by the active APCCdc20, which begins the process of APCHct1 activation. This stroke leads to full activation of APCHct1 and complete destruction of Clb2. However, it is reversed again in early S phase when early transcribed Clb3, Clb4, and Clb5 inactivate APCHct1, making way for Clb2 accumulation. These functional relationships are schematically depicted in Fig. 7.
FIG. 7.
A schematic depicting the interaction between APC and Clb proteins and the resulting oscillations in Clb2 abundance (see the text for details).
This study has taken a renewed look at the familiar oscillatory pattern in mitotic cyclins and has revealed the dynamic relationships that generate it. It also suggests that events which occur early in the division cycle (such as the inhibition of APCHct1 by Clb3, Clb4, and Clb5) have profound influence on the events in late mitosis. We suspect that many apparently simple patterns of cellular behavior emerge from such nonlinear functional interactions. These intertwined relationships are what make cells resilient and adaptable systems.
ACKNOWLEDGMENTS
We are grateful to Wolfgang Zachariae, Bruce Futcher, and David Morgan for various plasmids and constructs. We thank Shannon Allan for the CDC14-GFP construct and Bor Luen Tang for 12CA5 ascites.
This work was supported by the National Science and Technology Board, Singapore.
F.M.Y. and H.H.L. contributed equally to the work.
U.S. is an adjunct staff member of the Department of Pharmacology, National University of Singapore.
REFERENCES
- 1.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 2.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 3.Baumer M, Braus G H, Irniger S. Two different modes of cyclin Clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000;468:142–148. doi: 10.1016/s0014-5793(00)01208-4. [DOI] [PubMed] [Google Scholar]
- 4.Breeden L. Cyclin transcription: timing is everything. Curr Biol. 2000;10:R586–R588. doi: 10.1016/s0960-9822(00)00634-5. [DOI] [PubMed] [Google Scholar]
- 5.Fitch I, Dahman C, Surana U, Amon A, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast S. cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jasperson S L, Charles J F, Morgan D O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 7.Kramer E R, Scheuringer N, Podtelejnikov A V, Mann M, Peters J M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim H H, Goh P-Y, Surana U. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim H H, Goh P-Y, Surana U. Cdc20 is essential for APC-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 10.Lukas C, Sorensen C S, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters J M, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 11.Nougarede R, Della Seta F, Zarzov P, Schwob E. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol. 2000;20:3795–3806. doi: 10.1128/mcb.20.11.3795-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 13.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 14.Shirayama M, Toth A, Galova M, Nasmyth K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 15.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 16.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 17.Surana U, Amon A, Dowzer C, Mcgrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 19.Visintin R, Craig K, Hwang E S, Prinz S, Tyres M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 20.Visintin R, Hwang E S, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast Saccharomyces cerevisiae. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeong F M, Lim H H, Padmashree C G, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol Cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- 23.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 24.Zachariae W, Shevchenko A, Andrews P D, Stark M J R, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase complex from budding yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 25.Zachariae W. Progression into and out of mitosis. Curr Opin Cell Biol. 1999;11:708–716. doi: 10.1016/s0955-0674(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 26.Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]