Abstract
Long noncoding RNAs (lncRNAs) are involved in a variety of cancers, but the role of LncRNA DUBR in lung adenocarcinoma (LUAD), the most prevalent form of lung cancer, remains unclear. In this study we investigated the expression of DUBR in LUAD to ascertain its association with the clinical pathology and prognosis of LUAD. Analysis of mRNA expression in The Cancer Genome Atlas (TCGA) LUAD database and in-house LUAD cohort (n = 94) showed that DUBR was significantly downregulated in LUAD, and was associated with poor prognosis. In LUAD cell lines (H1975, A549), overexpression of DUBR significantly suppressed the migration and invasion of the LUAD cells. We demonstrated that c-Myc could bind to the promoter of DUBR, and transcriptionally suppressed its expression. Knockdown of c-Myc almost completely blocked the invasion and migration of LUAD cells, whereas knockdown of DUBR partially rescued c-Myc-knockdown suppressed cell migration and invasion. Furthermore, DUBR overexpression significantly increased the expression of a downstream protein of DUBR, zinc finger, and BTB domain containing 11 (ZBTB11), in H1975 and A549 cells; knockdown of ZBTB11 partially rescued the DUBR-overexpression suppressed cell migration and invasion; knockdown of c-Myc significantly upregulated the expression of ZBTB11 in LUAD cells. Finally, we revealed that DUBR/ZBTB11 axis suppressed oxidative phosphorylation in LUAD cells. In short, we demonstrate that c-Myc/DUBR/ZBTB11 axis suppresses migration and invasion of LUAD by attenuating cell oxidative phosphorylation, which provides new insights into the regulatory mechanism of DUBR.
Keywords: lung adenocarcinoma, metastasis, long noncoding RNAs, DUBR, c-Myc, ZBTB11, oxidative phosphorylation
Introduction
Lung cancer is the leading cause of cancer death worldwide, with a 5-year survival rate for advanced-stage patients of <5% [1, 2]. Lung adenocarcinoma (LUAD) accounts for the majority of lung cancer cases, and its clinicopathological characteristics have been extensively studied [2]. Tumor invasion and distant metastasis are the key factors related to prognosis, but the underlying molecular mechanisms have not been fully revealed [3]. Further understanding of these mechanisms is expected to contribute to developing therapeutic strategies to improve clinical outcomes.
Long noncoding RNAs (lncRNAs) are defined as noncoding RNAs (ncRNAs) with a length of more than 200 nucleotides [4]. Approximately 90% of noncoding sequences in the human genome are transcribed into ncRNAs, most of which are lncRNAs, and have moved to the forefront of ncRNA research [5]. Previous investigations have demonstrated that lncRNAs are involved in various biological processes and the occurrence of various diseases, such as fibrosis, aging, and cancer [6, 7]. A variety of lncRNAs have been reported to function in the tumorigenesis and progression of LUAD [8]. For example, several lncRNAs (ZXF1, MVIH, CCAT2, etc.) were found to be overexpressed in LUAD and were related to metastasis and poor survival [9–11]. In contrast, some lncRNAs (TARID, BANCR, etc.) were reported to have a tumor-suppressive function, with downregulated expression in LUAD [12, 13].
Although the roles of many lncRNAs have been clarified, to date, the functions of numerous lncRNAs have not been fully elucidated. The lncRNA DUBR, also called linc00883, was reported to be downregulated in recurrent neuroblastoma cell lines and was associated with unfavorable outcomes [14]. However, the role of DUBR in other cancers and its potential targets in LUAD are unclear; thus, further exploration is required to clarify the molecular mechanisms underlying its role. In this study, we investigated the expression of DUBR in LUAD to ascertain its association with the clinicopathological features and prognosis of LUAD. We further identified upstream regulators of DUBR, as well as downstream targets and signaling pathways regulated by DUBR, and investigated their contributions to LUAD metastasis.
Materials and methods
Bioinformatic analysis
Gene expression and prognostic data were obtained from The Cancer Genome Atlas (TCGA) at the online GEPIA website [15] (http://gepia.cancer-pku.cn/) and from Kaplan–Meier Plotter databases (http://kmplot.com/) [16], respectively. A list of genes related to DUBR and zinc finger and BTB domain containing 11 (ZBTB11) was downloaded from the cBioPortal website (http://www.cbioportal.org/). Gene Ontology (GO) biological process and KEGG pathway enrichment analyses were conducted by using the clusterProfiler package in R.
Patients and samples
A total of 94 patients with confirmed LUAD from Fudan University Shanghai Cancer Center (FUSCC) were included in the analysis. Pathological sections of tumor tissue and normal tissue were used to detect the expression level of DUBR. Clinicopathological characteristics and survival data were collected for subsequent analysis. All patients signed informed consent forms for donation of their samples to the tissue bank of FUSCC. None of the patients received preoperative therapy.
Cell culture and transfection
LUAD cell lines (H1975, A549, H23, H1299, and H1650) and the normal bronchial epithelial cell line BEAS-2B were purchased from the Cell Bank of the Chinese Academy of Sciences and cultured in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin solution. All cancer cells were maintained in a 37 °C incubator with 5% CO2. Cancer cells were seeded and transfected with DUBR overexpression vector, MYC overexpression vector, control vector, shRNA-MYC, siRNA-DUBR, siRNA-ZBTB11, and siRNA-control using Lipofectamine 2000 (Invitrogen, USA). After 48 h, the transfected cells were harvested, and the expression of the target genes was analyzed via qRT-PCR or Western blotting.
Plasmid construction
Recombinant plasmids containing full-length cDNA sequences of human DUBR (pENTER-DUBR) and c-Myc (pENTER-c-Myc) were purchased from Transheep (Shanghai, China). The cDNA sequences of DUBR and c-Myc were subcloned into the lentiviral vector pTSB-SH-copGFP-2A-PURO to generate the recombinant plasmids. The corresponding oligonucleotides used to generate shRNA against the open reading frames of c-Myc and ZBTB11 to silence their expression were as follows: c-Myc-1, 5′-CCGGCCTGAGACAGATCAGCAACAACTCGAGTTGTTGCTGATCTGTCTCAGGTTTTTT-3′; c-Myc-2, 5′-CCGGATCATCATCCAGGACTGTATGCTCGAGCAT ACAGTCCTGGATGATGATTTTTTT-3′; ZBTB11-1, 5′-CCGGGATACTTATAGAAGCAGGCTTCTCGAGAAGCCTGCTTCTATAAGTATCTTTTTTG-3′; and ZBTB11-1, 5′-CCGGGCTGAGGATATTTGTGCCGAACTCGAGTTCGGCACAAATATCCTCAGCTTTTTTG-3′. The small interfering RNA (siRNA), si-DUBR, and si-control constructs were purchased from Transheep.
Real-time quantitative PCR (qRT-PCR)
Briefly, all RNA was extracted with TRIzol reagent (Invitrogen, USA), and cDNA was synthesized using a PrimeScript RT Reagent Kit (Takara, Japan). Real-time PCR was performed in a ViiATM7Dx fluorescence quantitative PCR instrument (ABI, USA) using TB Green Premix Ex Taq II (Takara, Japan). PCR amplification was performed in a 96-well plate at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The relative expression levels of the target genes were normalized to that of β‐actin and analyzed using the 2−ΔΔCT method. The primer sequences used for PCR are listed in Supplementary Table 1.
Migration and invasion assays
Briefly, 105 LUAD cells in serum-free medium were added to the upper compartment of 24-well transwell plates, and serum containing 10% FBS was added to the lower compartment. For the invasion assay, the upper surface of the membrane was coated with Matrigel (BD Bioscience, USA). After incubation for 24 h, the cells remaining on the fibrous membrane were fixed with paraformaldehyde, stained with crystal violet, imaged, and counted.
Analysis of ATP production
An ENLITEN ATP Assay System (Promega; FF2000) was used according to the manufacturer’s instructions. Cells were harvested by scraping and resuspended in PBS. The cell suspension was divided into unequal aliquots. Part of the cell suspension was mixed with 5% trichloroacetic acid (TCA). The remaining cells were used for the cell counting. Tris-acetate buffer (pH 7.75) was then added to neutralize the TCA and to dilute the TCA to a final concentration of 0.1%. The diluted sample (40 mL) was added to an equal volume of rL/L reagent (Promega; FF2000). Then, luminescence was measured. The ATP standard (Promega; FF2000) was serially diluted to generate a standard curve for calculating the ATP concentration in individual samples. The relative ATP concentration was determined and normalized to that of the control cells, which was set at 1.
Oxygen consumption rate (OCR) measurement
Cellular mitochondrial function was measured using a Seahorse XF Cell Mito Stress Test Kit and a Bioscience XF96 Extracellular Flux Analyzer according to the manufacturer’s instructions. The glycolytic capacity was determined using a Glycolysis Stress Test Kit (ab222946, Abcam) according to the manufacturer’s instructions. Briefly, 4 × 104 cells were seeded in 96-well plates and incubated overnight. After washing the cells with Seahorse buffer (DMEM with phenol red containing 25 mmol/L glucose, 2 mmol/L sodium pyruvate, and 2 mmol/L glutamine), 175 mL of Seahorse buffer plus 25 mL each of 1 mmol/L oligomycin, 1 mmol/L FCCP, and 1 mmol/L rotenone was automatically injected to measure the OCR. The OCR values were calculated after normalization to the cell number and are plotted as the means ± SDs.
Western blot
Cells were lysed using RIPA buffer with protease inhibitors, and protein samples were then separated by SDS-PAGE and transferred to PVDF membranes (Roche, Switzerland). Next, the membranes were blocked and incubated with primary anti-ZBTB11 and anti-β-actin antibodies (ProteinTech Group, USA) at 4 °C overnight. Then, the membranes were incubated with a horseradish peroxidase (HRP)‐conjugated secondary antibody (1:10,000, Abclonal, China) at 37 °C for 1 h. An ECL detection system (Thermo Scientific, USA) was used for signal detection. The density of the β-actin band was used to normalize the density of the bands representing specific proteins.
Luciferase reporter assay
Luciferase reporter gene constructs containing the wild-type or mutant DUBR promoter were cloned into the promoter-less pGL3 enhancer plasmid. Briefly, luciferase reporter plasmids containing the wild-type or mutant DUBR promoter were co-transfected into 293T cells with various amounts of c-Myc overexpression plasmid. The cells were lysed 48 h after transfection, and the lysate was analyzed using a Dual Luciferase Assay System (Promega).
Chromatin-immunoprecipitation (ChIP)
The ChIP assay was performed using a ChIP kit (Millipore, USA) according to the manufacturer’s instructions 48 h after transfection. Anti-c-Myc and control IgG antibodies were used to immunoprecipitate the crosslinked protein-DNA complexes. The primer sequences used for real-time PCR were as follows: forward, 5′-GTTTGGGTTTGGCAGTGTCT-3′; and reverse, 5′-TTTTGGGCAGGATGAGTTTT-3′.
Statistical analysis
SPSS 22.0 software (SPSS, Chicago, USA) was used for data analysis. All data are expressed as the mean ± SD values, and P < 0.05 was considered statistically significant. All experiments were performed in triplicate, and two-tailed unpaired Student’s t test was used for comparisons between groups. Kaplan–Meier survival analysis was used to generate survival curves, and the log-rank test was used for analysis.
Results
DUBR is downregulated in LUAD and is associated with poor prognosis
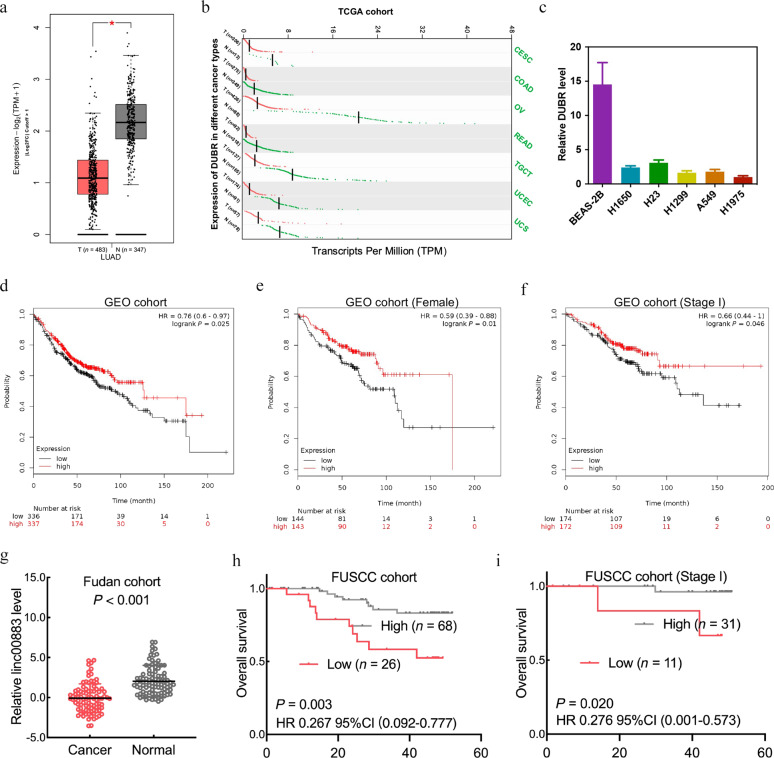
Analysis of TCGA LUAD dataset from the GEPIA online website [15] showed significantly decreased expression of DUBR in LUAD samples compared to the paired noncancerous lung tissues (Fig. 1a). The same pattern of downregulated expression was also observed in other cancer types (Fig. 1b). Consistent with these observations, the expression of DUBR was significantly reduced in LUAD cell lines compared with BEAS-2B normal bronchial epithelial cells (Fig. 1c). The online software Kaplan–Meier Plotter [16] was used to assess the relationship between DUBR expression and LUAD prognosis. We analyzed the expression data of DUBR in 673 patients with LUAD and found that patients with low expression of DUBR had short relapse-free survival times (Fig. 1d). DUBR expression showed similar trends in female patients (Fig. 1e) and patients with AJCC stage I disease (Fig. 1f). For further verification, we collected tissue samples from patients at FUSCC. Consistent with the above results, the expression level of DUBR in the tumor tissues of adenocarcinoma patients was lower than that in the adjacent tissues (Fig. 1g). Downregulation of DUBR was positively associated with advanced N stage (Table 1), patients with lower DUBR expression levels had shorter overall survival times (Fig. 1h), and DUBR expression showed similar tendencies in patients with AJCC stage I disease (Fig. 1i, Table 2). These data indicated that the expression of DUBR in LUAD was reduced and that downregulation of DUBR was associated with poor prognosis.
Fig. 1. Decreased DUBR expression is positively correlated with poor prognosis in LUAD.
a Box plots showing the downregulation of DUBR expression in the TCGA LUAD cohort downloaded from the GEPIA online database. T tumor tissues, N normal tissues. b DUBR expression was significantly decreased in the indicated TCGA cohorts downloaded from the GEPIA online database. c DUBR expression in five LUAD cell lines and one normal bronchial epithelial cell line using qRT-PCR. β‐Actin was used as the internal control. d Kaplan–Meier Plotter was used to analyze the relationship between DUBR expression and relapse-free survival in a group of 672 patients with LUAD. e Kaplan–Meier Plotter was used to analyze the relationship between DUBR expression and relapse-free survival in a group of 287 female patients with LUAD. f Kaplan–Meier Plotter was used to analyze the relationship between DUBR expression and relapse-free survival in a group of 346 patients with stage I LUAD. g Scatter plots showing the downregulation of DUBR expression in the FUSCC cohort. h Kaplan–Meier Plotter was used to analyze the relationship between DUBR expression and overall survival in the FUSCC cohort (n = 94). i Kaplan–Meier Plotter was used to analyze the relationship between DUBR expression and overall survival of 42 patients with stage I LUAD in the FUSCC cohort.
Table 1.
The relationship between linc00883 expression and clinicopathologic parameters.
| Characteristics | No. of patients (n = 94) |
Low expression (n = 26) |
High expression (n = 68) |
P value |
|---|---|---|---|---|
| Age (year) | 0.106 | |||
| ≤60 | 47 | 9 (19.1%) | 38 (80.9%) | |
| >60 | 47 | 17 (36.2%) | 30 (63.8%) | |
| Gender | 0.359 | |||
| Male | 46 | 15 (32.6%) | 31 (67.4%) | |
| Female | 28 | 11 (22.9%) | 27 (77.1%) | |
| Smoking status | 0.492 | |||
| No | 52 | 13 (25.0%) | 39 (75.0%) | |
| Yes | 40 | 13 (32.5%) | 27 (67.5%) | |
| Unknown | 2 | 0 (0.0%) | 2 (100%) | |
| Tumor size | 0.478 | |||
| ≤3 cm | 60 | 15 (25.0%) | 45 (75.0%) | |
| >3 cm | 34 | 11 (32.4%) | 23 (67.6%) | |
| T stage | 0.263 | |||
| T1 | 59 | 15 (25.4%) | 44 (74.6%) | |
| T2 | 23 | 6 (26.1%) | 17 (73.9%) | |
| T3 | 9 | 5 (55.6%) | 4 (44.4%) | |
| T4 | 1 | 0 (0.0%) | 1 (100.0%) | |
| N stage | 0.016 | |||
| N0 | 55 | 10 (18.2%) | 45 (81.8%) | |
| N1 | 8 | 5 (62.5%) | 3 (37.5%) | |
| N2 | 31 | 11 (35.5%) | 20 (64.5%) | |
| TNM stage | 0.374 | |||
| I | 42 | 8 (19.0%) | 34 (81.0%) | |
| II | 10 | 4 (40.0%) | 6 (60.0%) | |
| III | 37 | 12 (32.4%) | 25 (67.6%) | |
| IV | 5 | 2 (40.0%) | 3 (60.0%) |
TNM tumor–node–metastasis staging system.
Table 2.
Univariate and multivariate analysis of different prognostic factors for OS in patients with LUAD.
| Prognostic factors | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age (≤60/>60) | 1.005 | 0.960–1.052 | 0.846 | |||
| Gender (male/female) | 0.344 | 0.129–0.921 | 0.034 | |||
| Smoking status (no/yes) | 1.639 | 0.769–2.490 | 0.200 | |||
| Tumor size (≤3 cm/>3 cm) | 1.724 | 0.679–4.375 | 0.252 | |||
| T stage (T1 + 2/T3 + 4) | 1.847 | 1.016–3.356 | 0.044* | |||
| N stage (N0/N1–2) | 3.562 | 1.976–6.418 | 0.000* | 3.715 | 2.057–6.708 | 0.000* |
| TNM stage (I + II/III + IV) | 1.841 | 1.204–2.813 | 0.005* | |||
| Linc00883 (high/low) | 0.266 | 0.105–0.675 | 0.005* | |||
HR hazard ratio, CI confidence interval.
*P < 0.05.
DUBR suppresses LUAD cell migration and invasion
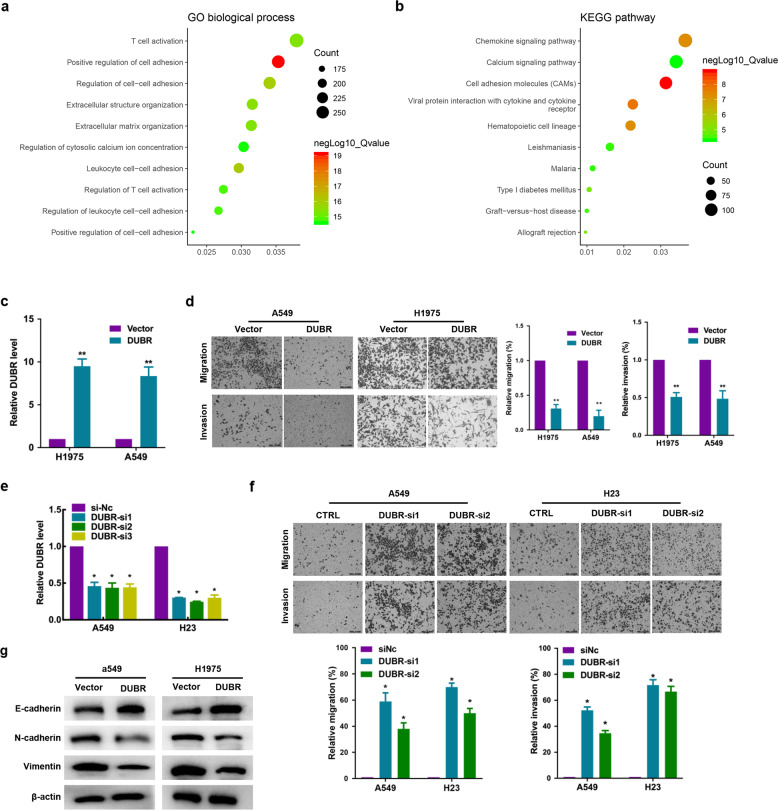
Given the clinical significance of DUBR downregulation in LUAD, we next identified 8646 genes whose expression was highly correlated with that of DUBR (Q value < 0.05; Supplementary Table S2) in the TCGA LUAD dataset (downloaded from the cBioPortal website) and explored the GO biological processes and KEGG pathways in which DUBR-correlated genes were enriched by using the clusterProfiler package in R. The functional enrichment analysis results showed that the overlapping genes were clustered in 1088 significant BP categories and 72 significant KEGG pathways (Supplementary Table S3). DUBR was closely related to cell adhesion (Fig. 2a, b); thus, we further evaluated the effect of DUBR on the migration and invasion capabilities of LUAD cells. We transfected two LUAD cell lines with a DUBR overexpression plasmid and verified the marked increase in expression by qRT-PCR (Fig. 2c). We found that DUBR overexpression markedly suppressed the migration and invasion of LUAD cells (Fig. 2d). EMT is one of the main mechanisms that promotes tumor invasion and metastasis. In DUBR-overexpressing cells, we observed significantly increased expression of the epithelial cell marker E-cadherin, while the expression of mesenchymal cell markers such as N-cadherin and vimentin was decreased (Fig. 2g). Moreover, knockdown of DUBR (Fig. 2e) significantly enhanced the ability of LUAD cells to pass through the chamber membrane (Fig. 2f). In summary, DUBR plays an important role in regulating the invasion and metastasis of LUAD cells.
Fig. 2. DUBR suppresses LUAD cell proliferation in vitro.
a, b GO enrichment and KEGG pathway analyses of DUBR-correlated genes (downloaded from the cBioPortal online database) in the biological process categories. The X-axis shows the enrichment levels. A larger value of the Rich factor indicates a higher level of enrichment. The colors of the dots represent different P values, and the sizes of the dots reflect the number of target genes enriched in the corresponding pathway. GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes. c qRT-PCR results showing the DUBR overexpression efficiency in A549 and H1975 cells. β-Actin was used as the internal control. **P < 0.01. d Representative images (left panel) and quantitative analysis (right panel) of the migration and invasion assay, which showed that overexpression of DUBR suppressed the migration and invasion of A549 and H1975 cells. **P < 0.01. e qRT-PCR results showing the efficiencies of DUBR knockdown efficiency in A549 and H23 cells. β-Actin was used as the internal control. *P < 0.05. f Representative images (upper panel) and quantitative analysis (lower panel) of the migration and invasion assay, which showed that knockdown of DUBR promoted the migration and invasion of A549 and H23 cells. *P < 0.05. g Western blot analysis showing upregulation of E-cadherin and downregulation of N-cadherin and Vimentin by DUBR overexpression in LUAD cells; β‐actin was used as the internal control.
Downregulation of DUBR expression is associated with c-Myc binding to the DUBR promoter
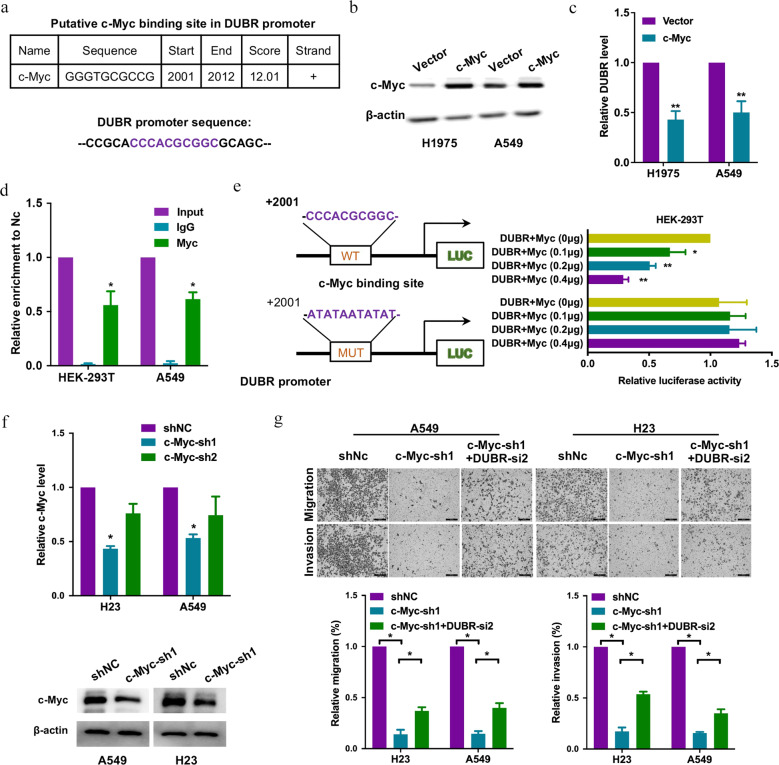
Since we discovered the downregulation of DUBR in LUAD, we next sought to explore the mechanisms that induced the phenomenon of DUBR downregulation in LUAD. It is well known that c-Myc is an important oncogenic transcription factor and is reported to play a regulatory role in the biogenesis of lncRNAs [17]. By bioinformatic analysis, we found a putative c-Myc binding site in the DUBR promoter (Fig. 3a). We also detected a significant decrease in DUBR levels in LUAD cell lines transfected with c-Myc (Fig. 3b, c). These results led us to speculate that c-Myc might function as a transcriptional suppressor of DUBR. To test this hypothesis, we performed ChIP assays to confirm the binding of c-Myc to the DUBR promoter and observed that the precipitate pulled down with the anti-c-Myc antibody was significantly enriched with DUBR compared to the precipitate pulled down with control IgG, confirming the effective binding of c-Myc to the putative binding site in the DUBR promoter c-Myc in LUAD cell lines (Fig. 3d). Additionally, we prepared wild-type and mutant DUBR promoter-reporter constructs to evaluate the regulation of DUBR transcription by c-Myc (Fig. 3e). As expected, the luciferase reporter assay showed that co-transfection of the c-Myc and wild-type DUBR promoter-reporter plasmids resulted in a decrease in luciferase activity, which further decreased with increasing c-Myc levels (Fig. 3e). However, transfection of the mutant DUBR promoter-reporter plasmid did not decrease luciferase activity and eliminated the response to changes in the c-Myc level. In conclusion, c-Myc inhibited the transcription of DUBR by binding to the DUBR transcriptional promoter. To further functionally verify the regulation of DUBR by c-Myc, we transfected LUAD cells with c-Myc shRNA and verified the knockdown efficiency by PCR and Western blotting (Fig. 3f). Knockdown of c-Myc almost completely abolished the invasion and migration of LUAD cells, while knockdown of DUBR restored the inhibitory effect (Fig. 3g), indicating that c-Myc promoted the migration and invasion of LUAD cells in part by transcriptionally suppressing DUBR.
Fig. 3. c-Myc transcriptionally inhibits DUBR.
a The putative c-Myc binding site in the DUBR promoter, as predicted by bioinformatics software. b Western blot showing the c-Myc overexpression efficiency in LUAD cells; β‐actin was used as the internal control. c qRT-PCR results showing that the overexpression of c-Myc suppressed the expression of DUBR in in LUAD cells; β‐actin was used as the internal control. **P < 0.01. d ChIP assays were performed in LUAD cells using anti-c-Myc antibodies or control IgG. The DUBR promoter fragment was detected by specific primers. *P < 0.05. e Wild-type or mutant DUBR promoter-reporter constructs and various amounts of c-Myc overexpression plasmids were co-transfected into 293T cells. *P < 0.05; **P < 0.01. f qRT-PCR and Western blot results showing the c-Myc knockdown efficiency c-Myc in H23 and A549 cells; β‐actin was used as the internal control. *P < 0.05. g Representative images (upper panel) and quantitative analysis (lower panel) of the migration and invasion assay, which showed that knockdown of DUBR partially reversed the MYC knockdown-induced suppression of A549 and H23 cell migration and invasion. *P < 0.05.
DUBR inhibits LUAD cell migration and invasion through ZBTB11
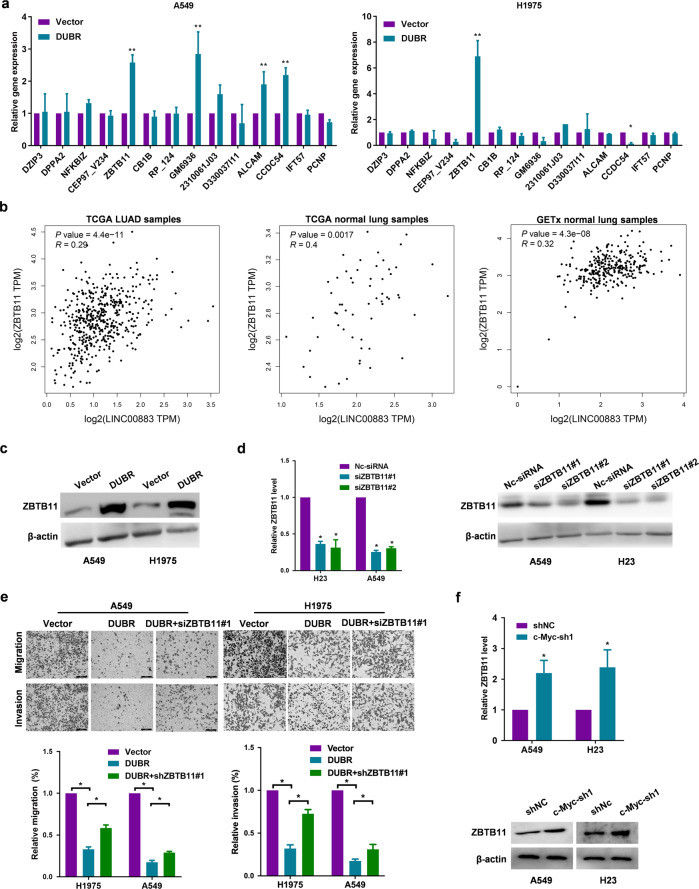
Prior studies have noted that lncRNAs may regulate the expression of their neighboring genes [18]. To identify the downstream target gene of DUBR in LUAD cells, we analyzed the expression of genes adjacent to DUBR by qRT-PCR. We found that the expression of ZBTB11 was significantly upregulated when DUBR was overexpressed in A549 and H1975 cells (Fig. 4a). Next, we verified the significant positive correlation between DUBR and ZBTB11 mRNA expression in LUAD and normal lung tissues by analyzing TCGA LUAD datasets and Genotype-Tissue Expression (GTEx) normal lung tissue datasets (Fig. 4b). The Western blot results also showed that ZBTB11 protein expression was upregulated by DUBR overexpression (Fig. 4c). To determine the role of ZBTB11 in DUBR-mediated suppression of cancer cell migration and invasion, siRNAs specifically targeting ZBTB11 were used to transfect LUAD cells, and the knockdown efficiency was confirmed by Western blotting (Fig. 4d). Knockdown of ZBTB11 by siRNAs partially restored the suppression of migration and invasion induced by DUBR (Fig. 4e). In addition, knockdown of the upstream gene c-Myc led to an increase in the expression of the downstream gene ZBTB11 (Fig. 4f), which further confirmed the regulatory relationship of the c-Myc/DUB/ZBTB11 axis.
Fig. 4. DUBR regulates ZBTB11 gene expression in cis.
a qRT-PCR results showing the effect of DUBR overexpression on the neighboring genes in A549 and H1975 cells; β‐actin was used as the internal control. **P < 0.01. b The correlation of DUBR and ZBTB11 expression in TCGA LUAD and Genotype-Tissue Expression (GTEx) normal lung tissue datasets downloaded from the GEPIA online database. c Western blot showing the upregulation of ZBTB11 by DUBR overexpression in LUAD cells; β‐actin was used as the internal control. d qRT-PCR and Western blot results showing the ZBTB11 knockdown efficiency in LUAD cells; β‐actin was used as the internal control. *P < 0.05. e Representative images (upper panel) and quantitative analysis (lower panel) of the migration and invasion assay, which showed that knockdown of ZBTB11 partially reversed the DUBR overexpression-induced suppression of A549 and H1975 cell migration and invasion. *P < 0.05. f qRT-PCR and Western blot results showing that knockdown of c-Myc upregulated the expression of ZBTB11 in LUAD cells; β‐actin was used as the internal control. *P < 0.05.
DUBR inhibits LUAD cell migration and invasion via ZBTB11-mediated oxidative phosphorylation
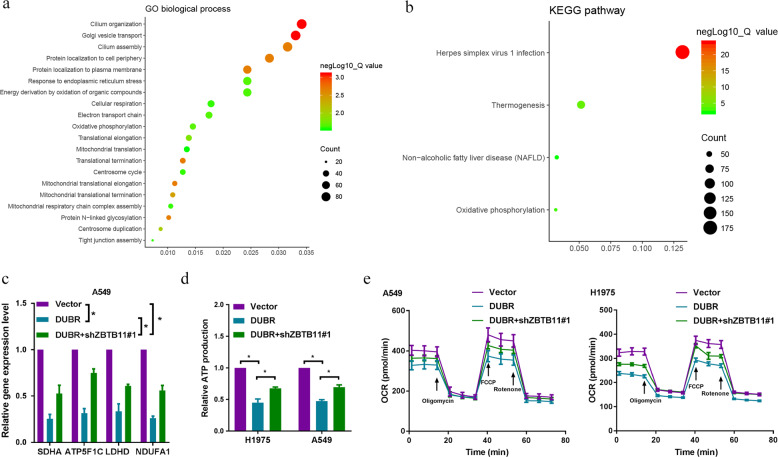
To explore the signaling pathway through which the DUBR/ZBTB11 axis regulates the invasion and metastasis of LUAD cells, we identified 7143 genes whose expression was highly correlated with that of ZBTB11 (Q value < 0.05; Supplementary Table 4) in the TCGA LUAD database and then took the intersection of DUBR-related genes and ZBTB11-related genes to identify 3149 overlapping genes (Supplementary Table 5). Next, we explored the GO biological processes and KEGG pathways in which those overlapping genes were enriched. The functional enrichment analysis results showed that the overlapping genes were clustered in 43 significant BP categories and 4 significant KEGG pathways (Supplementary Table 6). As shown in Fig. 5a, b (top 20 BP categories and all pathways), the overlapping genes were closely related to mitochondrial function and oxidative phosphorylation (OXPHOS). We further validated the bioinformatic analysis results by qRT-PCR, showing that overexpression of DUBR downregulated the expression of a list of OXPHOS-related genes (SDHA, ATP5F1C, LDHD, and NDUFA1), whereas knockdown of ZBTB11 partially reversed the DUBR overexpression-induced downregulation of these genes (Fig. 5c). Then, we investigated the effects of the DUBR/ZBTB11 axis on OXPHOS in LUAD cells. DUBR overexpression inhibited ATP production and decreased the oxygen consumption rate (OCR; reflecting mitochondrial respiration), and ZBTB11 knockdown partially restored the capacity for ATP production and the OCR of DUBR‐knockdown A549 and H1975 cells (Fig. 5d, e). Upregulating OXPHOS has been noted to induce the metastatic behavior of cancer cells [19]. Thus, these results suggested that the DUBR/ZBTB11 axis may play an antimetastatic role by targeting OXPHOS.
Fig. 5. The DUBR-ZBTB11 axis suppresses oxidative phosphorylation in LUAD.
a, b GO enrichment and KEGG pathway analyses of overlapping DUBR and ZBTB11-correlated genes (downloaded from the cBioPortal online database) in the biological process categories. The X-axis shows the enrichment levels. A larger value of the Rich factor indicates a higher level of enrichment. The colors of the dots represent different P values, and the sizes of the dots reflect the number of target genes enriched in the corresponding pathway. GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes. c qRT-PCR results showing that knockdown of ZBTB11 partially reversed the DUBR overexpression-induced suppression of SDHA, ATP5F1C, LDHD, and NDUFA1 expression in A549 cells. β‐Actin was used as the internal control. *P < 0.05. d ATP production was assessed after overexpression of DUBR with or without knockdown of ZBTB11 in A549 and H1975 cells. *P < 0.05. e OCR in A549 and H1975 cells treated as indicated.
Discussion
Recent studies on transcriptome and ncRNA expression profiles indicate that lncRNAs are involved in tumor growth, invasion, metastasis, and drug resistance in various cancers, including non-small cell lung cancer (NSCLC) [9–13]. However, only a few lncRNAs, such as ZXF1, MALAT1, and BANCR, have been identified as metastasis-associated in NSCLC [7, 13, 20]. DUBR is a novel lncRNA originally identified to be involved in the process of muscle cell differentiation and regeneration [21]. However, no previous experiments have revealed the biological functions and regulatory mechanisms of DUBR in lung cancer. Here, we analyzed the expression of DUBR in LUAD tissues and cell lines for the first time. In this study, we showed that DUBR was downregulated in LUAD tissues and LUAD cell lines. In addition, DUBR expression was negatively correlated with the prognosis of LUAD patients. We provided the first demonstration that DUBR plays a role as a tumor suppressor in LUAD by inhibiting migration and invasion via the DUBR/ZBTB11/OXPHOS signaling pathway.
Interestingly, an interaction between DUBR and c-Myc at the transcriptional level was observed in this study. c-Myc, an oncogenic transcription factor, is involved in various biological processes in cancer [17]. The results of ChIP and luciferase reporter assays indicated that c-Myc can specifically bind to the promoter region of DUBR because it contains a potential binding motif. c-Myc transcriptionally downregulated the expression of DUBR and thus promoted the invasion and migration of LUAD cells. Consistent with our results, previous evidence also indicated that c-Myc can transcriptionally repress the expression of lncRNAs with tumor suppressor functions and promote tumorigenesis [17, 22].
DUBR has been shown to have a strong cis-regulator in muscle cells [21]. In the present study, based on the results of qRT-PCR and Western blotting, we identified ZBTB11, as a downstream molecule upregulated by DUBR overexpression. Our findings suggest a novel mode of upregulating the ZBTB11 locus in which DUBR acts as a cis-acting lncRNA. ZBTB11 is a member of the ZBTB superfamily, which is involved in multiple biological processes, including tumorigenesis and lymphocyte differentiation, but it has not been fully studied [23, 24]. Previous studies verified that ZBTB11 is a direct transcriptional target of PU.1 and is involved in basal and emergency granulopoiesis [25]. It is still unclear to us whether DUBR is necessary for the regulation of ZBTB11 promoter activity. Further studies are needed to shed light on this aspect. TP53 has been identified as a direct target of ZBTB11 and regulates the cell death and cell cycle arrest signaling pathways [25]. Specific genes may be controlled through different mechanisms in specific physiopathological scenarios. In this study, we reported for the first time that DUBR inhibits LUAD cell invasion and migration partially by targeting ZBTB11, which indicated that ZBTB11 is possibly involved in tumor metastasis. Furthermore, DUBR-suppressed OXPHOS in LUAD cells by upregulating ZBTB11. Mitochondrial respiration is critical for cancer cell viability, and upregulation of OXPHOS has been noted to induce the metastatic behavior of cancer cells [19]. Thus, these results suggest that the DUBR/ZBTB11 axis may play an antimetastatic role by targeting OXPHOS. Although it must be pointed out that more effort is required to specifically connect DUBR-suppressed metastasis and oxidative phosphorylation, these results demonstrate that the invasive and migratory properties of cancer cells are dependent on mitochondrial proficiency and DUBR is identified as a potential target for therapeutic intervention.
Previous reports have indicated that lncRNAs can target DNA, RNA, or proteins to achieve transcriptional or posttranscriptional modulation of gene expression [26, 27]. In the present study, we identified a biochemical pathway connecting DUBR and ZBTB11 that merits consideration in normal and malignant tissues in which both DUBR and ZBTB11 are expressed. However, the precise mechanism of action of DUBR and ZBTB11 is historically diverse and elusive. They may act as signals, guides or scaffolds for chromatin to regulate the expression of their target genes and thus participate in diverse physiological and pathological processes in specific tumor types [28]. Therefore, more efforts are needed to clarify the details of the interaction between DUBR and its target genes and to explore the feasibility of clinical translation.
In summary, we first reported the downregulation of DUBR in LUAD, which was specifically mediated by c-Myc at the transcriptional level. Furthermore, we confirmed that ZBTB11 was a downstream molecule of DUBR and that its expression was positively correlated with that of DUBR. The DUBR/ZBTB11 axis repressed the invasion and migration of LUAD cells by targeting OXPHOS. This study indicated that the lncRNA DUBR might be a potential biomarker and new therapeutic target for LUAD.
Supplementary information
Acknowledgements
This research was financially supported by National Natural Science Foundation of China (No. 81601988), the Shanghai Science and Technology Development fund (No. 19MC1911000), Hospital Foundation of Fudan University Shanghai Cancer Center (No. YJMS201907), and Shanghai Chest Hospital Project of Collaborative Innovation (No. YJXT20190102).
Author contributions
BHH, MDX, and WN participated in the design of the study. JL, LLZ, SHC, and JWL carried out experiments and collected the data. MJH, FH, and CHL analyzed experimental data and conducted graphs and tables. FFQ, YW, QZ, and XYZ performed bioinformatics analysis. MJH and QZ wrote the paper. BHH, MDX, and WN reviewed the paper. All authors read and approved the final paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wei Nie, Min-juan Hu, Qin Zhang
Contributor Information
Xue-yan Zhang, Email: zhangxueyan570@163.com.
Mi-die Xu, Email: xumd27202003@sina.com.
Bao-hui Han, Email: 18930858216@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00624-5.
References
- 1.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–54. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed]
- 3.Greenawalt EJ, Edmonds MD, Jain N, Adams CM, Mitra R, Eischen CM. Targeting of SGK1 by miR-576-3p inhibits lung adenocarcinoma migration and invasion. Mol Cancer Res. 2019;17:289–98. doi: 10.1158/1541-7786.MCR-18-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Jiang S, Shang J, Jiang Y, Dai Y, Xu B, et al. LncRNA: shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res Rev. 2019;52:17–31. doi: 10.1016/j.arr.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M, et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35:7587–94. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 10.Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Zhou XF, Pan GF, Zhao JP. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68:401–7. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–14. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utnes P, Løkke C, Flægstad T, Einvik C. Clinically relevant biomarker discovery in high-risk recurrent neuroblastoma. Cancer Inform. 2019;18:1176935119832910. doi: 10.1177/1176935119832910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick M, et al. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019;450:63–75. doi: 10.1016/j.canlet.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Yan P, Luo S, Lu JY, Shen X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:170–8. doi: 10.1016/j.gde.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Porporato PE, Payen VL, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 2: Mitochondria, lipid and amino acid metabolism. Cell Mol Life Sci. 2016;73:1349–63. doi: 10.1007/s00018-015-2100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt LH, Görlich D, Spieker T, Rohde C, Schuler M, Mohr M, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol. 2014;9:1294–304. doi: 10.1097/JTO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Zhao Y, Bao X, Zhu X, Kwok YK, Sun K, et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–50. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T, Cui R, Jeon YJ, Fadda P, Alder H, Croce CM. MYC-repressed long noncoding RNAs antagonize MYC-induced cell proliferation and cell cycle progression. Oncotarget. 2015;6:18780–9. doi: 10.18632/oncotarget.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siggs OM, Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–69. [DOI] [PMC free article] [PubMed]
- 24.Chevrier S, Corcoran LM. BTB-ZF transcription factors, a growing family of regulators of early and late B-cell development. Immunol Cell Biol. 2014;92:481–8. doi: 10.1038/icb.2014.20. [DOI] [PubMed] [Google Scholar]
- 25.Keightley MC, Carradice DP, Layton JE, Pase L, Bertrand JY, Wittig JG, et al. The Pu.1 target gene Zbtb11 regulates neutrophil development through its integrase-like HHCC zinc finger. Nat Commun. 2017;8:14911. doi: 10.1038/ncomms14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 27.Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. OncoTargets Ther. 2016;9:4411–24. doi: 10.2147/OTT.S121785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, Huang Z, Sheng W, Xu MD. Emerging roles of long non-coding RNAs in tumor metabolism. J Hematol Oncol. 2018;11:106. doi: 10.1186/s13045-018-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.