Abstract
Alzheimer’s disease (AD) is a progressive and irreversible neurodegenerative disorder clinically characterized by cognitive impairment, abnormal behavior, and social deficits, which is intimately linked with excessive β-amyloid (Aβ) protein deposition along with many other misfolded proteins, neurofibrillary tangles formed by hyperphosphorylated tau protein aggregates, and mitochondrial damage in neurons, leading to neuron loss. Currently, research on the pathological mechanism of AD has been elucidated for decades, still no effective treatment for this complex disease was developed, and the existing therapeutic strategies are extremely erratic, thereby leading to irreversible and progressive cognitive decline in AD patients. Due to gradually mental dyscapacitating of AD patients, AD not only brings serious physical and psychological suffering to patients themselves, but also imposes huge economic burdens on family and society. Accordingly, it is very imperative to recapitulate the progress of gene editing-based precision medicine in the emerging fields. In this review, we will mainly focus on the application of CRISPR/Cas9 technique in the fields of AD research and gene therapy, and summarize the application of CRISPR/Cas9 in the aspects of AD model construction, screening of pathogenic genes, and target therapy. Finally, the development of delivery systems, which is a major challenge that hinders the clinical application of CRISPR/Cas9 technology will also be discussed.
Keywords: gene therapy, CRISPR/Cas9, cognitive function, Alzheimer’s disease, delivery system
Introduction
Alzheimer’s disease (AD) is a progressive and irreversible neurodegenerative disorder that is characterized by cognitive dysfunction, abnormal behavior, social deficits, and an eventual inability to perform daily tasks. It is predicted that by 2050, the number of Alzheimer’s patients aged 65 years and older in the U.S. may reach 13.8 million (No authors listed, 2020). In China, more than 15.07 million people aged 60 years or older have dementia, and approximately 9.83 million of them suffer from AD (Jia et al., 2020). Accordingly, so many AD patients will bring major public health challenges. And to make matters worse, the pathogenesis of AD is currently still unclear. Until now, β-amyloid protein deposition and tau protein hyperphosphorylation are widely regarded as key contributions to the neurobiological mechanisms underlying the pathogenesis of AD (Mila-Aloma et al., 2020; Zetterberg and Bendlin, 2021). Besides, other age-related, protective, and disease-promoting factors probably interact with the core mechanisms of AD and may be involved in the onset of AD. For instance, emerging extensive research has shown that some disregarded partners such as vascular dysfunction (Sweeney et al., 2019), oxidative stress (Tonnies and Trushina, 2017), proteinopathy (Sharma and Kim, 2021), metal ions (Huang et al., 2015; Liu et al., 2019b), neuroinflammation (Calsolaro and Edison, 2016), mitochondrial dysfunction (Mantzavinos and Alexiou, 2017; Perez Ortiz and Swerdlow, 2019), and microbiota-gut-brain axis (Cryan et al., 2019) are also closely associated with the AD progress.
Due to the complexity of the pathogenesis, the clinical manifestations of AD vary greatly between individuals. In addition to cognitive impairment, it also involves many neuropsychiatric symptoms including apathy, aggression, depression, anxiety, sleep disorders, hallucination, and irritability. These symptoms require active treatment when they cause serious adverse effects, however, the presently clinical drugs available merely delay the progression of AD even though some have certain efficacy, but the ultimate misfortune outcome is inevitable. Meanwhile, the irreversibility of the disease process also brought great challenges to developing more effective therapeutic interventions in AD. At present, the global researchers in the field are committed to seeking potential biomarkers for early diagnosis of AD and has sought to expand the mechanism of AD progress, and attempts to reverse the progress of AD through efficient gene editing therapy.
Recently, CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats, CRISPR associated nuclease, CRISPR/Cas) technology is developing rapidly and showing a great potential in the field of basic research and disease therapeutics. Also, the gene editing technology was evaluated as a promising approach in AD research and treatment (Giau et al., 2018; Hanafy et al., 2020). This review first will introduce the development and advantages of CRISPR/Cas9 technology, and then focus on the application of CRISPR/Cas9 in the aspects of AD model construction, screening of pathogenic genes, and target therapy. Finally, we have also summarized the development of delivery systems, which is a major challenge that hinders the clinical application of CRISPR/Cas9 technology (Figure 1).
FIGURE 1.
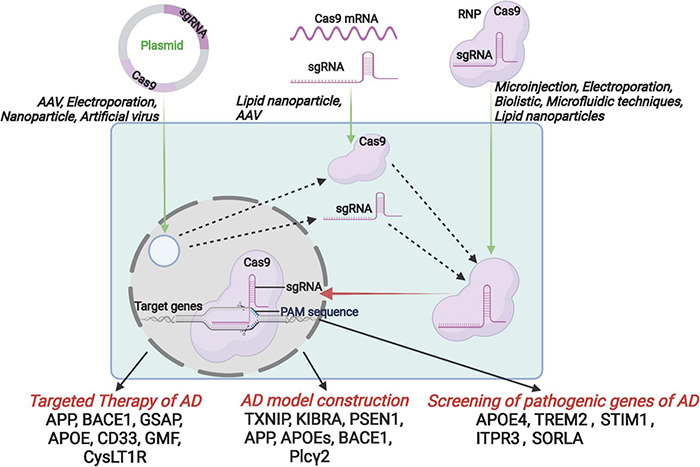
Schematic diagram of the introduction of CRISPR/Cas9 components into host cells by the format of CRISPR/Cas9 plasmid, mRNA and RNP, and further application for the targeted therapy, AD model construction and the screening of pathogenic genes.
CRISPR/Cas9 Gene-Editing Technology
The Composition and Principle of CRISPR/Cas9 System
The CRISPR/Cas9 system is an adaptive bacterial and archaeal defense mechanism that recognizes and disables invading bacteriophages or other foreign nucleic acids (Giau et al., 2018). In general, the gene-editing systems are divided into two classes. The class 1 system contains types I, III and IV, and the class 2 system contains types II, V, and VI (Gupta et al., 2019). Type II CRISPR/Cas9 is the most routinely used for CRISPR gene-editing system and usually refer to CRISPR. The CRISPR/Cas9 system is composed of the Cas9 protein and the sgRNA (single guide RNA, sgRNA). In this system, the sgRNA guides the system to the target, and the Cas9 can cleave the double strands of DNA (Liu et al., 2017). The sgRNA is essential for precise gene editing as its 5′-terminal 20-nucleotide sequence interacts with a target sequence of host DNA via Watson and Crick base pairing rules, while its 3′ duplex structure allows binding to Cas9 proteins (Jinek et al., 2012). The sgRNA is composed of the trans-activating crRNA (tracrRNA) and crRNA. The crRNA contains a 20-nt protospacer element and an additional sequence that is complementary to the tracrRNA. The tracrRNA hybridizes to the crRNA and binds the Cas9 protein, forming the Cas9-sgRNA complex to create double-stranded breaks (DSBs) at target sites in the genome (Cong et al., 2013; Yang et al., 2013; Dow et al., 2015). The Cas9 protein has six domains including REC I, REC II, Bridge Helix, PAM-interacting, HNH and RuvC. The protospacer adjacent motif (PAM) is a short sequence of nucleotides directly adjacent to the target DNA sequence. The Cas9 nucleases from different bacterial species recognize different PAM sequences for seeking targets, with SpCas9 using “NGG” PAM as a binding target while SaCas9 employing “NNGRRT” PAM (Lino et al., 2018; Xie et al., 2018). The HNH and RuvC domains are nuclease domains that cut single-stranded DNA. The RuvC domain cleaves non-complementary DNA strands, while the HNH domain cleaves complementary DNA strands (Jinek et al., 2012). This system presumably functions via the high nuclease activity of CRISPR/Cas9 that induces highly efficient, targeted double-stranded breaks (DSBs). Following a DSB, there are multiple fates for the broken chromosomal DNA. Primary DSB repair mechanisms include non-homologous end joining (NHEJ) or homology directed repair (HDR) pathways (Kaulich et al., 2015). NHEJ involves direct ligation of the two broken chromosomal DNA strands to one another and is the main cellular DSB repair mechanism. This process is error-prone owing to random insertions or deletions (indels) of nucleotides to assist in ligation, which can cause gene disruption via frameshift or nonsense mutations (Lieber, 2010). The HDR pathway performs DNA repair through homologous recombination and rarely mismatches in replicating DNA.
The Development of CRISPR/Cas9 System and Its Advantages Over Other Gene-Editing Technologies
The first CRISPR was cloned from E. coli by accident in 1987. Ishino et al. (1987) noted the presence of a 29-nucleotide repeat sequence in Escherichia coli, which was interrupted by unrelated, non-repetitive short sequences (spacers). These regularly spaced motifs were clustered next to the lap gene, which encodes an aminopeptidase in Escherichia coli K12. Two years later, a second array was found in the same genome, and hybridization assays suggested the presence of similar sequences in very close relatives (Shigella and Salmonella species) (Nakata et al., 1989). In 1991, interspaced direct repeats (DR) were identified in strains of an evolutionarily distant group of bacteria, the Mycobacterium tuberculosis complex (MTBC) (Hermans et al., 1991). The DR-intervening sequences, known as spacers, were found to differ among isolates, and hence were harnessed for strain typing (Jeffreys et al., 1991; Groenen et al., 1993). Later the similar CRISPR sequences were also cloned from other bacteria and Archaea (Hermans et al., 1991; Mojica et al., 1993; Jansen et al., 2002). However, at the first almost one decade, the scientists did not know the function of these special repeat sequences and just thought they are a special sequence among different species and used them as strain typing.
In 2000, the presence of similar repeats was reported in most prokaryotes and was then named “CRISPR” by Ruud Jansen (Mojica et al., 2000; Jansen et al., 2002). The significance of the spacers being derived from foreign genetic elements was established in 2005 (Mojica et al., 2005). In 2012, two independent laboratories reported that in vitro reconstructed CRISPR/Cas systems had biological functions and were capable of cleaving an individual DNA sequence (Gasiunas et al., 2012; Jinek et al., 2012). This provides foundation for using CRISPR/Cas as a genome editing tool. By January 2013, three independent US teams led by Luciano Marraffini (Jiang et al., 2013), Feng Zhang (Cong et al., 2013) and George Church (Mali et al., 2013) succeed in editing the bacterial and mammalian genomes using Cas9.
In recent years, based on the discovery of a variety of high-efficiency nucleases, gene editing technology has been rapidly developed and widely used. The artificial endonuclease-mediated genome editing technology developed most rapidly (Gupta and Shukla, 2017), mainly including 4 types: meganuclease technology (meganuclease), zinc finger nuclease technology (zinc finger nuclease, ZFN), transcription activator-like effector nuclease technology (transcription activator-like effector nuclease, TALEN) and clustered regularly interspaced short palindromic repeats (CRISPR-associated proteins, Cas) system. Different from ZFNs, TALENs and Meganucleases identifying and linking to the DNA to create a double-strand break by the proteins, the binding domain of the CRISPR/Cas9 deriving from RNA implies that the system is more straightforward, efficient and convenient (Khadempar et al., 2019). In this review, we summarized the advantages and limitations of these gene-editing methods (see Table 1). With the development of this technology, CRISPR/Cas9 gene editing system has been extensively applied to gene engineering such as genes knocking-in of mammalian cells (Banan, 2020), human diseases modeling (Torres-Ruiz and Rodriguez-Perales, 2017) and gene therapy, etc. In recent years, the CRISPR/Cas9 technology has improved dramatically over the past decade, and the efficiency of CRISPR/Cas9 system can be greatly increased by lipid-encapsulated gold nanoparticles as a multifunctional vehicle enabling a more effective therapy for tumor (Wang et al., 2018a). By means of the application of artificial virus, CRISPR/Cas9 system has many advantages such as minimum side effects, high efficiency and security over other superinfect and lipofectamine methods (Li et al., 2017). In conclusion, CRISPR/Cas9 technology is increasingly mature and it has the potential to become an effective tool for gene editing in clinical therapy.
TABLE 1.
Comparison of different gene-editing methods.
| CRISPR/Cas9 | ZFNs | TALENs | Meganucleases | RNAi | |
| Nuclease | Cas9 | FOKI | FOKI | I-SceI | Dicer and Argonaute proteins |
| Recognition mechanism | PAM sequences and the complementary sgRNA site-DNA | Zinc finger proteins-DNA | RVDs- DNA | Protein-DNA | RNA |
| Cycle | Short | Long | Long | Long | Long |
| Cytotoxicity | Low | High | Low | Low | Variable to high |
| Versatility | High | High | High | Limited | Not mentioned |
| Cost | Low | High | High | Low | Low |
| Delivery | Easily | Limited | Limited | Limited | Limited |
| RNA editing | Yes | No | No | No | Yes |
| Specificity | High | High | High | High | High |
| Stability | High | High | High | High | Low |
| Application case | AD target therapy | CCR5 gene mutagenesis resisting HIV | Cystic Fibrosis-Gene Therapy | Embed artificial gene networks | Antiviral therapy |
CRISPR/Cas9 in Alzheimer’s Disease
In recent years, due to the short experimental period and relatively low consumption of CRISPR/Cas9 technology, CRISPR/Cas9 is currently widely used in the AD field including construction of AD model, screening pathogenic genes, and target therapy.
Alzheimer’s Disease Model Construction
Cell models are widely used in studies of various neurological diseases including AD because this does not involve ethical concerns, and the experimental period is relatively short, and the cost is low as well. In the past decades, numerous in vitro cell models of AD were established. To date, cell lines commonly used in AD research include human neuroblastoma cells SH-SY5Y and SK-N-SH, mouse hippocampal neuron cell lines HT22 and glial cells BV2, and mouse glioblastoma cells N2a. The emergence of CRISPR/Cas9 gene editing technology can facilitate more efficiently developing AD cell models. Wang et al. (2019) reported that down-regulation of Thioredoxin-interacting protein (Txnip) level in HT22 cells via CRISPR/Cas9 system can effectively attenuate amyloid-β-induced protein cysteine oxidative modification. The findings indicated that Txnip may be a therapeutic target for the treatment of AD. Meanwhile, Song et al. (2019) also found that knockdown of KIBRA (KIdney and BRAin expressed protein) in HT22 cells suppressed its growth and caused apoptosis while treatment with Aβ1–42 oligomers. In addition, Sun et al. (2017) revealed that knock-out of Presenilin 1 (PSEN1) genes in N2a cells by using CRISPR/Cas9 system can eliminate the background of endogenous γ-secretase, and found that exogenous addition of recombinant protein derived from PSEN1 mutations can decrease the production of Aβ42 and Aβ40. However, owing to the differences in genome and physiology characteristics between these cell lines and genuine neural cells, the constructed AD model using these cells still cannot precisely elucidate the molecular mechanism responsible for AD occurrence. Therefore, patient-derived induced pluripotent stem cells (iPSCs) have received more and more attention due to their unique physiological profiles. In 2016, Paquet et al. (2016) introduced mutations of amyloid precursor protein (APP) and PSEN1 which cause AD, into human iPSCs. Subsequently, the higher Aβ levels were detected in the converted neurons from the mutant homozygous and heterozygous human iPSCs compared with isogenic controls, implying the AD-associated mutations could be modeled in human neural cells by CRISPR/Cas9 technology.
Apart from building AD cell models, CRISPR/Cas9 technology can also be used to build AD animal models. In 2020, Serneels et al. (2020) created a novel model named mouse and rat Apphu/hu by using CRISPR/Cas9 strategy to generate a humanized Aβ sequence (G676R, F681Y, and R684H) in APP gene of mouse and rat. And by inserting the three amino acids in the rodent Aβ sequence, the level of Aβ increased more than three times compared to original WT strain. Apolipoprotein E (APOE) has different variants including APOE2, APOE3, APOE3r, and APOE4. Komor et al. (2016) transfer APOE3r into APOE4 in mouse astrocytes by converting C in codon 158 to T via CRISPR/Cas9 system, which implicates the point mutations being modified by CRISPR/Cas9 method. Until 2019, Cas9 nanocomplexes with sgRNAs targeting tyrosine hydroxylase (Th) and β-secretase 1 (BACE1) genes were delivered into mouse primary neurons to assess the efficiency of Cas9 nanocomplexes by Park et al. (2019). Excitingly, almost none of off-target was found by using a variety of survey assays, Sanger sequencing, and deep sequencing. Concomitantly, they injected Cas9-BACE1 nanocomplexes into the hippocampal of 6-month-old AD mouse. Four weeks later, a remarkable decrease of BACE1 expression and β-cleavage products of APP was found in the hippocampal of the AD mice (Park et al., 2019). Moreover, Takalo et al. (2020), designed a Plcγ2-P522R knock-in mouse model by using CRISPR/Cas9 gene editing strategy and assessed the protective effects of Plcγ2-P522R variant. Their results revealed that the microglial functions were enhanced in Plcγ2-P522R knock-in mice, implying that the gene editing strategy is likely to be a new treatment approach for AD (Takalo et al., 2020).
Non-human primates are one of the most suitable models for human diseases owing to their genetic, physiological and pathological similarities with human beings. In 2014, several researchers from China succeeded in achieving two gene modifications (Rag1 and Ppar-γ) of cynomolgus monkeys by CRISPR/Cas9 system at one step. Inspiringly, no off-target mutations were found. Soon afterward, the gene modification of both ear punch tissues and placenta are also conducted, demonstrating the success of global gene modification (Niu et al., 2014). Next year, the Cas9-mediated gene modifications were substantiated in monkey germline, implying the possibility that the artificial modifications are transmitted to their offspring (Chen et al., 2015). For more thorough gene knockout, the injection of various sgRNAs is more effective than single type of high concentration sgRNAs. The thorough gene knock-out by CRISPR/Cas9 system is conducive to reduce the test times of non-human primates and guarantee the accuracy, which is strongly supported by ethics (Zuo et al., 2017). Although no AD primate models were designed via CRISPR/Cas9 technology, however, to further study on the pathogenesis of AD and more efficiently build AD primate models, it is necessary to construct a primate AD model by CRISPR/Cas9 strategy.
Screening of Pathogenic Genes for Sporadic Alzheimer’s Disease
Alzheimer’s disease is classified into sporadic (sporadic AD, SAD) and familial (familiar AD, FAD). APP, PSEN1, and PSEN2 are the main pathogenic genes caused FAD, and the mutation in each gene could contribute to the onset of FAD. However, more than 95% of AD cases are sporadic (Zhang et al., 2020). The pathogenesis is very complicated, involving genetic factors (such as APOE), and is associated with age, environmental factors and many diseases including diabetes (Baglietto-Vargas et al., 2016) and vascular diseases (Zetterberg and Mattsson, 2014). Therefore, the screening of SAD pathogenic genes is crucial for understanding the molecular pathogenesis, early diagnosis, risk prediction, and treatment of AD.
The development of high-throughput sequencing and CRISPR/Cas9 has provided great help for the screening of SAD pathogenic genes. The APOE gene is currently recognized as a susceptibility gene for SAD. Wadhwani et al. (2019) used CRISPR/Cas9 system to correct the E4 allele into E3/E3 genotype in iPSCs from 2 AD patients, and found that E3 neurons are more resistant to ionomycin-induced cytotoxicity. However, E4 cells exhibited an increase in tau phosphorylation and contributed to the calcium dysregulation and ultimately cell death (Wadhwani et al., 2019). This finding intensifies our understanding of the relationship between APOE4 and the pathogenesis of SAD. In addition, Kolay and Diamond (2020) reported that knock-out of the 22 genes (derived from GWAS) associated with AD via CRISPR/Cas9 in HEK293T cells had no effect on the tau metabolism. A recent study conducted by the lab of the University of California Irvine reveals the crucial roles of TREM2 in the pathogenesis of AD (McQuade et al., 2020). They found that knock-out of TREM2 in iPSCs via CRISPR system, the survival of microglia, the clearance of APOE and SDF-1α/CXCR4-mediated chemotaxis was seriously impacted, which ultimately caused the impairment response to beta-amyloid plaques. As for the function of AD-associated TREM2 variant, Cheng-Hathaway et al. (2018) generate a mouse model of AD encoding the one copy R47H variant in TREM2 via CRISPR/Cas9 method, and found the heterozygous AD mouse showed reduced expression of TREM2 in cells and decreased myeloid cell responses to amyloid deposition, indicating the relationship between TREM2 R47H variant and increased risk of AD. Beyond that, researchers recently also found that p.S1038C variant may strengthen the risk of AD when a homozygous mutation (rs377155188, C > G, p.S1038C) was introduced in TTC3 gene via CRISPR/Cas9 system (Laverde-Paz et al., 2021). Furthermore, in order to investigate the function of STIM1 gene in neurodegeneration, Pascual-Caro et al. (2019) establish a STIM1 knock-out SH-SY5Y neuroblastoma cells model by CRISPR/Cas9 system, and demonstrated that once knock-out of STIM1 gene, mitochondrial function abnormality and calcium homeostasis imbalance rapidly occurred, ultimately leading to cell death. Notably, the expression of STIM1 is significantly reduced in the brain of patients with AD, and STIM1-deficient SH-SY5Y cells further substantiated the relationship between STIM1 gene and the onset of AD in vitro. In 2020, Pascual-Caro et al. (2020) found a remarkable reduction of ITPR3 following the knock-out of STIM1 gene, which caused the mitochondrial function abnormality. This further confirmed the potential role of STIM1-ITPR3 axis in the pathogenesis of AD (Pascual-Caro et al., 2020). Similarly, Knupp et al. (2020) found that the depletion of SORLA in hiPSCs by using CRISPR/Cas9 can cause early endosome enlargement in hiPSCs-derived neurons rather than hiPSC microglia, and inhibition of BACE is not able to rescue the endosome enlargement phenotype in hiPSC neurons, indicating the independent function of SORLA in AD pathogenesis. More convenient is a design of Duan et al. (2021) which is imaging based arrayed CRISPR to investigate the genes related to AD features. This will also provide a platform to explore the AD biology and an opportunity for drug discovery (Duan et al., 2021).
Targeted Therapy
AD is the most common neurodegenerative disease in the elderly. Cortical neuritic plaques and the neurofibrillary tangles are two neuropathological hallmarks of AD (Glenner and Wong, 1984; Masters et al., 1985). The neurofibrillary tangles are composed of hyperphosphorylated tau protein. The neuritic plaques are generally the result of the excessive cleavage of the APP by BACE1. Herein, we will mainly discuss the gene therapy strategies for AD (see Table 2), which focus on the regulation of Aβ expression by using CRISPR/Cas9 technology.
TABLE 2.
Targeted therapy of AD via CRISPR/Cas9 system.
| Target genes | Main results | Model system | Delivery system | References |
| Amyloid precursor protein (APP) | APP↓ Aβ↓ | Tg2576 mice as mutant models of APP familiar form of Alzheimer | Adeno-associated viral (AAV)-1 vectors | Gyorgy et al., 2018 |
| 3′-UTR APP | APP↓ Aβ↓ | C57BL/6 mice | px330 plasmid | Nagata et al., 2018 |
| beta-secretase 1 (Bace1) | Bace1↓ Aβ↓ Memory impairment↓ | 5XFAD as Alzheimer mouse model and wild-type mice | Micelle | Park et al., 2019 |
| γ-Secretase activating protein (GSAP) | GSAP↓ γ-Secretase activity↓ Aβ↓ | HEK-APP cell lines | Plasmid | Wong et al., 2019 |
| APOE | Turning APOE4 to APOE3↑ Hyper-phosphorylation Tau protein↓ Deposition of amyloid ↓ | Induced pluripotent stem cells (iPSCs) | Electroporation with three episomal plasmids | Wadhwani et al., 2019 |
| CD33 | hCD33m+ hCD33M- Aβ1–42 phagocytosis in microglia↑ | U937 cells | Not mentioned | Bhattacherjee et al., 2021 |
| Glia maturation factor (GMF) | GMF↓ p38 MAPK ↓ | BV2 microglial cell line | AAVpro | Raikwar et al., 2019 |
| CysLT1R | CysLT1R–/– hippocampal synaptic plasticity↑ amyloidogenesis↓ neuroinflammation in the hippocampus↓ | APP/PS1 mice | Not mentioned | Chen et al., 2021 |
CRISPR/Cas9 Targets APP Gene Mutations
The mutation in the APP gene causes dominantly inherited AD as a result of increased β-secretase cleavage of the amyloid-β (Aβ) precursor protein. The KM670/671NL APP mutation, indigenous to Sweden (APPsw for the mutation and APPsw for the mutant allele), results in an increase in enzymatic cleavage by β-secretase, and thereby increased Aβ protein levels. Gyorgy et al. (2018) reported that the expression of Aβ protein decreases when APP alleles were knocked out using CRISPR/Cas9 technology. Therefore, the CRISPR/Cas9 system may provide gene therapy strategies for AD patients with APP mutations. Moreover, Nagata et al. (2018) identified possible protective deletion mutations in the 3′-UTR of the APP gene in mice. They found a drastic reduction of Aβ accumulation when ∼700-bp of the 891-bp APP 3′-UTR in the mouse model zygotes was deleted using CRISPR/Cas9 technology. Interestingly, the A673T mutation is the reason for an Icelandic population that did not show symptoms of AD at an advanced age. This mutation can reduce β-secretase cleavage by 40% (Jonsson et al., 2012). Accordingly, Guyon et al. (2021) hypothesized that the insertion of this mutation in patients’ neurons could be an effective and sustainable method of slowing down or even hindering the progression of AD. To this end, they introduced a new mutation using CRISPR/Cas9-based strategy to modify the APP gene by converting the alanine codon to a threonine in HEK293T cells and SH-SY5Y cells (containing the APP gene with deaminated cytosine1 and cytosine2 positions). The accumulation of Aβ peptide have further reduced owing to a successful introduction of the A673T mutation in 53% of HEK293T cells alongside a new mutation (E674K). Likewise, in Sun’s laboratory (Sun et al., 2019), they also selectively edited endogenous APP at the extreme C-terminus using a CRISPR/Cas9-based strategy in cell and animal models, and reciprocally manipulated the amyloid pathway. Therefore, the Aβ production have been reduced by attenuating APP-β-cleavage, while elevating neuroprotective APP-α-cleavage.
CRISPR/Cas9 Targets Key Enzymes of Aβ Protein
CRISPR/cas9 Nanocomplex Targets BACE1
Aβ protein is formed by sequential modification of APP through BACE1 and γ-secretase. Thereby targeting BACE1 is a potential therapeutic strategy for the treatment of AD. Park et al. (2019) reported that the expression of BACE1 has been successfully reduced by employing Cas9 nanocomplexes, which was prepared by adding amphiphilic R7L10 peptide to Cas9-sgRNA, in two mouse models of AD.
CRISPR/cas9 Targets γ-Secretase Protease
Another target of gene therapy in AD is a large intramembrane protein complex known as γ-secretase protease which is regulated by γ-secretase activating protein (GSAP). There is evidence to suggest that reduction in GSAP expression decreases Aβ levels significantly (He et al., 2010; Ghosh and Surolia, 2011). Based on the claims, Wong et al. (2019) also knocked out GSAP with CRISPR-Cas9 technology in HEK293 cells that stably express APP (HEK-APP), leading to a remarkable reduction in Aβ secretion and γ-secretase activity.
Given that γ-secretase is regulated by the expression of the GSAP, and PSEN1 and PSEN2 are the key components of the γ secretase complex. Therefore, the mutations of PSEN1 would cause AD and be associated with most familial AD (Raux et al., 2005; Wu et al., 2011; Chavez-Gutierrez et al., 2012; Wang et al., 2018b). Most of these mutations impair amyloid metabolism, resulting in an elevation in Aβ42/40 ratio and Aβ42 levels, and/or reduced production of Aβ40 (Hung and Livesey, 2018). Also, PSEN1 gene mutations have been proven to be linked with the majority of early-onset familial AD. The statement was confirmed by Fang et al. (2006) who found a novel V97L missense mutation at codon 97 (Val97Leu) of the PSEN1 gene in a Chinese familial AD pedigree. To verify whether this mutation is pathologically functional, they established a mutation type of SH-SY5Y cell line via CRISPR/Cas9 system and detected Aβ production. Strikingly, the level of Aβ42 was significantly elevated both intracellularly and extracellularly in mutation type SH-SY5Y cells at 48 h when compared to wild type cells. This indicates that the mutation of PSEN1 is a potential factor of AD pathogenesis. In addition, Ortiz-Virumbrales et al. (2017) reported that cell lines harboring the PSEN2 N141I mutation displayed an increase in the Aβ42/40 in iPSC-derived basal forebrain cholinergic neurons (BFCNs). More importantly, increased Aβ42/40 was normalized following CRISPR/Cas-mediated correction of the N141I mutation in PSEN2.
CRISPR/Cas9 Targets Editing of APOE Genotype
The APOE4 isoform is the strongest genetic risk factor for SAD (Huang et al., 2017). As we know, APOE is mainly expressed by astrocytes in the central nervous system. However, APOE expression in neurons will imply occurrence of these events including the age-related cognitive impairment, neurological injury, and neurodegeneration. A study regarding therapeutic target for APOE4 by Wadhwani et al. (2019) showed that when the E4 allele was corrected to E3/E3 genotype in iPSCs from two patients with AD via CRISPR/Cas9 method, E3 neurons were less susceptible to ionomycin-induced cytotoxicity, and exhibited a decrease in tau phosphorylation. Furthermore, Lin et al. (2018) identified APOE4 function by using hiPSC and CRISPR/Cas9 technology, their results showed that APOE4 had an impact on the Aβ metabolism in a cell-type-specific manner by different ways. More exciting results showed that isogenic conversion of APOE4 to APOE3 can attenuate multiple AD-related pathologies (Lin et al., 2018). These findings further revealed that APOE4 is a promising target for the treatment of AD.
CRISPR/Cas9 Targets Proinflammatory Molecules
CRISPR/cas9 Targets CD33
Human genetic association studies indicate that immune response is also the major pathway of AD etiology. The importance of chronic neuroinflammation in AD, has been demonstrated by accumulating evidence. CD33, an immunomodulatory receptor, is expressed at high levels on neutrophils and low levels on microglia and has divergent roles in regulating phagocytosis responsible for AD pathology (Griciuc et al., 2013; Estus et al., 2019). Recent research by Bhattacherjee et al. (2019) showed that the genetic ablation of mCD33 enhanced microglia phagocytosis to increase Aβ clearance via CRISPR/Cas9 technology in U937 (highly expressing hCD33M) and attenuated the pathological phenotype of AD. Furthermore, their team disrupted CD33 gene using CRISPR/Cas9 and complemented with different variants of hCD33, leading to the alleviation of Aβ pathology and neurodegeneration. On the contrary to hCD33M that represses phagocytosis, hCD33m is a variant of hCD33 and improve Aβ phagocytosis (Bhattacherjee et al., 2021). These results provided strong support that AD-protective CD33 allele will facilitate AD therapeutics targeting these receptors.
CRISPR/cas9 Targets Glia Maturation Factor
Glia maturation factor (GMF), a newly discovered pro-inflammatory molecule, is predominantly expressed in the reactive glial cells surrounding the amyloid plaques and highly expressed in various AD brain regions (Ahmed et al., 2017). Overexpression of GMF usually leads to neuronal cell death by activating the p38 MAPK signaling pathway and oxidative toxicity. Raikwar et al. (2019) successfully reduced GMF expression in BV2 cells via CRISPR/Cas9 method, resulting in inhibition of pp38 MAPK to regulate GMF-induced proinflammation in microglia.
CRISPR/cas9 Targets cysLT1R
Cysteinyl leukotrienes (Cys-LTs) are a group of the inflammatory lipid molecules and initiate inflammatory signaling cascades by two major G-protein coupled receptors (CysLT1R and CysLT2R). In the recent years, mounting evidence shows that CysLT1R is intimately associated with the occurrence and development of AD, and can mediate inflammatory response via the NF-κB pathway (Yu et al., 2005; Wang et al., 2011). Reportedly, the high level of Aβ1–42 significantly causes the elevation of CysLT1R expression. The CysLT1R antagonist can remarkably suppress the overexpression of CysLT1R-mediated inflammatory response via the NF-κB pathway. Chen et al. (2021) also demonstrated that the deletion of CysLT1R via CRISPR/Cas9 system reduces amyloid pathology and alleviates neuroinflammation in APP/PS1 mice. As a result, Aβ1-42-induced cognitive and hippocampal synaptic impairments in APP/PS1 mice were ameliorated. In summary, this study will provide an insight into elucidation of the mechanism underlying CysLT1R-mediated AD pathology.
Delivery System of CRISPR/Cas9 in Alzheimer’s Disease
For therapeutic application, the CRISPR components must be delivered to mammalian cells to allow gene alteration in the host cell. Although considerable advances have been made in the in vivo administration of CRISPR/Cas9, the biocompatibility, safety and tissue specificity remain challenging. Relying on the genetic modification desired, different components of CRISPR/Cas9 are delivered. Amongst the simplest implementation, the Cas9/sgRNA pair is sufficient to disrupt genes (such as knock-out), however, the delivery of an additional piece of DNA is required for advanced bio-functions such as gene repair or insertion (knock-in). The CRISPR/Cas9 components can be delivered into cells in various forms: viral, mRNA, plasmid, and protein-based approaches. In the subsequent sections, we will further discuss the strengths and limitations regarding these potentially clinically translatable approaches, as well as their own separate potential for clinical applications.
Viral Delivery Methods
Viral methods are the most broadly used approaches for delivery of CRISPR/Cas9. At the present, many virus delivery systems have been utilized to deliver CRISPR/Cas9 including adeno-associated viruses (AAV), Adenoviral Vectors (AdV) and Lentiviral Vectors (LV) (Xu et al., 2019).
Adeno-Associated Viruses
Adeno-associated virus (AAV), one of the smallest viruses, is originally discovered as a contaminant of purified adenovirus preparation, and its replication is dependent on adenovirus due to a failure to encode a polymerase, thereby they have been called AAV (Rose et al., 1966). Along with numerous classes of viral vectors, AAVs have been widely used for CRISPR genome editing, the specific reasons are multiple. Firstly, after entering the cell, most of AAV is free from the host cell genome. Once integrated into the host genome, the provirus will be able to stably and continuously express for 1–2 years, which is relatively beneficial for the treatment of diseases. Secondly, AAV can widely infect different tissues due to the distinct capsids. Weinmann et al. (2020) has identified a myotropic AAV-9 variant by massively parallel in vivo evaluation of barcoded capsid variants, which exhibits superior efficiency and specificity in the musculature including skeletal muscle, heart and diaphragm following peripheral delivery. Thirdly, AAV can tolerate pH and temperature changes, and constantly keep its stable activity (Rayaprolu et al., 2013). To date, the only limitation seems to be the formulated concentration for delivery (Wright, 2008).
The robust stability of these vectors provides ample options for different administration routes and specialized delivery strategies. Researchers have delivered gene-editing components to mdx mice (a model of DMD) after birth by adeno-associated virus-9 (AAV9), finally leading to modification of the mutant dystrophin gene, and achieved success in treating DMD in vivo (Long et al., 2016). In addition, an open-label clinical trial proved that bilateral stereotactic administration of AAV2-NGF (CERE-110) to the nucleus basalis of Meynert could produce long-term, biologically active NGF expression, and AAV2-NGF was safe and well-tolerated for 2 years. Hence, AAV2-NGF may become an effective target for the treatment of AD (Rafii et al., 2014). CERE-120, also known as an adeno-associated virus type-2 (AAV2) vector encoding NTN, is currently being developed as a potential therapy for neurodegenerative disease. It was reported that when monkeys received bilateral injections of CERE-120 across a different range of doses, caused a dose-related increase in NTN protein expression within the striatum and substantia nigra (SN) pars compacta including nigrostriatal tyrosine hydroxylase (Th). Besides, the phosphorylated signal-regulated kinase responsible for common neurotrophic signaling event was activated (Herzog et al., 2008). Furthermore, Marks et al. (2016) provide evidence for the long-term safety of CERE-120 gene transfer.
One of the major limitations of AAVs is the small genome-packaging capacity of ∼4.7 kb. Since many diseases are caused by genes whose coding sequence exceeds this capacity, packaging into a single AAV capsid is currently not feasible for these genes. To overcome the size limit of AAV vector, one strategy is to split large transgenes into two or three parts, generating dual or triple AAV vectors (Akil, 2020); the other strategy is to use a smaller Cas9 variants that allow for the packaging of genes encoding both Cas9 and sgRNA into a single vector (Ran et al., 2015). Nevertheless, no method is currently perfect, and there are still many limitations to the application of AAV. As for dual or triple AAV vector strategy, the accumulation of mRNA (Xu et al., 2004) and low transduction levels (Maddalena et al., 2018) have become new obstacles. In addition, a smaller Cas9 variant from streptococcus aureus Cas9 (saCas9) could cause the body to produce humoral and cell-mediated adaptive immunity, again hindering its therapeutic application (Charlesworth et al., 2019).
Adenoviral Vectors
Adenoviridae is a medium-sized virus, about 90–100 nm large, is an icosahedral DNA virus without a mantle, and has a nucleocapsid. The genetic material of adenovirus is linear double-stranded DNA with a total length of about 30,000–42,000 bp (Wold and Toth, 2013). There are at least 57 serotypes of human Ads (Adenoviruses), Ad1-Ad57, that form seven “species” A–G (Wold and Toth, 2013). Ads mainly infect a variety of vertebrates, including humans, leading to lifelong immunity. In addition, there are no drugs approved specifically to treat Ad infections. Although Ad is dangerous, AdV was found to have many advantages. For instance, AdV can grow into high titer stable stocks and persist in their expression extrachromosomally rather than by integrating into the host genome (Wold and Toth, 2013). Furthermore, gene delivery vectors that do not rely on host cell genome integration could avoid insertion mutation and position effect variegation (Athanasopoulos et al., 2017).
Nonetheless, AdV also have certain shortcomings, for instance, a well-known highly immunogenicity. Early clinical trials for gene correction using AdV did not yield several successes in clinical therapy, and one trial resulted in a tragic fatality (Raper et al., 2003). In order to avoid this serious situation from happening again, Capasso et al. (2014) further demonstrated that making chemical modifications in AdV facilitated overcoming some of the early challenges regarding liver targeting and host immunity. Nevertheless, things are two-sided, as for cancer gene therapy, the strongly immune responses are beneficial for therapeutic outcomes. Therefore, AdV have recently been used in cancer treatment as oncolytic viruses. Many clinical trials use AdV to target a number of different cancers, such as prostate, ovarian, bladder, and refractory solid tumors (Burke et al., 2012; Kim et al., 2013; Freytag et al., 2014; Hemminki et al., 2015).
Lentiviral Vectors
Retroviruses are single-stranded RNA viruses whose genome is reverse transcribed into double-stranded DNA and integrated into the infected cell genome. Genomic integration leads to stable maintenance and potentially sustained expression. These features make them particularly appreciated when the stable, long-term expression was sought. The basic genes required for retroviral and lentiviral survival and function are the gag, pol, and env genes (Escors and Breckpot, 2010). In addition, the lentivirus genus of retroviruses includes a variety array of accessory genes (Delenda, 2004). The lentiviruses that infect various mammals have been transformed into lentiviral vectors, but the most used are those based on HIV-1 (Naldini et al., 1996). With the continuous development of lentiviral vectors, the safety of the third-generation lentiviral vectors is greatly improved (Milone and O’Doherty, 2018).
Retrovirus shows a very important role in clinical gene therapy. For instance, direct clinical benefit with chimeric antigen receptor T (CAR-T) cells is a promising novel therapy for many malignancies, such as leukemia and lymphoma (Schuster et al., 2019; Sterner et al., 2019). CAR-T cells are produced by ex vivo transduction of T cells with lentiviral vectors. Schuster et al. (2019) have found an exciting result that high rates of durable responses were produced with the use of tisagenlecleucel in relapsed or refractory diffuse large B-cell lymphoma in adults. However, because CD19 is a pan-B cell marker, one side effect is normal B-cell depletion. Nevertheless, CAR-T cell therapy is still an attractive alternative to treating or preventing leukemia and lymphoma. Following CAR-T cell therapy, another sensational study recently demonstrated the infinite possibilities of lentiviral vectors. The researchers used a lentiviral-mediated gene therapy program-LentiGlobinBB305 to treat patients with β-thalassemia and achieved significant curative effects. Meanwhile, they showed that gene therapy with lentiviral vectors is safe. Therefore, this new gene therapy is likely to rescue or reduce long-term blood transfusion for patients with transfusion-dependent beta thalassemia (TDT) (Thompson et al., 2018).
Plasmid-Based Approaches
Delivery of DNA encoding the Cas9 protein is an attractive way to introduce the CRISPR/Cas9 machinery into the cells. This method has advantages as below: firstly, gene synthesis is relatively simple; secondly, the synthesized gene does not need integrating into the host genome after being transferred into the host cell through a plasmid, and can be continuously expressed. Furthermore, the organ specific delivery of the CRISPR/Cas9 system is very important for the further application. One potential advantage of plasmid-based delivery is that tissue or cell-specific targeting can also be integrated into the plasmid itself.
In 2020, Lu et al. reported a first-in-human phase I clinical trial of CRISPR/Cas9 PD-1-edited T cells in patients with advanced non-small-cell lung cancer (Lu et al., 2020). In this clinical trial, the plasmids expressing two sgRNAs and Cas9 edited hPD-1 plasmids were co-transfected into T cells by electroporation. After infusion of edited T cells, the treatment-related adverse events were grade 1/2, indicating the safety and feasibility of therapeutic application of CRISPR/Cas9 in non-small-cell lung cancer.
Despite promising reports of the utility of CRISPR/Cas9 for in vivo gene editing, a principal problem in implementing process is how to deliver high molecular weight DNA to cells. At the present, Jo et al. (2020) designed a PLGA nanoparticle fluorescently labeled with the fluorophore 6, 13-bis (triisopropylsilylethynyl) pentacene (TIPS pentacene) to delivered Cas9 into primary bone marrow derived macrophages. Inspiringly, the expression of Cas9 protein was initially found after 24 h, and TIPS fluorescence was detected in most cells (Jo et al., 2020). In addition, Wang et al. (2018a) reported a strategy to deliver Cas9-sgPlk-1 plasmids (CPs) which is condensed on TAT peptide-modified Au nanoparticles (AuNPs/CP, ACP) via electrostatic interactions, following lipid-encapsulated and laser-controlled. This method for CRISPR/Cas9 delivery is highly efficient (Wang et al., 2018a). These studies indicate that the method for nanoparticles coating plasmids is likely to effective in the in vivo therapeutic application of CRISPR/Cas9. Besides, researchers have constructed a multifunctional nucleus-targeting “core-shell” artificial virus (RRPHC) to delivery CRISPR/Cas9 plasmid. The system can induce a higher targeted gene disruption than traditional transfection reagent (such as Lipofectamine 3000). More excitingly, the artificial virus can effectively target ovarian cancer via dual-receptor-mediated endocytosis (Li et al., 2017). Therefore, this will provide an ideal idea for efficient delivery and targeting of CRISPR/Cas9.
RNA-Based Delivery
Delivery of Cas9-encoded mRNA is another commonly used approach for the introduction of CRISPR machinery into the cells. Unlike gene-based delivery methods, mRNA-based strategies are transient in functioning, and circumventing the risks associated with integration into the host genome (Nelles et al., 2016). mRNA-based methods also benefit from more rapid effect since mRNA is transcribed in a matter of minutes (Zetsche et al., 2015). However, the obvious challenges regarding the maintenance of RNA stability and delivery of each component of CRISPR/Cas9 individually remains.
Recently, Beijing National Laboratory for Molecular Science has reported an CRISPR/Cas9 delivery nanocarrier called BAMEA-O16B that can efficiently deliver Cas9 mRNA and sgRNA into target cells. After intravenous injection of BAMEA-O16B/Cas9 mRNA/sgRNA nanoparticle, the lipid nanoparticle can efficiently accumulate in the liver, leading to a significant decrease of the proprotein convertase subtilisin/kexin type 9 in mouse serum (Liu et al., 2019a). Library of engineered LNPs (lipid nanoparticles) containing barcoded mRNA (b-mRNA) has been designed by Guimaraes et al. (2019) which enables direct barcoding and subsequent quantification of a functional mRNA, can accelerate the in vivo screening and design of LNPs for mRNA therapeutic applications such as CRISPR/Cas9 gene editing. These further broaden the therapeutic prospects of mRNA-based CRISPR/Cas9 technology. In addition, a previous study reported a combinatorial delivery method. In this study, researchers used lipid nanomaterials to deliver Cas9 mRNA, and used adeno-associated virus to deliver sgRNA and a repair template. Finally, treatment alleviated the disease symptoms in the mouse model of the human hereditary tyrosinemia (Yin et al., 2016). This result further shows the joint multiple delivery methods may be an effective strategy for the clinical application of CRISPR/Cas9 technology in the future, which can remedy the shortcomings of a single delivery method.
To improve genome-editing efficiency by increasing sgRNA stability, further studies reported the benefits of synthetic modifications to the sgRNA (Yin et al., 2017). This represents an effective method to overcome the stability issues associated with RNA-based CRISPR/Cas9 delivery.
Protein-Based CRISPR/Cas9 Strategies
In CRISPR/Cas9 gene editing technology, ribonucleoprotein (RNP) consisting of Cas9 protein and sgRNA is a powerful method for genome editing, which has various advantages including fast and safe, lower off-targeting, and higher editing efficiency. In addition, the Cas9 RNP system can be applied to various model organisms and cell types, such as stem cells (Wang et al., 2021), immune cells (Schumann et al., 2015), primary cells (Seki and Rutz, 2018), etc.
At the present, there are many options including physical approaches (such as microinjection, electroporation, biolistic, and microfluidic techniques) and synthetic carriers (lipid nanoparticles) can be used for the delivery Cas9 RNP system. Nevertheless, microinjection has strict requirements, cumbersome process and high cost. Hence the method needs to be further improved. More encouragingly, a recent work conducted by Chen et al. (2016) showed a simple and economic electroporation-based strategy to delivery Cas9 RNP system. Compared to microinjection, CRISPR RNP Electroporation of Zygotes (CRISPR-EZ) can efficiently increase the embryo viability (Chen et al., 2016). As for lipid nanoparticles to delivery Cas9 RNP, the lab of Hokkaido University has reported a lipid nanoparticle (LNP)-based CRISPR/Cas ribonucleoprotein (RNP) combined microfluidic techniques to delivery Cas9 and Cpf1 RNPs. The delivery technique following optimized formula, can efficiently suppress both HBV DNA and covalently closed circular DNA (cccDNA) in HBV-infected human liver cells (Suzuki et al., 2021). These progressively findings have made significant contributions to the development of the CRISPR/Cas9 delivery system and its therapeutic and clinical applications.
The above-mentioned delivery strategies are not actually independent of each other. Each delivery strategy has its own merits and limitations (see Table 3). Lattanzi et al. (2019) compared the different delivery systems-based CRISPR/Cas9 into HSPCs (hematopoietic stem and progenitor cells). Results showed that plasmid-based method has high editing efficiency but was associated with significant cell toxicity. RNA-mediated way has similar cell toxicity to plasmid-based method, and has less editing efficiency. Although LVs-based delivery system has minimal cell toxicity, the genome-editing efficiency is low. By contrast, RNP-based delivery of CRISPR/Cas9 exhibits a good balance between cytotoxicity and editing efficiency. Above all, it is often necessary to jointly apply these delivery methods to achieve desired result.
TABLE 3.
Generalized comparison of different CRISPR/Cas9 delivery format.
| Delivery format | Delivery vehicle | Advantages | Editing efficiency | Immunogenicity | Limitations | References |
| Plasmid | Electroporation; nanoparticle; AAV; artificial virus | Gene synthesis is simple; no need to integrate into the genome; tissue or cell-specific targeting | Moderate | Moderate | Low capacity | Li et al., 2017; Thompson et al., 2018; Wang et al., 2018a; Lu et al., 2020 |
| mRNA | Lipid nanoparticle; AAV | Transient in function; No need to integrate into the genome | Moderate | Moderate | Low RNA stability; delivery component individually | Zetsche et al., 2015; Yin et al., 2016; Guimaraes et al., 2019; Liu et al., 2019a |
| Protein | Microinjection; electroporation; biolistic; microfluidic techniques; lipid nanoparticles | Fast; lower off-targeting | High | Low | Non-specific | Chen et al., 2016; Seki and Rutz, 2018; Suzuki et al., 2021 |
Conclusion and Future Perspectives
Genome editing has entered a blooming period of development in recent years due to its extensive and effective application promise for scientific researches and disease treatment. The CRISPR/Cas9 system has been successfully applied to modify the genomes of different animals including mouse (Komor et al., 2016; Serneels et al., 2020; Takalo et al., 2020), rat (Serneels et al., 2020), monkey (Niu et al., 2014; Chen et al., 2015), cell lines including HT22 (Wang et al., 2019), N2a (Sun et al., 2017), stem cells (Paquet et al., 2016). More recently, there are already many publications on AD-related research mediated by CRISPR/Cas9, which mainly involves the use of this technology to construct AD model, screen pathogenic genes and treat AD via specific target genes (such as APP, BACE1, APOE4, CD33, GMF, and CysLT1R). However, considering potential off-target mismatches and specific tissue targeting in this technology, there are still numerous challenges to the eventual application of CRISPR-Cas9 to the clinical treatment of AD.
At present, a large number of researches are mainly focused on finding more efficient delivery systems. Protein-based CRISPR/Cas9 methods received more and more attention due to its low off-target effects, and higher editing efficiency. The development of CRISPR/Cas ribonucleoprotein (RNP) technology based on lipid nanoparticles (LNP) (Suzuki et al., 2021) has laid the foundation for the application of CRISPR/Cas9 technology in AD research and further clinical treatment. Notwithstanding these achievements, the numerous problems still exist. For instance, the AD animal models currently constructed are mostly rodents, however, the rodent model is not able to effectively and truly mimic the situation in the human brain, especially the occurrence of AD is closely related to the age factor since rodents generally have a short lifespan. Therefore, the application of results from animal models to the treatment of AD requires great care. Although the use of primates to build AD models by CRISPR/Cas9 strategy is likely to be a good strategy for simulating the age-dependent characteristics of AD, the long reproductive cycle of primates and the strict requirements for the growth environment also limit its application.
In summary, CRISPR/Cas9 is a masterpiece in gene editing technology this century. The gene scissors open new avenues in clinical gene therapy. Compared to other gene editing technologies, it has the advantages of short cycle, low cytotoxicity, low price, simple delivery, etc. Hence, all these characteristics make CRISPR-Cas9 system endowed with a broader application prospect in the clinical therapy of AD albeit taking into consideration of some drawbacks.
Author Contributions
LL and XY wrote the manuscript with support from YC, MS, and HY. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We try to provide readers with comprehensive and updated reviews of recent literature. However, some studies may have been omitted involuntarily. We apologize for the inconvenience caused to you, and hope our colleagues can forgive us for neglecting their work. We also thank the reviewers for their work.
Funding
This work was supported by grants from the National Key R&D Program of China (2019YFA0802600), the National Natural Science Foundation of China (81974244, 82071551, and 81570960), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (5832013521).
References
- Ahmed M. E., Iyer S., Thangavel R., Kempuraj D., Selvakumar G. P., Raikwar S. P., et al. (2017). Co-Localization of glia maturation factor with NLRP3 inflammasome and autophagosome markers in human Alzheimer’s disease brain. J. Alzheimers Dis. 60 1143–1160. 10.3233/JAD-170634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil O. (2020). Dual and triple AAV delivery of large therapeutic gene sequences into the inner ear. Hear. Res. 394:107912. 10.1016/j.heares.2020.107912 [DOI] [PubMed] [Google Scholar]
- Athanasopoulos T., Munye M. M., Yanez-Munoz R. J. (2017). Nonintegrating gene therapy vectors. Hematol. Oncol. Clin. North Am. 31 753–770. 10.1016/j.hoc.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Baglietto-Vargas D., Shi J., Yaeger D. M., Ager R., LaFerla F. M. (2016). Diabetes and Alzheimer’s disease crosstalk. Neurosci. Biobehav. Rev. 64 272–287. [DOI] [PubMed] [Google Scholar]
- Banan M. (2020). Recent advances in CRISPR/Cas9-mediated knock-ins in mammalian cells. J. Biotechnol. 308 1–9. 10.1016/j.jbiotec.2019.11.010 [DOI] [PubMed] [Google Scholar]
- Bhattacherjee A., Jung J., Zia S., Ho M., Eskandari-Sedighi G., St Laurent C. D., et al. (2021). The CD33 short isoform is a gain-of-function variant that enhances Abeta1-42 phagocytosis in microglia. Mol. Neurodegener. 16:19. 10.1186/s13024-021-00443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee A., Rodrigues E., Jung J., Luzentales-Simpson M., Enterina J. R., Galleguillos D., et al. (2019). Repression of phagocytosis by human CD33 is not conserved with mouse CD33. Commun. Biol. 2:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Lamm D. L., Meng M. V., Nemunaitis J. J., Stephenson J. J., Arseneau J. C., et al. (2012). A first in human phase 1 study of CG0070, a GM-CSF expressing oncolytic adenovirus, for the treatment of nonmuscle invasive bladder cancer. J. Urol. 188 2391–2397. 10.1016/j.juro.2012.07.097 [DOI] [PubMed] [Google Scholar]
- Calsolaro V., Edison P. (2016). Neuroinflammation in Alzheimer’s disease: current evidence and future directions. Alzheimers Dement. 12 719–732. 10.1016/j.jalz.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Capasso C., Garofalo M., Hirvinen M., Cerullo V. (2014). The evolution of adenoviral vectors through genetic and chemical surface modifications. Viruses 6 832–855. 10.3390/v6020832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth C. T., Deshpande P. S., Dever D. P., Camarena J., Lemgart V. T., Cromer M. K., et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25 249–254. 10.1038/s41591-018-0326-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Gutierrez L., Bammens L., Benilova I., Vandersteen A., Benurwar M., Borgers M., et al. (2012). The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 31 2261–2274. 10.1038/emboj.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Fang S., Du Y., Ghosh A., Reed M. N., Long Y., et al. (2021). CRISPR/Cas9-mediated CysLT1R deletion reverses synaptic failure, amyloidosis and cognitive impairment in APP/PS1 mice. Aging (Albany NY) 13 6634–6661. 10.18632/aging.202501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Lee B., Lee A. Y., Modzelewski A. J., He L. (2016). Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 291 14457–14467. 10.1074/jbc.M116.733154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cui Y., Shen B., Niu Y., Zhao X., Wang L., et al. (2015). Germline acquisition of Cas9/RNA-mediated gene modifications in monkeys. Cell Res. 25 262–265. 10.1038/cr.2014.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng-Hathaway P. J., Reed-Geaghan E. G., Jay T. R., Casali B. T., Bemiller S. M., Puntambekar S. S., et al. (2018). The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol. Neurodegener. 13:29. 10.1186/s13024-018-0262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J. F., O’Riordan K. J., Cowan C. S. M., Sandhu K. V., Bastiaanssen T. F. S., Boehme M., et al. (2019). The microbiota-gut-brain axis. Physiol. Rev. 99 1877–2013. [DOI] [PubMed] [Google Scholar]
- Delenda C. (2004). Lentiviral vectors: optimization of packaging, transduction and gene expression. J. Gene Med. 6 (Suppl. 1), S125–S138. 10.1002/jgm.501 [DOI] [PubMed] [Google Scholar]
- Dow L. E., Fisher J., O’Rourke K. P., Muley A., Kastenhuber E. R., Livshits G., et al. (2015). Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Hu M., Tamm J. A., Grinberg Y. Y., Shen F., Chai Y., et al. (2021). Arrayed CRISPR reveals genetic regulators of tau aggregation, autophagy and mitochondria in Alzheimer’s disease model. Sci. Rep. 11:2879. 10.1038/s41598-021-82658-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Breckpot K. (2010). Lentiviral vectors in gene therapy: their current status and future potential. Arch. Immunol. Ther. Exp. (Warsz) 58 107–119. 10.1007/s00005-010-0063-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estus S., Shaw B. C., Devanney N., Katsumata Y., Press E. E., Fardo D. W. (2019). Evaluation of CD33 as a genetic risk factor for Alzheimer’s disease. Acta Neuropathol. 138 187–199. 10.1007/s00401-019-02000-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B., Jia L., Jia J. (2006). Chinese Presenilin-1 V97L mutation enhanced Abeta42 levels in SH-SY5Y neuroblastoma cells. Neurosci. Lett. 406 33–37. 10.1016/j.neulet.2006.06.072 [DOI] [PubMed] [Google Scholar]
- Freytag S. O., Stricker H., Lu M., Elshaikh M., Aref I., Pradhan D., et al. (2014). Prospective randomized phase 2 trial of intensity modulated radiation therapy with or without oncolytic adenovirus-mediated cytotoxic gene therapy in intermediate-risk prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 89 268–276. 10.1016/j.ijrobp.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U S A. 109 E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Surolia A. (2011). gamma-secretase activating protein is a therapeutic target for alzheimer ’s disease: a path-breaking discovery finally paving way towards therapeutic intervention. Ann. Neurosci. 18 1–2. 10.5214/ans.0972.7531.1118101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giau V. V., Lee H., Shim K. H., Bagyinszky E., An S. S. A. (2018). Genome-editing applications of CRISPR-Cas9 to promote in vitro studies of Alzheimer’s disease. Clin. Interv. Aging 13 221–233. 10.2147/CIA.S155145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenner G. G., Wong C. W. (1984). Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120 885–890. 10.1016/s0006-291x(84)80190-4 [DOI] [PubMed] [Google Scholar]
- Griciuc A., Serrano-Pozo A., Parrado A. R., Lesinski A. N., Asselin C. N., Mullin K., et al. (2013). Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78 631–643. 10.1016/j.neuron.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen P. M., Bunschoten A. E., van Soolingen D., van Embden J. D. (1993). Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10 1057–1065. 10.1111/j.1365-2958.1993.tb00976.x [DOI] [PubMed] [Google Scholar]
- Guimaraes P. P. G., Zhang R., Spektor R., Tan M., Chung A., Billingsley M. M., et al. (2019). Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control Release 316 404–417. 10.1016/j.jconrel.2019.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D., Bhattacharjee O., Mandal D., Sen M. K., Dey D., Dasgupta A., et al. (2019). CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci. 232:116636. 10.1016/j.lfs.2019.116636 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Shukla P. (2017). Gene editing for cell engineering: trends and applications. Crit. Rev. Biotechnol. 37 672–684. 10.1080/07388551.2016.1214557 [DOI] [PubMed] [Google Scholar]
- Guyon A., Rousseau J., Begin F. G., Bertin T., Lamothe G., Tremblay J. P. (2021). Base editing strategy for insertion of the A673T mutation in the APP gene to prevent the development of AD in vitro. Mol. Ther. Nucleic Acids 24 253–263. 10.1016/j.omtn.2021.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy B., Loov C., Zaborowski M. P., Takeda S., Kleinstiver B. P., Commins C., et al. (2018). CRISPR/Cas9 mediated disruption of the swedish APP allele as a therapeutic approach for early-onset Alzheimer’s disease. Mol. Ther. Nucleic Acids 11 429–440. 10.1016/j.omtn.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafy A. S., Schoch S., Lamprecht A. (2020). CRISPR/Cas9 delivery potentials in Alzheimer’s disease management: a mini review. Pharmaceutics 12:801. 10.3390/pharmaceutics12090801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Luo W., Li P., Remmers C., Netzer W. J., Hendrick J., et al. (2010). Gamma-secretase activating protein is a therapeutic target for Alzheimer’s disease. Nature 467 95–98. 10.1038/nature09325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki O., Parviainen S., Juhila J., Turkki R., Linder N., Lundin J., et al. (2015). Immunological data from cancer patients treated with Ad5/3-E2F-Delta24-GMCSF suggests utility for tumor immunotherapy. Oncotarget 6 4467–4481. 10.18632/oncotarget.2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P. W., van Soolingen D., Bik E. M., de Haas P. E., Dale J. W., van Embden J. D. (1991). Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 59 2695–2705. 10.1128/iai.59.8.2695-2705.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog C. D., Dass B., Gasmi M., Bakay R., Stansell J. E., Tuszynski M., et al. (2008). Transgene expression, bioactivity, and safety of CERE-120 (AAV2-neurturin) following delivery to the monkey striatum. Mol. Ther. 16 1737–1744. 10.1038/mt.2008.170 [DOI] [PubMed] [Google Scholar]
- Huang H. C., Hong L., Chang P., Zhang J., Lu S. Y., Zheng B. W., et al. (2015). Chitooligosaccharides attenuate Cu2+-induced cellular oxidative damage and cell apoptosis involving Nrf2 activation. Neurotox Res. 27 411–420. 10.1007/s12640-014-9512-x [DOI] [PubMed] [Google Scholar]
- Huang Y. A., Zhou B., Wernig M., Sudhof T. C. (2017). ApoE2, ApoE3, and ApoE4 differentially stimulate App transcription and abeta secretion. Cell 168 42 427–441.e21. 10.1016/j.cell.2016.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. O. Y., Livesey F. J. (2018). Altered gamma-Secretase processing of APP disrupts lysosome and autophagosome function in monogenic Alzheimer’s disease. Cell Rep. 25 3647–3660.e2. 10.1016/j.celrep.2018.11.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 5429–5433. 10.1128/jb.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Embden J. D., Gaastra W., Schouls L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43 1565–1575. 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., MacLeod A., Tamaki K., Neil D. L., Monckton D. G. (1991). Minisatellite repeat coding as a digital approach to DNA typing. Nature 354 204–209. 10.1038/354204a0 [DOI] [PubMed] [Google Scholar]
- Jia L., Du Y., Chu L., Zhang Z., Li F., Lyu D., et al. (2020). Prevalence, risk factors, and management of dementia and mild cognitive impairment in adults aged 60 years or older in China: a cross-sectional study. Lancet Public Health. 5 e661–e671. 10.1016/S2468-2667(20)30185-7 [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A., Ringel-Scaia V. M., McDaniel D. K., Thomas C. A., Zhang R., Riffle J. S., et al. (2020). Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR-Cas9 plasmid. J. Nanobiotechnol. 18:16. 10.1186/s12951-019-0564-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., Bjornsson S., et al. (2012). A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488 96–99. 10.1038/nature11283 [DOI] [PubMed] [Google Scholar]
- Kaulich M., Lee Y. J., Lonn P., Springer A. D., Meade B. R., Dowdy S. F. (2015). Efficient CRISPR-rAAV engineering of endogenous genes to study protein function by allele-specific RNAi. Nucleic Acids Res. 43:e45. 10.1093/nar/gku1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadempar S., Familghadakchi S., Motlagh R. A., Farahani N., Dashtiahangar M., Rezaei H., et al. (2019). CRISPR-Cas9 in genome editing: its function and medical applications. J. Cell Physiol. 234 5751–5761. 10.1002/jcp.27476 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Dmitriev I. P., Saddekni S., Kashentseva E. A., Harris R. D., Aurigemma R., et al. (2013). A phase I clinical trial of Ad5/3-Delta24, a novel serotype-chimeric, infectivity-enhanced, conditionally-replicative adenovirus (CRAd), in patients with recurrent ovarian cancer. Gynecol. Oncol. 130 518–524. 10.1016/j.ygyno.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knupp A., Mishra S., Martinez R., Braggin J. E., Szabo M., Kinoshita C., et al. (2020). Depletion of the AD risk gene SORL1 selectively impairs neuronal endosomal traffic independent of amyloidogenic APP processing. Cell Rep. 31:107719. 10.1016/j.celrep.2020.107719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolay S., Diamond M. I. (2020). Alzheimer’s disease risk modifier genes do not affect tau aggregate uptake, seeding or maintenance in cell models. FEBS Open Bio. 10 1912–1920. 10.1002/2211-5463.12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattanzi A., Meneghini V., Pavani G., Amor F., Ramadier S., Felix T., et al. (2019). Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol. Ther. 27 137–150. 10.1016/j.ymthe.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverde-Paz M. J., Nuytemans K., Wang L., Vance J. M., Pericak-Vance M. A., Dykxhoorn D. M., et al. (2021). Derivation of stem cell line UMi028-A-2 containing a CRISPR/Cas9 induced Alzheimer’s disease risk variant p.S1038C in the TTC3 gene. Stem Cell Res. 52:102258. 10.1016/j.scr.2021.102258 [DOI] [PubMed] [Google Scholar]
- Li L., Song L., Liu X., Yang X., Li X., He T., et al. (2017). Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano 11 95–111. 10.1021/acsnano.6b04261 [DOI] [PubMed] [Google Scholar]
- Lieber M. R. (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79 181–211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Seo J., Gao F., Feldman H. M., Wen H. L., Penney J., et al. (2018). APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-Derived brain cell types. Neuron 98:1294. 10.1016/j.neuron.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino C. A., Harper J. C., Carney J. P., Timlin J. A. (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25 1234–1257. 10.1080/10717544.2018.1474964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhang L., Liu H., Cheng K. (2017). Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 266 17–26. 10.1016/j.jconrel.2017.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L., et al. (2019a). Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 31:e1902575. 10.1002/adma.201902575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Nguyen M., Robert A., Meunier B. (2019b). Metal ions in Alzheimer’s disease: a key role or not? Acc. Chem. Res. 52 2026–2035. 10.1021/acs.accounts.9b00248 [DOI] [PubMed] [Google Scholar]
- Long C., Amoasii L., Mireault A. A., McAnally J. R., Li H., Sanchez-Ortiz E., et al. (2016). Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351 400–403. 10.1126/science.aad5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., et al. (2020). Publisher correction: safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26:1149. 10.1038/s41591-020-0973-6 [DOI] [PubMed] [Google Scholar]
- Maddalena A., Tornabene P., Tiberi P., Minopoli R., Manfredi A., Mutarelli M., et al. (2018). Triple vectors expand AAV transfer capacity in the retina. Mol. Ther. 26 524–541. 10.1016/j.ymthe.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzavinos V., Alexiou A. (2017). Biomarkers for Alzheimer’s disease diagnosis. Curr. Alzheimer Res. 14 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks W. J., Jr., Baumann T. L., Bartus R. T. (2016). Long-Term safety of patients with Parkinson’s disease receiving rAAV2-Neurturin (CERE-120) gene transfer. Hum. Gene Ther. 27 522–527. 10.1089/hum.2015.134 [DOI] [PubMed] [Google Scholar]
- Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. (1985). Amyloid plaque core protein in Alzheimer disease and down syndrome. Proc. Natl. Acad. Sci. U S A. 82 4245–4249. 10.1073/pnas.82.12.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A., Kang Y. J., Hasselmann J., Jairaman A., Sotelo A., Coburn M., et al. (2020). Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer’s disease. Nat Commun. 11:5370. 10.1038/s41467-020-19227-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mila-Aloma M., Salvado G., Gispert J. D., Vilor-Tejedor N., Grau-Rivera O., Sala-Vila A., et al. (2020). Amyloid beta, tau, synaptic, neurodegeneration, and glial biomarkers in the preclinical stage of the Alzheimer’s continuum. Alzheimers Dement. 16 1358–1371. 10.1002/alz.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone M. C., O’Doherty U. (2018). Clinical use of lentiviral vectors. Leukemia 32 1529–1541. 10.1038/s41375-018-0106-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J., Diez-Villasenor C., Garcia-Martinez J., Soria E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60 174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Mojica F. J., Diez-Villasenor C., Soria E., Juez G. (2000). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36 244–246. 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- Mojica F. J., Juez G., Rodriguez-Valera F. (1993). Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 9 613–621. 10.1111/j.1365-2958.1993.tb01721.x [DOI] [PubMed] [Google Scholar]
- Nagata K., Takahashi M., Matsuba Y., Okuyama-Uchimura F., Sato K., Hashimoto S., et al. (2018). Generation of App knock-in mice reveals deletion mutations protective against Alzheimer’s disease-like pathology. Nat. Commun. 9:1800. 10.1038/s41467-018-04238-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata A., Amemura M., Makino K. (1989). Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J. Bacteriol. 171 3553–3556. 10.1128/jb.171.6.3553-3556.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F. H., et al. (1996). In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272 263–267. 10.1126/science.272.5259.263 [DOI] [PubMed] [Google Scholar]
- Nelles D. A., Fang M. Y., O’Connell M. R., Xu J. L., Markmiller S. J., Doudna J. A., et al. (2016). Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165 488–496. 10.1016/j.cell.2016.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156 836–843. 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- No authors listed (2020). 2020 Alzheimer’s disease facts and figures. Alzheimers Dement. Online ahead of print. 10.1002/alz.12068 [DOI] [PubMed] [Google Scholar]
- Ortiz-Virumbrales M., Moreno C. L., Kruglikov I., Marazuela P., Sproul A., Jacob S., et al. (2017). CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer’s PSEN2 (N141I) neurons. Acta Neuropathol. Commun. 5:77. 10.1186/s40478-017-0475-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533 125–129. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- Park H., Oh J., Shim G., Cho B., Chang Y., Kim S., et al. (2019). In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 22 524–528. 10.1038/s41593-019-0352-0 [DOI] [PubMed] [Google Scholar]
- Pascual-Caro C., Berrocal M., Lopez-Guerrero A. M., Alvarez-Barrientos A., Pozo-Guisado E., Gutierrez-Merino C., et al. (2019). Correction to: STIM1 deficiency is linked to Alzheimer’s disease and triggers cell death in SH-SY5Y cells by upregulation of L-type voltage-operated Ca(2+) entry. J. Mol. Med. (Berl) 97 1215–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Caro C., Orantos-Aguilera Y., Sanchez-Lopez I., de Juan-Sanz J., Parys J. B., Area-Gomez E., et al. (2020). STIM1 deficiency leads to specific down-regulation of ITPR3 in SH-SY5Y cells. Int. J. Mol. Sci. 21:6598. 10.3390/ijms21186598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Ortiz J. M., Swerdlow R. H. (2019). Mitochondrial dysfunction in Alzheimer’s disease: role in pathogenesis and novel therapeutic opportunities. Br. J. Pharmacol. 176 3489–3507. 10.1111/bph.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii M. S., Baumann T. L., Bakay R. A., Ostrove J. M., Siffert J., Fleisher A. S., et al. (2014). A phase1 study of stereotactic gene delivery of AAV2-NGF for Alzheimer’s disease. Alzheimers Dement. 10 571–581. 10.1016/j.jalz.2013.09.004 [DOI] [PubMed] [Google Scholar]
- Raikwar S. P., Thangavel R., Dubova I., Selvakumar G. P., Ahmed M. E., Kempuraj D., et al. (2019). Targeted gene editing of glia maturation factor in microglia: a novel Alzheimer’s disease therapeutic target. Mol. Neurobiol. 56 378–393. 10.1007/s12035-018-1068-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Cong L., Yan W. X., Scott D. A., Gootenberg J. S., Kriz A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper S. E., Chirmule N., Lee F. S., Wivel N. A., Bagg A., Gao G. P., et al. (2003). Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 80 148–158. 10.1016/j.ymgme.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Raux G., Guyant-Marechal L., Martin C., Bou J., Penet C., Brice A., et al. (2005). Molecular diagnosis of autosomal dominant early onset Alzheimer’s disease: an update. J. Med. Genet. 42 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayaprolu V., Kruse S., Kant R., Venkatakrishnan B., Movahed N., Brooke D., et al. (2013). Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 87 13150–13160. 10.1128/JVI.01415-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Hoggan M. D., Shatkin A. J. (1966). Nucleic acid from an adeno-associated virus: chemical and physical studies. Proc. Natl. Acad. Sci. U S A. 56 86–92. 10.1073/pnas.56.1.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann K., Lin S., Boyer E., Simeonov D. R., Subramaniam M., Gate R. E., et al. (2015). Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U S A. 112 10437–10442. 10.1073/pnas.1512503112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. J., Bishop M. R., Tam C. S., Waller E. K., Borchmann P., McGuirk J. P., et al. (2019). Tisagenlecleucel in adult relapsed or refractory diffuse large B-Cell lymphoma. N. Engl. J. Med. 380 45–56. [DOI] [PubMed] [Google Scholar]
- Seki A., Rutz S. (2018). Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 215 985–997. 10.1084/jem.20171626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serneels L., T’Syen D., Perez-Benito L., Theys T., Holt M. G., De Strooper B. (2020). Modeling the beta-secretase cleavage site and humanizing amyloid-beta precursor protein in rat and mouse to study Alzheimer’s disease. Mol. Neurodegener. 15:60. 10.1186/s13024-020-00399-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Kim S. R. (2021). Linking oxidative stress and proteinopathy in Alzheimer’s disease. Antioxidants (Basel) 10:1231. 10.3390/antiox10081231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Tang S., Dong L., Han X., Cong L., Dong J., et al. (2019). The neuroprotection of KIBRA in promoting neuron survival and against amyloid beta-induced apoptosis. Front. Cell Neurosci. 13:137. 10.3389/fncel.2019.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R. M., Sakemura R., Cox M. J., Yang N., Khadka R. H., Forsman C. L., et al. (2019). GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood 133 697–709. 10.1182/blood-2018-10-881722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Carlson-Stevermer J., Das U., Shen M., Delenclos M., Snead A. M., et al. (2019). CRISPR/Cas9 editing of APP C-terminus attenuates beta-cleavage and promotes alpha-cleavage. Nat. Commun. 10:53. 10.1038/s41467-018-07971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhou R., Yang G., Shi Y. (2017). Analysis of 138 pathogenic mutations in presenilin-1 on the in vitro production of Abeta42 and Abeta40 peptides by gamma-secretase. Proc. Natl. Acad. Sci. U S A. 114 E476–E485. 10.1073/pnas.1618657114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Onuma H., Sato R., Sato Y., Hashiba A., Maeki M., et al. (2021). Lipid nanoparticles loaded with ribonucleoprotein-oligonucleotide complexes synthesized using a microfluidic device exhibit robust genome editing and hepatitis B virus inhibition. J. Control Release 330 61–71. 10.1016/j.jconrel.2020.12.013 [DOI] [PubMed] [Google Scholar]
- Sweeney M. D., Montagne A., Sagare A. P., Nation D. A., Schneider L. S., Chui H. C., et al. (2019). Vascular dysfunction-the disregarded partner of Alzheimer’s disease. Alzheimers Dement. 15 158–167. 10.1016/j.jalz.2018.07.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takalo M., Wittrahm R., Wefers B., Parhizkar S., Jokivarsi K., Kuulasmaa T., et al. (2020). The Alzheimer’s disease-associated protective Plcgamma2-P522R variant promotes immune functions. Mol. Neurodegener. 15:52. 10.1186/s13024-020-00402-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. A., Walters M. C., Kwiatkowski J., Rasko J. E. J., Ribeil J. A., Hongeng S., et al. (2018). Gene therapy in patients with transfusion-dependent beta-thalassemia. N. Engl. J. Med. 378 1479–1493. 10.1056/NEJMoa1705342 [DOI] [PubMed] [Google Scholar]
- Tonnies E., Trushina E. (2017). Oxidative stress, synaptic dysfunction, and Alzheimer’s disease. J. Alzheimers Dis. 57 1105–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz R., Rodriguez-Perales S. (2017). CRISPR-Cas9 technology: applications and human disease modelling. Brief Funct. Genomics 16 4–12. 10.1093/bfgp/elw025 [DOI] [PubMed] [Google Scholar]
- Wadhwani A. R., Affaneh A., Van Gulden S., Kessler J. A. (2019). Neuronal apolipoprotein E4 increases cell death and phosphorylated tau release in alzheimer disease. Ann. Neurol. 85 726–739. 10.1002/ana.25455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Alinaghi S., Tafakhori A., Sikora E., Azcona L. J., Karkheiran S., et al. (2018a). Genetic screening in two Iranian families with early-onset Alzheimer’s disease identified a novel PSEN1 mutation. Neurobiol. Aging 62 244.e15–244.e17. 10.1016/j.neurobiolaging.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Du C., Lv J., Wei W., Cui Y., Xie X. (2011). Antiasthmatic drugs targeting the cysteinyl leukotriene receptor 1 alleviate central nervous system inflammatory cell infiltration and pathogenesis of experimental autoimmune encephalomyelitis. J. Immunol. 187 2336–2345. 10.4049/jimmunol.1100333 [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang L., Zheng W., Cong L., Guo Z., Xie Y., et al. (2018b). Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew Chem. Int. Ed Engl. 57 1491–1496. 10.1002/anie.201708689 [DOI] [PubMed] [Google Scholar]
- Wang R., Graham S., Gao L., Tam J., Levesque M. C. (2021). Editing the immune system in vivo in mice using CRISPR/Cas9 ribonucleoprotein (RNP)-mediated gene editing of transplanted hematopoietic stem cells. Methods 194 30–36. 10.1016/j.ymeth.2021.01.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y., Bharti V., Zhou H., Hoi V., Tan H., et al. (2019). Upregulation of thioredoxin-interacting protein in brain of amyloid-beta protein precursor/presenilin 1 transgenic mice and amyloid-beta treated neuronal cells. J. Alzheimers Dis. 72 139–150. 10.3233/JAD-190223 [DOI] [PubMed] [Google Scholar]
- Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., et al. (2020). Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 11:5432. 10.1038/s41467-020-19230-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Toth K. (2013). Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 13 421–433. 10.2174/1566523213666131125095046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E., Liao G. P., Chang J. C., Xu P., Li Y. M., Greengard P. (2019). GSAP modulates gamma-secretase specificity by inducing conformational change in PS1. Proc. Natl. Acad. Sci. U S A. 116 6385–6390. 10.1073/pnas.1820160116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. F. (2008). Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther. 15 840–848. 10.1038/gt.2008.65 [DOI] [PubMed] [Google Scholar]
- Wu Y. Y., Cheng I. H., Lee C. C., Chiu M. J., Lee M. J., Chen T. F., et al. (2011). Clinical phenotype of G206D mutation in the presenilin 1 gene in pathologically confirmed familial Alzheimer’s disease. J. Alzheimers Dis. 25 145–150. 10.3233/JAD-2011-102031 [DOI] [PubMed] [Google Scholar]
- Xie H., Tang L., He X., Liu X., Zhou C., Liu J., et al. (2018). SaCas9 requires 5′-NNGRRT-3′ PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol. J. 13:e1700561. [DOI] [PubMed] [Google Scholar]
- Xu C. L., Ruan M. Z. C., Mahajan V. B., Tsang S. H. (2019). Viral delivery systems for CRISPR. Viruses 11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Yue Y., Lai Y., Ye C., Qiu J., Pintel D. J., et al. (2004). Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum. Gene Ther. 15 896–905. 10.1089/hum.2004.15.896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., Jaenisch R. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Song C. Q., Dorkin J. R., Zhu L. J., Li Y., Wu Q., et al. (2016). Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34 328–333. 10.1038/nbt.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Song C. Q., Suresh S., Wu Q., Walsh S., Rhym L. H., et al. (2017). Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35 1179–1187. 10.1038/nbt.4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Wei E. Q., Zhang S. H., Xu H. M., Chu L. S., Zhang W. P., et al. (2005). Montelukast, a cysteinyl leukotriene receptor-1 antagonist, dose- and time-dependently protects against focal cerebral ischemia in mice. Pharmacology 73 31–40. 10.1159/000081072 [DOI] [PubMed] [Google Scholar]
- Zetsche B., Volz S. E., Zhang F. (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33 139–142. 10.1038/nbt.3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H., Bendlin B. B. (2021). Biomarkers for Alzheimer’s disease-preparing for a new era of disease-modifying therapies. Mol. Psychiatry 26 296–308. 10.1038/s41380-020-0721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H., Mattsson N. (2014). Understanding the cause of sporadic Alzheimer’s disease. Expert Rev. Neurother. 14 621–630. 10.1586/14737175.2014.915740 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen C., Mak M. S., Lu J., Wu Z., Chen Q., et al. (2020). Advance of sporadic Alzheimer’s disease animal models. Med. Res. Rev. 40 431–458. 10.1002/med.21624 [DOI] [PubMed] [Google Scholar]
- Zuo E., Cai Y. J., Li K., Wei Y., Wang B. A., Sun Y., et al. (2017). One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 27 933–945. 10.1038/cr.2017.81 [DOI] [PMC free article] [PubMed] [Google Scholar]


