Abstract
Intrinsically disordered proteins (IDPs) are proteins that lack rigid 3D structure but exist as conformational ensembles. Because of their structural plasticity, they can interact with multiple partners. The protein interactions between IDPs and their partners form scale-free protein interaction networks (PINs) that facilitate information flow in the cell. Because of their plasticity, IDPs typically occupy hub positions in cellular PINs. Furthermore, their conformational dynamics and propensity for post-translational modifications contribute to “conformational” noise which is distinct from the well-recognized transcriptional noise. Therefore, upregulation of IDPs in response to a specific input, such as stress, contributes to increased noise and, hence, an increase in stochastic, “promiscuous” interactions. These interactions lead to activation of latent pathways or can induce “rewiring” of the PIN to yield an optimal output underscoring the critical role of IDPs in regulating information flow. We have used PAGE4, a highly intrinsically disordered stress-response protein as a paradigm. Employing a variety of experimental and computational techniques, we have elucidated the role of PAGE4 in phenotypic switching of prostate cancer cells at a systems level. These cumulative studies over the past decade provide a conceptual framework to better understand how IDP conformational dynamics and conformational noise might facilitate cellular decision-making.
Keywords: Protein conformational dynamics, Intrinsically disordered proteins, Phenotypic switching, PAGE4, Conformational noise, MRK hypothesis
Introduction
Despite the initial skepticism regarding the existence of proteins that lacked structure, mainly because of the dominance of the structure/function paradigm that was predicated on the “lock-and-key” hypothesis formulated in the late nineteenth century by Emil Fischer (Uversky, 2021), it is generally held that intrinsically disordered proteins (IDPs) constitute a significant fraction of the proteomes of organisms across all three kingdoms of life (Ward et al, 2004; Xue et al, 2012a; Peng et al, 2015). Although IDPs, and intrinsically disordered regions (IDRs) within structured proteins, may lack rigid 3D structure, they can populate different conformations and, hence, exist as conformational ensembles (Tompa, 2011; Uversky, 2017; Wright and Dyson, 1999; Uversky & Dunker, 2010). Indeed, the unusually high degree of malleability facilitates their interactions with multiple partners (Uversky, 2015), and such interactions constitute a network referred to as protein interaction network (PIN). The PIN configuration defines a cell’s phenotype and its ability to “make” decisions.
Although it was tacitly assumed that PINs are “wired” randomly, seminal work beginning in the late 1990s by Barabási and colleagues (Barabási and Albert, 1999; Barabási, 2009; Dehmamy et al, 2018) revealed that, indeed, cellular PINs are organized following a “scale-free” architecture. In such networks, the degree distribution P(k) exhibits a power-law behavior as a function of the degree k. A salient feature of scale-free networks is that they are robust to failure of random nodes. However, they are vulnerable to failure of hubs (Barabási and Albert, 1999). Furthermore, the fact that the organization and properties of the PINs are conserved during evolution (Rangarajan et al., 2015a) underscores their functional significance.
Consistent with their unique ability to interact with multiple partners by virtue of their plasticity, IDPs are typically found in hub positions defined as nodes with multiple interactions (defined as edges) in PINs (Dosztányi et al., 2006; Haynes et al., 2006; Gsponer & Babu, 2009; Patil et al, 2010; Hu et al., 2017) and play critical roles in many biological processes including transcription, splicing, translation, and signaling (Wright & Dyson, 2015; Bürgi et al., 2016; Shammas, 2017). Furthermore, they also regulate several key processes such as cell division (Galea et al, 2008; Yoon et al, 2012; Mitrea et al, 2012), circadian rhythmicity (Baggio et al., 2013; Hurley et al, 2013, 2016; Dong et al, 2016; Michael et al, 2017), and phenotypic plasticity (Xue et al, 2012b; Mooney et al, 2016). Therefore, it is not surprising that, under physiological conditions, the levels of IDPs are tightly regulated from transcript synthesis to protein degradation (Gsponer et al, 2008; Edwards et al, 2009; Babu et al, 2011; Uversky VN, 2014). Indeed, when dysregulated, IDPs have the potential to engage in multiple “promiscuous” interactions resulting in pathological states (Vavouri et al, 2009; Marcotte and Tsechansky, 2009). Remarkably, several proteins that are dysregulated in disease pathology such as the oncogenes MYC, c-Jun, c-Fos, and the cancer/testis antigens are IDPs (Iakoucheva et al, 2002; Uversky et al 2008; Rajagopalan et al, 2011; Babu MM, 2016). Yet, the molecular mechanisms by which IDPs accomplish their functions, and engage in promiscuous interactions, are not fully understood. When compared to number of IDPs, for example, in the human, where ~ 50% of the proteome is estimated to be comprised of IDPs (Dunker et al, 2001; Dyson and Wright, 2021), only a tiny fraction of IDPs have been characterized in significant detail.
Nonetheless, these studies have revealed that IDPs can transition from disorder to order upon binding to their cognate partners, a phenomenon referred to as, “coupled folding and binding” (Dyson & Wright, 2002; Sugase et al, 2007). However, while in some IDPs, such as the GTPase-binding domain (GBD) of the Wiskott–Aldrich syndrome protein (WASP) and the phosphorylated kinase-inducible domain (pKID) of the cAMP-response element binding (CREB) protein, which interacts with the KIX domain of the transcriptional coactivator CREB-binding protein (CBP), an ordered conformation is induced by the interacting partner – the “induced fit” hypothesis, the opposite may be true in other instances such as the α-MoRE located within the intrinsically disordered C-terminal domain of the measles virus (MeV) nucleoprotein called the NTAIL, wherein the IDP ensembles populate multiple conformations a priori, and the ligand selects the most favored prefolded state from these conformations (Boehr et al, 2009). Nonetheless, it appears that, in many cases, a combination of the two extremes underlies the transition (Wang et al, 2013; Arai et al, 2015), suggesting that the intrinsic secondary structure propensities of the IDPs determine their binding mechanisms.
In contrast to the above scenarios, some IDPs can stochastically switch among distinct conformational states suggesting that IDPs can alter the conformation of their ensembles while remaining disordered (Choi et al, 2011; Choi et al., 2019). Together, these observations suggest that despite being disordered, many IDPs are only marginally unstable and can easily transition to active conformations. On the other hand, it has also been observed that several IDPs (Chakrabortee et al, 2010; He et al, 2015; Kulkarni et al, 2017; Borgia et al, 2018) appear to remain largely disordered even while interacting with their biological targets to form what are known as “fuzzy” complexes (Sharma et al., 2015; Fuxreiter, 2018). Fuzzy binding is seen when the degree of disorder in the bound state of the IDP varies with the partner or cellular conditions such as polymorphic bound structures, conditional folding, and dynamic binding highlighting the structural continuum of complexes as well as their context-dependent interaction behaviors (Fuxreiter, 2020).
A recent report suggested that, in fact, frustration in such fuzzy complexes contributes to the versatility (one-to-many interactions), and high specificity but low affinity interactions, associated with IDPs (Freiberger et al., 2021). Complexes of IDP exhibit a high degree of local frustration, especially at the binding interface. However, the authors noted that the suboptimal interactions can potentially lead to multiple bound substates, each displaying distinct frustration patterns, which are differently populated in complexes with different partners. Therefore, IDPs appear to achieve specificity without a single common bound conformation, and the conflict between different interactions is leveraged to control the binding to multiple partners. From the foregoing, it may be summarized that IDPs may explore myriad interaction mechanisms, ranging from induced folding to formation of fuzzy complexes where significant levels of disorder are preserved to polyvalent stochastic interactions (Uversky, 2018; Fuxreiter, 2020).
Prostate-associated gene 4 (PAGE4) is a remarkably prostate-specific protein in the normal human adult and is overexpressed in prostate cancer (PCa). It is also an IDP that appears to remain disordered when interacting with its partner (see below). Therefore, using PAGE4 as a paradigm, here, we discuss how its conformational dynamics, and consequently, conformational noise, can influence a PCa cell’s decision to switch between an androgen-dependent and androgen-independent phenotype. These findings shed new light on how non-genetic mechanisms may contribute to phenotypic heterogeneity in the population and highlight important therapeutic implications.
Conformational noise
The term noise in biology implies random variability in quantities arising in biological systems including isogenic systems. Therefore, cells in an isogenic population can display very different phenotypes in response to the same stimulus by switching their phenotypes (Huang S, 2009; Brock et al, 2009). In fact, phenotypic switching due to noise has been observed in development, stress response, pathological states such as cancer, and evolution (Mahmoudabadi et al, 2013; Jia et al., 2017).
Presently, noise in biology typically implies transcriptional noise mainly because gene expression is an intrinsically stochastic process which results in variability in protein levels between individual cells in a population (Raj and van Oudenaarden, 2009; Hansen et al, 2018). However, information transduced in cellular signaling networks also appears to be significantly affected by noise (Ladbury and Arold, 2012), particularly, noise contributed by the “non-functional” interactions of proteins (Kuwahara and Gao, 2013). This noise results from the intrinsic promiscuity of protein–protein interactions that modulate cellular signal transduction (Kontogeorgaki et al, 2017). Since a majority of the transcription factors and signaling molecules are IDPs that can engage in promiscuous interactions when dysregulated, they play a significant role in generating noise enhancing the potential to switch phenotypes. Furthermore, the overexpression of IDPs is observed to correlate with altered physiological (Vavouri et al, 2009) and pathological states (Iakoucheva et al, 2002; Uversky et al 2008; Rajagopalan et al, 2011; Babu, 2016).
The MRK hypothesis
Almost a decade ago, we (Mahmoudabadi et al, 2013) proposed a model (the MRK model, Kulkarni and Kulkarni, 2019) to account for noise contributed by the conformational dynamics of IDPs. This noise referred to as “conformational noise” is defined as the random variability in the various confirmations sampled by the IDP ensemble which results in stochastic promiscuous interactions with other proteins. Although interconversions of conformations of the IDPs are in fast exchange, we postulated that the conformational preferences of the ensemble are impacted by post-translational modifications and, therefore, can have significant half-lives (in the order of several minutes to hours) contributing to conformational noise. Furthermore, the model showed that conformational noise can be an integral part of transcriptional noise, and therefore, IDPs could potentially amplify total noise in the system in response to intrinsic or extrinsic perturbations. Thus, conformational noise arising from the stochasticity of the promiscuous interactions initiated by the IDPs in response to a specific input allows the system to sample the network interaction space. This heuristic rewires the network and drives phenotypic switching to generate phenotypic heterogeneity (Fig. 1). Stated differently, the model suggests that IDPs uncover network configurations that are causal in phenotypic switching but are latent under normal conditions (Mahmoudabadi et al, 2013). Indeed, such stochasticity in phenotypic switching has been linked to cellular differentiation (Eldar & Elowitz, 2010; Nichol et al., 2016; Safdari et al, 2020), generation of induced pluripotent stem cells (iPS cells) via reprogramming (McArthur et al., 2008; Yamanaka, 2009; Wakao et al, 2013; Chung et al, 2014; Smith et al., 2015; Lin et al., 2018), emergence of cancer stem cells from non-stem cancer cells (Gupta et al, 2011; Sehl et al., 2015; Rambow et al., 2019), and emergence of chemoresistance (Kumar et al., 2019).
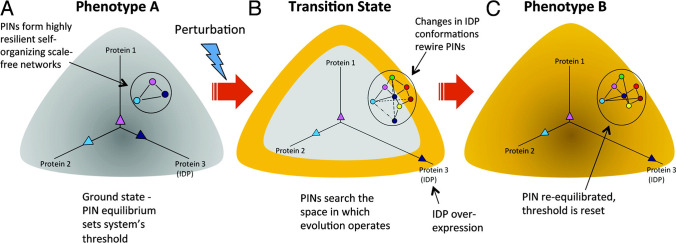
Fig. 1.
Rewiring of protein networks facilitates state-switching by activating latent pathways. (A) The state of a cell with phenotype A is depicted in grey and shows a simple protein network with three proteins (1‒3), of which one is an IDP (indicated in dark blue), and expressed at different levels represented by the three vectors. This configuration represents the protein network’s ground state threshold. (B) Depicts a transition state. A perturbation causes increased IDP expression (protein 3). Overexpression of the IDP results in promiscuity and the protein network explores the network search space shown by the various dashed lines. This transition state is depicted state in yellow around the grey area. (C) The state of the cell after it has transitioned to phenotype B from phenotype A represented in yellow. A particular configuration of the protein network that increased its fitness is “selected,” which now represents the new ground state.
Reproduced with permission from Mahmoudabadi et al. 2013
Several important points characterize the MRK hypothesis. First, according to the model, the information that specifies the cell’s phenotype resides in the configuration of its PIN. Second, cell fate is not determined a priori (is not deterministic), and hence, it is likely that each cell in the population has the potential to undergo specific phenotypic transition in response to the given input. Third, in response to a specific input, IDPs can rewire the network to uncover latent configurations and, thus, actuate a phenotypic switch. Fourth, the model proposes that, at least in some cases, upon withdrawal of the input, the PIN can rewire itself to the normal (default) network configuration, thereby reversing the phenotypic switch. Fifth, information can operate across spatiotemporal timescales. Thus, while information that operates over relatively short timescales maybe retained within the PIN, information operating over longer periods such as cellular transformation, development, and evolution is transferred to the genome to ensure it is heritable (Sonnenschein et al., 2014),
Therefore, contrary to the prevailing wisdom that phenotype specification is deterministic, the MRK hypothesis advocates that stochasticity contributed by IDP conformational noise may be a confounding factor in specifying cell fate. Consistent with this line of thinking, several studies have shown that cells can reversibly switch phenotypes, such as, epithelial to mesenchymal transition (EMT), a drug-sensitive cell developing resistance and switching back to becoming drug sensitivity (Sharma et al, 2010; Al Emran et al, 2017; Su et al, 2017; Hammerlindl & Schaider, 2018; Sahoo et al, 2021) or a normal cell transforming into a malignant one and its reversal to normalcy (dormancy) (Shachaf et al, 2004; Shachaf & Felsher, 2005). A theoretical perspective (Rangarajan et al., 2015b) demonstrating how the oncogene c-Myc, an IDP, lends further credence to the MRK hypothesis.
IDP conformational dynamics, noise, and cell decisions
PAGE4 is a small protein of 102 amino acids that is highly intrinsically disordered (Zeng et al., 2011; Rajagopalan et al, 2014; He et al, 2015) (Fig. 2). It primarily resides in the cytoplasm where it functions as a stress-response protein (Zeng et al, 2013). In response to stress, PAGE4 is upregulated and translocates to the mitochondrion and appears to suppress production of reactive oxygen species (ROS). Thus, overexpression of PAGE4 decreases the phosphorylation of MAP2K4, JNK, and c-JUN while increasing phosphorylation of ERK1/2. Taken together, these data indicate that under stress, PAGE4 appears to promote the survival of PCa cells by regulating MAPK pathway (Lv et al, 2019). In addition to serving as a stress response factor, PAGE4 is also a transcriptional regulator and potentiates transactivation by c-Jun (Rajagopalan et al, 2014).
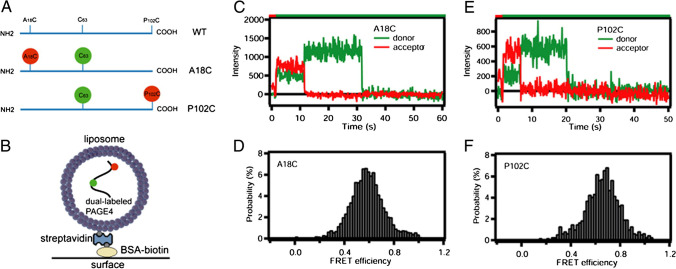
Fig. 2.
Single molecule FRET indicates that PAGE4 is an intrinsically disordered protein. (A) Schematic of the PAGE4 constructs with the native cysteine (green) and the introduced cysteine (red). Single PAGE4 protein molecules were encapsulated inside 100 nm diameter liposomes tethered to a quartz surface. (B) Shows a cartoon of this immobilization scheme (not to scale). Fluorescence emission time courses in the donor and acceptor spectral bands were collected and those indicating exactly 1 donor and 1 acceptor were further analyzed. Example intensity time courses showing anti-correlated donor/acceptor behavior upon photobleaching, which is characteristic of single molecules, are shown for the A18C/63C (C) and P102C/63C (E) FRET mutants. The color bar at the top indicates the illumination color. Red illumination at the start driving only acceptor fluorescence allows identification of molecules containing an active acceptor. The disappearance of red emission (with anticorrelated recovery of green) is photobleaching of the acceptor, and disappearance of green emission is photobleaching of the donor. Histograms assembled from all FRET active data points of over 300 molecules are shown for A18C/63C (D) and P102C/63C (F) PAGE4 mutants. These FRET signals agree with expectations based upon modeling PAGE4 as a highly flexible IDP.
Reproduced with permission from Rajagopalan et al. 2014
The latter function of PAGE4 is modulated by the conformational dynamics of its differentially phosphorylated ensembles by kinases, namely HIPK1 and CLK2. HIPK1 is a stress-response kinase which phosphorylates PAGE4 at T51 and, to a minor extent, S9 (Mooney et al, 2014; He et al, 2015). Employing multidimensional NMR, small angle X-ray scattering, and single molecule Förster resonance energy transfer microscopy (Fig. 3), we determined that threonine phosphorylation, predominantly at T51, leads to compaction of the PAGE4 ensemble (radius of gyration, Rg, 34.7 ± 1.2 Å compared to non-phosphorylated where the Rg is 36 ± 1.1 Å) which is facilitated by the looping of the N-terminal region (He et al, 2015; Kulkarni et al, 2017; Lin et al., 2018; Lin et al., 2019). HIPK1-phosphorylated PAGE4 (HIPK1-PAGE4) acts as a strong potentiator of c-Jun which heterodimerizes with c-Fos to form the AP-1 transcription factor complex. AP-1 is a negative regulator of the androgen receptor (AR) activity in PCa cells (Sato et al, 1997; Tilman et al., 1998), and AR, in turn, is a negative regulator of the CDC-like kinase 2 (CLK2) (Kulkarni et al, 2017). Therefore, inhibiting AR de-represses CLK2. CLK2 hyperphosphorylates PAGE4 (CLK2-PAGE4) at all S/T residues in the molecule leading to remodeling of the PAGE4 ensemble which now prefers to assume a more random coil-like confirmation (Rg, 49.8 ± 1.9 Å) (Table 1). Furthermore, in contrast to HIPK1-PAGE4, CLK2-PAGE4 attenuates c-Jun potentiation (Kulkarni et al, 2017).
Fig. 3.
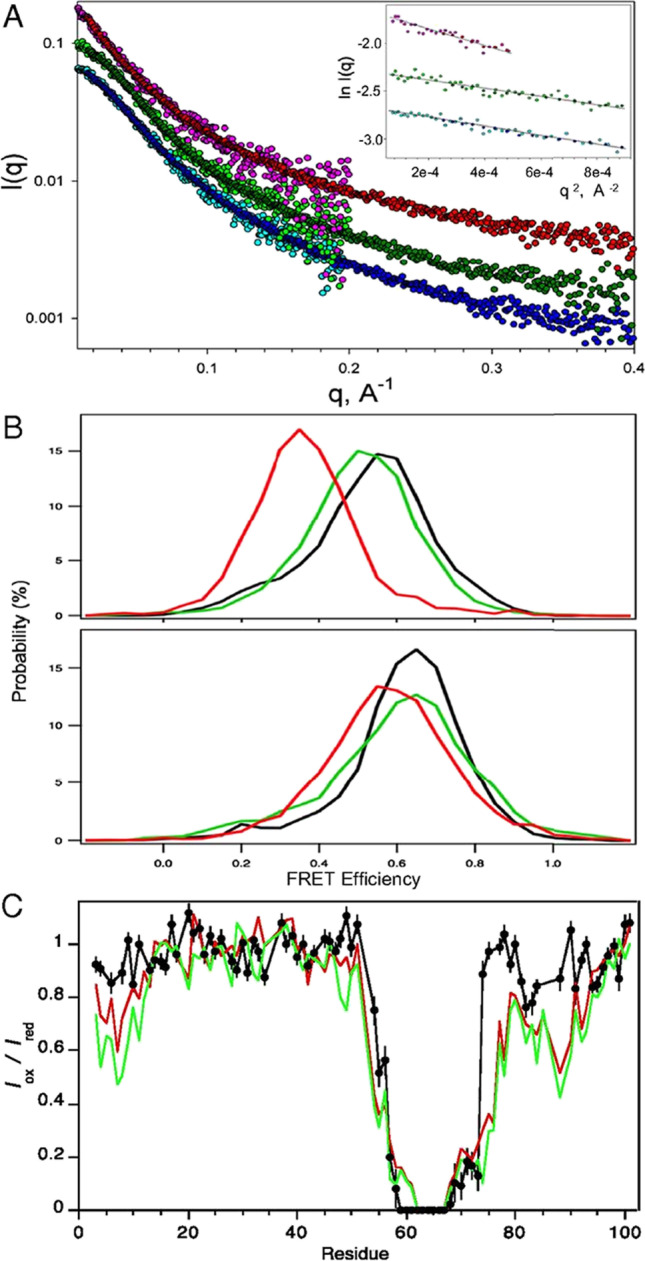
Conformational expansion of PAGE4 upon hyperphosphorylation by CLK2. (A) Experimental X-ray scattering data for the WT-PAGE4 (bottom curve, cyan/blue), HIPK1-PAGE4 (middle curve, light green/dark green), and CLK2-PAGE4 (top curve, pink/red). For each of the variants, the two colors denote independent data collections probing lower-q and medium-q ranges of the scattering data. The curves are offset for clarity. (Inset) Guinier fits of the lowest q data that yield model-free estimates of the ensemble-averaged radii of gyration for the three variants. (B) smFRET measurements. (Upper) Distributions of smFRET efficiency measurements for PAGE4 with donor and acceptor sites at positions 18 and 63 WT-PAGE4 (black), HIPK1-PAGE4 (green), and CLK2-PAGE4 (red). (Lower) Donor and acceptor sites are at positions 63 and 102 for WT-PAGE4 (black), HIPK1-PAGE4 (green), and CLK2-PAGE4 (red). (C) PRE data for CLK2-PAGE4 (black) with an MTSL spin label at C63. Results are compared with earlier observations for WT-PAGE4 (red) and HIPK1-PAGE4 (green).
Reproduced with permission from Kulkarni et al. 2017
Table 1.
A summary of the size measurements of the PAGE4 phosphoforms from both the AAWSEM simulations and SAXA and smFRET experiments
| SAXS (Rg) (Å) | FRET efficiency (E) | FRET RMS Dist (Å) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Res. 18–63 | Res. 63–102 | Res. 18–63 | Res. 63–102 | |||||||
| EXPa | SIMb | EXPa | SIMb | EXPa | SIMb | EXP | SIM | EXP | SIM | |
| Wild-type form | 36 ± 1.1 | 32.9 | 0.55 | 0.48 | 0.64 | 0.60 | 56 | 57.4 | 50 | 51.2 |
| HIPK1form | 34.7 ± 1.2 | 32.1 | 0.52 | 0.53 | 0.63 | 0.60 | 59 | 55.6 | 50 | 51.4 |
| CLK2 form | 39.8 ± 1.9 | 41.8 | 0.35 | 0.22 | 0.58 | 0.52 | 75 | 73.4 | 55 | 54.8 |
aEXP, experimental results [16]
bSIM, simulation results (this study)
Reproduced with permission from Lin et al. 2018
Given the enormity of the conformational space of PAGE4, deriving an ensemble average picture representing its conformational plasticity represents a challenge. Therefore, to further understand the mechanisms driving the conformational diversity among different PAGE4 ensembles, we analyzed their simulated atomistic trajectories using the associative memory, water-mediated, structure and energy Model (AWSEM) forcefield, and the energy landscapes were elucidated using the energy landscape visualization (ELViM) method. This method identifies and compares the population distributions of different PAGE4 ensembles using the same effective phase space. The results showed a conformational ensemble with an extended C-terminal segment of non-phosphorylated “wild-type” (WT) PAGE4 to be predominant. Interestingly, this conformation exposes the T51 residue, underscoring its potential of undertaking a fly-casting mechanism while binding to its cognate partner (Fig. 4). In contrast, in the case of HIPK1-PAGE4, the compacted ensemble populates a conformation that sequesters the phosphorylated T51 which is consistent with the experimentally observed weaker affinity of HIPK1-PAGE4 for c-Jun (Mooney et al, 2014).
Fig. 4.
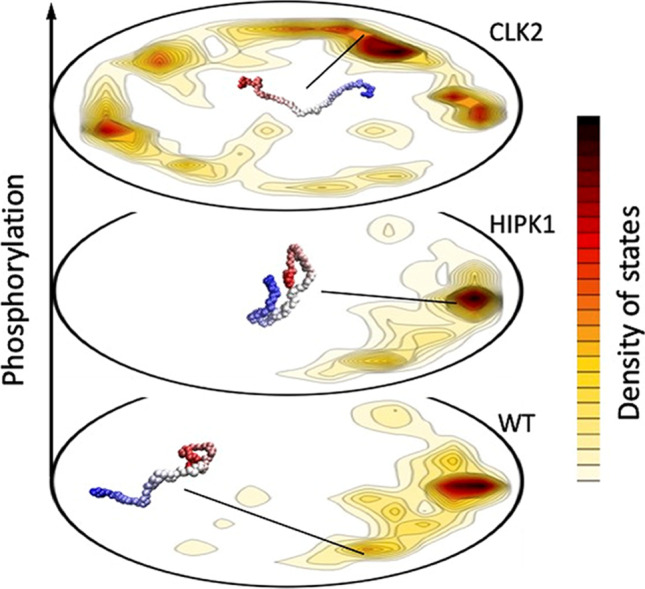
Employing the energy landscape visualization method (ELViM), different PAGE4 ensembles are represented in one single conformational phase space. The density of states, shown in the contour plots, varies according to the physical–chemical conditions, which in this case is the PAGE4 phosphorylation state. Each free energy valley can be characterized by specific conformations that entail particular binding affinities, typical of the promiscuous behavior of IDPs. For WT-PAGE4, through a fly-casting mechanism, the C-terminal region is extended, allowing the binding to its cognate partner. For the HIPK1-PAGE4, the lower free energy of the compact state decreases the affinity for c-Jun. Finally, the dominant extended conformations of CLK2-PAGE4 inhibit any binding affinity
Oscillatory dynamics of the HIPK1-PAGE4-AP1-AR-CLK2 circuit guides cellular decisions
Mathematical modeling of the HIPK1-PAGE4-AP1-AR-CLK2 circuit in PCa cells (Kulkarni et al, 2017; Lin et al, 2018; Salgia et al., 2018; Lin et al., 2019) suggested that these interactions between HIPK1, PAGE4, the AP-1 transcription factor complex, androgen receptor (AR), and CLK2 form a negative feedback loop which may give rise to oscillations in intracellular levels of the different conformational ensembles of PAGE4 as well as AR activity (Kulkarni et al, 2017) (Fig. 5). Therefore, in cells that express both HIPK1 and CLK2, the feedback loop can lead to “dynamic” regulatory circuits due to changes in PINs. Thus, conformational noise that is contributed by differential phosphorylation of PAGE4 can result in cell-to-cell variability due to rewiring of the network circuit and promote phenotypic heterogeneity in a population of androgen-dependent PCa cells. Thus, it follows that due to the oscillatory dynamics, a cell can exhibit a varying degree of androgen dependence at different time points. Thus, even non-synchronous oscillations can generate heterogeneity in an isogenic population by a non-genetic, IDP conformation-based mechanism. These oscillations can be dampened by depriving the system of androgen but may be reinstituted if deprivation is withheld or administered intermittently, suggesting that PCa cells can potentially transition from an androgen-independent to an androgen-dependent phenotype reversibly (Lin et al., 2018).
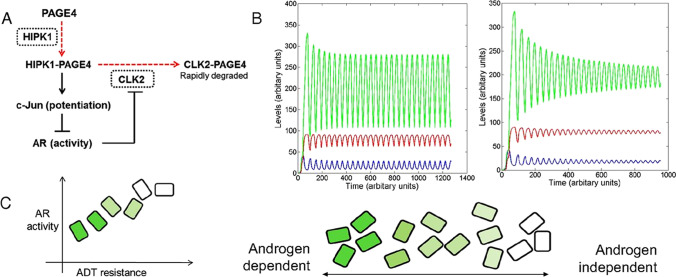
Fig. 5.
Modeling the PAGE4/AP-1/AR/CLK2 regulatory circuit. (A) Regulatory circuit for PAGE4/AP-1/AR/CLK2 interactions. Dashed red lines denote enzymatic reactions, and solid black lines denote non-enzymatic reactions. CLK2 and HIPK1, the two enzymes involved, are shown in dotted rectangles. (B) Dynamics of the circuit showing sustained and damped oscillations for HIPK1-PAGE4 (PAGE4M, shown in blue), CLK2-PAGE4 (PAGE4H, shown in red), and CLK2 (shown in green). (C) Distribution of androgen dependence for an isogenic population over a spectrum, as indicated by the shade of green. Dark green boxes denote highly androgen-dependent (i.e., ADT-sensitive) cells, and white boxes denote androgen-independent cells.
Reproduced with permission from Kulkarni et al. 2017
Corroborating these observations, a recent study (Lv et al, 2019) reported that PAGE4 overexpression in androgen-dependent (LNCaP) and independent (DU145) cells treated with hydrogen peroxide (H2O2) suppressed production of reactive oxygen species (ROS) in response to stress. However, co-expressing PAGE4 and CLK2 in these cells attenuated the ability of PAGE4 to suppress ROS suggesting that hyperphosphorylation inactivates PAGE4. On the other hand, co-expression of HIPK1 with PAGE4 reduced ROS production after H2O2 treatment in LNCaP. But in DU145 cells, co-transfection of HIPK1 and PAGE4 increased ROS suggesting that HIPK1 may impact PAGE4 function in a cell type-dependent manner.
Coupled feedback loops involving PAGE4, EMT, and Notch signaling, and non-genetic heterogeneity in PCa cells
It is now well-recognized that non-genetic mechanisms can give rise to functional heterogeneity. However, the design principles of the regulatory networks are not fully understood. Therefore, we (Singh et al., 2021) examined coupled dynamics of feedback loops involving oscillations in and AR signaling mediated through PAGE4, multistability in EMT, and Notch-Delta-Jagged signaling mediated cell–cell communication, each of which can generate non-genetic heterogeneity through multistability and/or oscillations. Interestingly, we found that different coupling strengths between AR and EMT signaling can lead to monostability, bistability, or oscillations in the levels of AR, as well as propagation of oscillations to EMT dynamics (Fig. 6). More specifically, we observed that depending on the relative strengths of the effect of ZEB1, an EMT inducer, on AR and vice-versa, the stand-alone dynamical features of EMT and AR circuits (multistability and oscillations) could percolate to the other circuit. In other words, the EMT circuit exhibits oscillations, and/or the AR circuit exhibits multistability. While multistability in EMT has been reported previously (George et al., 2017; Karacosta et al., 2019; Ruscetti et al., 2016), this is the first report to suggest oscillations in EMT.
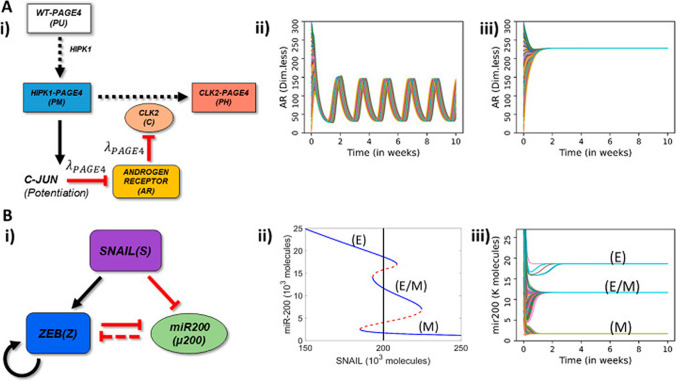
Fig. 6.
Schematic representation of PAGE4-AR and EMT circuits and their stand-alone dynamics. (A) (i) Schematic representation of PAGE4-Androgen Receptor (AR) circuit: The enzyme HIPK1 double phosphorylates WT-PAGE4 and forms the HIPK1-PAGE4 complex which can be further hyperphosphorylated by CLK2 enzyme. Solid arrows show activation, dotted arrows show phosphorylation and red hammer heads show inhibition. In turn, the HIPK1-PAGE4 complex regulates CLK2 levels via the intermediates c-JUN and AR. A strong inhibition of AR by c-JUN and that of CLK2 by AR leads to oscillations (λPAGE4 = 0.1) (ii) or a single steady state (mono-stability) (λPAGE4 = 0.9) (iii). (B) (i) EMT circuit: ZEB and microRNA-200 form a mutually inhibiting loop while SNAIL acts as an external EMT inducer. Solid arrows show transcriptional activation, dashed line show microRNA-mediated inhibition, and solid hammerheads show transcriptional inhibition. (ii) Bifurcation diagram of microRNA (miR)-200 as a function of SNAIL shows tristability, bistability or mono-stability depending on SNAIL levels. Blue and red curves show stable and unstable states respectively. The vertical black line depicts the SNAIL level (= 200,000 molecules) used in panel (iii). (iii) Dynamics of miR-200 for SNAIL = 200 K showing the existence of three states-epithelial (high miR-200; 20 K molecules), mesenchymal (low miR-200; ~ 12 K molecules). In panels A—ii, A—iii, B—iii, different curves depict AR and miR-200 dynamics starting from multiple randomized initial conditions.
Reproduced with permission from Singh et al. Entropy (Basel). 2021 Feb 26;23(3):288
The bidirectional coupling between AR signaling and EMT suggests a potential link between progression towards a partial or full EMT with significant therapeutic implications. Thus, while the epithelial phenotype usually co-occurs with PAGE4 oscillations, transitions to hybrid E/M or mesenchymal phenotypes quench these oscillations and promote low AR levels. Therefore, EMT induction can potentially promote therapy resistance by stabilizing an androgen-independent PCa phenotype through the ZEB1-AR signaling axis. Similarly, coupling between AR and ZEB1 implies that EMT can promote a drug-resistant state (Zheng et al., 2015; Fischer et al., 2015). Conversely, a switch from drug-sensitive to drug-resistant state can also trigger EMT. Taken together, these results reveal the emergent dynamics of coupled oscillatory and multistable systems and shed new light on potential mechanisms of non-genetic heterogeneity and cellular decision-making that are actuated by IDP conformational dynamics.
Conclusions and future directions
From the foregoing, it appears plausible that conformational noise contributed by IDP conformational dynamics is an additional source of noise that has hitherto remained unappreciated. As propagators of conformational and transcriptional noise, IDPs rewire PINs and unmask latent interactions in response to perturbations. Further, it may also be noted that IDPs could likely relay, and even amplify, other types of intrinsic and extrinsic noise and perturbations in the system. Therefore, noise-driven activation of latent pathways actuated by IDPs drives phenotypic switching and, thus, generates heterogeneity via non-genetic mechanisms as postulated by the MRK hypothesis.
Our cumulative efforts have provided empirical evidence for many aspects of the MRK hypothesis. However, a quantitative measure of conformational noise that is implied to originate from conformational dynamics of an IDP ensemble is still lacking. Nonetheless, the identification of the conformational preferences of the various phosphorylated forms of PAGE4 could be leveraged to authenticate conformational noise. For example, hyperphosphorylation of all 8 S/T residues in PAGE4 by CLK2 results in an almost random coil-like conformation that is non-functional. However, it is conceivable, perhaps highly plausible, that some S/T residues are more critical in unfolding the polypeptide than others. Hence, the relative abundance of these phosphoforms, the dynamics of CLK2, and the as yet unidentified phosphatase that dephosphorylates these S/T residues can impinge on the half-lives and the activity of the differentially phosphorylated ensembles of PAGE4 and, therefore, contribute to conformational noise. Interestingly, in the case of the Elk-1 transcription factor, multisite phosphorylation of 8 S/T residues also leads to opposing effects on its transcriptional activation potential. However, time-resolved NMR spectroscopy revealed that phosphorylation proceeds at significantly different rates (with differences ranging from > 30 min to 3 h), and while phosphorylation at the fast and intermediate sites promoted transactivation by Elk-1, phosphorylation at the slow site opposed it (Mylona et al., 2016), lending further support to the concept of conformational noise. Further research on PAGE4 and phenotypic switching in PCa that is currently under way in our respective laboratories should help gain deeper insight into IDP conformational noise and cellular decision-making.
Acknowledgements
PK thanks Dr. Amita Behal for her thoughtful comments on a previous version of this manuscript.
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest/Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al Emran A, Marzese DM, Menon DR, Stark MS, Torrano J, Hammerlindl H, Zhang G, Brafford P, Salomon MP, Nelson N, Hammerlindl S, Gupta D, Mills GB, Lu Y, Sturm RA, Flaherty K, Hoon DSB, Gabrielli B, Herlyn M, Schaider H. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2017;9(9):8206–8222. doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M, Sugase K, Dyson HJ, Wright PE. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci U S A. 2015;112(31):9614–9619. doi: 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21(3):432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44(5):1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggio F, Bozzato A, Benna C, Leonardi E, Romoli O, Cognolato M, Tosatto SC, Costa R, Sandrelli F. 2mit, an intronic gene of Drosophila melanogaster timeless2, is involved in behavioral plasticity. PLoS One. 2013;8(9):e76351. doi: 10.1371/journal.pone.0076351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabási AL. Scale-free networks: a decade and beyond. Science. 2009;325(5939):412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, Kragelund BB, Best RB, Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555(7694):61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A, Chang H, Huang S. Non-genetic heterogeneity–a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10(5):336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- Bürgi J, Xue B, Uversky VN, van der Goot FG. Intrinsic disorder in transmembrane proteins: roles in signaling and topology prediction. PLoS One. 2016;11(7):e0158594. doi: 10.1371/journal.pone.0158594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Meersman F, Kaminski Schierle GS, Bertoncini CW, McGee B, Kaminski CF, Tunnacliffe A. Catalytic and chaperone-like functions in an intrinsically disordered protein associated with desiccation tolerance. Proc Natl Acad Sci U S A. 2010;107(37):16084–16089. doi: 10.1073/pnas.1006276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure. 2011;19(4):566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi UB, Sanabria H, Smirnova T, Bowen ME, Weninger KR. Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules. 2019;9(3):114. doi: 10.3390/biom9030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Kolling FW, 4th, Gajdosik MD, Burger S, Russell AC, Nelson CE. Single cell analysis reveals the stochastic phase of reprogramming to pluripotency is an ordered probabilistic process. PLoS One. 2014;9(4):e95304. doi: 10.1371/journal.pone.0095304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmamy N, Milanlouei S, Barabási AL. A structural transition in physical networks. Nature. 2018;563(7733):676–680. doi: 10.1038/s41586-018-0726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Fan Y, Sun J, Lv M, Yi M, Tan X, Liu S. A dynamic interaction process between KaiA and KaiC is critical to the cyanobacterial circadianoscillator. Sci Rep. 2016;6:25129. doi: 10.1038/srep25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5(11):2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. NMR illuminates intrinsic disorder. Curr Opin Struct Biol. 2021;2(70):44–52. doi: 10.1016/j.sbi.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards YJ, Lobley AE, Pentony MM, Jones DT. Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol. 2009;10(5):R50. doi: 10.1186/gb-2009-10-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467(7312):167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberger MI, Wolynes PG, Ferreiro DU, Fuxreiter M. Frustration in fuzzy protein complexes leads to interaction versatility. J Phys Chem B. 2021;125(10):2513–2520. doi: 10.1021/acs.jpcb.0c11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M. Towards a stochastic paradigm: from fuzzy ensembles to cellular functions. Molecules. 2018;23(11):pii E3008. doi: 10.3390/molecules23113008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M. Fuzzy protein theory for disordered proteins. Biochem Soc Trans. 2020;48(6):2557–2564. doi: 10.1042/BST20200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47(29):7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JT, Jolly MK, Xu S, Somarelli JA, Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77(22):6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Babu MM. The rules of disorder or why disorder rules. Prog Biophys Mol Biol. 2009;99(2–3):94–103. doi: 10.1016/j.pbiomolbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322(5906):1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hammerlindl H, Schaider H. Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J Cell Commun Signal. 2018;12(1):133–141. doi: 10.1007/s12079-017-0435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MMK, Desai RV, Simpson ML, Weinberger LS. Cytoplasmic amplification of transcriptional noise generates substantial cell-to-cell variability. Cell Syst. 2018;7(4):384–397. doi: 10.1016/j.cels.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen Y, Mooney SM, Rajagopalan K, Bhargava A, Sacho E, Weninger K, Bryan PN, Kulkarni P, Orban J. Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J Biol Chem. 2015;290(41):25090–25102. doi: 10.1074/jbc.M115.658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wu Z, Uversky VN, Kurgan L. Functional analysis of human hub proteins and their interactors involved in the intrinsic disorder-enriched interactions. Int J Mol Sci. 2017;18(12):pii: E2761. doi: 10.3390/ijms18122761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol Cell. 2013;52(6):832–843. doi: 10.1016/j.molcel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 2016;41(10):834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Jia D, Jolly MK, Kulkarni P, Levine H. Phenotypic plasticity and cell fate decisions in cancer: insights from dynamical systems theory. Cancers (basel) 2017;9(7):70. doi: 10.3390/cancers9070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karacosta LG, Anchang B, Ignatiadis N, Kimmey SC, Benson JA, Shrager JB, Tibshirani R, Bendall SC, Plevritis SK. Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat Commun. 2019;10(1):5587. doi: 10.1038/s41467-019-13441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontogeorgaki S, Sánchez-García RJ, Ewing RM, Zygalakis KC, MacArthur BD. Noise-Processing by Signaling Networks Sci Rep. 2017;7(1):532. doi: 10.1038/s41598-017-00659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P, Jolly MK, Jia D, Mooney SM, Bhargava A, Kagohara LT, Chen Y, Hao P, He Y, Veltri RW, Grishaev A, Weninger K, Levine H, Orban J. Phosphorylation-induced conformational dynamics in an intrinsically disordered protein and potential role in phenotypic heterogeneity. Proc Natl Acad Sci U S A. 2017;114(13):E2644–E2653. doi: 10.1073/pnas.1700082114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni V, Kulkarni P. Intrinsically disordered proteins and phenotypic switching: implications in cancer. Prog Mol Biol Transl Sci. 2019;166:63–84. doi: 10.1016/bs.pmbts.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Kumar N, Cramer GM, Dahaj SAZ, Sundaram B, Celli JP, Kulkarni RV. Stochastic modeling of phenotypic switching and chemoresistance in cancer cell populations. Sci Rep. 2019;9(1):10845. doi: 10.1038/s41598-019-46926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara H, Gao X. Stochastic effects as a force to increase the complexity of signaling networks. Sci Rep. 2013;3:2297. doi: 10.1038/srep02297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury JE, Arold ST. Noise in cellular signaling pathways: causes and effects. Trends Biochem Sci. 2012;37(5):173–178. doi: 10.1016/j.tibs.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kulkarni P, Bocci F, Schafer NP, Roy S, Tsai MY, He Y, Chen Y, Rajagopalan K, Mooney SM, Zeng Y, Weninger K, Grishaev A, Onuchic JN, Levine H, Wolynes PG, Salgia R, Rangarajan G, Uversky V, Orban J, Jolly MK. Structural and dynamical order of a disordered protein: molecular insights into conformational switching of PAGE4 at the systems level. Biomolecules. 2019;9(2):77. doi: 10.3390/biom9020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Hufton PG, Lee EJ, Potoyan DA. A stochastic and dynamical view of pluripotency in mouse embryonic stem cells. PLoS Comput Biol. 2018;14(2):e1006000. doi: 10.1371/journal.pcbi.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C, Fu S, Dong Q, Yu Z, Zhang G, Kong C, Fu C, Zeng Y. PAGE4 promotes prostate cancer cells survive under oxidative stress through modulating MAPK/JNK/ERK pathway. J Exp Clin Cancer Res. 2019;38(1):24. doi: 10.1186/s13046-019-1032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur BD, Please CP, Oreffo RO. Stochasticity and the molecular mechanisms of induced pluripotency. PLoS One. 2008;3(8):e3086. doi: 10.1371/journal.pone.0003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudabadi G, Rajagopalan K, Getzenberg RH, Hannenhalli S, Rangarajan G, Kulkarni P. Intrinsically disordered proteins and conformational noise: implications in cancer. Cell Cycle. 2013;12(1):26–31. doi: 10.4161/cc.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Tsechansky M. Disorder, promiscuity, and toxic partnerships. Cell. 2009;138(1):16–18. doi: 10.1016/j.cell.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Fribourgh JL, Van Gelder RN, Partch CL. Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond. Photochem Photobiol. 2017;93(1):128–140. doi: 10.1111/php.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea DM, Yoon MK, Ou L, Kriwacki RW. Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem. 2012;393(4):259–274. doi: 10.1515/hsz-2011-0254. [DOI] [PubMed] [Google Scholar]
- Mooney SM, Jolly MK, Levine H, Kulkarni P. Phenotypic plasticity in prostate cancer: role of intrinsically disordered proteins. Asian J Androl. 2016;18(5):704–710. doi: 10.4103/1008-682X.183570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Qiu R, Kim JJ, Sacho EJ, Rajagopalan K, Johng D, Shiraishi T, Kulkarni P, Weninger KR. Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry. 2014;53(10):1670–1679. doi: 10.1021/bi500013w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona A, Theillet FX, Foster C, Cheng TM, Miralles F, Bates PA, Selenko P, Treisman R. Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science. 2016;354(6309):233–237. doi: 10.1126/science.aad1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol D, Robertson-Tessi M, Jeavons P, Anderson AR. Stochasticity in the genotype-phenotype map: implications for the robustness and persistence of bet-hedging. Genetics. 2016;204(4):1523–1539. doi: 10.1534/genetics.116.193474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Kinoshita K, Nakamura H. Hub promiscuity in protein-protein interaction networks. Int J Mol Sci. 2010;11(4):1930–1943. doi: 10.3390/ijms11041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72(1):137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow F, Marine JC, Goding CR. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 2019;33(19–20):1295–1318. doi: 10.1101/gad.329771.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan N, Fox Z, Singh A, Kulkarni P, Rangarajan G. Disorder, oscillatory dynamics and state switching: the role of c-Myc. J Theor Biol. 2015;386:105–114. doi: 10.1016/j.jtbi.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Rangarajan N, Kulkarni P, Hannenhalli S. Evolutionarily conserved network properties of intrinsically disordered proteins. PLoS One. 2015;10(5):e0126729. doi: 10.1371/journal.pone.0126729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–270. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem. 2011;112(11):3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan K, Qiu R, Mooney SM, Rao S, Shiraishi T, Sacho E, Huang H, Shapiro E, Weninger KR, Kulkarni P. The stress-response protein prostate-associated gene 4, interacts with c-Jun and potentiates its transactivation. Biochim Biophys Acta. 2014;1842(2):154–163. doi: 10.1016/j.bbadis.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti M, Dadashian EL, Guo W, Quach B, Mulholland DJ, Park JW, Tran LM, Kobayashi N, Bianchi-Frias D, Xing Y, Nelson PS, Wu H. HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene. 2016;35(29):3781–3795. doi: 10.1038/onc.2015.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdari H, Kalirad A, Picioreanu C, Tusserkani R, Goliaei B, Sadeghi M. Noise-driven cell differentiation and the emergence of spatiotemporal patterns. PLoS One. 2020;15(4):e0232060. doi: 10.1371/journal.pone.0232060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S, Mishra A, Kaur H, Hari K, Muralidharan S, Mandal S, Jolly MK. A mechanistic model captures the emergence and implications of non-genetic heterogeneity and reversible drug resistance in ER+ breast cancer cells. NAR Cancer. 2021;3(3):zcab027. doi: 10.1093/narcan/zcab027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, Sato S, Lange PH, Gleave ME. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272(28):17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- Salgia R, Jolly MK, Dorff T, Lau C, Weninger K, Orban J, Kulkarni P. Prostate-associated gene 4 (PAGE4): leveraging the conformational dynamics of a dancing protein cloud as a therapeutic target. J Clin Med. 2018;7(6):156. doi: 10.3390/jcm7060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Felsher DW. Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy. Cancer Res. 2005;65(11):4471–4474. doi: 10.1158/0008-5472.CAN-05-1172. [DOI] [PubMed] [Google Scholar]
- Shammas SL. Mechanistic roles of protein disorder within transcription. Curr Opin Struct Biol. 2017;42:155–161. doi: 10.1016/j.sbi.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS. Modeling of cancer stem cell state transitions predicts therapeutic response. PLoS One. 2015;10(9):e0135797. doi: 10.1371/journal.pone.0135797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Raduly Z, Miskei M, Fuxreiter M. Fuzzy Complexes: specific binding without complete folding. FEBS Lett. 2015;589(19 Pt A):2533–42. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Singh D, Bocci F, Kulkarni P, Jolly MK. Coupled feedback loops involving PAGE4, EMT and Notch signaling can give rise to non-genetic heterogeneity in prostate cancer cells. Entropy (basel) 2021;23(3):288. doi: 10.3390/e23030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Q, Stukalin E, Kusuma S, Gerecht S, Sun SX. Stochasticity and spatial interaction govern stem cell differentiation dynamics. Sci Rep. 2015;31(5):12617. doi: 10.1038/srep12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenschein C, Soto AM, Rangarajan A, Kulkarni P. Competing views on cancer. J Biosci. 2014;39(2):281–302. doi: 10.1007/s12038-013-9403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Wei W, Robert L, Xue M, Tsoi J, Garcia-Diaz A, Homet Moreno B, Kim J, Ng RH, Lee JW, Koya RC, Comin-Anduix B, Graeber TG, Ribas A, Heath JR. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A. 2017;114(52):13679–13684. doi: 10.1073/pnas.1712064115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Tillman K, Oberfield JL, Shen XQ, Bubulya A, Shemshedini L. c-Fos dimerization with c-Jun represses c-Jun enhancement of androgen receptor transactivation. Endocrine. 1998;9(2):193–200. doi: 10.1385/ENDO:9:2:193. [DOI] [PubMed] [Google Scholar]
- Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci. 2014;1:6. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 2015;589(19 Pt A):2498–506. doi: 10.1016/j.febslet.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Paradoxes and wonders of intrinsic disorder: stability of instability. Intrinsically Disord Proteins. 2017;5(1):e1327757. doi: 10.1080/21690707.2017.1327757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsic disorder, protein-protein interactions, and disease. Adv Protein Chem Struct Biol. 2018;110:85–121. doi: 10.1016/bs.apcsb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Per aspera ad chaos: a personal journey to the wonderland of intrinsic disorder. Biochem J. 2021;478(15):3015–3030. doi: 10.1042/BCJ20210146. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138(1):198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Wakao S, Kitada M, Dezawa M. The elite and stochastic model for iPS cell generation: multilineage-differentiating stress enduring (Muse) cells are readily reprogrammable into iPS cells. Cytometry A. 2013;83(1):18–26. doi: 10.1002/cyto.a.22069. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chu X, Longhi S, Roche P, Han W, Wang E, Wang J. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci U S A. 2013;110(40):E3743–E3752. doi: 10.1073/pnas.1308381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8(1):134–150. doi: 10.1039/c1mb05163f. [DOI] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Yoon MK, Mitrea DM, Ou L, Kriwacki RW. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans. 2012;40(5):981–988. doi: 10.1042/BST20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, He Y, Yang F, Mooney SM, Getzenberg RH, Orban J, Kulkarni P. The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem. 2011;286(16):13985–13994. doi: 10.1074/jbc.M110.210765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Gao D, Kim JJ, Shiraishi T, Terada N, Kakehi Y, Kong C, Getzenberg RH, Kulkarni P. Prostate-associated gene 4 (PAGE4) protects cells against stress by elevating p21 and suppressing reactive oxygen species production. Am J Clin Exp Urol. 2013;1(1):39–52. [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.