Abstract
Coronavirus disease 2019 (COVID-19) is a new respiratory illness caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and now spreads globally. Currently, therapeutics and effective treatment options remain scarce and there is no proven drug to treat COVID-19. Targeting the positive-sense RNA genome and viral mRNAs of SARS-CoV-2 to simultaneously degrade viral genome templates for replication and viral mRNAs for essential gene expression would be a strategy to completely realize virus elimination. Type VI CRISPR enzymes Cas13 have recently been identified as programmable RNA-guided, RNA-targeting Cas proteins with nuclease activity that allows for RNA cleavage and degradation. The precise viral RNA detection and antiviral application of the CRISPR/Cas13 system depend on high-efficient and minimal off-target crRNAs. Although a computer-based algorithm has been applied for the design of crRNAs targeting SRAS-CoV-2, the experimental screening system to identify optimal crRNA is not available. We develop a one-step experimental screening system to identify high-efficient crRNAs with minimal off-target effects for CRISPR/Cas13-based SARS-CoV-2 elimination. This platform provides the foundation for CRISPR/Cas13-based diagnostics and therapeutics for COVID-19. This platform is versatile and could also be applied for crRNAs screening for other RNA viruses.
Abbreviations: COVID-19, Coronavirus disease 2019; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; scRNA-seq, Single-cell RNA-seq; ACE2, Angiotensin-converting enzyme 2
Keywords: COVID-19, SARS-CoV-2, CRISPR/Cas13, scRNA-seq
Introduction
Coronavirus disease 2019 (COVID-19) is a new respiratory illness caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and now spreads globally [1], [2]. SARS-CoV-2 is classified in the Coronaviridae family and betacoronavirus genus and the seventh coronavirus known to infect humans and it is an enveloped, positive-sense, single-stranded RNA virus with mammalian and avian hosts [3]. The SARS-CoV-2 virus enters the human cell, releases its RNA genome into the cytoplasm, and synthesizes negative-sense RNA intermediates which serve as the templates for the synthesis of sub-genomic RNAs and positive-sense viral genome for new virus production [4]. A critical feature of SARS-CoV-2 is the fast transmission by human-to-human contact, or by touching surfaces contaminated by the infected person, or by breathing aerosol that quickly led to a worldwide public health emergency [5]. The clinical spectrum of SARS-CoV-2 infection ranges from asymptomatic infection to fatal disease [5]. Some people may suffer from fever, cough, fatigue, shortness of breath, occasionally watery diarrhea, or worsened symptoms such as pneumonia, acute respiratory distress syndrome, and death, which resulted in serious public and healthcare-related casualties worldwide [5]. Unfortunately, therapeutics and effective treatment options remain scarce and there is no proven drug to treat COVID-19 [6].
To combat the SARS-CoV-2 virus, many drugs are repurposed to treat COVID-19 by targeting the SARS-CoV-2 virus, such as chloroquine, hydroxychloroquine, lopinavir/ritonavir, and ribavirin [7]. Immunomodulatory drugs such as IL-6 inhibitors (eg, sarilumab or tocilizumab) or anti-GM-CSF compounds (eg, lenzilumab and gimsilumab) are clinically tested to prevent or reverse SARS-CoV-2-caused pneumonia and acute respiratory distress syndrome [8]. In addition, various novel drugs are developed to inhibit SARS-CoV-2 virus replication and infection [9]. Prasad et al found annonaceous acetogenins showed a good inhibition activity against the SARS-CoV-2 spike protein through computational approaches [10]. Nevertheless, these drugs are designed to inhibit the replication and infection of SARS-CoV-2 but not eliminate it in the infected lung. Moreover, we do not yet know which drugs would be showed effective against SARS-CoV-2. The devastating impact of the SARS-CoV-2 pandemic on society and economics emphasizes an unprecedentedly urgent need to find effective drugs to reduce the clinical consequence of COVID-19.
The hypothesis
Type VI CRISPR (clustered regularly interspaced short palindromic repeats) enzymes Cas13 have recently been identified as programmable RNA-guided, RNA-targeting Cas proteins with nuclease activity that allows for RNA cleavage and degradation [11], [12], [13], [14], [15], [16]. Cas13 proteins are guided to the target RNA by a single CRISPR RNA (crRNA) composed of a direct repeat (DR) stem-loop and a spacer sequence (gRNA) that mediates target recognition by RNA–RNA hybridization and then causes targeted RNA cleavage and degradation. CRISPR/Cas13-based methods have been developed for COVID-19 diagnosis (SHERLOCK and CREST) and therapeutics (PAC-MAN and ABACAS) [17]. CRISPR/Cas13-based diagnostic platforms seem rapid, sensitive, and specific for the detection of SARS-CoV-2, but their efficiency in clinical use remains unknown [17]. The major shortcoming of these methods is the off-target effects that led to the poor signaling and misinterpretation of results [17]. CRISPR/Cas13-based therapeutic platforms are more dependent on the high specificity and low off-target effects for clinical use [18]. To improve the specificity and avoid the off-target effects of CRISPR/Cas13 system, we need to design high-efficient and minimal off-target crRNAs. A computer-based algorithm has been developed for the design of crRNAs to target SRAS-CoV-2 [19]. However, sequence analysis by GUIDE-seq revealed that many off-target sites cannot be predicted by in silico methods [20]. It implies that crRNA design cannot completely depend on the computer-based algorithms and the experimental screening system to identify optimal crRNA is necessary. So far, there is no such experimental crRNA screening system to be reported.
We hypothesize that targeting the positive-sense RNA genome and viral mRNAs of SARS-CoV-2 by CRISPR/Cas13 system to simultaneously degrade viral genome templates for replication and viral mRNAs for essential gene expression would be a strategy to completely realize SARS-CoV-19 virus elimination. To reach this purpose, we develop a one-step experimental screening system to identify high-efficient crRNAs with minimal off-target for CRISPR/Cas13-based SARS-CoV-2 elimination. This platform provides the foundation for CRISPR/Cas13-based diagnostics and therapeutics for COVID-19. This platform is versatile and could also be applied for crRNAs screening for other RNA viruses.
Experimental procedure
The design of crRNAs targeting SARS-CoV-2
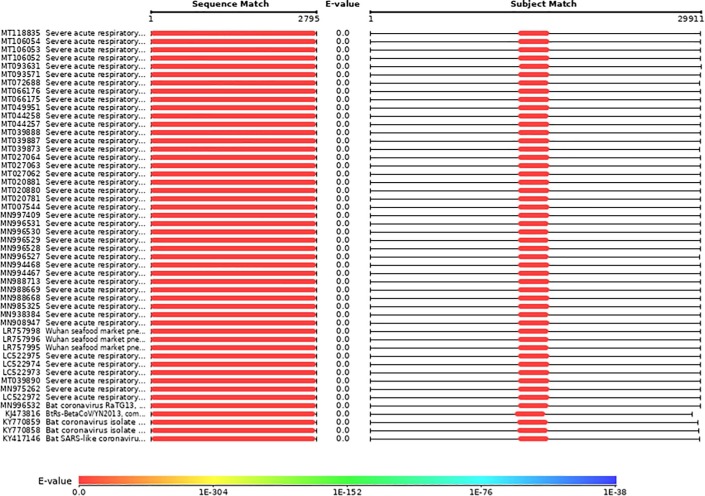
Theoretically, Cas13 can target anywhere of the RNA sequence because no protospacer flanking sequence (PFS) restriction has been documented for the Cas13 orthologs, including Cas13a, Cas13b, and Cas13d, in mammalian cells [11], [12], [14], [16]. Because there is no available SARS-CoV-2 virus strain for conventional laboratory application, a conserved genomic region coding the RNA-dependent RNA polymerase (RdRp) is selected as the target sequence to design crRNAs for one-step platform assessment. RdRp is an essential gene for viral replication and conserved among human coronaviruses and a versatile drug target for anti-coronavirus [21], [22], [23], [24]. Sequence alignment analysis demonstrated that the genomic region of SARS-CoV-2 RdRp is rarely mutated among COVID-19 patients (Fig. 1 ). In addition, there is no the similar sequence in the human transcriptome except a 19-nt sequence (CCTGTTGTAGATTCTTATT) that localizing in the KMT2C mRNA. Therefore, the RdRp is an ideal target for designing crRNAs. Considering that the secondary structure of RNA will reduce the efficiency of the CRISPR/Cas13 system, the RNAxs platform is used to find regions of SARS-CoV-2 RdRp that have good accessibility for crRNAs [25].
Fig. 1.
The RdRp genomic region of SARS-COV-2 is rarely mutated in various virus strains isolated world widely.
LentiCRISPR library construction
The CRISPR library is constructed according to a protocol from Wessels et al. [19]. Briefly, pooled crRNA libraries are synthesized as single-stranded oligonucleotides (Twist Biosciences), amplified using NEBNext High-Fidelity 2X PCR Master Mix (M0541S) and subcloned into lentiGuide-Puro (Addgene # 52963) along with the replacement of gRNA scaffold with direct repeat (DR). All constructs are confirmed by Next Generation Sequence and subsequently pooled using equal amounts. Complete library representation with minimal bias (90th percentile: 10th percentile crRNA read ratio: 1.68 – 2.17) is verified by Illumina sequencing (MiSeq). Lentiviruses are produced via transfection of library plasmid with appropriate packaging plasmids (psPAX2: Addgene, Cat# 12260; pMD2.G: Addgene, Cat# 12259). At 3 days post-transfection, the viral supernatant are collected and passed through a 0.45-μm filter and stored at −80 °C until use.
Lentivirus transduction and single-cell RNA-seq (scRNA-seq)
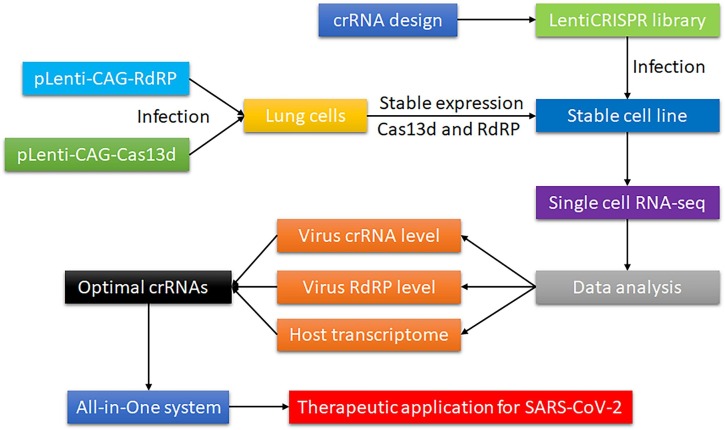
A human lung cell line stably expressing SARS-CoV-2 RdRP and Cas13 nuclease is generated for crRNA screening. The Cas13d is preferred to use as a CRISPR nuclease for this experiment because Cas13d has the most robust and substantial RNA knockdown in mammalian cells among Cas13 subtypes and orthologs [15], [26]. The synthesized fragment of the SARS-CoV-2 RdRp sequence and Cas13d cDNA sequence are subcloned into a lentivirus vector for cell transduction to generate a stable lung cell line expressing SARS-CoV-2 RdRp and Cas13d in human lung epithelial A549 cells (ATCC CCL-185). This lung cell line is infected with lentivirus CRISPR library at a low multiplicity of infection (approximately 0.3) to ensure that most cells receive only 1 viral construct with high probability. Three independent infections are conducted, and then 3 independent libraries are produced. After a 2-day infection, cells are selected with puromycin. After a 7-day infection, cells are collected for scRNA-seq. Prior to analysis, cells are diluted to the final concentration and loaded on the 10X Chromium system (8,000 cells/channel) and scRNA-seq libraries are generated following the manufacturer’s instructions. A digital expression matrix is obtained for each experiment using 10X’s CellRanger pipeline with default parameters. Data are analyzed by a bioinformatician. The expression profile of host lung cells and SARS-CoV-2 RdRp expression in each single cell with a certain crRNA expression is compared to that in a single cell without crRNA expression. The single cell showing maximal suppression of SARS-CoV-2 RdRp expression and minimal change of expression profile of the host lung cells will contain an applicable candidate crRNA that will be the highly efficient and selective crRNAs with minimal off-target effects (Fig. 2 ).
Fig. 2.
Flow chart of the one-step screening platform for high-efficient and minimal off-target CRISPR/Cas13 crRNAs targeting SARS-COV-2. Virus crRNA level indicates that the single cell is infected by a certain crRNA. Virus RdRp level indicates the knockdown efficiency induced by the crRNA in a single cell. Host transcriptome change indicates off-target effects induced by the crRNA in a single cell.
All-in-One system construction for therapeutic application
Considering that angiotensin-converting enzyme 2 (ACE2) is the receptor of SARS-CoV-2, the expression of Cas13d is controlled by a human ACE2 promotor for a unique expression in lung ACE2+ cells, where SARS-CoV-2 infects lung epithelial cells and enters the human body [27], [28], [29]. The crRNAs and direct repeat sequence are subcloned to the Cas13d-expressed vector and the U6 promoter is used to drive crRNA expression. Combinations of several optimal crRNAs are used to form a crRNA array for multiple editing to enhance the targeting efficiency. Thus, a plasmid containing all components for SARS-CoV-2 elimination is generated. To test the efficiency of the CRISPR/Cas13 system for SARS-CoV-2 RdRp knockdown in cultured human cells, the synthesized fragment of SARS-CoV-2 RdRp is subcloned into pcDNA3 expression vector, and this vector is transfected into human primary bronchial/tracheal epithelial cells (ATCC Cat#PCS-300-010). The expression levels of SARS-CoV-2 RdRp are determined by RT-qPCR. The All-in-One CRISPR/Cas13 system containing the high-efficient and minimal off-target crRNA array is co-transfected with the pcDNA3-RdRp expression vector to evaluate the SARS-CoV-2 RdRp knockdown efficiency. The off-target effects are analyzed by scRNA-seq. The All-in-One CRISPR/Cas13 with scramble crRNAs is co-transfected with the pcDNA3-RdRp expression vector as a control group. Because of the lack of laboratory strains of SARS-CoV-2, the synthesized DNA fragment of SARS-CoV-2 RdRp is subcloned into an AAV-based expression vector and the constructed RdRp-AAV virus particles are delivered into the mouse lung for transduction via intranasal administration. When SARS-CoV-2 RdRp is highly expressed (e.g., at 21 days post-transduction [30]), the All-in-One CRISPR/Cas13 vector containing the identified crRNAs targeting SARS-CoV-2 RdRp or scramble crRNAs is delivered into the lung intranasally by the nanoparticle technology for highly efficient delivery. The SARS-CoV-2 RdRP knockdown efficiency and off-target effects are determined by scRNA-seq.
Discussion
The guide RNA, including single-guide RNA (sgRNA) for CRISPR/Cas9 system and CRISPR RNA (crRNA) for the CRISPR/Cas13 system, is a key component of CRISPR/Cas system because the quality of guide RNA is critical for the efficacy and specificity of CRISPR/Cas-mediated DNA/RNA editing [31]. Therefore, more attention is paid to design an efficient and functional guide RNA with high on-target efficacy and low off-target effects [32]. Three different computation-based genres appear for guide RNA designing: 1. pattern recognition genre; 2. feature rule genre; 3. machine learning genre [33]. In theory, the CRISPR/Cas system searches PAM sequence in genome and guide RNA recognizes target site to activate endonuclease activity to cut specific locus. However, sequence features of guide RNA, epigenetic features of the host genome, energetics features of guide RNA and the host genome affect guide RNA efficacy, which make guide RNA design become extraordinarily complex [33]. Therefore, guide RNAs need to be experimentally validated for the efficacy after designing by the software. To avoid off-target effects, in silico methods are developed to evaluate off-target sites by aligning short guide RNA sequence to reference genome to detect mismatch number [32]. However, sequence analysis by GUIDE-seq revealed that numerous off-targets with high-mismatch or even with one-mismatch cannot be predicted by sequence alignment algorithm [20]. So, off-target effects need to be validated by established methods such as CIRCLE-seq [34], GUIDE-seq [20], DISCOVER-seq [35], and Digenome-seq [36]. This one-step guide RNA screening platform is a combination of CRISPR screening and single-cell RNA sequencing and is the first method for simultaneously screening guide RNAs with high on-target efficacy and low off-target effects.
RdRp is selected as a target to design crRNA for the application of CRISPR/Cas13-based therapeutics based on several reasons: 1) RdRp is a versatile enzyme for viral RNA replication and transcription of viral genome in host cells, which makes RdRp essential for viral survival and spread [3], [24]; 2) RdRp is considered to be a highly conserved enzyme across all RNA viruses and some DNA viruses, such as coronavirus, influenza virus, zika virus, hepatitis C virus [37]; 3) RdRp has a very low mutation rate among COVID-19 patients (Fig. 1); 4) RdRp has no homologs in human cells and its RNA sequence shows an extremely low similarity with human transcriptome [38]; 5) The essential role of RdRp for viral RNA replication and transcription makes it become a promising therapeutic target and many drugs are repurposed and developed to target it for COVID-19 treatment, such as remdesivir [22], favipiravir [39], ribavirin [40], sofosbuvir [41], and galidesivir [42].
Conclusion
We develop a one-step experimental screening platform to identify high-efficient and selective crRNAs with minimal off-target effects for therapeutic eradication of SARS-CoV-2 virus in vivo laying a foundation for effective treatment of COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This research was funded by the National Natural Science Foundation of China (grant number: 81901485).
Ethical approval
The study was approved by the Institutional Review Board of the University of Henan University of Science and Technology (HUST).
References
- 1.Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., et al. COVID-19: towards controlling of a pandemic. Lancet. 2020;395(10229):1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chowell G., Mizumoto K. The COVID-19 pandemic in the USA: what might we expect? Lancet. 2020;395(10230):1093–1094. doi: 10.1016/S0140-6736(20)30743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberfeld B., Achanta A., Carpenter K., Chen P., Gilette N.M., Langat P., et al. SnapShot: COVID-19. Cell. 2020;181(4):954–954.e1. doi: 10.1016/j.cell.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganesh B., Rajakumar T., Malathi M., Manikandan N., Nagaraj J., Santhakumar A., et al. Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: an updated overview of current knowledge and future perspectives. Clin Epidemiol Glob Health. 2021;10:100694. doi: 10.1016/j.cegh.2020.100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baden L.R., Rubin E.J. Covid-19 - The search for effective therapy. N Engl J Med. 2020;382(19):1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijayvargiya P., Esquer Garrigos Z., Castillo Almeida N.E., Gurram P.R., Stevens R.W., Razonable R.R. Treatment considerations for COVID-19: a critical review of the evidence (or Lack Thereof) Mayo Clin Proc. 2020;95(7):1454–1466. doi: 10.1016/j.mayocp.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stasi C., Fallani S., Voller F., Silvestri C. Treatment for COVID-19: an overview. Eur J Pharmacol. 2020;889:173644. doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternberg A., McKee D.L., Naujokat C. Novel drugs targeting the SARS-CoV-2/COVID-19 machinery. Curr Top Med Chem. 2020;20(16):1423–1433. doi: 10.2174/1568026620999200517043137. [DOI] [PubMed] [Google Scholar]
- 10.Prasad S.K., Pradeep S., Shimavallu C., Kollur S.P., Syed A., Marraiki N., et al. Evaluation of Annona muricata Acetogenins as potential anti-SARS-CoV-2 agents through computational approaches. Front Chem. 2020;8 doi: 10.3389/fchem.2020.62471610.3389/fchem.2020.624716.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299) doi: 10.1126/science:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smargon A.A., Cox D.B.T., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., et al. Cas13b Is a type VI-B CRISPR-associated RNA-guided rnase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630.e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safari F., Afarid M., Rastegari B., Borhani-Haghighi A., Barekati-Mowahed M., Behzad-Behbahani A. CRISPR systems: novel approaches for detection and combating COVID-19. Virus Res. 2021;294:198282. doi: 10.1016/j.virusres.2020.198282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straiton J. CRISPR vs COVID-19: how can gene editing help beat a virus? Biotechniques. 2020;69(5):327–329. doi: 10.2144/btn-2020-0145. [DOI] [PubMed] [Google Scholar]
- 19.Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 2020;38(6):722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M et al: Structural Basis for the Inhibition of the RNA-Dependent RNA Polymerase from SARSCoV-2 by Remdesivir. 2020. [DOI] [PMC free article] [PubMed]
- 23.Lung J., Lin Y.-S., Yang Y.-H., Chou Y.-L., Shu L.-H., Cheng Y.-C., et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J Med Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tafer H., Ameres S.L., Obernosterer G., Gebeshuber C.A., Schroeder R., Martinez J., et al. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol. 2008;26(5):578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 26.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santry L.A., Ingrao J.C., Yu D.L., de Jong J.G., van Lieshout L.P., Wood G.A., et al. AAV vector distribution in the mouse respiratory tract following four different methods of administration. BMC Biotechnol. 2017;17(1) doi: 10.1186/s12896-017-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanna R.E., Doench J.G. Design and analysis of CRISPR-Cas experiments. Nat Biotechnol. 2020;38(7):813–823. doi: 10.1038/s41587-020-0490-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu G., Zhang Y., Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput Struct Biotechnol J. 2020;18:35–44. doi: 10.1016/j.csbj.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Zhao G., Ahmed F.Y.H., Yi T., Hu S., Cai T., et al. In silico method in CRISPR/Cas system: an expedite and powerful booster. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.584404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14(6):607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J., et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364(6437):286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 37.Venkataraman S., Prasad B., Selvarajan R. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses. 2018;10(2):76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naydenova K, Muir KW, Wu LF, Zhang Z, Coscia F, Peet MJ, Castro-Hartmann P, Qian P, Sader K, Dent K et al: Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc Natl Acad Sci USA 2021; 118(7). [DOI] [PMC free article] [PubMed]
- 40.Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y.i. Novel coronavirus treatment with ribavirin: Groundwork for an evaluation concerning COVID-19. J Med Virol. 2020;92(7):740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbass S., Kamal E., Salama M., Salman T., Sabry A., Abdel‐Razek W., et al. Efficacy and safety of sofosbuvir plus daclatasvir or ravidasvir in patients with COVID-19: a randomized controlled trial. J Med Virol. 2021;93(12):6750–6759. doi: 10.1002/jmv.27264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celik I., Erol M., Duzgun Z. In silico evaluation of potential inhibitory activity of remdesivir, favipiravir, ribavirin and galidesivir active forms on SARS-CoV-2 RNA polymerase. Mol Divers. 2021 doi: 10.1007/s11030-021-10215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]