Abstract
Long non-coding RNAs (lncRNAs) can exhibit cell-type and cancer-type specific expression profiles, making them highly attractive as therapeutic targets. Pan-cancer RNA sequencing data revealed broad expression of the SAMMSON lncRNA in uveal melanoma (UM), the most common primary intraocular malignancy in adults. Currently, there are no effective treatments for UM patients with metastatic disease, resulting in a median survival time of 6–12 months. We aimed to investigate the therapeutic potential of SAMMSON inhibition in UM. Antisense oligonucleotide (ASO)-mediated SAMMSON inhibition impaired the growth and viability of a genetically diverse panel of uveal melanoma cell lines. These effects were accompanied by an induction of apoptosis and were recapitulated in two uveal melanoma patient derived xenograft (PDX) models through subcutaneous ASO delivery. SAMMSON pulldown revealed several candidate interaction partners, including various proteins involved in mitochondrial translation. Consequently, inhibition of SAMMSON impaired global, mitochondrial and cytosolic protein translation levels and mitochondrial function in uveal melanoma cells. The present study demonstrates that SAMMSON expression is essential for uveal melanoma cell survival. ASO-mediated silencing of SAMMSON may provide an effective treatment strategy to treat primary and metastatic uveal melanoma patients.
Subject terms: Targeted therapies, Non-coding RNAs
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, with an incidence of 5−7.4 cases per million annually [1–3]. Current treatments consist of radiotherapy and enucleation, but despite the advances in local therapy towards eye-preserving therapeutic choices, no substantial progress in overall survival has been achieved. The main cause of death of UM patients is due to the metastatic dissemination, mainly to the liver, in about 50% of the patients. Patients with metastatic UM have extremely poor survival, with a median survival time of 6−12 months [4]. Uveal melanoma is a genetically and biologically distinct type of melanoma that arises from choroidal melanocytes in the choroidal plexus, ciliary body and iris of the eye. In addition, UM and cutaneous melanoma differ in their chromosomal aberrations and mutational signature. Recurrent genomic aberrations in UM include loss of 1p, monosomy of chromosome 3, loss of 6q and 8p and gain of 6p and 8q, of which loss of chromosome 3 and gain of 8q have been associated with a high mortality rate [5, 6]. Tumor cells are characterized by activated G protein-coupled receptor (GPCR) signaling, which occurs in almost all UM tumors by specific mutations that have been related to UM, such as mutations in GNAQ or GNA11 (90% of UM tumors). These mutations result in the activation of the pathways downstream of Gαq and Gα11 such as the Ras Homolog Family Member/Ras-related C3 botulinum toxin/Yes Associated Protein (Rho/Rac/YAP) pathway, Phosphatidylinositol (4,5)-bisphosphate 3-kinase (PI3K)/AKT pathway and Phospholipase C (PLC) which subsequently activates the Mitogen-Activated Protein Kinase (MAPK) pathway (RAF/MEK/ERK) [7]. Although GNAQ and GNA11 mutations occur in most UM, they are not associated with metastasis [8]. In addition, inactivating somatic mutations in BRCA1-associated protein 1 (BAP1) have been frequently detected in UM tumors and are associated with the development of metastasis [9]. Tumor suppressor gene BAP1, located on 3p21.1, is frequently mutated on the remaining allele present in monosomy 3 tumors, which might explain the poor prognosis of monosomy 3 tumors [10]. Loss of BAP1 affects several pathways, including DNA damage repair, cell cycle regulation, cell differentiation and cell growth [11, 12]
Long non-coding RNAs are an emerging class of regulatory RNA molecules that interact with proteins, DNA, and other RNA molecules. Compared to protein-coding mRNAs, lncRNAs show a more tissue restricted expression profile, making them attractive targets for therapy. LncRNAs are regulating a variety of cellular functions such as transcription, splicing, mRNA stability and translation, and do so via different mechanisms. Recently, Survival Associated Mitochondrial Melanoma Specific Oncogenic Non-coding RNA (SAMMSON) was discovered on chromosome 3p13 as a lineage survival oncogene in skin melanoma [13]. Interaction of SAMMSON with proteins involved in ribosomal RNA (rRNA) maturation and protein synthesis such as p32 (C1QBP), CARF and XRN2 and sequestration of CARF in the cytoplasm result in an elevated cytosolic and mitochondrial rRNA processing and protein synthesis [14]. In this study, we show that SAMMSON is consistently expressed in UM and conjunctival melanoma (CM) cells, the latter of which are phenotypically and genetically more related to skin melanoma. SAMMSON silencing, by means of locked nucleid acid (LNA) antisense oligonucleotides (ASOs), revealed an essential role for SAMMSON in UM survival in vitro and in vivo and provides perspectives for RNA-targeted therapy.
Results
SAMMSON is consistently expressed in uveal melanoma tumors
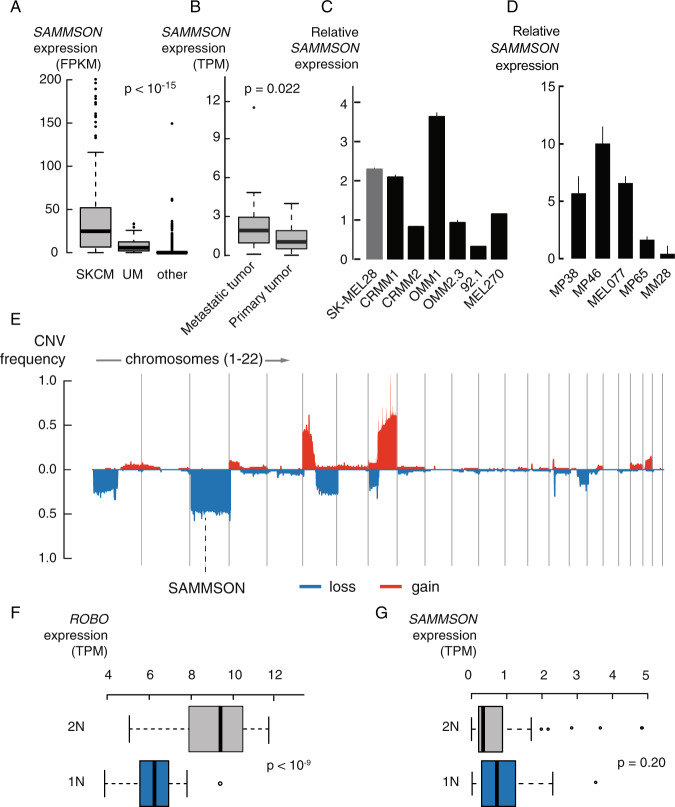
Pan cancer RNA sequencing data from more than 10 000 tumor samples representing 32 cancer types (The Cancer Genome Atlas, TCGA) showed the highest and most consistent SAMMSON expression in skin melanoma (SKCM, SAMMSON expression in >90% of tumor samples) followed by uveal melanoma (SAMMSON expression in >80% of tumor samples) (Fig. 1A, p < 10−15, Mann–Whitney test). While SAMMSON expression in uveal melanoma tumors is independent from patient survival, tumor stage, primary tumor localization site (choroid, ciliary body or iris) and metastatic stage of the patient, SAMMSON expression is elevated in metastatic tumors compared to matched primary tumors (Fig. 1B, p = 0.022, Wilcoxon matched-pairs signed rank test, Supplemental Fig. 1). SAMMSON expression was further verified by RT-qPCR in various UM cell lines originating from primary tumors (92.1 and MEL270) and metastatic tumors (OMM2.3 and OMM1) (Fig. 1C), as well as in UM PDX-derived cell lines (MP38 (BAP1 negative, monosomy 3), MP46 (BAP1 negative, monosomy 3), MEL077, MP65 (BAP1 negative, monosomy 3), and MM28) (Fig. 1D). SAMMSON expression was also detected in the conjunctival melanoma (CM) cell lines CRMM1 and CRMM2 that are genetically and phenotypically more related to skin melanoma [15] (Fig. 1C). In contrast to skin melanoma tumors, where SAMMSON is invariably co-amplified with Melanocyte Inducing Transcription Factor (MITF) on chromosome 3, uveal melanoma cells are characterized by frequent (50−60%) loss of an entire copy of chromosome 3 (ref. [6, 16]) (Fig. 1E). While the majority of genes expressed from chromosome 3, for example Roundabout Guidance Receptor 1 (ROBO1) (Fig. 1F, p < 10−9, Mann-Whitney test), are significantly downregulated in monosomy 3 UM tumors (Supplemental Fig. 2, 89% downregulated genes (n = 581), 81% of those genes are significantly downregulated (n = 472/581)), SAMMSON expression is independent of chromosome 3 copy number (Fig. 1G, p = 0.20, Mann–Whitney test). This suggests a compensation mechanism by which UM tumors cells maintain high SAMMSON levels in the presence of chromosome 3 loss.
Fig. 1. LncRNA SAMMSON is consistently expressed in UM tumors.
A RNA sequencing data from >10,000 tumor samples and 32 cancer types (TCGA) showing SAMMSON expression in skin melanoma (SKCM), uveal melanoma (UM) and other cancer types (p < 10−15, Mann–Whitney test). B RNA sequencing data showing elevated SAMMSON expression levels in metastatic UM tumors compared to matching primary tumors (n = 21, p = 0.022, Wilcoxon matched-pairs signed rank test). C Relative SAMMSON expression in CM cell lines (CRMM1, CRMM2) and UM cell lines (OMM1, OMM2.3, 92.1 and MEL270) compared to skin melanoma cell line SK-MEL28. Error bars represent ± standard error (SE) of qPCR replicates. D Relative SAMMSON expression in UM PDX lines. Error bars represent ± standard error (SE) of qPCR replicates. E Frequent chromosomal aberrations in UM tumors and the location of SAMMSON on chromosome 3, which is frequently lost. F Significantly lower ROBO1 expression levels in monosomy 3 (1 N, n = 36) tumors compared to disomy 3 (2 N, n = 44) tumors (p < 10−9, Mann–Whitney test). G No difference in SAMMSON expression levels between disomy 3 (2 N, n = 44) and monosomy 3 (1 N, n = 36) tumors (p = 0.20, Mann–Whitney test).
SAMMSON expression is required for UM and CM cell survival in vitro
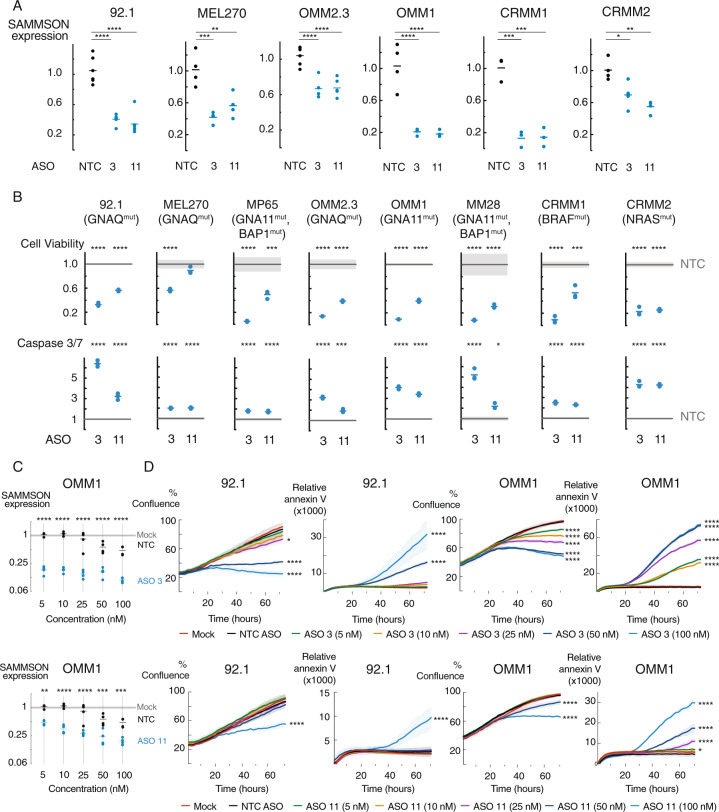
To evaluate the importance of SAMMSON in UM and CM, we studied the effects of SAMMSON knockdown in various UM and CM cell lines using two independent SAMMSON targeting ASOs (ASO 3 and ASO 11). Both ASOs (ASO 3 i.e. GapmeR3 and ASO 11 i.e. GapmeR11) have previously been validated by Leucci et al. [13] for its efficient knockdown performance without toxic effects in SAMMSON negative cell lines. Transfection of 100 nM of both SAMMSON inhibiting ASOs significantly decreased SAMMSON expression in four UM (92.1, MEL270, OMM2.3 and OMM1) and two CM cell lines (CRMM1 and CRMM2) (Fig. 2A, p < 0.05, one-way ANOVA). SAMMSON knock-down resulted in a strong and significant decrease in cell viability (based on ATP measurements) in multiple UM and two CM cell lines (Fig. 2B, p≤0.001 for all except ASO 11 in MEL270, one-way ANOVA). These effects were accompanied by the induction of apoptosis, as evidenced by a significant increase in caspase-3/7 levels, observed for both SAMMSON inhibiting ASOs in 8 different UM and CM cell lines (Fig. 2B, p < 0.05, one-way ANOVA). In line with earlier observations in skin melanoma cells [13], these effects were independent of the mutational status (GNAQ, GNA11, BAP1, BRAF and NRAS) of the UM and CM cell lines.
Fig. 2. SAMMSON knockdown reduces cell viability and confluence and induces apoptosis.
A Relative SAMMSON expression 48 h after transfection in four UM cell lines (92.1, MEL270, OMM2.3, OMM1) and two CM cell lines (CRMM1, CRMM2) transfected with 100 nM of a scrambled ASO (NTC), ASO 3 or ASO 11. The individual data points and mean are presented. B Cell viability (Cell Titer Glo) reduction and apoptosis (Caspase-Glo 3/7) induction in ASO 3 and ASO 11 treated UM and CM (CRMM1 and CRMM2) cells compared to NTC ASO treated cells (72 h post-transfection). The individual data points and mean of ASO 3 and ASO11 are presented. The NTC ASO treated data are presented as the mean of three replicates ± s.d. P values in (A) and (B) were calculated for each ASO 3/ASO 11 concentration and compared to NTC ASO using one-way ANOVA with Dunnett’s multiple testing correction. C Relative SAMMSON expression in UM cell line OMM1 48 h after transfection with 5, 10, 25, 50, or 100 nM of NTC ASO, ASO 3 or ASO 11 and scaled to untreated cells (Mock). The individual data points and mean of NTC ASO, ASO 3 and ASO11 are presented. The mock treated data are presented as the mean of four replicates ± s.d. P values were calculated for each ASO 3/ASO 11 concentration and compared to its corresponding NTC ASO concentration using one-way ANOVA with Dunnett’s multiple testing correction. D Reduction of proliferation (% confluence) and induction of apoptosis (annexin V) in 92.1 and OMM1 upon transfection with 5, 10, 25, 50, or 100 nM ASO 3 or ASO 11 compared to NTC ASO treatment or untreated cells (mock) measured by time-lapse microscopy (every 2–3 h) using the IncuCyte device. Data represents mean ± s.d. of three replicates. NTC ASO data represents the mean ± s.d. of all NTC ASO concentrations (5, 10, 25, 50, and 100 nM). P values were calculated at the 72 h time point compared to NTC ASO using one-way ANOVA with Dunnett’s multiple testing correction. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
To evaluate whether SAMMSON knockdown depends on ASO dosing, we performed a dose response experiment with both ASOs in the UM cell line OMM1 and CM cell line CRMM1. We observed a dose dependent decrease in SAMMSON expression for both ASOs (Fig. 2C and Supplemental Fig. 3A, p ≤ 0.0001 and p ≤ 0.001 for all tested concentrations of ASO 3 in OMM1 and CRMM1, respectively and p ≤ 0.01 for all tested concentrations of ASO 11 in OMM1 and CRMM1, except 5 nM in CRMM1, one-way ANOVA). We also observed a dose dependent reduction in cell growth and proliferation in UM cell lines OMM1, 92.1 and CM cell line CRMM1, measured via real-time cell imaging (Fig. 2D and Supplemental Fig. 3B, p < 0.05 for the three highest ASO 3 concentrations and the highest ASO 11 concentration in 92.1, p ≤ 0.0001 for all ASO 3 concentrations and the 2 highest ASO 11 concentrations in OMM1 and p ≤ 0.0001 for the three highest ASO three and two highest ASO 11 concentrations in CRMM1, one-way ANOVA). These effects were accompanied by a dose dependent induction in apoptosis, measured by annexin V, in both UM cell lines (Fig. 2D, p ≤ 0.0001 for the two highest ASO 3 and highest ASO 11 concentration in 92.1 and p < 0.05 for all ASO 3 concentrations and the four highest ASO 11 concentrations in OMM1, one-way ANOVA).
To verify that the observed effects were not related to the lipid-based delivery of the ASOs, we evaluated an alternative non-lipid-based delivery method using the TransIT-X2 transfection reagent. Transfection of 100 nM of ASO 3 significantly decreased SAMMSON expression in 92.1 and OMM1 (Supplemental Fig. 4A, p < 0.0001, one-way ANOVA). Consequently, SAMMSON knockdown was associated with a significant decrease in cell viability in 92.1 and OMM1 (Supplemental Fig. 4B, p < 0.0001, one-way ANOVA). Taken together, these results reveal an important role for SAMMSON in maintaining UM and CM cell survival in vitro. Due to the genetic similarity between CM and SKCM, where SAMMSON has been extensively studied [13, 14], this research will further mainly focus on the role of SAMMSON in UM.
SAMMSON inhibition affects protein synthesis and mitochondrial function in UM
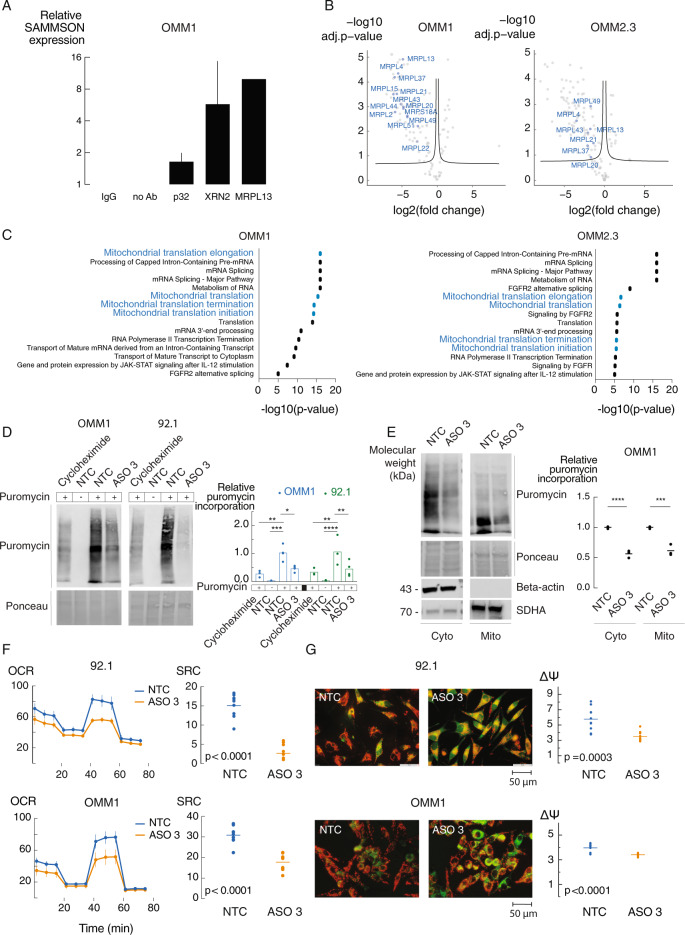
To investigate the mechanism by which SAMMSON contributes to UM tumor cell survival, we studied SAMMSON interaction partners in UM cell lines. First, we evaluated SAMMSON binding to p32 and XRN2, 2 interaction partners previously identified in skin melanoma [13, 14], using RIP-qPCR. Immunoprecipitation of both p32 and XRN2 in 92.1 and OMM1 cells revealed a 2- and 6-fold enrichment in OMM1 and a 4- and 64-fold enrichment in 92.1 of SAMMSON RNA, respectively, indicating that SAMMSON binding to these factors is conserved in uveal melanoma cells (Fig. 3A and Supplemental Fig. 5A). We then applied chromatin isolation by RNA purification and mass spectrometry (ChIRP-MS) in two UM cell lines (OMM1 and OMM2.3) to identify additional interaction partners of SAMMSON in UM cells. We verified enrichment of SAMMSON RNA upon SAMMSON pull down with biotinylated probes (Supplemental Fig. 5B) and subsequently applied mass spectrometry to quantify protein abundance in both the SAMMSON and lacZ pull-down samples. Of note, the non-SAMMSON binding control LacZ did not result in SAMMSON RNA enrichment (data not shown). We detected 83 and 84 proteins that were significantly enriched upon SAMMSON pull down in OMM1 and OMM2.3, respectively, of which 57 were found in both cell lines (Fig. 3B, Supplemental Fig. 5C and Supplemental Table 1). We next performed pathway enrichment analysis and found that pathways involved in mitochondrial translation were highly enriched among the candidate interaction partners in both cell lines (Fig. 3C and Supplemental Table 1). Closer inspection of the candidate interaction partners revealed 13 (OMM1) and 7 (OMM2.3) mitochondrial ribosomal proteins (MRPs) (Fig. 3B) that are all part of the 39 S large mitoribosomal subunit involved in mitochondrial translation. One of the identified interaction partners, Mitochondrial Ribosomal Protein L13 (MRPL13), was validated in OMM1 cells by means of immunoprecipitation with a 10-fold enrichment of SAMMSON RNA (Fig. 3A). The importance of three MRPs (MRPL4, MRPL13 and MRPL37, all identified as SAMMSON interaction partners in both UM cell lines), in maintaining mitochondrial function was assessed by measuring the oxygen consumption rate (OCR) as a proxy for mitochondrial respiration. Differences in OCR after injections of oligomycin, fluoro-carbonyl cyanide phenylhydrazone (FCCP) and rotenone/antimycin A were measured [17]. Knockdown of those MRPs (Supplemental Fig. 5D, p < 0.001 for all tested MRP siPOOLs in 92.1 and OMM1, unpaired t-test) resulted in a significant decrease in mitochondrial spare respiratory capacity (SRC) of 49% in 92.1 and 32% in OMM1 (Supplemental Fig. 5E, p = 0.0008 (92.1) and p = 0.0063 (OMM1), unpaired t-test). Together with validated p32 and XRN2 interactions, these results suggest a role for SAMMSON in regulating translation and mitochondrial function in uveal melanoma. To further investigate the effect of SAMMSON knockdown on translation, we quantified global, mitochondrial and cytosolic translation levels using a puromycin incorporation assay (SUnSET [18]). SAMMSON knockdown significantly impaired global translation rates by 56% and 61% in UM cell lines OMM1 and 92.1, respectively (Fig. 3D and Supplemental Fig 6A, p = 0.0155 (OMM1) and p = 0.0077 (92.1), two-way ANOVA). Fractionation revealed a reduction of both mitochondrial and cytosolic translation rates by 38% and 44%, respectively in OMM1 (Fig. 3E and Supplemental Fig 6B, p = 0.0002 (mito) and p < 0.0001 (cyto), one-way ANOVA).
Fig. 3. SAMMSON inhibition affects translation and mitochondrial function.
A P32, XRN2 and MRPL13 were identified as SAMMSON interacting proteins by means of RIP-qPCR in OMM1. Data presented as the mean ± standard error (SE) of qPCR replicates. B Volcano plot depicting SAMMSON interacting proteins identified by ChIRP-MS (left). Probes targeting LacZ were included as a control (right). Mitochondrial ribosomal proteins (MRPs) are indicated on the graph. Significance was calculated using two-sided t-test. C Pathway enrichment analysis for ChIRP results showed participation of SAMMSON in mitochondrial translation pathways. D Representative images of WB-SUnSET analysis of UM cells treated with cycloheximide (translation inhibitor, positive control), scrambled ASO (NTC) (without puromycin, negative control), NTC ASO or ASO 3 (50 nM). Quantification of protein synthesis measured by calculating the intensity of the puromycin signal on WB. The individual data points (n = 3 biological replicates) and mean are presented. P values were calculated using two-way ANOVA with Tukey’s multiple comparisons test. E Representative images of WB-SUnSET analysis of UM cells treated with scrambled ASO (NTC) or ASO 3 (100 nM) followed by mitochondrial (mito) and cytosolic (cyto) fractionation. Quantification as described in D. The individual data points (n = 3 biological replicates) and mean are presented. P values were calculated using one-way ANOVA with Tukey’s multiple comparisons test. F Oxygen Consumption Rate (OCR) measurements over time after sequential injections of oligomycin, fluoro-carbonyl cyanide phenylhydrazone (FCCP) and rotenone/antimycin A in UM cell lines treated for 24 h with NTC ASO or ASO 3 (100 nM). Data are represented as the mean of three replicates ± s.d. Spare respiratory capacity (SRC) was obtained by subtracting the basal respiration from the maximal respiration. The individual data points and mean are presented. P values were calculated using unpaired two-tailed t-test. G 5,5’,6,6’-Tetraethylbenzimidazolyl-carbocyanine iodide (JC-1) staining in UM cells treated with NTC ASO or ASO 3 (100 nM) (magnification of the images x400). Quantification of the electric membrane potential (ΔΨ) as the red over green fluorescence of 10 random selected fields. The individual data points and mean are presented. P values were calculated using unpaired two-tailed t-test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
We then explored the importance of SAMMSON for proper mitochondrial function by measuring the OCR. Upon SAMMSON knockdown, we observed a significant decrease in mitochondrial spare respiratory capacity (SRC) of 82%, 42% and 72% in 92.1, OMM1 and OMM2.3, respectively (Fig. 3F and Supplemental Fig 7A, p < 0.0001, unpaired t-test). This was further verified by a 5,5′,6,6′-tetraethylbenzimidazolyl-carbocyanine iodide (JC-1) fluorescence staining, where the electric membrane potential (ΔΨ) is quantified based on the relative red over green fluorescence [19]. SAMMSON knockdown decreases ΔΨ with 39%, 14% and 31% in 92.1, OMM1 and OMM2.3, respectively, indicating oxidative phosphorylation (OXPHOS) impairment (Fig. 3G and Supplemental Fig 7B, p = 0.0003 (92.1) and p < 0.0001 (OMM1 and OMM2.3), unpaired t-test). These data suggest that the mechanism by which SAMMSON affects translation is conserved between UM and SKCM, resulting in an impairment of mitochondrial function. In addition, knockdown of MRPL13, together with Mitochondrial Ribosomal Protein L4 and L37 (MRPL4 and MRPL37), three SAMMSON interaction partners identified in both OMM1 and OMM2.3, resulted in a significant decrease in spare respiratory capacity of 49% in 92.1 and 32% in OMM1, demonstrating their importance in mitochondrial function.
To assess the importance of mitochondrial translation in UM cell survival, SAMMSON expressing UM cell lines 92.1 and OMM1 and non-SAMMSON expressing cell lines CT5.3hTERT and HEK293T were treated with tigecycline, a bacteriostatic antibiotic of the tetracyclines family [20–23]. In both UM cell lines, we observed a tigecycline dose-dependent reduction in cell confluence and induction of apoptosis (Supplemental Fig 8A, p < 0.0001 (confluence) and p < 0.0001 for all concentrations, except 3.125 µM in 92.1 and p≤0.01 for the two highest concentrations in OMM1 (apoptosis), one-way ANOVA), verifying that UM cells strongly depend on mitochondrial translation. While the growth of non-SAMMSON expressing cells HEK293T and CT5.3hTERT is also affected by tigecycline treatment (Supplemental Fig 8B, p < 0.0001 (HEK293T) and p ≤ 0.05 for all concentrations, except 3.125 µM (CT5.3hTERT), two-way ANOVA), tigecycline was markedly less effective in CT5.3hTERT cells (cell viability reduction of 35% using 50 µM of tigecycline compared to ≥55% in 92.1, OMM1 and HEK293T).
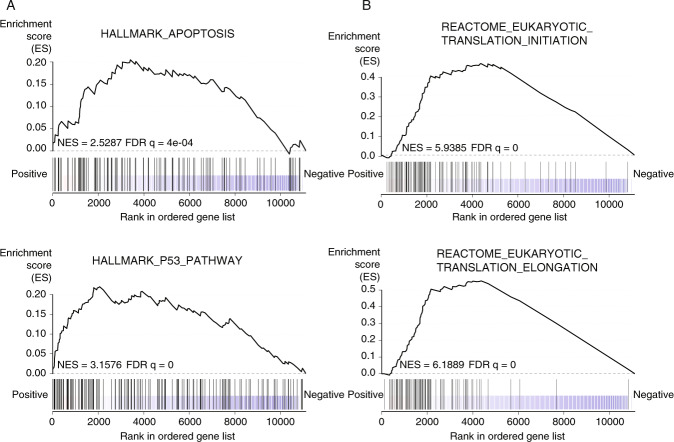
To assess the impact of SAMMSON on the UM transcriptome, we performed shallow RNA sequencing of UM cell lines 92.1 and OMM1 treated with either NTC ASO or ASO 3. Differential gene expression analysis revealed 378 up- and 255 downregulated genes (adjusted p value <0.05). Gene set enrichment analysis (GSEA) revealed a significant upregulation of gene sets associated with apoptosis and p53 response upon SAMMSON silencing (Fig. 4A, FDR q = 4e–04 (apoptosis) and FDR q = 0 (p53) and Supplemental Table 2), supporting our phenotypic observations. Surprisingly, SAMMSON inhibition also resulted in a transcriptional activation of genes involved in translation (Fig. 4B, FDR q = 0).
Fig. 4. Enrichment of apoptosis, p53 and translation gene sets upon SAMMSON knockdown.
A, B GSEA results for Hallmark gene sets apoptosis and p53 (MsigDB) (A) and c2 curated gene sets translation initiation and translation elongation (MsigDB) (B) that are enriched upon ASO 3 treatment (50 nM). Normalized enrichment score (NES) and false discovery rate (FDR) are depicted on the enrichment plots.
SAMMSON inhibition suppresses UM PDX growth
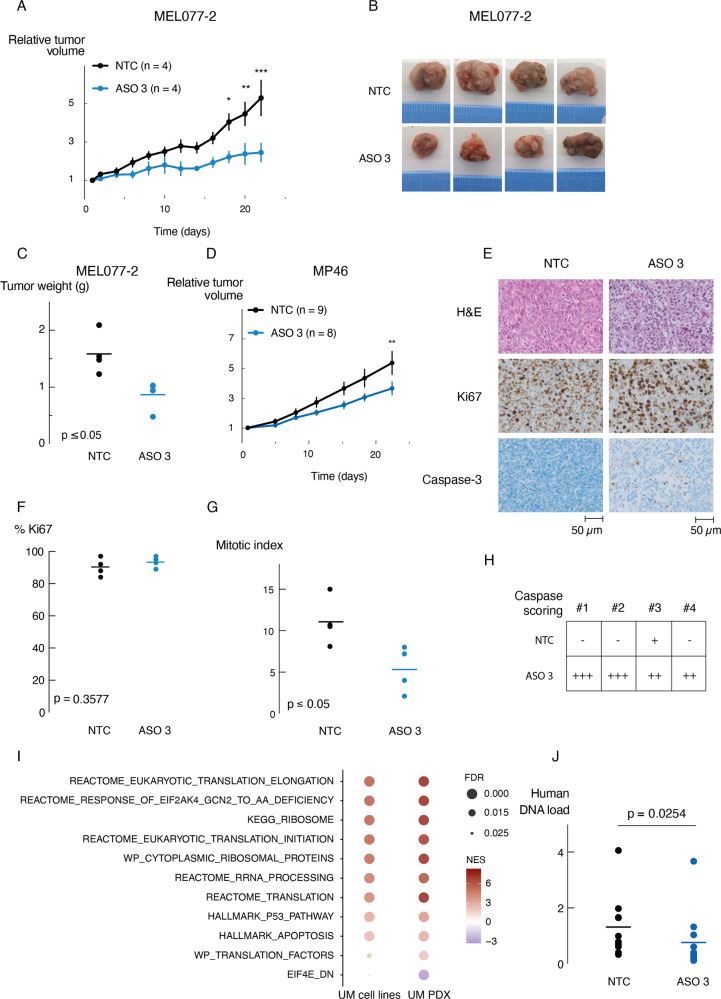
In vivo anti-tumor effects of SAMMSON knockdown were evaluated in three independent experiments using two UM patient derived xenografts (PDX) models (MP46 (GNAQQ209L) and MEL077 (derived from a patient progressing on the immune checkpoint inhibitor pembrolizumab)) showing high SAMMSON expression levels (Fig. 1D). When the tumor xenografts reached a volume of 60–180 mm3, the mice were randomly separated into two groups and were subcutaneously injected with either NTC ASO or ASO 3 (10 mg/kg). Tumor growth, monitored for 22 days, was significantly delayed (MEL077-2, Fig. 5A, p = 0.0001 at endpoint, two-way ANOVA) and tumor weight significantly reduced in MEL077 PDX mice treated with ASO 3 compared to NTC ASO (MEL077-2, Fig. 5B, C, p = 0.0183, unpaired t-test). A similar trend could be observed in an independent repeat in PDX model MEL077 (MEL077-1, Supplemental Fig 9A) and a significant delay in tumor growth was also observed in UM PDX model MP46 (Fig. 5D, p = 0.0050 at endpoint, two-way ANOVA). Notably, the mice of both PDX models did not suffer from any weight loss after ASO 3 treatment (Supplemental Fig 9B-D).
Fig. 5. SAMMSON inhibition reduces tumor growth in vivo.
A Relative tumor volume of MEL077 PDX mice (MEL077-2) subcutaneously injected with NTC ASO or ASO 3 (10 mg/kg). Data are mean ± s.e.m. of multiple replicates (n = 4). P values were calculated using two-way ANOVA with Sidak’s multiple testing correction. B Representative tumors of MEL077 PDX mice (MEL077-2) 22 days after treatment (n = 4/treatment group). C Tumor weight of tumors as shown in B. Individual data points and mean are presented. P value was calculated using unpaired two-tailed t-test. D Relative tumor volume of MP46 PDX mice subcutaneously injected with scrambled ASO (NTC) or ASO 3 (10 mg/kg). Data are mean ± s.e.m. of multiple replicates (n = 8–9/treatment group). P values were calculated using two-way ANOVA with Sidak’s multiple testing correction. E Representative images of H&E, Ki67 and Caspase-3 staining of MEL077 tumor sections (MEL077-2) 22 days after treatment with NTC ASO or ASO 3. F Percentage of Ki67 positive cells in 500 counted cells per tumor sample (MEL077-2, n = 4/treatment group) (magnification x100). Individual data points and mean are presented. P value was calculated using unpaired two-tailed t-test. G Number of mitotic cells based on H&E staining (MEL077-2) in E of 10 random high power fields (HPFs) (magnification x400). Each data point represents the mean of 10 HPFs per tumor sample (n = 4/treatment group). P value was calculated using unpaired two-tailed t-test. H Caspase-3 scoring based on morphology and staining intensity as shown in E of four tumor samples per treatment group (MEL077-2). I Selected GSEA results of RNA seq data from UM cell lines OMM1 and 92.1 and UM MEL077 PDX tumors (MEL077-2) demonstrating overlapping enrichment of gene sets involved in apoptosis, p53 and translation upon ASO 3 treatment. The depth of the color represents the normalized enrichment score (NES). The area of the circle represents the false discovery rate (FDR). J Human DNA load measured in lung tissues of MEL077-2 (n = 4/treatment group) and MP46 (n = 10–11/treatment group) mice by means of qPCR of Alu-Sq repetitive sequence. Individual data points and mean are presented. P value was calculated using unpaired one-tailed t-test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Immunohistochemistry (IHC) analysis with anti-Ki67 of the MEL077 model revealed highly aggressive tumors, with over 80% of proliferating cells in all tumor samples (Fig. 5E). While the difference in Ki67 expression between the ASO 3 and NTC ASO treated mice was too small to be statistically significant (Fig. 5E, F, p = 0.3577, unpaired t-test), further investigation of the mitotic index showed a significant reduction in the number of mitotic cells in the ASO 3 treated tumor samples (Fig. 5E, G, p = 0.0278, unpaired t-test). The discrepancy between Ki67 and MI results might be explained by the selected regions and cell cycle phase coverage of both methods, which has also been described in other tumors [24–26]. In addition, caspase-3 activity was elevated in tumors from mice treated with ASO 3 compared to NTC ASO, indicating SAMMSON knockdown induces apoptosis in vivo (Fig. 5E, H). Moreover, all tumor samples were correctly labeled as ‘ASO 3-treated’ or ‘NTC ASO-treated’, independently by two pathologists and blinded to treatment.
To further investigate these tumors at the molecular level, we applied RNA-sequencing to generate transcriptome profiles to define pathways significantly enriched or repressed upon PDX ASO treatment. Notably, we observed a strong overlap (p < 0.0001, Fisher exact test) between pathways induced in the PDX model and pathways induced in the UM cell lines upon SAMMSON knockdown. Gene sets related to apoptosis and p53 response were significantly upregulated (Fig. 5I, FDR q < 0.05, Supplemental Table 2). Similarly, pathways related to translation were also found transcriptionally upregulated. Median SAMMSON expression was decreased in tumors from ASO 3 treated mice however this difference was not significant (Supplemental Figure 9E).
Although no macro-metastatic lesions were observed in both PDX models, we investigated the presence of tumoral DNA in blood and various murine tissues resected from both PDX models [27]. To this end, we extracted genomic DNA (gDNA) from blood, liver and lung of PDX mice that received ASO 3 or NTC ASO treatment and performed quantitative PCR using the human specific Alu-Sq, SVA and LINE-1 repetitive sequences as an indicator of tumor load in these tissues. Copy number levels of the murine Hprt1 and Pthlh genes were used for normalization. Tumor DNA was only detected in the murine lung tissue samples and compared to NTC ASO treated mice, mice treated with ASO 3 had significantly reduced levels of tumor DNA in lung tissue (Fig. 5J and Supplemental Fig 9F, G, p = 0.0254 (Alu-Sq), p = 0.0357 (LINE-1), p = 0.0202 (SVA), unpaired t-test).
Discussion
Long non-coding RNAs are crucial players in many cellular and biological processes including regulation of gene expression, cell growth, differentiation and development. Aberrant expression of lncRNAs is observed in virtually all tumor types and some lncRNAs appear to be cell type- and tissue-specific [28, 29]. The lncRNA SAMMSON was discovered as a melanoma specific lncRNA [13], but several studies have also shown occasional expression in gastric cancer (GC), hepatocellular carcinoma (HCC), glioblastoma (GBM) and papillary thyroid carcinoma (PTC) [30–34]. In the present study, we found that SAMMSON is expressed in more than 80% of uveal melanoma tumors at levels that are substantially higher compared to non-melanoma tumors. Furthermore, SAMMSON is significantly higher expressed in metastatic UM tumors compared to matching primary tumors. In addition, SAMMSON expression could be observed in conjunctival melanoma (CM) cells that are genetically and phenotypically more related to skin melanoma. While 50% of uveal melanoma tumors are characterized by loss of an entire copy of chromosome 3, SAMMSON expression levels are not reduced in monosomy 3 tumors whereas the majority of genes on chromosome 3 do show a clear gene-dosage effect. These observations suggest the existence of a compensation mechanism that requires further investigation. Potentially, monosomy 3 tumors upregulate one or multiple transcription factors that drive SAMMSON expression from the remaining allele.
The lack of treatments for UM patients with metastatic dissemination that occurs in 50% of the patients and is accompanied by extremely poor survival [4] demonstrates the high unmet need for new treatment modalities. Therapeutic nucleic acid-based approaches like siRNA and antisense oligonucleotides (ASO) hold enormous potential to target RNA molecules, including lncRNAs. Advances in therapeutic ASO and siRNA technology research, such as the development of locked nucleid acid (LNA) and S-constrained ethyl (cEt) modified ASOs and 2’-OMe and 2’-fluoro modified siRNAs, enabled the development and approval of several ASO and siRNA drugs to treat diseases such as spinal muscular atrophy (SMA) and hereditary transthyretin-mediated amyloidosis (hATTR) [35–37]. Our work demonstrates that, similar to skin melanoma, ASO-mediated SAMMSON knockdown in various in vitro and in vivo UM models induces a potent anti-tumor response including growth reduction, induction of apoptosis, and reduced levels of tumor-derived DNA in lung tissue. Whether this DNA is cell-free or derived from tumor cells invading these tissues remains to be investigated. The fact that we did not observe tumor DNA in blood may be indicative of the latter. Additional studies in uveal melanoma xenograft models that metastasize [38–40] should further assess the impact of SAMMSON knockdown on metastatic disease. Furthermore, the in vivo kinetics of SAMMSON expression upon ASO treatment should be explored in more detail. While we observed a trend of SAMMSON knockdown in vivo 3 weeks after start of treatment, the difference was not significant. Differences in baseline SAMMSON expression levels and (time-dependent) variability in knockdown efficiency between mice may explain these observations. SAMMSON expression quantification at earlier treatment time points could provide more insights in knockdown dynamics. Our in vitro data, based on multiple cell lines with a different genetic background, and in vivo data, based on two genetically different UM PDX models, suggest that the genetic background of the tumor does not influence the response to SAMMSON inhibition. Furthermore, cell lines originating from primary and metastatic UM tumors showed similar responses to SAMMSON knockdown, including viability reduction and induction of apoptosis. Since SAMMSON expression levels are elevated in metastatic tumors, these results highlight the broad relevance of SAMMSON inhibition as a potential therapeutic strategy, which may be primarily of interest for metastatic UM patients, where the therapeutic need is high. On-target toxicity of the ASO treatment could not be investigated due to the absence of a SAMMSON homologue in the mouse genome, which is a limitation of the current study. On the other hand, the fact that SAMMSON is expressed in most uveal melanoma tumors (irrespective of genetic background or tumor stage) and absent in differentiated human tissues [13] suggests that therapeutic targeting of SAMMSON could selectively kill tumor cells without on-target toxicity in other tissues.
Earlier studies in SKCM demonstrated SAMMSON interaction with proteins involved in mitochondrial and cytosolic translation, such as p32, XRN2 and CARF [14]. In UM, interactions with p32 and XRN2 were confirmed and novel candidate interactions with multiple proteins belonging to the 39S large mitoribosomal subunit were identified, of which interaction with MRPL13 was validated, further supporting the role of SAMMSON in regulating mitochondrial and cytosolic translation. Additional mechanistic studies are required to further elucidate the role of these novel candidate interactors. Of note, XRN2 and p32 were not identified by mass-spectrometry analysis, suggesting that we may have missed other interaction partners. In support of these interactions and the established role of SAMMSON in SKCM, SAMMSON inhibition effectively impairs mitochondrial and cytosolic translation rates. Since all proteins required for mitochondrial translation, including the mitoribosomal proteins and the mitochondrial translation factors, are translated by cytosolic ribosomes [41], further investigation is needed to determine whether the observed inhibition on mitochondrial translation is a direct or indirect effect. Furthermore, all 13 polypeptides synthesized by the mitochondrial ribosomes (mitoribosomes) are essential components of the oxidative phosphorylation machinery [42]. SAMMSON inhibition indeed impairs mitochondrial function, as shown in three UM cell lines. Moreover, knockdown of the identified SAMMSON interaction partners MRPL4, MRPL13 and MRPL37 also impaired mitochondrial respiration, demonstrating their involvement in mitochondrial function. This results in mitochondrial precursor overaccumulation stress (mPOS), characterized by the toxic accumulation and aggregation of unimported mitochondrial proteins in the cytosol [43]. Cells need to balance between import of mitochondrial proteins and the cytosolic capacity to handle unimported mitochondrial proteins. Induction of mPOS can tip that balance, hereby compromising cell viability. Although SAMMSON inhibition affects global translation, transcriptome analysis in both UM cell lines and PDX models revealed a transcriptional upregulation of various components involved in the translational machinery. These observations have also been observed in yeast when using translation inhibitors such as cycloheximide (CHX) [44] and upregulation in ribosomal biogenesis (RiBi) gene expression has been observed when inducing ribosomal stress which lowers the translational capacity [45]. Ribosomal biogenesis is one of the most energy consuming processes, which requires a coordinated de-repression and active induction of RiBi gene expression [46]. Transcriptional upregulation of these genes while global translation is affected, suggest the existence of a feedback loop to compensate for the loss in translational capacity.
In conclusion, our work shows that inhibition of SAMMSON impairs cytosolic and mitochondrial translation, which consequently affects mitochondrial function and ultimately results in decreased cell viability and induction of apoptosis, both in vitro and in vivo. Together, our findings suggest that SAMMSON is an attractive therapeutic target for UM and that SAMMSON can be targeted in vivo using ASO technology.
Material and methods
Cell culture
For all human cell lines, an ethical approval was obtained from the Ghent University commission for medical ethics. The human uveal melanoma cell lines 92.1 [47], OMM1 [16], OMM2.3, MEL270, MP38 [48], MP46 [48], MM28 [48] and MP65 [48] and conjunctival melanoma cell lines CRMM1 and CRMM2 [49] were obtained from the Leiden University Medical Center, The Netherlands. OMM2.3 and MEL270 were a kind gift from Bruce Ksander. MM28, MP65, MP46 and MP38 were a kind gift from Sergio Roman-Roman. Uveal melanoma cell line MEL077 and skin melanoma cell line SK-MEL28 was obtained from the Laboratory for Molecular Cancer Biology, VIB-KU Leuven, Belgium. SAMMSON negative cell lines HEK293T and CT5.3hTERT were obtained from ATCC (Manassas, Virginia, USA) and the Department of Radiation oncology and experimental cancer research, Ghent University, Belgium, respectively. The cell lines 92.1, OMM1, OMM2.3 and MEL270 were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco)/F12––Roswell Park Memorial Institute (RPMI, Gibco) 1640 (1:1) medium. SK-MEL28 was grown in DMEM/F-12 GlutaMAX medium (Gibco), CT5.3hTERT in DMEM medium, CRMM1 and CRMM2 in Ham’s F-12K (Kaighn’s) medium (Gibco), MM28, MP65, MP46 and MP38 in Iscove’s Modified Dulbecco’s Medium (IMDM) and HEK293T and MEL077 in RPMI 1640 medium. Media of MM28, MP65, MP46 and MP38 were supplemented with 20% fetal bovine serum (FBS) and all other media with 10% FBS, 2 mM L-glutamine (Gibco) and 100 IU/ml penicillin/streptomycin (Gibco) and all cell lines were incubated in a humidified atmosphere containing 5% CO2 at 37 °C. For executing experiments, cells were grown in L-glutamine and penicillin/streptomycin free media. Short tandem repeat (STR) genotyping was used to validate cell line authenticity and absence of mycoplasma was verified on a monthly basis for all cell lines in culture.
Supplementary information
Supplemental Fig 1. SAMMSON expression is independent from patient survival, tumor stage, tumor localization site and metastatic state of the UM patient.
Supplemental Fig 2. Expressed genes in monosomy 3 UM tumors compared to disomy 3 UM tumors.
Supplemental Fig 3. SAMMSON knockdown affects cell growth in CM cell line CRMM1.
Supplemental Fig 4. Non-lipid-based delivery of ASOs in UM cells.
Supplemental Fig 5. Identification of SAMMSON interaction partners.
Supplemental Fig 6. Uncropped images of WB-SUnSET analysis.
Supplemental Fig 7. Impairment of mitochondrial function upon SAMMSON knockdown in UM cell line OMM2.3.
Supplemental Fig 8. Phenotypic results using tigecycline are comparable to SAMMSON inhibition in UM cells.
Supplemental Fig 9. SAMMSON inhibition slows down tumor growth in vivo.
Acknowledgements
We would like to thank the VIB proteomics core for performing LC-MS/MS analysis and Prof. Olivier De Wever for providing the CT5.3hTERT cell line. The authors would also like to thank the Animal PlatformCRP2-UMS 3612 CNRS-US25 Inserm-IRD (Faculty of Pharmacy, Paris University, Paris), and the Animal Platform of Institut Curie. They also thank Justine Fleury for her technical assistance (Laboratory of preclinical Investigation, Institut Curie). This work was supported by a Foundation Against Cancer grant (Stichting tegen Kanker), a Standup Against Cancer grant (Kom op tegen Kanker), a Ghent University Industrial Research Fund (IOF), a Ghent University GOA research grant and Fund for Scientific Research Flanders (FWO Vlaanderen). EL is the recipient of the Melanoma Research Alliance (MRA) Amanda and Jonathan Eilian young investigator award. SD, LD and JDW are recipients of a grant from the Fund of Scientific Research Flanders (FWO Vlaanderen).
Author contributions
PM conceived and supervised the project; SD, LD, BDP, EDB, JDW, KV, NY, JN, EVE, JS, MV, and SE designed and performed experiments; PM, SD, LD, EDB, JDW, JA analyzed the data; FN, EL performed PDX experiments; MR and PZ provided and analyzed RNA seq data; AJ provided cell lines; DD, JVD, CM, JV, RVC, SE contributed technical support and resources; SD and PM wrote the paper. All authors contributed to manuscript editing and approved the final draft.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/15/2021
A Correction to this paper has been published: 10.1038/s41388-021-02051-6
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-021-02006-x.
References
- 1.Bakalian S, Marshall J-C, Logan P, Faingold D, Maloney S, di Cesare S, et al. Molecular Pathways Mediating Liver Metastasis in Patients with Uveal Melanoma. Clin Cancer Res: Off J Am Assoc Cancer Res. 2008;14:951–6. doi: 10.1158/1078-0432.CCR-06-2630. [DOI] [PubMed] [Google Scholar]
- 2.Papastefanou VP, Cohen VML. Uveal Melanoma. J Ski Cancer. 2011;2011:1–13. doi: 10.1155/2011/573974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez A, Dueñas-Gonzalez A, Delgado-Pelayo S. Clinical presentation and management of uveal melanoma. Mol Clin Oncol. 2016;5:675–7. doi: 10.3892/mco.2016.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krantz BA, Dave N, Komatsubara KM, Marr BP, Carvajal RD. Uveal melanoma: epidemiology, etiology, and treatment of primary disease. Clin Ophthalmol. 2017;ume 11:279–89. doi: 10.2147/OPTH.S89591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer cell. 2017;32:204–20. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoiom V, Helgadottir H. The genetics of uveal melanoma: current insights. Application Clin Genet. 2016;ume 9:147–55. doi: 10.2147/TACG.S69210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JJ, Diefenbach RJ, Joshua AM, Kefford RF, Carlino MS, Rizos H. Oncogenic signaling in uveal melanoma. Pigment Cell Melanoma Res. 2018;31:661–72. doi: 10.1111/pcmr.12708. [DOI] [PubMed] [Google Scholar]
- 8.van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbour JW, Onken MD, Roberson EDO, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues M, Ait Rais K, Salviat F, Algret N, Simaga F, Barnhill R, et al. Association of Partial Chromosome 3 Deletion in Uveal Melanomas with Metastasis-Free Survival. JAMA Ophthalmol. 2020;138:182–8. doi: 10.1001/jamaophthalmol.2019.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang A, Papneja A, Hyrcza M, Al-Habeeb A, Ghazarian D. BAP1: Gene of the month. J Clin Pathol. 2016;69:750–3. doi: 10.1136/jclinpath-2016-203866. [DOI] [PubMed] [Google Scholar]
- 12.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013;13:1–12. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–22. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 14.Vendramin R, Verheyden Y, Ishikawa H, Goedert L, Nicolas E, Saraf K, et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat Struct Mol Biol. 2018;324:1021–9. doi: 10.1038/s41594-018-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, Stei MM, Fröhlich H, Holz FG, Loeffler KU, Herwig-Carl MC. Genetic and epigenetic insights into uveal melanoma. Clin Genet. 2018;93:952–61. [DOI] [PubMed]
- 16.Luyten GPM, Naus NC, Mooy CM, Hagemeijer A, Kan-Mitchell J, van Drunen E, et al. Establishment and characterization of primary and metastatic uveal melanoma cell lines. Int J Cancer. 1996;66:380–7. [DOI] [PubMed]
- 17.Nicholas D, Proctor EA, Raval FM, Ip BC, Habib C, Ritou E, et al. Advances in the quantification of mitochondrial function in primary human immune cells through extracellular flux analysis. PLOS ONE. 2017;12:e0170975–19. doi: 10.1371/journal.pone.0170975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat methods. 2009;6:275–7. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 19.de Paepe B, Smet J, Vanlander A, Seneca S, Lissens W, de Meirleir L, et al. Fluorescence imaging of mitochondria in cultured skin fibroblasts: a useful method for the detection of oxidative phosphorylation defects. Pediatr Res. 2012;72:232–40. doi: 10.1038/pr.2012.84. [DOI] [PubMed] [Google Scholar]
- 20.Roger AJ, Muñoz-Gómez SA, Kamikawa R. The origin and diversification of mitochondria. Curr Biol. 2017;27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Xie F, Chen D, Wang L. Inhibition of mitochondrial respiration by tigecycline selectively targets thyroid carcinoma and increases chemosensitivity. Clin Exp Pharmacol Physiol. 2019;46:890–7. doi: 10.1111/1440-1681.13126. [DOI] [PubMed] [Google Scholar]
- 22.Noskin GA. Tigecycline: a new glycylcycline for treatment of serious infections. Clin Infect Dis. 2005;41:S303–S314. doi: 10.1086/431672. [DOI] [PubMed] [Google Scholar]
- 23.Doan PT-L, PharmD BHBF, Mehta PD, Riska MDPF. Tigecycline: a glycylcycline antimicrobial agent. Clin Therapeutics. 2006;28:1079–106. doi: 10.1016/j.clinthera.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Khan MS, Luong TV, Watkins J, Toumpanakis C, Caplin ME, Meyer T. A comparison of Ki-67 and mitotic count as prognostic markers for metastatic pancreatic and midgut neuroendocrine neoplasms. Br J Cancer. 2013;108:1838–45. doi: 10.1038/bjc.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mooy CM, de Jong PTVM, van der Kwast TH, et al. Immunostaining in uveal melanoma: the effect of pre-enucleation radiotherapy. Ophthalmology. 1990;97:1275–80. doi: 10.1016/S0161-6420(90)32420-X. [DOI] [PubMed] [Google Scholar]
- 26.Rossi L, Laas E, Mallon P, Vincent-Salomon A, Guinebretiere JM, Lerebours F, et al. Prognostic impact of discrepant Ki67 and mitotic index on hormone receptor-positive, HER2-negative breast carcinoma. Br J Cancer. 2015;113:996–1002. doi: 10.1038/bjc.2015.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Maerken T, Ferdinande L, Taildeman J, Lambertz I, Yigit N, Vercruysse L, et al. Antitumor activity of the selective MDM2 antagonist nutlin-3 against chemoresistant neuroblastoma with wild-type p53. JNCI: J Natl Cancer Inst. 2009;101:1562–74. doi: 10.1093/jnci/djp355. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Wen L, Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5:1–10. [DOI] [PMC free article] [PubMed]
- 29.Bhan A, Soleimani M, Mandal S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed]
- 30.Xie J, Wang X, Liu S, Chen C, Jiang F, Mao K, et al. LncRNA SAMMSON overexpression distinguished glioblastoma patients from patients with diffuse neurosarcoidosis. NeuroReport. 2019;30:817–21. doi: 10.1097/WNR.0000000000001278. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Cai H, Hu B, Tu J. LncRNA SAMMSON negatively regulates miR-9-3p in hepatocellular carcinoma cells and has prognostic values. Biosci Rep. 2019;39:BSR20190615–7. doi: 10.1042/BSR20190615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Li M, Chen J, Dai H, Wang L, Xiong Y, et al. SAMMSON drives the self-renewal of liver tumor initiating cells through EZH2-dependent Wnt/β-catenin activation. Oncotarget. 2017;8:103785–96. doi: 10.18632/oncotarget.21792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Ni H, Wang K, Xie P, Zuo J, Liu W, Liu C. LncRNA SAMMSON knockdown inhibits the malignancy of glioblastoma cells by inactivation of the PI3K/Akt pathway. Cell Mol Neurobiol. 2020;41:1–12. [DOI] [PMC free article] [PubMed]
- 34.Shao L, Sun W, Wang Z, Dong W, Qin Y. Long noncoding RNA SAMMSON promotes papillary thyroid carcinoma progression through p300/Sp1 axis and serves as a novel diagnostic and prognostic biomarker. IUBMB Life. 2019;72:237–46. doi: 10.1002/iub.2158. [DOI] [PubMed] [Google Scholar]
- 35.Verma A. Neuromuscular diseases: recent advances in antisense oligonucleotide therapy. Astrocyte. 2018;5:81–5. doi: 10.4103/astrocyte.astrocyte_55_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7:314ra185–314ra185. doi: 10.1126/scitranslmed.aac5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saw PE, Song E-W. siRNA therapeutics: a clinical reality. Science China. Life Sci. 2019;63:485–500. doi: 10.1007/s11427-018-9438-y. [DOI] [PubMed] [Google Scholar]
- 38.Sugase T, Lam BQ, Danielson M, Terai M, Aplin AE, Gutkind JS, et al. Development and optimization of orthotopic liver metastasis xenograft mouse models in uveal melanoma. J Transl Med. 2020;18:1–14. [DOI] [PMC free article] [PubMed]
- 39.Richards JR, Yoo JH, Shin D, Odelberg SJ. Mouse models of uveal melanoma: strengths, weaknesses, and future directions. Pigment Cell &. Melanoma Res. 2020;33:264–78. doi: 10.1111/pcmr.12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Fang G, Huang X, Yu J, Hsieh C-L, Grossniklaus HE. In-vivo xenograft murine human uveal melanoma model develops hepatic micrometastases. Melanoma Res. 2008;18:95–103. doi: 10.1097/CMR.0b013e3282f628df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koripella RK, Sharma MR, Risteff P, Keshavan P, Agrawal RK. Structural insights into unique features of the human mitochondrial ribosome recycling. PNAS. 2019;116:8283–8. doi: 10.1073/pnas.1815675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Brien TW, Denslow ND, Anders JC, Courtney BC. The translation system of mammalian mitochondria. Biochimica et Biophysica Acta. 2002;1050:174–8. [DOI] [PubMed]
- 43.Coyne LP, Chen XJ. mPOS is a novel mitochondrial trigger of cell death - implications for neurodegeneration. FEBS Lett. 2017;592:759–75. doi: 10.1002/1873-3468.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng Z, Brar GA. Global translation inhibition yields condition-dependent de-repression of ribosome biogenesis mRNAs. Nucleic Acids Res. 2019;47:1–13. [DOI] [PMC free article] [PubMed]
- 45.Shamsuzzaman M, Rahman N, Gregory B, Bruno VM, Lindahl L. Mosaic changes to the global transcriptome in response to inhibiting ribosome formation versus inhibition of ribosome function. bioRxiv. 2020:2020.10.15.341230.
- 46.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: Functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Waard Siebinga I, Blom DJR, Griffioen M, Schrier PI, Hoogendoorn E, Beverstock G, et al. Establishment and characterization of an uveal‐melanoma cell line. Int J Cancer. 1995;62:155–61. doi: 10.1002/ijc.2910620208. [DOI] [PubMed] [Google Scholar]
- 48.Amirouchene-Angelozzi N, Némati F, Gentien D, Nicolas A, Dumont A, Carita G, et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Molecular. Oncology. 2014;8:1508–20. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nareyeck G, Wuestemeyer H, von der Haar D, Anastassiou G. Establishment of two cell lines derived from conjunctival melanomas. Exp Eye Res. 2005;81:361–2. doi: 10.1016/j.exer.2005.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig 1. SAMMSON expression is independent from patient survival, tumor stage, tumor localization site and metastatic state of the UM patient.
Supplemental Fig 2. Expressed genes in monosomy 3 UM tumors compared to disomy 3 UM tumors.
Supplemental Fig 3. SAMMSON knockdown affects cell growth in CM cell line CRMM1.
Supplemental Fig 4. Non-lipid-based delivery of ASOs in UM cells.
Supplemental Fig 5. Identification of SAMMSON interaction partners.
Supplemental Fig 6. Uncropped images of WB-SUnSET analysis.
Supplemental Fig 7. Impairment of mitochondrial function upon SAMMSON knockdown in UM cell line OMM2.3.
Supplemental Fig 8. Phenotypic results using tigecycline are comparable to SAMMSON inhibition in UM cells.
Supplemental Fig 9. SAMMSON inhibition slows down tumor growth in vivo.