Abstract
Many novel tigecycline-inactivating enzymes encoded by tet(X) variants from different bacteria were discovered since the plasmid-mediated tet(X3) and tet(X4) genes conferring high-level resistance to tigecycline in Enterobacterales and Acinetobacter were reported. However, there have been no comprehensive studies of the prevalence of different tet(X) variants in poultry farms. In this study, we collected 45 chicken fecal samples, isolated tet(X)-positive strains, and performed antimicrobial susceptibility testing, conjugation assay, whole-genome sequencing, and bioinformatics analysis. A total of 15 tet(X)-bearing strains were isolated from 13 samples. Species identification and tet(X) subtyping analysis found that the 15 strains belonged to eight different species and harbored four different tet(X) variants. Genomic investigation showed that transmission of tet(X) variants was associated with various mobile genetic elements, and tet(X4) was the most prevalent variant transferred by conjugative plasmids. Meanwhile, we characterized a plasmid co-harboring tet(X6) and blaOXA–58 in Acinetobacter baumannii. In summary, we demonstrated that different tet(X) variants were widely disseminated in the chicken farming environment and dominated by tet(X4). This finding expands the understanding of the prevalence of tet(X) among different animal sources, and it was advocated to reduce the usage of antibiotics to limit the emergence and transmission of novel tet(X) variants in the poultry industry.
Keywords: tigecycline resistance, tet(X), plasmids, chickens, whole-genome sequencing
Introduction
Tigecycline is a broad-spectrum antibiotic of glycylcyclines and is one of the last-resort antibiotics to treat serious infections caused by multidrug-resistant (MDR) Gram-negative bacteria (Sun et al., 2013). The mechanisms of tigecycline resistance were mainly the overexpression of non-specific active efflux pumps or mutations within the drug-binding sites in the ribosome, which were limited by less capability of horizontal transfer among bacteria (Pournaras et al., 2016). However, the emergence and dissemination of plasmid-mediated high-level tigecycline resistance genes tet(X3) and tet(X4) are bringing formidable threats to public health (He et al., 2019; Sun J. et al., 2019). A variety of tet(X) variants containing tet(X3.2) (Li et al., 2019), tet(X5) (Wang et al., 2019), tet(X6) (He et al., 2020; Liu et al., 2020; Peng et al., 2020), and tet(X14) (Cheng et al., 2020) have been detected in Empedobacter, Enterobacterales, and Acinetobacter so far. These widespread tet(X) variants will limit treatment options for MDR bacteria infections.
The livestock industry has been a critical reservoir of resistance genes due to the abuse and misuse of antibiotics in animal agriculture. Many clinically significant resistance genes, such as mcr-1 (Liu et al., 2016), tet(X3), and tet(X4) (He et al., 2019), were first detected in bacteria of animal origin. According to a retrospective screening project, the prevalence of tet(X)-positive isolates of animal source (6.9%) was much higher than that of human source (0.07%) (He et al., 2019). Recent studies also showed that the detection rate of tet(X) genes in isolates from animals (Cui et al., 2020; Li et al., 2020b,c) was higher than that from humans (Wang et al., 2019). Hence, it is critical to enrich more information about the animal source associated with tet(X)-bearing pathogens. The prevalence of tet(X) genes in swine farms and slaughterhouses has been systematically investigated (Li et al., 2020b,c), but the comprehensive molecular characterization of tet(X)-bearing bacteria of chicken was unexplored. In this study, we focused on the prevalence of tet(X) variants in cultivable bacteria among chicken fecal microbiota and demonstrated that tet(X) genes in diverse bacteria are worthy of continuous surveillance among different sources.
Materials and Methods
Sample Collection and Bacterial Isolates
A total of 45 chicken fecal samples were collected from a chicken farm in Jiangsu Province, China, in May 2020. We incubated 0.5 g feces in 5 ml of Tryptic Soy Broth (TSB) for 6 h to perform bacteria enrichment. The tet(X)-positive isolates were screened by Tryptic Soy Agar (TSA) plates supplemented with tigecycline (4 mg/L) and further identified by PCR using primers previously described (He et al., 2019). The species of all tet(X)-positive isolates were determined by 16S rRNA gene sequencing.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of all tet(X)-positive isolates were tested by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2018). Escherichia coli ATCC25922 was used for quality control. The resistance breakpoint for tigecycline was interpreted as >0.5 mg/L according to European Committee on Antimicrobial Susceptibility Testing (EUCAST)1.
Conjugation Experiments
In order to verify the transferability of tet(X) genes, we conducted conjugation experiments using E. coli C600 and a clinical carbapenem-resistant Acinetobacter baumannii 5AB as recipients. Briefly, the donor and recipient strains were cultured to the logarithmic growth phase with an optical density at 600 nm (OD600) of 0.4 in LB broth, mixed at a ratio of 1:1, and cultured overnight on TSB agar plates at 37°C. For the tet(X)-positive Acinetobacter, we also conducted the conjugation assay at 30°C. Then, the transconjugants were selected using TSA plates containing tigecycline (2 mg/L) and rifampin (300 mg/L) or meropenem (2 mg/L). And we further confirmed the recovered transconjugants by PCR for tet(X) and 16S rRNA genes. The frequencies of conjugation transfer were calculated by the number of transconjugants per recipient.
Genomic DNA Extraction and Whole-Genome Sequencing
Genomic DNA of tet(X)-positive isolates were extracted using FastPure Bacteria DNA Isolation Mini Kit (Vazyme™, Nanjing, China) following the manufacturer’s instruction. The quality and purity of genomic DNA were evaluated by Qubit 4 Fluorometer (Thermo Fisher Scientific™, Hennigsdorf, Germany) and Titertek-Berthold Colibri (Berthold™, Bad Wildbad, Germany). The genomic DNA of all tet(X)-positive isolates was subjected to the short-read sequencing (2 × 150 bp) by Illumina Hiseq 2500 platform. According to the assembly result of short-read sequencing, the genomic DNA of isolates with different tet(X) genetic contexts was further sequenced by long-read sequencing platform Oxford Nanopore Technologies MinION with a rapid barcoding library preparation strategy.
Bioinformatics Analysis
De novo short-read assembly was performed using SPAdes (Bankevich et al., 2012). The complete bacterial genomes were obtained using a hybrid assembly strategy combining long-read Nanopore and short-read Illumina sequencing data (Wick et al., 2017; Li R. et al., 2018). Antibiotic resistance genes, insertion sequence (IS) elements, and plasmid replicon types were identified by CGE services.2 The draft genomes were annotated by Prokka (Seemann, 2014). Functional annotation of the complete genome sequences was annotated automatically using the RAST3 and modified manually. Multilocus sequence typing (MLST) of assembled bacterial genomes was performed using the mlst tool4 and Pubmlst.5 The complete genomes of tet(X4)-bearing E. coli were downloaded from nr database in the National Center for Biotechnology Information (NCBI). The phylogenetic tree of strains was constructed using Roary and FastTree based on single-nucleotide polymorphisms (SNPs) of core genomes with default parameters (Price et al., 2009; Page et al., 2015). BRIG and Easyfig tools were used to visualize plasmid comparisons (Alikhan et al., 2011) and genetic context comparisons (Sullivan et al., 2011).
Results
Characterization of Tigecycline-Resistant Strains
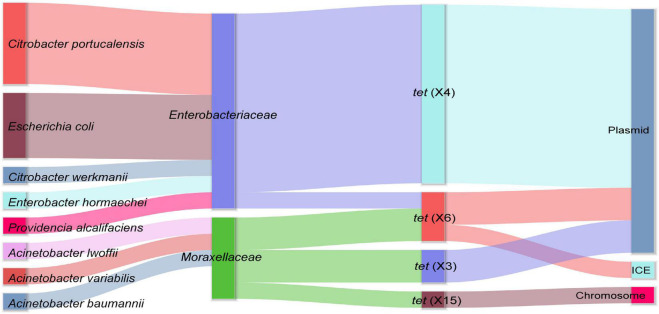
Out of 45 chicken fecal samples, a total of 15 tet(X)-positive strains were isolated from 13 samples (13/45, 28.89%). These tet(X)-positive strains consisted of eight different species including five Citrobacter portucalensis, four E. coli, one Enterobacter hormaechei, one Citrobacter werkmanii, one Acinetobacter variabilis, one Acinetobacter lwoffii, one A. baumannii, and one Providencia alcalifaciens. Meanwhile, four different tet(X) variants were detected in these strains, containing tet(X3), tet(X4), and tet(X6) reported previously and a novel tet(X) variant, designated as tet(X15) in another study (Li et al., 2021; Figure 1). Among them, tet(X4) carried by Enterobacteriaceae was the most pervasive. Although three different tet(X) variants, tet(X3), tet(X6), and the novel tet(X15), were found in Acinetobacter, all of them showed low prevalence. Notably, the phenomenon that such a number of tet(X) variants were distributed in bacteria with different species within a farm was not observed in other studies. The extensive prevalence of tet(X) genes in this chicken farm suggested that poultry may be an important reservoir of tet(X), and the tet(X) genes are likely to be transmitted to humans through environmental interactions and chicken consumption (Sun et al., 2020).
FIGURE 1.
The distribution of tet(X)-positive strains and the location of different tet(X) variants. The sankey diagram shows the host range diversity and genetic structure features of tet(X) variants investigated in this study.
Although tet(X4)-harboring E. coli was the most dominant in other research (He et al., 2019; Sun C. et al., 2019; Li et al., 2020a,b; Mohsin et al., 2021), tet(X4)-harboring E. coli of chicken source was rarely reported previously (Mohsin et al., 2021). To investigate the clonal relationship of tet(X4)-harboring E. coli between chicken and other sources, the genomes of 26 tet(X4)-positive E. coli with different hosts including pig, dog, chicken, cow, and human were downloaded from NCBI database and analyzed. Phylogenetic analysis based on the core genome indicated that the prevalence feature of tet(X4)-harboring E. coli has no clear clonal relationship with their sources (Supplementary Figure 1). It is worth noting that a tet(X4)-harboring E. coli we detected showed a close relationship with a tet(X4)-positive E. coli detected in human gut microbiota (Ding et al., 2020; Supplementary Figure 1). Hence, the serious prevalence of tet(X) of animal source has a potential threat to human health.
Antimicrobial Susceptibility Testing and Transfer of Different tet(X) Variants
Resistance phenotype analysis found that 15 tet(X)-positive strains showed resistance to multiple antibiotics and were all resistant to tigecycline and florfenicol (Table 1). In addition, most of them also conferred resistance to amoxicillin and enrofloxacin, but all strains were still susceptible to meropenem. Subsequently, we analyzed the distribution of resistance genes in tet(X)-positive strains according to draft genome sequences constructed by Illumina sequencing data. These strains contained multiple antibiotic resistance genes ranging from 8 to 19 (Supplementary Table 1). Besides, an extended-spectrum beta-lactamase (ESBL) gene blaCTX–M–55 and a carbapenemase gene blaOXA–58 were detected in some tet(X)-positive strains.
TABLE 1.
Antimicrobial susceptibility testing (MICs, mg/L) of 15 tet(X)-positive strains and their transconjugants.
| Strain | Source | Conjugation frequency to C600 | ST type | Species | Antimicrobials |
|||||||
| AMX | ENR | CFF | MEM | CL | KAN | TIG | FFC | |||||
| SC2-6 | Feces | / | 284 | Citrobacter portucalensis | >256 | 16 | 1 | ≤0.25 | ≤0.25 | >128 | 32 | >128 |
| cSC2-6 | Transconjugant | 2.8 × 10–9 | / | Escherichia coli | >256 | 16 | 1 | ≤0.25 | ≤0.25 | 4 | 16 | >128 |
| LHC3 | Feces | / | 284 | C. portucalensis | >256 | 16 | 1 | ≤0.25 | ≤0.25 | >128 | 64 | >128 |
| cLHC3 | Transconjugant | 1.9 × 10–9 | / | E. coli | >256 | 16 | 1 | ≤0.25 | ≤0.25 | 8 | 32 | >128 |
| LHC31-1 | Feces | / | 284 | C. portucalensis | >256 | 32 | 1 | ≤0.25 | ≤0.25 | >128 | 32 | >128 |
| cLHC31-1 | Transconjugant | 3.9 × 10–11 | / | E. coli | >256 | 4 | 1 | ≤0.25 | ≤0.25 | 4 | 8 | >128 |
| XMY1F802-7 | Feces | / | 284 | C. portucalensis | >256 | 16 | 0.5 | ≤0.25 | ≤0.25 | 4 | 64 | 128 |
| cXMY1F802-7 | Transconjugant | 3.9 × 10–11 | / | E. coli | >256 | 4 | 0.5 | ≤0.25 | ≤0.25 | 4 | 64 | >128 |
| XM10F302-7 | Feces | / | 284 | C. portucalensis | >256 | 32 | 1 | ≤0.25 | ≤0.25 | >128 | 128 | >128 |
| cXM10F302-7 | Transconjugant | 1.4 × 10–10 | / | E. coli | >256 | 32 | 1 | ≤0.25 | ≤0.25 | ≤0.25 | 64 | >128 |
| LHC5-1 | Feces | / | novel | Citrobacter werkmanii | >256 | 16 | 16 | ≤0.25 | ≤0.25 | 32 | 64 | >128 |
| cLHC5-1 | Transconjugant | 1.3 × 10–9 | / | E. coli | >256 | 0.5 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | 32 | >128 |
| XM3F402-1 | Feces | / | 93 | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | 8 | 32 | >128 |
| cXM3F402-1 | Transconjugant | 4 × 10–11 | / | E. coli | >256 | 0.5 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | 16 | >128 |
| XMC1F102-2 | Feces | / | 93 | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | 32 | >128 |
| cXMC1F102-2 | Transconjugant | NA | / | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | 16 | >128 |
| XM3F402-7 | Feces | / | 1,286 | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | >128 | 64 | >128 |
| cXM3F402-7 | Transconjugant | 4 × 10–11 | / | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 | 16 | >128 |
| XM7F102 | Feces | / | 155 | E. coli | >256 | 32 | >64 | ≤0.25 | ≤0.25 | 8 | 32 | >128 |
| cXM7F102 | Transconjugant | NA | / | E. coli | >256 | 2 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | 16 | >128 |
| LHC3-2 | Feces | / | 327 | Enterobacter hormaechei | >256 | 4 | 32 | ≤0.25 | ≤0.25 | 8 | 32 | >128 |
| cLHC3-2 | Transconjugant | NA | / | E. coli | >256 | 4 | ≤0.25 | ≤0.25 | ≤0.25 | 4 | 16 | 128 |
| LHC2-1 | Feces | / | / | Providencia alcalifaciens | >256 | 8 | 32 | ≤0.25 | >128 | >128 | >128 | >128 |
| XM9F202-2 | Feces | / | / | Acinetobacter variabilis | 2 | 4 | 4 | ≤0.25 | ≤0.25 | >128 | 32 | >128 |
| XMC5X702 | Feces | / | / | Acinetobacter lwoffii | 128 | 0.5 | 1 | ≤0.25 | ≤0.25 | 8 | 32 | >128 |
| LHC22-2 | Feces | / | 1,459 | Acinetobacter baumannii | >256 | 32 | 32 | 1 | ≤0.25 | 16 | 64 | 128 |
NA, not available. The transfer frequencies of these samples were too low to be calculated accurately.
MICs, minimum inhibitory concentrations; ST, sequence typing; AMX, amoxicillin; ENR, enrofloxacin; CFF, ceftiofur; MEM, meropenem; CL, colistin; KAN, kanamycin; TIG, tigecycline; FFC, florfenicol.
To investigate the transmissibility of different tet(X) variants, all strains were performed by conjugation assay. All tet(X4) genes in this study were successfully transferred into the recipient E. coli C600 with low frequencies, resulting in resistance to tigecycline in transconjugants. The remaining tet(X)-positive strains failed in conjugation assay. The higher horizontal transfer percentage of tet(X4) might explain its high prevalence.
The Genetic Contexts of tet(X) Variants
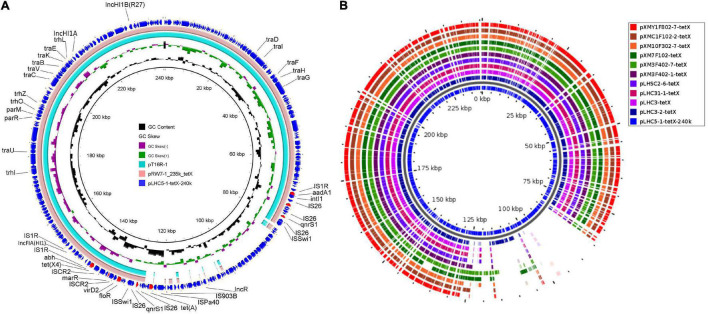
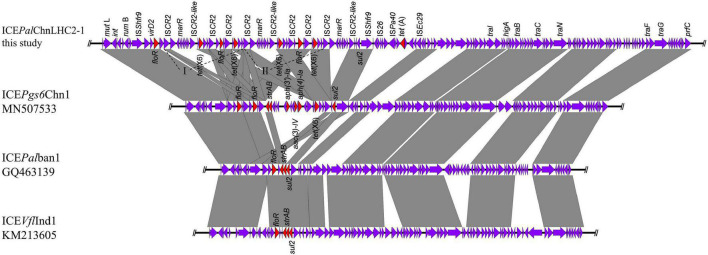
In order to investigate the genetic contexts of different tet(X) variants, five strains (one C. werkmanii LHC5-1, one P. alcalifaciens, and three Acinetobacter) were performed with Nanopore long-read sequencing to obtain complete genomes together with short-read data (Supplementary Table 1). Genetic analysis of strain LHC5-1 found that tet(X4) gene was located in a 240-kb IncFIA(HI1)/IncHI1A/IncHI1B(R27)/IncR hybrid plasmid, named pLHC5-1-tetX-240k. Many plasmids with a similar structure to pLHC5-1-tetX-240k were found in the NCBI nr database (Figure 2A), and most of these plasmids were positive for tet(X4) and harbored by E. coli. The emergence of tet(X4)-bearing IncFIA(HI1)/IncHI1A/IncHI1B(R27)/IncR plasmid in Citrobacter spp. exacerbated the transmission of tet(X4) among different species of bacteria. Comparative analysis of plasmid pLHC5-1-tetX-240k and other similar hybrid plasmids found that a ca. 190-kb backbone region with replicons IncFIA(HI1)/IncHI1A/IncHI1B(R27) in these hybrid plasmids was conserved (Figure 2A). Some small plasmids with replicons IncX1, IncX4, and IncR could integrate into a hybrid plasmid with replicons IncFIA(HI1)/IncHI1A/IncHI1B(R27) and form larger and more complex plasmids, such as pRW7-1_235k_tetX (Li et al., 2020b). Subsequently, we investigated the genetic feature of tet(X4) in other strains in this study. The result showed that all tet(X4) genes in this study were carried by plasmids with a similar backbone to pLHC5-1-tetX-240k and located in a conserved ca. 190-kb region harboring IncFIA(HI1)/IncHI1A/IncHI1B(R27) plasmid replicons (Figure 2B). In addition, we found that these hybrid plasmids were widely distributed in different species of bacteria. Therefore, the diffusion of tet(X4) was strongly associated with the IncFIA(HI1)/IncHI1A/IncHI1B(R27) hybrid plasmids and their evolved complex plasmids. Apart from these tet(X4)-bearing plasmids in Enterobacteriaceae, one tet(X6) gene was detected in a P. alcalifaciens of Enterobacteriaceae for the first time. tet(X6) gene was located in variable region III (VRIII) of a chromosomal integrative and conjugative element (ICE), designated as ICEPalChnLHC2-1. A total of four tet(X6) genes were detected in VRIII within two tandem repeat units (Figure 3). Although tandem repeats of different tet(X) variants were frequently observed, two tet(X6) in one repeat unit have not been reported. The molecular mechanism of tet(X) tandem repeat deserved further investigations. Then, we searched for homologous ICEs with ICEPalChnLHC2-1 in the NCBI database, and three tet(X)-negative ICEs from Proteus genomosp, P. alcalifaciens, and Vibrio fluvialis were downloaded and compared. The four ICEs showed high similarity with each other, which implied that they originated from one ancestor and were popular because of horizontal transfer between different bacterial chromosomes. Meanwhile, we observed an evolution of genetic context in VRIII of the four ICEs, which was a manifestation of the adaptation of bacteria to changes in the external environment.
FIGURE 2.
Structure analysis of tet(X4)-bearing plasmids. (A) Comparison analysis of the plasmid pLHC5-1-tetX-240k with other similar plasmids including pRW7-1_235k_tetX (GenBank accession_number: MT219825) and pT16R-1 (CP046717). (B) Structure features of tet(X4)-bearing plasmids carried by Enterobacteriaceae in this study. The structural diversity of these plasmids existed within a multidrug-resistant (MDR) region. Resistance genes in plasmid pLHC5-1-tetX-240k were highlighted in red arrows. The reference sequence in panel (B) is pLHC5-1-tetX-240k, and colored circles indicate the sequences in draft genomes, which are mapped to the reference sequence.
FIGURE 3.
Linear comparison of the tet(X6)-bearing integrative and conjugative element (ICE) ICEPalChnLHC2-1 with other similar ICEs. The multidrug-resistant (MDR) region encoding tet(X6) was inserted into variable region III conserved in ICEs.
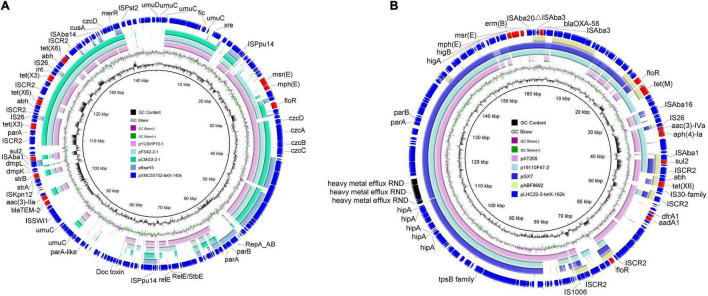
Although only three tet(X)-positive strains belonging to Moraxellaceae were identified, complex genetic contexts of tet(X) variants were found in the three strains. Co-occurrence of two different tet(X) variants in one strain was detected in strains XM9F202-2 and XMC5X702. A plasmid-mediated tet(X3) and a chromosomal novel tet(X) variant, designated as tet(X15), were found in A. variabilis XM9F202-2, which has been investigated in detail in our previous study (Li et al., 2021). In A. lwoffii XMC5X702, two different tet(X) variants corresponded to tet(X3) and tet(X6). Genetic analysis found that tet(X3) and tet(X6) were located on a 145-kb plasmid pXMC5X702-tetX-145k with unknown replicon types. Multiple plasmids co-harboring tet(X3) and tet(X6) carried by Acinetobacter were found in the nr database, and they share more than 50% coverage and more than 95% identify to pXMC5X702-tetX-145k (Figure 4A). However, the replicon gene in pXMC5X702-tetX-145k differed from those plasmids co-harboring tet(X3) and tet(X6). The plasmids that harbor the same replicon as pXMC5X702-tetX-145k showed low identity to pXMC5X702-tetX-145k. Hence, the structure of pXMC5X702-tetX-145k was novel, and it enriched the profile of tet(X)-bearing plasmids in Acinetobacter. tet(X6) gene in A. baumannii LHC22-2 was carried by a 162-kb plasmid pLHC22-2-tetX-162k. What is noteworthy is that a carbapenemase gene blaOXA–58 was found in the tet(X6)-bearing plasmid. Although many plasmids co-harboring tet(X3) and blaOXA–58 have been reported in other species of Acinetobacter (Cui et al., 2020; Ma et al., 2020), tet(X6)- and blaOXA–58-bearing plasmid was rarely reported (Zheng et al., 2020). As a clinically critical opportunistic pathogen, the emergence of carbapenem- and tigecycline-resistant A. baumannii poses a great threat to public health. Phenotype analysis of antimicrobial resistance showed that LHC22-2 was resistant to imipenem but susceptible to meropenem. The expression of blaOXA–58 could be enhanced by an intact upstream ISAba3 and result in resistance to meropenem (Hamidian and Nigro, 2019), but ISAba3 in plasmid pLHC22-2-tetX-162k was truncated. Subsequently, one plasmid pABF9692 co-harboring tet(X6)- and blaOXA–58 from A. baumannii and three plasmids with different sizes from Acinetobacter towneri showing similar backbone with pLHC22-2-tetX-162k were retrieved from the nr database and analyzed (Figure 4B). Notably, the backbone of pABF9692 was different with pLHC22-2-tetX-162k. In contrast, the three plasmids co-harboring tet(X3) and blaOXA–58 showed similar backbone with pLHC22-2-tetX-162k (Figure 4B). Two different tet(X) variants, tet(X3) and tet(X6), were detected in these plasmids, which indicated that such plasmids played a vital role in capturing and propagating the tet(X) genes.
FIGURE 4.
Circular comparisons between tet(X)-positive plasmids of Acinetobacter origin in this study and similar plasmids in the National Center for Biotechnology Information (NCBI) database. (A) Comparative analysis of pXMC5X702-tetX-145k with four closely related plasmids including pBspH3 (GenBank accession_number: CP055285), pCMG3-2-1 (CP044446), pFS42-2-1 (CP046596), and pYUSHP10-1 (MT107270). pXMC5 × 102-tetX-145k was used as the reference plasmid. (B) Comparative analysis of pLHC22-2-tetX-162k with four closely related plasmids including pABF9692 (CP048828), pGX7 (CP071772), p19110F47-2 (CP046044), and pAT205 (CP048015). Plasmid pBspH3 in panel (A) shared the same replicon gene with plasmid pXMC5X702-tetX-145k. Plasmid pABF9692 co-harbored tet(X6) and blaOXA–58 but shared limited homologous regions with plasmid pLHC22-2-tetX-162k.
The Core Genetic Structures of tet(X) Variants in This Study
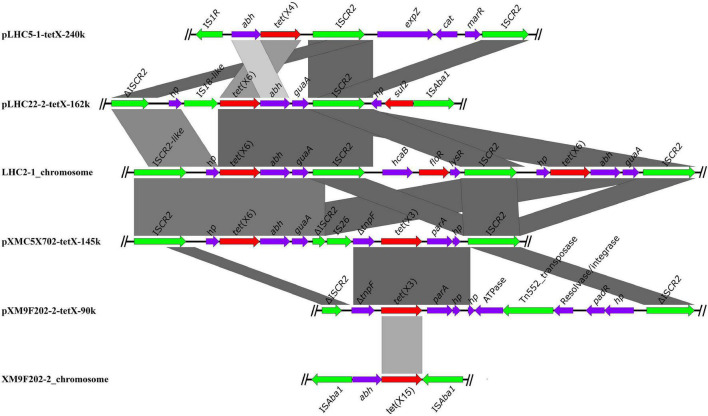
The different tet(X) variants were harbored by various genetic structures and distributed in different bacteria in the chicken farm. However, ISCR2 was always associated with different tet(X) variants except for the novel tet(X15) (Figure 5). This phenomenon was consistent with previous studies (He et al., 2019; Li et al., 2020b,c), implying that ISCR2 was a major driving factor for the dissemination of tet(X) variants. We also found many other IS elements in the surroundings of different tet(X) variants, such as ISAba1 in the downstream of tet(X6) in plasmid pLHC22-2-tetX-162k and IS26 in the upstream of tet(X3) in plasmid pXMC5X702-tetX-145k. These IS elements will probably be involved in the transfer of tet(X) variants and hereby have evolved novel genetic context of tet(X) variants. Apart from ISCR2-associated tet(X)-bearing genetic contexts, we identified a novel tet(X15) located in an ISAba1-bound composite transposon Tn6866 (Li et al., 2021). The ISAba1 in the composite transposon Tn6866 is directly responsible for the movement of tet(X15), which differs from that of tet(X6). Therefore, monitoring the genetic context of tet(X) variants is important for understanding their transmission and evolution destiny.
FIGURE 5.
The core genetic structures of tet(X) investigated in this study. The resistance genes are shown as red arrows, and the mobile elements are shown as green arrows.
Discussion
The emergence of high-level tigecycline resistance genes tet(X3) and tet(X4) has caused great concern throughout the world. A large number of tet(X) variants, from tet(X3) to tet(X44), were identified from different bacteria in humans and animals within 2 years (Wang et al., 2019; Cheng et al., 2020; Gasparrini et al., 2020; Peng et al., 2020; Zheng et al., 2020; Umar et al., 2021). The current outbreak and widespread situation of tet(X) is rapidly diminishing the effectiveness of tetracycline antibiotics, including tigecycline and the US Food and Drug Administration (FDA) newly approved eravacycline and omadacycline. Tetracyclines have been used in livestock farms for many years in China. However, few studies investigated the epidemiological and genetic features of tet(X) in livestock farms, with limited research focusing on the tet(X)-bearing Enterobacterales or Acinetobacter (Cui et al., 2020; Li et al., 2020c). Meanwhile, the prevalence of tet(X) in bacteria of chicken origin has not been investigated fully to date. In this work, we systematically explored the distribution and genetic characteristics of different tet(X) variants and their host bacteria in a chicken farm. We found that the prevalence of mobilizable tet(X4) was the highest and more worrisome than that of other variants. Apart from E. coli, Citrobacter spp. was also an emerging host for tet(X4). The high prevalence of tet(X4) might be associated with their host plasmids. Although only three tet(X)-positive strains belonging to Acinetobacter spp. were identified, three different tet(X) variants were identified in them. The epidemic pattern of tet(X) in Acinetobacter differed from that in Enterobacterales, and the relationship between them warrants further investigations.
Genetic analysis found that plasmids are an important vector for the dissemination of tet(X). However, some chromosomal mobile elements, such as ICEs and transposons, also contribute to the transfer of tet(X). According to transfer experiments, all tet(X4)-positive plasmids in Enterobacterales could transfer to E. coli C600, and the other tet(X)-bearing genetic structures in Enterobacterales and Acinetobacter failed to transfer in conjugation assay. The phenomenon explained the high prevalence of tet(X4) in the chicken farm and demonstrated that the prevalence of tet(X) genes was positively related to the horizontal transferability of their vectors within specific bacterial hosts. Notably, the transmission of tet(X4) was associated with various plasmids reported in our previous study (Li et al., 2020b). In this study, we first noticed that serious prevalence of tet(X4) in different bacteria mediated by IncFIA(HI1)/IncHI1A/IncHI1B(R27) plasmids occurred in the chicken farm. Currently, the worldwide dissemination of critical resistance genes was possible with the help of some common types of plasmids, such as blaNDM–5-positive IncX3 plasmid (Li X. et al., 2018; Liu et al., 2019; Zhao et al., 2021) and mcr-1-positive IncI2 plasmid (Elbediwi et al., 2019; Gelbicova et al., 2019; Lu et al., 2020). Hence, the emergence of tet(X4)-positive common plasmids with high mobility might cause an increasing prevalence of tet(X4). In addition, we found that all tet(X)-positive plasmids in Acinetobacter in the chicken farm had no ability of horizontal transfer, which is consistent with the previous reports (Cui et al., 2020; Ma et al., 2020; Wang et al., 2020). Genetic structure analysis found that those tet(X) genes in plasmids harbored by Acinetobacter were adjacent to ISCR2, indicating that ISCR2-mediated mobilization of tet(X) also deserved concerns among bacteria of different genus.
In conclusion, we comprehensively investigated the prevalence of tet(X) in a chicken farm first and identified multiple tet(X) variants from diversified bacteria. The prevalence of tet(X4) in the chicken farm was mainly determined by their host plasmids. The Acinetobacter spp. is an important reservoir for other tet(X) variants. Apart from ISCR2, ISAba1 might also be an important element for the mobilization of tet(X). Therefore, we propose that effective measures should be formulated to decelerate the dissemination of tet(X) in animal- and human-associated environments.
Data Availability Statement
The sequences obtained in this article have been deposited in the GenBank database under BioProject number: PRJNA750704.
Author Contributions
RL and ZW conceived and designed the experiments, and manuscript reviewing and editing. YL and KP conducted the experiments, analyze the data, and wrote the draft. YY, XS, and WZ conducted long-read sequencing and bioinformatics analysis. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported in part by the National Natural Science Foundation of China (31872526 and 31872523) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.751006/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Chen Y., Liu Y., Guo Y., Zhou Y., Xiao T., et al. (2020). Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg. Microbes Infect. 9 1843–1852. 10.1080/22221751.2020.1803769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. CLSI supplement M100. [Google Scholar]
- Cui C. Y., Chen C., Liu B. T., He Q., Wu X. T., Sun R. Y., et al. (2020). Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob. Agents Chemother. 64:e02502-19. 10.1128/AAC.02502-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Saw W. Y., Tan L. W. L., Moong D. K. N., Nagarajan N., Teo Y. Y., et al. (2020). Emergence of tigecycline- and eravacycline-resistant tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J. Antimicrob. Chemother. 75 3480–3484. 10.1093/jac/dkaa372 [DOI] [PubMed] [Google Scholar]
- Elbediwi M., Li Y., Paudyal N., Pan H., Li X., Xie S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms 7:461. 10.3390/microorganisms7100461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparrini A. J., Markley J. L., Kumar H., Wang B., Fang L., Irum S., et al. (2020). Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun. Biol. 3:241. 10.1038/s42003-020-0966-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbicova T., Barakova A., Florianova M., Jamborova I., Zelendova M., Pospisilova L., et al. (2019). Dissemination and comparison of genetic determinants of mcr-mediated colistin resistance in Enterobacteriaceae via retailed raw meat products. Front. Microbiol. 10:2824. 10.3389/fmicb.2019.02824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Nigro S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant. Acinetobacter baumannii. Microb. Genom. 5:e000306. 10.1099/mgen.0.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Wang L., Zhao S., Liu L., Liu J., Hu G., et al. (2020). A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J. Antimicrob. Chemother. 75 1159–1164. 10.1093/jac/dkaa012 [DOI] [PubMed] [Google Scholar]
- He T., Wang R., Liu D., Walsh T. R., Zhang R., Lv Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4 1450–1456. 10.1038/s41564-019-0445-2 [DOI] [PubMed] [Google Scholar]
- Li R., Lu X., Peng K., Liu Z., Li Y., Liu Y., et al. (2020b). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-Bearing plasmidome among bacteria. mSystems 5:e00134-20. 10.1128/mSystems.00134-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Peng K., Li Y., Liu Y., Wang Z. (2020c). Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci. Total Environ. 733:139306. 10.1016/j.scitotenv.2020.139306 [DOI] [PubMed] [Google Scholar]
- Li R., Lu X., Liu Z., Liu Y., Xiao X., Wang Z. (2020a). Rapid detection and characterization of tet(X4)-positive Escherichia coli strains with nanopore sequencing. J. Antimicrob. Chemother. 75 1068–1070. 10.1093/jac/dkz528 [DOI] [PubMed] [Google Scholar]
- Li R., Peng K., Xiao X., Wang Y., Wang Z. (2021). Characterization of novel ISAba1-bounded tet(X15)-bearing composite transposon Tn6866 in Acinetobacter variabilis. J. Antimicrob. Chemother. 76 2481–2483. [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Wong M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Fu Y., Shen M., Huang D., Du X., Hu Q., et al. (2018). Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7:59. 10.1186/s13756-018-0349-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liu Z., Peng K., Liu Y., Xiao X., Wang Z. (2019). Co-occurrence of two tet(X) variants in an Empedobacter brevis of shrimp origin. Antimicrob. Agents Chemother. 63:e01636-19. 10.1128/AAC.01636-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zhai W., Song H., Fu Y., Schwarz S., He T., et al. (2020). Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J. Antimicrob. Chemother. 75 1428–1431. 10.1093/jac/dkaa037 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Li Y., Liu Y., Li R., Wang Z. (2019). Emergence of IncX3 Plasmid-Harboring bla NDM-5 Dominated by Escherichia coli ST48 in a Goose Farm in Jiangsu. China. Front. Microbiol. 10:2002. 10.3389/fmicb.2019.02002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Xiao X., Liu Y., Huang S., Li R., Wang Z. (2020). Widespread prevalence of plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli from Pere David’s Deer in China. mSphere 5:e01221-2. 10.1128/mSphere.01221-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wang J., Feng J., Liu Y., Yang B., Li R., et al. (2020). Characterization of three porcine Acinetobacter towneri strains co-harboring tet(X3) and bla OXA-58. Front. Cell Infect. Microbiol. 10:586507. 10.3389/fcimb.2020.586507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsin M., Hassan B., Martins W., Li R., Abdullah S., Sands K., et al. (2021). Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia. Sci. Total Environ. 787:147613. 10.1016/j.scitotenv.2021.147613 [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Li R., He T., Liu Y., Wang Z. (2020). Characterization of a porcine Proteus cibarius strain co-harbouring tet(X6) and cfr. J. Antimicrob. Chemother. 75 1652–1654. 10.1093/jac/dkaa047 [DOI] [PubMed] [Google Scholar]
- Pournaras S., Koumaki V., Spanakis N., Gennimata V., Tsakris A. (2016). Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int. J. Antimicrob. Agents 48 11–18. 10.1016/j.ijantimicag.2016.04.017 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Cui M., Zhang S., Wang H., Song L., Zhang C., et al. (2019). Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008-2018. Emerg. Microbes Infect. 8 1524–1527. 10.1080/22221751.2019.1678367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen C., Cui C. Y., Zhang Y., Liu X., Cui Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4 1457–1464. 10.1038/s41564-019-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liao X. P., D’souza A. W., Boolchandani M., Li S. H., Cheng K., et al. (2020). Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 11:1427. 10.1038/s41467-020-15222-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Cai Y., Liu X., Bai N., Liang B., Wang R. (2013). The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 41 110–116. [DOI] [PubMed] [Google Scholar]
- Umar Z., Chen Q., Tang B., Xu Y., Wang J., Zhang H., et al. (2021). The poultry pathogen Riemerella anatipestifer appears as a reservoir for Tet(X) tigecycline resistance. Environ. Microbiol. 10.1111/1462-2920.15632 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Wu H., Wang Z. Y., Shen P. C., Tian Y. Q., et al. (2020). Coexistence of blaOXA-58 and tet(X) on a Novel Plasmid in Acinetobacter sp. from Pig in Shanghai, China. Front. Microbiol. 11:578020. 10.3389/fmicb.2020.578020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu D., Lv Y., Cui L., Li Y., Li T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 64:e01326-19. 10.1128/AAC.01326-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Berglund B., Zou H., Zhou Z., Xia H., Zhao L., et al. (2021). Dissemination of blaNDM-5 via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ. Pollut. 273:116370. 10.1016/j.envpol.2020.116370 [DOI] [PubMed] [Google Scholar]
- Zheng X. R., Zhu J. H., Zhang J., Cai P., Sun Y. H., Chang M. X., et al. (2020). A novel plasmid-borne tet(X6) variant co-existing with blaNDM-1 and blaOXA-58 in a chicken Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 75 3397–3399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained in this article have been deposited in the GenBank database under BioProject number: PRJNA750704.