Abstract
As more uses for biomarkers are sought after for an increasing number of disease targets, single-target biomarkers are slowly giving way for biomarker panels. These panels incorporate various sources of biomolecular and clinical data to guarantee a higher robustness and power of separation for a clinical test. Multifactorial diseases such as psychiatric disorders show great potential for clinical use, assisting medical professionals during the analysis of risk and predisposition, disease diagnosis and prognosis, and treatment applicability and efficacy. More specific tests are also being developed to assist in ruling out, distinguishing between, and confirming suspicions of multifactorial diseases, as well as to predict which therapy option may be the best option for a given patient’s biochemical profile. As more complex datasets are entering the field, involving multi-omic approaches, systems biology has stepped in to facilitate the discovery and validation steps during biomarker panel generation. Filtering biomolecules and clinical data, pre-validating and cross-validating potential biomarkers, generating final biomarker panels, and testing the robustness and applicability of those panels are all beginning to rely on machine learning and systems biology and research in this area will only benefit from advances in these approaches.
Keywords: Proteomics, Biomarkers, Biomarker panels, Post-translational modifications, Bioinformatics
Introduction
In the ongoing quest for personalized medicine and ever more precise medical tests, molecules referred to as biomarkers have shown great potential. Life at a very fundamental level is a complex mixture of DNA, RNA, proteins, lipids, and metabolites. With the activation and suppression of biological processes, both beneficial and detrimental to an organism’s wellbeing, these biological molecules can also vary, not only in quantity, but also regarding their shape, interactions, location, and activity. More concisely, the World Health Organization has defined a biomarker to be “any substance, structure, or process that can be measured in the body or its products and influence or predict the incidence of outcome or disease” (Organization and Safety 2001). According to an early definition (Frank and Hargreaves 2003), for a biological molecule to be useful as a biomarker, it should fit into one of the three following types: type 0, which represents a patient’s natural biology with perturbations being directly correlated with symptoms, though this may include correlation and not necessarily causation; type 1, which represents the effects of treatment, being directly associated with a biological mechanism affected by the therapeutic molecule; and type 2, which represents a molecule that is directly and unambiguously associated with a patient’s feelings, biological functions, or chance of survival and which can be objectively used as a clinical data point. Type 2 biomarkers are also referred to as surrogate endpoints, as they step in for traditional clinical endpoints, in either a responsive or predictive sense. While these three types of biomarkers still encompass the field and the overall definition has not changed much, several subcategories with more precise definitions have evolved over the last two decades. A remapping of data points as predictive, diagnostic, prognostic, or therapeutic biomarkers has gained popularity (Fong and Winter 2012; Carlomagno et al. 2017; Ankeny et al. 2018; Verstockt et al. 2019; Chang and Ladame 2020; Nguyen et al. 2021; Tian et al. 2021), clearly categorizing them by their purpose and function.
Modern science has brought about many new biomarkers that can be used in research and in the clinic; however, various factors have slowed or even inhibited their entry into the health sector, and have been extensively discussed (Mayeux 2004; Agache and Rogozea 2017; Hampel et al. 2018; Locke et al. 2019). Cost is one such factor, compounded by several sources such as purchasing and maintaining the equipment used for identifying and quantifying biomolecules, various reagents, and hiring trained specialists in that specific technique. Also, along these lines is profitability, since funding agencies look at performance, patentability, market fit, and overall need and demand when identifying which biomarkers to support. Next, variations in populations and subpopulations, resulting both from geography and non-regional environmental and lifestyle factors, can also limit scalability; only biomarkers that stand up to rigorous testing conditions can progress to the clinic. Moreover, the source of a particular biomarker also has to be considered for clinical implementability. Non-invasive sources of biological material such as saliva, urine, and epithelial cells (i.e. cheek swab), as well as imaging such as fMRI, are easily obtained. Minimally invasive techniques like drawing blood are also commonly used for biomarkers, though other sources like internal organ tissue and cerebrospinal fluid are sometimes the only possibility. All sources of biological material possess distinct sets of molecules, each able to answer different biological questions. Genetic markers can easily be identified with non-invasive buccal swabbing and some metabolic biomarkers can be collected from urine or saliva, though a proteomic marker might need to be measured from blood or other tissues. Other classes of biomarkers, such as imaging data from MRI scans, though non-invasive, may be costly and time-consuming. Even when covering all these topics, regulatory agencies must see a clear benefit for the use and implementation.
Despite these drawbacks and limitations, biomarkers as a whole show great potential in the health sector by introducing new clinical tests and offering alternatives to methods that rely on phenotype and symptoms alone. Especially in psychiatry, patients often exhibit symptoms that overlap with multiple disorders and classification by symptoms alone can also create very heterogeneous groups of patients, all expected to receive the same treatment options (García-Gutiérrez et al. 2020). Of course, biomarker tests are not intended to replace medical professionals, but rather assist them by integrating genetics, behavioral science, proteomics, and other areas with the phenotypic data that they analyze when calculating risks, benefits, prognoses, and treatment options (Venigalla et al. 2017).
Biomarkers in human diseases
One of the primary compounding factors when trying to discover biomarkers for human diseases is the inherent variability between individuals and within the same individual over time due to genetic, environmental, and endogenous factors (Aylward et al. 2014; Enroth et al. 2014; Thyagarajan et al. 2016; Aziz et al. 2019). Differences in biological molecules can arise from ethnicity, sex, and lifestyle choices like smoking and alcohol use. As such, any biomarker must pass stringent testing and be replicable, reliable, and robust to ensure the lowest rate possible of false negatives and false positives. Several categories of data have entered the field to predict the risk of an individual to develop a disease, detect the disease at an earlier stage, identify or confirm a disease, and predict a patient’s response to a particular treatment and their prognosis.
The first biomarker tests were originally possible by studying a single biological molecule, and in their elegant simplicity, are still widely used to this day. In 2000, specific types of DNA mutations were linked to their environmental causes (Shugart 2000); a year later, mutations in certain protein-encoding DNA sequences increased the risk for an individual to develop cancer, and an array of genetic, epigenetic, and protein data points were suggested as potential biomarkers to identify cancer (Srinivas et al. 2001); and over the following few years, other biomarkers had been identified to measure the progression of Alzheimer’s disease in a drug trial (Jack et al. 2003), predict atherothrombotic events (Ridker et al. 2004), measure cellular senescence (Krishnamurthy et al. 2004), identify metabolic syndrome (Ryo et al. 2004), and assess oxidative stress (Dalle-Donne et al. 2006). Since these initial studies, the number of publications involving biomarkers has skyrocketed, having reached a total of over one million publications with the keyword biomarker in PubMed (NCBI Resource Coordinators 2018). The path taken to research biomarkers, however, shifted with the confirmation that many diseases and disorders cannot be boiled down to a single dysregulated gene, protein, or metabolite, which is especially true with psychiatric disorders, since a combination of environmental factors and genetic factors often leads to their development (Kubota et al. 2012; Meyer-Lindenberg and Tost 2012; Klengel and Binder 2015).
Each class of biomarker calls for a different technique for analysis, which can also vary depending on the properties of the biomarker itself. RNA in all its forms, proteins and polypeptides, lipids, and metabolites can all be analyzed differently (see Fig. 1). More relevant to the patient, each type of biomarker is well-suited to respond to certain physiological questions (see Table 1). Genomics can be used for predictive, prognostic, and diagnostic biomarkers, which may allow a medical professional to predict disease risk; however, it usually cannot be used for therapeutic biomarkers, since DNA rarely varies in response to treatment or medication, excluding gene therapy. In contrast, RNA and protein levels are dynamic and are associated with a vast array of biological processes, making them suitable for all four classes of biomarkers. Metabolites also show great potential as diagnostic, prognostic, and therapeutic biomarkers (Tolstikov et al. 2020). Lastly, non-molecular biomarkers, such as visual phenotyping via radiographic capture, circulating tumor cells (Punnoose et al. 2010; Cabel et al. 2017; Pantel et al. 2019; Yang et al. 2019), physiological parameters like blood pressure and resting heart rate, as well as the human microbiota (Manor et al. 2020; Julie et al. 2021), all have their niches in various aspects of biomarker use.
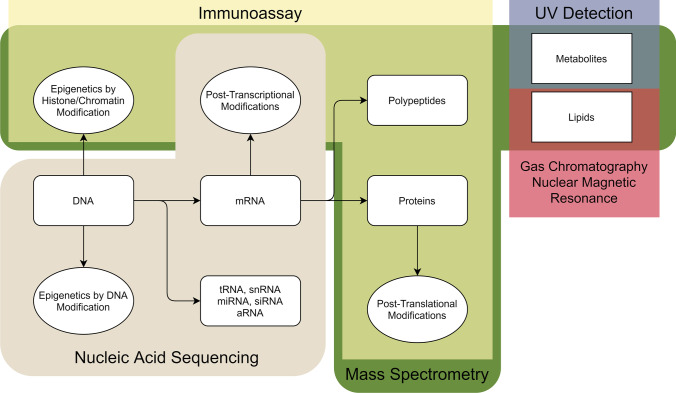
Fig. 1.
Possible sources for molecular biomarkers with each molecule type categorized depending on the method(s) by which they are most commonly detected. Created using open
source software at Diagrams.net. Different classes of biomarkers are separated based on the method(s) available for detection. Nucleic acid sequencing can be used for DNA, epigenetics by DNA modification, mRNA, post-transcriptional modifications, along with tRNA, snRNA, miRNA, siRNA, and aRNA. Immunoassays can detect epigenetics by histone/chromatin modification, polypeptides, proteins, and post-translational modifications. Mass spectrometry can detect all the biomarkers in immunoassays with the addition of metabolite and lipids. UV detection can also be used for metabolites. Gas chromatography and nuclear magnetic resonance can be used for lipids
Table 1.
Different classes of molecular biomarkers, subclasses, and some published applications
| Molecule | Biomarker | Common applications |
|---|---|---|
| Nucleic acids | Genetic | -Risk factors and predisposition |
| -DNA | -Diagnosis and prognosis | |
| -Therapy efficacy | ||
| -Drug dosing (absorption and metabolism) | ||
| Transcriptomic | -Diagnosis and prognosis | |
| -RNA | -Determining physiological states | |
| -Identifying infection states | ||
| Epigenetic | -Diagnosis and prognosis | |
| -DNA | -Risk factors and predisposition | |
| -Histones | -Therapy | |
| -Chromatin | ||
| Amino acids | ||
| Proteomic | -Diagnosis and prognosis | |
| -Proteins | -Therapy | |
| -Polypeptides | ||
| -Post-translational modifications | ||
| Metabolites | Metabolomic | -Diagnosis and prognosis |
| -Hormones | -Treatment response | |
| -Free amino acids | -Forensic toxicology | |
| -Drug metabolites | -Drug efficacy/toxicity | |
| -Vitamins | ||
| Lipids | Lipidomic | -Diagnosis and prognosis |
| -Phospholipids | -Treatment response | |
| -Glycerides | -Risk factors and predisposition | |
| -Sterols | ||
| -Fatty acids |
During the ongoing quest for biomarkers, new data points are constantly being investigated, both in terms of identity and in origin. Despite a great deal of studies regarding protein levels in diseases, as well as the well-established use of protein-based biomarkers in the clinic, protein post-translational modifications (PTMs) have entered the clinic without much fanfare (García-Giménez et al. 2017). PTMs affect several aspects of protein function and properties, modifying localization, binding, activity, and stability, among other characteristics (Ramazi and Zahiri 2021). Each modification can be added and removed in different ways, is reversible or irreversible, enzymatically regulated or spontaneous, site-specific or global, and is controlled by various regulatory pathways (Walsh et al. 2005). PTM data is an untapped source of information for biomarkers and proteomic studies overall since PTMs can respond rapidly to stimuli, as PTMs can occur more quickly than protein translation, simultaneously offering a more phenotypic profile than transcriptomic data. The different mechanisms of addition and removal and their associated biological pathways make PTMs even more attractive for biomarker studies, with the enzymes themselves also being potential biomarkers for some diseases and disorders. There are hundreds of known protein modifications and, though some are more consistently found, such as sulfide bridges between cysteine residues, many can play roles both upstream and downstream to protein-based dysregulations in diseases.
Not all applications have been implemented in the clinic; this is not an exhaustive list of applications or references. Documented uses of biomarkers that are genetic (Novelli et al. 2008; Ziegler et al. 2012; Coppedè et al. 2014; Zhang et al. 2019; Center for Drug Evaluation and Research 2021), transcriptomic (Heidecker et al. 2011; Pedrotty et al. 2012; van Rensburg and Loxton 2015; Xi et al. 2017; Zhang et al. 2019), epigenetic (Kubota et al. 2012; Coppedè et al. 2014; Li et al. 2014; García-Giménez et al. 2017; Soler-Botija et al. 2019), proteomic (Hewitt et al. 2004; Theodorescu et al. 2006; Egerer et al. 2009; Humphries et al. 2014; Raemdonck et al. 2014; Thelin et al. 2017; Zhang et al. 2019; Zhao et al. 2020), metabolomic (Helander and Beck 2005; Chen et al. 2011; Zhang et al. 2015; Tam et al. 2017; Wang et al. 2020; Long et al. 2020), and lipidomic (Chandler et al. 2016; Kim et al. 2017; Yan et al. 2017, 2018; Aquino et al. 2018; El-Ansary et al. 2020; Liu et al. 2020).
Phosphorylation, for example, is a widely studied regulator of protein activity and has been studied in relation to multiple diseases including identifying tumors (Carter et al. 2020) and neurofibrillary tangles in Alzheimer’s disease (Buerger et al. 2006; Henriques et al. 2016). Other targets include multiple PTMs potentially related to Parkinson’s disease (Schmid et al. 2013) and the complex field of glycoproteins, which has shown promise in identifying neoplastic diseases (Díaz-Fernández et al. 2018). PTMs are also known to play roles in aging and age-related diseases (Santos and Lindner 2017) and inflammatory processes (Yang et al. 2017), and dysregulated modification of tubulin has been suggested to be behind a wide range of human diseases including cancer, cardiac diseases, and bleeding disorders (Magiera et al. 2018). As not all modifications are performed enzymatically, cellular conditions can also affect how proteins are modified; succinylation, malonylation, formylation, succination, and acetylation are all hypothesized—or have been proven—to be possible without any transferase (Lin et al. 2012). Documenting these changes has thus far led to insight into the metabolic (de)regulation and metastasis that occurs in gastric cancer (Song et al. 2017) and GAPDH malonylation has been suggested to play a role in inflammation in macrophages (Galván-Peña et al. 2019). Overall, PTMs show great potential as markers of phenotypes while still maintaining flexibility and dynamicity due to their fast and often reversible nature.
Unfortunately, due to variations between individuals (Enroth et al. 2014) and within individuals over time (Cicognola et al. 2015), the infeasibility of measuring biomarkers directly in more sensitive tissue like the brain or other internal organs, and the fact that some more complex, multifactorial diseases like psychiatric disorders stem from a vast number of small changes (2009; Genovese et al. 2016; Selzam et al. 2018), not every potential biomarker can exhibit such stark differences as those seen in the some of the first clinical biomarkers. In response, the scientific community has found ways to use biomarkers with lower robustness. In one approach, a biomarker test can be prescribed a complementary technique in parallel with other analyses to distinguish between similar diseases, confirm suspicions, or objectively measure disease progression or treatment efficacy. Another approach is biomarker panels, a set of independent biomarkers that, together, provide a more robust and reproducible answer to a given medical question. As computer processing capacity increases, so does the complexity of problems that can be solved, and systems biology has now made it feasible to sift through tens of thousands of data points to return a set of potential biomarkers that strengthen one another. Overall, biomarker panels are able to perform a slightly different function from individual biomarkers, since the panel is additionally able to help exclude compounding factors and work with multifactorial disorders with more certainty.
In one example, a panel of metabolites has been used to concisely map a profile for the healthy metabolic response to physical activity (Netzer et al. 2011). In another study, over 1600 plasma and urine protein levels were studied for associations with dozens of human diseases (Dudley and Butte 2009). Such studies have not been limited to being reactive, but have also been proposed to be applicable for proactive, long-term health monitoring and other facets of personalized medicine (Miller et al. 2019). In conjunction with magnetic resonance imaging, transcriptomic data of 30 different genes were used to predict a progression from clinically isolated syndrome to multiple sclerosis (Tossberg et al. 2013). Trends in fasting plasma glucose levels can be predicted with a panel of 9 metabolites, despite neither the individual metabolites nor a panel of standard risk factors such as BMI and age being able to accurately model fasting plasma glucose levels (Hische et al. 2012). Another panel of metabolites can predict dysregulation in fasting plasma glucose levels, type-2 diabetes, and insulin resistance (Menni et al. 2013), sometimes even a decade or more before developing a disorder (Tabák et al. 2009; Rhee et al. 2011; Wang et al. 2011). A three-part panel for detecting colorectal cancer and pre-malignant colorectal neoplasia has also been approved using genetic, epigenetic, and protein assays (Imperiale et al. 2014a, b). Using NMR-based metabolomic data and multivariate statistics, Song et al. compiled 82 references to form a list of potential and identified biomarkers in over 10 disease categories before classifying the biomarkers for categories like diagnosis, treatment, and prognosis (Song et al. 2019) and Dhama et al. compiled a comprehensive list of several types of potential biomarkers for physiological and psychological stress, along with their sources and possible clinical significance (Dhama et al. 2019). Volatile organic compounds are also being investigated using gas chromatography mass spectrometry, with the potential to identify cancers, genetic and metabolic disorders, infectious diseases, and various other conditions (Buljubasic and Buchbauer 2015). Lastly, comprehensive tissue-based DNA sequencing panels have been approved by the FDA as companion diagnostic tools to identify cancer types and suggest compatible therapy options (U.S. Food & Drug Administration 2021) and new panels are being tested and approved every year.
Among the many classes of human diseases, psychiatric disorders are among those that most need biomarkers, since the clinically observable symptoms and genetic background for many disorders are often similar and have significant overlap (Doherty and Owen 2014). This makes an early, fast, and precise diagnosis through clinical observation alone extremely difficult. Moreover, antipsychotics, which are used to alleviate symptoms in conditions such as schizophrenia and bipolar disorder, do not have a universally positive response. While one patient might respond well to a given medication, another patient may not respond, or may even have a negative response (Leucht et al. 2013). Repeated negative responses to treatment paired with debilitating side effects increase patient stress and can lead to treatment dropout (Wahlbeck et al. 2001; Rabinowitz and Davidov 2008; Rabinowitz et al. 2009), reducing the patient’s quality of life and incurring more medical costs. Another important point is that, for many psychiatric disorders, by the time symptoms appear and can be effectively diagnosed and treated, the patient’s quality of life may have already been negatively, potentially irreversibly, affected (Guest et al. 2016). Biomarker panels for diagnosing neurological and psychological conditions in the prodromal phase or earlier, distinguishing between two or more similar psychiatric disorders, and predicting a patient’s response to a given treatment option without needing to perform a trial-and-error testing method would all be extremely beneficial to patients and healthcare providers alike.
Generating biomarker panels
Unlike the less fruitful searches for molecular signatures related to psychiatric disorders, diagnostic biomarkers for other diseases, such as cancer, have been established with relative success (Parkes et al. 1995; Cramer et al. 2011; Fontecha et al. 2016). Potential diagnostic biomarkers for ovarian cancer, for example, were discovered after the analysis of five predictive models across three combined stages of validation and discovery, compared against an already well-established biomarker for some types of cancers, the CA-125 protein (Zhu et al. 2011). Although this is a reliable method for establishing new biomarkers, in the absence of any previously established biomarkers, mathematical modeling has paved new avenues in biomarker development.
Establishing biomarkers is a multistep and multidisciplinary process that can be split into three major stages before passing through regulatory bodies into the clinic: discovery, development and validation, and implementation (Fig. 2) (Martins-de-Souza et al. 2011; Menetski et al. 2019). During the discovery and development/validation stages, the selection criteria for patient eligibility for a predictive model form a key step for efficiently achieving statistical significance. Thus, establishing biomarkers requires not only an understanding of the biochemical pathways involved in the disease or biological state by analyzing molecular data but also properly collecting clinical data from patients for selection and stratification, as well as for establishing exclusion and validation criteria (Laifenfeld et al. 2012; Drucker and Krapfenbauer 2013; Fröhlich et al. 2018; Golubnitschaja et al. 2018). Overall, the discovery step seeks to do just that: collect potentially vast amounts of data from various biomolecular and clinical assays to analyze and filter for potentially relevant data points. Molecular data from different omics and clinical information are used to compose molecular signatures after increasingly advanced statistical processing steps involving advanced computational methods, algorithms, and machine learning. When using single omics methods, molecular signature investigations benefit from multivariate approaches with penalties for classification of features or regression. However, in the case of higher complexity multi-omics, the lasso or elastic net selection methods help to efficiently reduce data dimensionality (Bravo-Merodio et al. 2019; Freue et al. 2019). Moreover, algorithms such as tree ensembles can combine different models to identify potential molecular signatures using distinct techniques (Chen et al. 2013; Toth et al. 2019; Shuwen et al. 2020; Acharjee et al. 2020). Mathematical models are being successfully applied to a small portion of omic datasets, and without them, the search for biomarkers would take a prohibitive amount of time or even find dead ends. Despite great advances, however, there are still no computational methods or data interpretation protocols that are so well established that they have become state-of-the-art for molecular signatures.
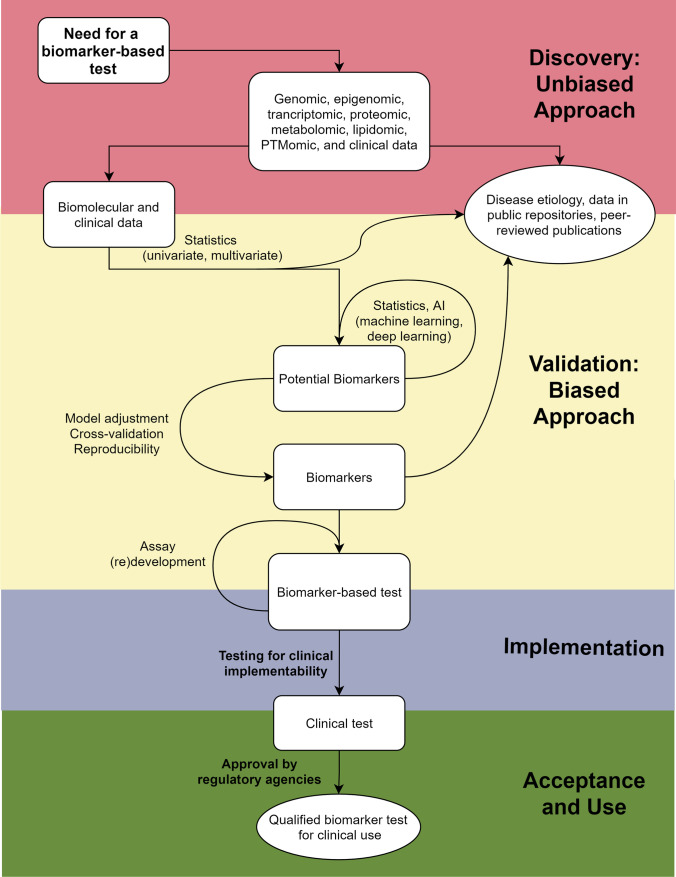
Fig. 2.
A workflow describing the steps going from a need for a biomarker-based test to a qualified biomarker test for clinical use, passing through an unbiased discovery stage, a biased development and validation stage, and an implementation stage before being proposed for acceptance and clinical use. Created using the open
source software at Diagrams.net. A flow chart detailing the process from a need for a biomarker-based test, through genomic, epigenomic transcriptomic, proteomic, metabolomic, lipidomic, PTMomic, and clinical data in a discovery-based, unbiased approach into biomolecular and clinical data. Using univariate and multivariate statistics, this and the previously mentioned data can both be published in publicly available repositories and peer-reviewed articles. Statistically filtered data can then pass into potential biomarkers that loop through more statistics and AI-based filtering using machine and deep learning before passing through a model adjustment, cross-validation, and reproducibility tests to reach biomarker status. This then passes on to a biomarker-based test, all of which occurred in a validation-based, biased approach. During implementation, the assay may be redeveloped and is eventually tested for clinical implementability and reaches clinical test status. In the acceptance and use phase, approval by regulatory agencies must be obtained to reach a final, qualified biomarker test for clinical use
After the discovery phase, the development and validation phase is often composed of two verification rounds to further narrow down the data points highlighted during the first phase. The first, cross-validation round reduces the number of potentially false biomarkers through sets of algorithm retraining, which may be based on similar samples to those used in the discovery phase or based on a new set of samples in an omics or multi-omics experiment (Smit et al. 2007; Singh et al. 2019; Chierici et al. 2020). During this round, one common method of pre-validation is to randomize and divide the retraining samples into a number k of groups of similar sizes, leading to its name k-fold cross-validation. In this method, one group is set aside as a validation set, while the computer models a fit to the training set. During cross-validation, the number of potential targets in the biomarker panel is reduced as the model is adjusted, contributing to increased specificity and sensitivity thresholds, along with a greater power of discrimination between biological states (Harris et al. 2009).
Next, a second validation round is where the greatest challenge lies when establishing biomarkers due to both the greater number of samples required for analysis and a dependence on long-term resources to carry out studies over a potentially great period of time (Bonassi et al. 2001). During this stage, many new samples must be collected to analyze the potential biomarkers. A biomarker or panel can undergo different levels of validation to be applicable for more restricted or broader populations, as well as conditions under which it is valid. Furthermore, biomarker validation must also include confirmatory clinical endpoint analyses to prevent misleading conclusions (Strimbu and Tavel 2010). During development, statistical methods can be used to predict a model's error when applied to new cases, such as in the double cross-validation method (Smit et al. 2007; Szász et al. 2016). After this phase, the rigor of the methods used to establish the potential biomarkers and the accessibility of the tool for clinical use are considered. To ensure smooth translatability to the clinic, a great deal of care must be taken at every step of the discovery process, especially when creating sample groups, working with missing values, selecting assays, and testing for robustness and scalability across subpopulations (Mnatsakanyan et al. 2018). Meeting the criteria for a particular regulatory and implementing body, such as the FDA in the United States or the EMA in Europe, is a complex and time-consuming procedure as well, as these clinical trials must carefully weigh the risks and benefits of the use of the biomarker panel. Robustness must be proven, and time must be taken to determine the precision of the test; false negatives and false positives must be at a minimum or the panel may be sent back for statistical refinement, requiring a new round of clinical trials and once the panel of biomarkers has passed these trials, it may then be qualified for eventual implementation (Manolis et al. 2015; Menetski et al. 2019). To ensure a robust test, future biomarker investigations should be carried out with multi-omics techniques on several layers of biological systems, also providing a better understanding of the molecular mechanisms involved and adding extra dimensions to the mathematical model being developed. While this makes the process of building a panel of biomarkers more challenging, it creates a more useful test with a higher power of separation.
Conclusions
The early days of biomarker research brought important advances for a few select cases, dominated by individual biomolecules. As different clinical targets received focus, the field of biomarker research quickly shifted once single-molecule biomarkers began to slow and new target classes were scrutinized through transcriptomics, lipidomics, PTMomics, and metabolomics. New targets for disease risk and predisposition, diagnosis, prognosis, and treatment are constantly being discovered before passing through rigorous testing steps to ensure reproducibility and population-wide applicability. Though the era of individual biomarkers has not come to a close, biomarker panels have entered the scene to increase the robustness of tests for clinical implementation. Biomarker panels bring together various data points, even using multi-omic approaches, to compose tests to complement current clinical practices. By using the tests in a controlled manner, medical professionals can efficiently diagnose, rule out, confirm, or distinguish between various possible diseases, allowing a patient to be more quickly and accurately diagnosed and therefore treated quickly and effectively. Multifactorial diseases, such as psychiatric disorders, with a culmination of both genetic and environmental factors have already benefited greatly from such biomarker panels and show great potential for further development. Tests to facilitate the diagnosis and treatment of such disorders are of great importance to doctors and patients alike, improving the quality of life of the patient and simultaneously reducing the cost of medical care.
Propelling this new chapter of biomarker research is advanced computational approaches, able to process and filter data in ways that are simply not feasible to be performed manually or with simple algorithms. Machine learning has found a niche in biomarker research, performing vital steps during both the discovery and validation phases of panel generation. Pre-validation, selection methods, and cross-validation of data all contribute to a high-quality panel of potential biomarkers with higher certainty of achieving implementable results and a lower chance of having confounding factors influencing the results. As computational methods and processing speed advance, increasingly large datasets involving multiple data sources and collection methods will be accessible. Though this field is still in its infancy with no widely standardized methods for data processing or validation, it is nonetheless paving the way for new biomarker-based tests with various clinical applications.
Funding
This research is funded by the Coordination for the Improvement of Higher Education Personnel (CAPES; grant number 88887.495565/2020–00) and The São Paulo Research Foundation (FAPESP; grant numbers 2016/07948–8, 2017/25588–1, 2018/03422–7, 2019/25957–2, and 2020/04746–0).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Acharjee A, Larkman J, Xu Y, et al. A random forest based biomarker discovery and power analysis framework for diagnostics research. BMC Med Genomics. 2020;13:178. doi: 10.1186/s12920-020-00826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agache I, Rogozea L. Asthma biomarkers: do they bring precision medicine closer to the clinic? Allergy Asthma Immunol Res. 2017;9:466–476. doi: 10.4168/aair.2017.9.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankeny JS, Labadie B, Luke J, et al. Review of diagnostic, prognostic, and predictive biomarkers in melanoma. Clin Exp Metastasis. 2018;35:487–493. doi: 10.1007/s10585-018-9892-z. [DOI] [PubMed] [Google Scholar]
- Aquino A, Alexandrino GL, Guest PC, et al. Blood-based lipidomics approach to evaluate biomarkers associated with response to olanzapine, risperidone, and quetiapine treatment in schizophrenia patients. Front Psychiatry. 2018;9:209. doi: 10.3389/fpsyt.2018.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward LL, Hays SM, Smolders R, et al. Sources of variability in biomarker concentrations. J Toxicol Environ Health B Crit Rev. 2014;17:45–61. doi: 10.1080/10937404.2013.864250. [DOI] [PubMed] [Google Scholar]
- Aziz N, Detels R, Quint JJ, et al. Biological variation of immunological blood biomarkers in healthy individuals and quality goals for biomarker tests. BMC Immunol. 2019;20:33. doi: 10.1186/s12865-019-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonassi S, Neri M, Puntoni R. Validation of biomarkers as early predictors of disease. Mutat Res-Fund Mol M. 2001;480–481:349–358. doi: 10.1016/S0027-5107(01)00194-4. [DOI] [PubMed] [Google Scholar]
- Bravo-Merodio L, Williams JA, Gkoutos GV, Acharjee A. Omics biomarker identification pipeline for translational medicine. J Transl Med. 2019;17:155. doi: 10.1186/s12967-019-1912-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger K, Ewers M, Pirttilä T, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- Buljubasic F, Buchbauer G. The scent of human diseases: a review on specific volatile organic compounds as diagnostic biomarkers. Flavour Fragr J. 2015;30:5–25. doi: 10.1002/ffj.3219. [DOI] [Google Scholar]
- Cabel L, Proudhon C, Gortais H, et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol. 2017;22:421–430. doi: 10.1007/s10147-017-1105-2. [DOI] [PubMed] [Google Scholar]
- Carlomagno N, Incollingo P, Tammaro V, et al (2017) Diagnostic, predictive, prognostic, and therapeutic molecular biomarkers in third millennium: a breakthrough in gastric cancer. BioMed Research International 2017:e7869802. 10.1155/2017/7869802 [DOI] [PMC free article] [PubMed]
- Carter AM, Tan C, Pozo K, et al. Phosphoprotein-based biomarkers as predictors for cancer therapy. PNAS. 2020;117:18401–18411. doi: 10.1073/pnas.2010103117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Drug Evaluation and Research (2021) Table of pharmacogenomic biomarkers in drug labeling. FDA
- Chandler PD, Song Y, Lin J, et al. Lipid biomarkers and long-term risk of cancer in the Women’s Health Study. Am J Clin Nutr. 2016;103:1397–1407. doi: 10.3945/ajcn.115.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JYH, Ladame S (2020) Chapter 1.1 - Diagnostic, prognostic, and predictive biomarkers for cancer. In: Ladame S, Chang JYH (eds) Bioengineering Innovative Solutions for Cancer. Academic Press, pp 3–21
- Chen T, Cao Y, Zhang Y, et al. Random forest in clinical metabolomics for phenotypic discrimination and biomarker selection. eCAM. 2013;2013:e298183. doi: 10.1155/2013/298183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Xie G, Wang X, et al (2011) Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma *. Molecular & Cellular Proteomics 10. 10.1074/mcp.M110.004945 [DOI] [PMC free article] [PubMed]
- Chierici M, Bussola N, Marcolini A, et al (2020) Integrative network fusion: a multi-omics approach in molecular profiling. Front Oncol 0: 10.3389/fonc.2020.01065 [DOI] [PMC free article] [PubMed]
- Cicognola C, Chiasserini D, Parnetti L. Preanalytical confounding factors in the analysis of cerebrospinal fluid biomarkers for Alzheimer’s disease: the issue of diurnal variation. Front Neurol. 2015;6:143. doi: 10.3389/fneur.2015.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20:943–956. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer DW, Bast RC, Berg CD, et al. Ovarian cancer biomarker performance in prostate, lung, colorectal, and ovarian cancer screening trial specimens. Cancer Prev Res. 2011;4:365–374. doi: 10.1158/1940-6207.CAPR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo R, et al. Biomarkers of oxidative damage in human disease. Clin Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- Dhama K, Latheef SK, Dadar M, et al (2019) Biomarkers in stress related diseases/disorders: diagnostic, prognostic, and therapeutic values. Front Mol Biosci 6. 10.3389/fmolb.2019.00091 [DOI] [PMC free article] [PubMed]
- Díaz-Fernández A, Miranda-Castro R, de-Los-Santos-Álvarez N, Lobo-Castañón MJ (2018) Post-translational modifications in tumor biomarkers: the next challenge for aptamers? Anal Bioanal Chem 410:2059–2065. 10.1007/s00216-018-0861-9 [DOI] [PubMed]
- Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6:29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA Journal. 2013;4:7. doi: 10.1186/1878-5085-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley JT, Butte AJ (2009) Identification of discriminating biomarkers for human disease using integrative network biology. Pac Symp Biocomput 27–38 [PMC free article] [PubMed]
- Egerer K, Feist E, Burmester G-R. The serological diagnosis of rheumatoid arthritis: antibodies to citrullinated antigens. Dtsch Arztebl Int. 2009;106:159–163. doi: 10.3238/arztebl.2009.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ansary A, Chirumbolo S, Bhat RS, et al. The Role of Lipidomics in Autism Spectrum Disorder. Mol Diagn Ther. 2020;24:31–48. doi: 10.1007/s40291-019-00430-0. [DOI] [PubMed] [Google Scholar]
- Enroth S, Johansson Å, Enroth SB, Gyllensten U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat Commun. 2014;5:4684. doi: 10.1038/ncomms5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong ZV, Winter JM. Biomarkers in pancreatic cancer: diagnostic, prognostic, and predictive. Cancer J. 2012;18:530–538. doi: 10.1097/PPO.0b013e31827654ea. [DOI] [PubMed] [Google Scholar]
- Fontecha N, Basaras M, Hernáez S, et al. Assessment of human papillomavirus E6/E7 oncogene expression as cervical disease biomarker. BMC Cancer. 2016;16:852. doi: 10.1186/s12885-016-2885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- Freue GVC, Kepplinger D, Salibián-Barrera M, Smucler E. Robust elastic net estimators for variable selection and identification of proteomic biomarkers. Ann Appl Stat. 2019;13:2065–2090. doi: 10.1214/19-AOAS1269. [DOI] [Google Scholar]
- Fröhlich H, Patjoshi S, Yeghiazaryan K, et al. Premenopausal breast cancer: potential clinical utility of a multi-omics based machine learning approach for patient stratification. EPMA Journal. 2018;9:175–186. doi: 10.1007/s13167-018-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván-Peña S, Carroll RG, Newman C, et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun. 2019;10:338. doi: 10.1038/s41467-018-08187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Giménez JL, Seco-Cervera M, Tollefsbol TO, et al. Epigenetic biomarkers: current strategies and future challenges for their use in the clinical laboratory. Crit Rev Clin Lab Sci. 2017;54:529–550. doi: 10.1080/10408363.2017.1410520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gutiérrez MS, Navarrete F, Sala F, et al (2020) Biomarkers in psychiatry: concept, definition, types and relevance to the clinical reality. Front Psychiatry 0: 10.3389/fpsyt.2020.00432 [DOI] [PMC free article] [PubMed]
- Genovese G, Fromer M, Stahl EA, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubnitschaja O, Polivka J, Yeghiazaryan K, Berliner L. Liquid biopsy and multiparametric analysis in management of liver malignancies: new concepts of the patient stratification and prognostic approach. EPMA Journal. 2018;9:271–285. doi: 10.1007/s13167-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest FL, Guest PC, Martins-de-Souza D. The emergence of point-of-care blood-based biomarker testing for psychiatric disorders: enabling personalized medicine. Biomark Med. 2016;10:431–443. doi: 10.2217/bmm-2015-0055. [DOI] [PubMed] [Google Scholar]
- Hampel H, O’Bryant SE, Molinuevo JL, et al. Blood-based biomarkers for Alzheimer disease: mapping the road to the clinic. Nat Rev Neurol. 2018;14:639–652. doi: 10.1038/s41582-018-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Girolami M, Mischak H, et al. Definition of valid proteomic biomarkers: a Bayesian solution. In: Kadirkamanathan V, Sanguinetti G, Girolami M, et al., editors. Pattern Recognition in Bioinformatics. Berlin, Heidelberg: Springer; 2009. pp. 137–149. [Google Scholar]
- Heidecker B, Kittleson MM, Kasper EK, et al. Transcriptomic biomarkers for the accurate diagnosis of myocarditis. Circulation. 2011;123:1174–1184. doi: 10.1161/CIRCULATIONAHA.110.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander A, Beck O. Ethyl sulfate: a metabolite of ethanol in humans and a potential biomarker of acute alcohol intake. J Anal Toxicol. 2005;29:270–274. doi: 10.1093/jat/29.5.270. [DOI] [PubMed] [Google Scholar]
- Henriques AG, Müller T, Oliveira JM, et al. Altered protein phosphorylation as a resource for potential AD biomarkers. Sci Rep. 2016;6:30319. doi: 10.1038/srep30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SM, Dear J, Star RA. Discovery of protein biomarkers for renal diseases. JASN. 2004;15:1677–1689. doi: 10.1097/01.ASN.0000129114.92265.32. [DOI] [PubMed] [Google Scholar]
- Hische M, Larhlimi A, Schwarz F, et al. A distinct metabolic signature predicts development of fasting plasma glucose. Journal of Clinical Bioinformatics. 2012;2:3. doi: 10.1186/2043-9113-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JM, Penno MAS, Weiland F, et al (2014) Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1844:1051–1058. 10.1016/j.bbapap.2014.01.018 [DOI] [PubMed]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;371:187–188. doi: 10.1056/NEJMc1405215. [DOI] [PubMed] [Google Scholar]
- Jack CR, Slomkowski M, Gracon S, et al. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003;60:253–260. doi: 10.1212/01.WNL.0000042480.86872.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julie V, Romain V, Nicolas B, Mathilde B. Gut microbiota as potential biomarker and/or therapeutic target to improve the management of cancer: focus on colibactin-producing Escherichia coli in colorectal cancer. Cancers (basel) 2021;13:2215. doi: 10.3390/cancers13092215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-Y, Lee H, Kim S-H, et al. Discovery of potential biomarkers in human melanoma cells with different metastatic potential by metabolic and lipidomic profiling. Sci Rep. 2017;7:8864. doi: 10.1038/s41598-017-08433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Binder EB. Epigenetics of Stress-Related Psychiatric Disorders and Gene × Environment Interactions. Neuron. 2015;86:1343–1357. doi: 10.1016/j.neuron.2015.05.036. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Miyake K, Hirasawa T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: a new concept of clinical genetics. Clin Epigenet. 2012;4:1. doi: 10.1186/1868-7083-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laifenfeld D, Drubin DA, Catlett NL, et al. Early patient stratification and predictive biomarkers in drug discovery and development. In: Goryanin II, Goryachev AB, et al., editors. Advances in Systems Biology. New York, NY: Springer; 2012. pp. 645–653. [DOI] [PubMed] [Google Scholar]
- Leucht S, Cipriani A, Spineli L, et al. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. The Lancet. 2013;382:951–962. doi: 10.1016/S0140-6736(13)60733-3. [DOI] [PubMed] [Google Scholar]
- Li J, Jin H, Wang X. Epigenetic biomarkers: potential applications in gastrointestinal cancers. ISRN Gastroenterology. 2014;2014:e464015. doi: 10.1155/2014/464015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Meister M, Zhang S, et al. Identification of lipid biomarker from serum in patients with chronic obstructive pulmonary disease. Respir Res. 2020;21:242. doi: 10.1186/s12931-020-01507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke WJ, Guanzon D, Ma C, et al. DNA methylation cancer biomarkers: translation to the clinic. Front Genet. 2019;10:1150. doi: 10.3389/fgene.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Yang Z, Wang L, et al. Metabolite biomarkers of type 2 diabetes mellitus and pre-diabetes: a systematic review and meta-analysis. BMC Endocr Disord. 2020;20:174. doi: 10.1186/s12902-020-00653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiera MM, Singh P, Gadadhar S, Janke C. Tubulin posttranslational modifications and emerging links to human disease. Cell. 2018;173:1323–1327. doi: 10.1016/j.cell.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Manolis E, Koch A, Deforce D, Vamvakas S. The European Medicines Agency Experience with Biomarker Qualification. In: Vlahou A, Makridakis M, editors. Clinical Proteomics: Methods and Protocols. New York, NY: Springer; 2015. pp. 255–272. [DOI] [PubMed] [Google Scholar]
- Manor O, Dai CL, Kornilov SA, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206. doi: 10.1038/s41467-020-18871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Vanattou-Saifoudine N, et al. Chapter 4 - Proteomic Technologies for Biomarker Studies in Psychiatry: Advances and needs. In: Guest PC, Bahn S, et al., editors. International Review of Neurobiology. Academic Press; 2011. pp. 65–94. [DOI] [PubMed] [Google Scholar]
- Mayeux R. Biomarkers: Potential Uses and Limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski JP, Hoffmann SC, Cush SS, et al. The foundation for the National Institutes of Health Biomarkers Consortium: past accomplishments and new strategic direction. Clin Pharmacol Ther. 2019;105:829–843. doi: 10.1002/cpt.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C, Fauman E, Erte I, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Tost H. Neural mechanisms of social risk for psychiatric disorders. Nat Neurosci. 2012;15:663–668. doi: 10.1038/nn.3083. [DOI] [PubMed] [Google Scholar]
- Miller IJ, Peters SR, Overmyer KA, et al (2019) Real-time health monitoring through urine metabolomics. npj Digit Med 2:1–9. 10.1038/s41746-019-0185-y [DOI] [PMC free article] [PubMed]
- Mnatsakanyan R, Shema G, Basik M, et al. Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry. Expert Rev Proteomics. 2018;15:515–535. doi: 10.1080/14789450.2018.1483340. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer M, Weinberger KM, Handler M, et al. Profiling the human response to physical exercise: a computational strategy for the identification and kinetic analysis of metabolic biomarkers. J Clin Bioinformatics. 2011;1:34. doi: 10.1186/2043-9113-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Kacimi SEO, Nguyen TL, et al. MiR-21 in the cancers of the digestive system and its potential role as a diagnostic, predictive, and therapeutic biomarker. Biology. 2021;10:417. doi: 10.3390/biology10050417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli G, Ciccacci C, Borgiani P, et al. Genetic tests and genomic biomarkers: regulation, qualification and validation. Clin Cases Miner Bone Metab. 2008;5:149–154. [PMC free article] [PubMed] [Google Scholar]
- Organization WH, Safety IP on C (2001) Biomarkers in risk assessment : validity and validation. World Health Organization
- Pantel K, Hille C, Scher HI. Circulating tumor cells in prostate cancer: from discovery to clinical utility. Clin Chem. 2019;65:87–99. doi: 10.1373/clinchem.2018.287102. [DOI] [PubMed] [Google Scholar]
- Parkes C, Wald NJ, Murphy P, et al. Prospective observational study to assess value of prostate specific antigen as screening test for prostate cancer. BMJ. 1995;311:1340–1343. doi: 10.1136/bmj.311.7016.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotty DM, Morley MP, Cappola TP. Transcriptomic biomarkers of cardiovascular disease. Prog Cardiovasc Dis. 2012;55:64–69. doi: 10.1016/j.pcad.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnoose EA, Atwal SK, Spoerke JM, et al. Molecular biomarker analyses using circulating tumor cells. PLoS ONE. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J, Davidov O. The association of dropout and outcome in trials of antipsychotic medication and its implications for dealing with missing data. Schizophr Bull. 2008;34:286–291. doi: 10.1093/schbul/sbm161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J, Levine SZ, Barkai O, Davidov O. Dropout rates in randomized clinical trials of antipsychotics: a meta-analysis comparing first- and second-generation drugs and an examination of the role of trial design features. Schizophr Bull. 2009;35:775–788. doi: 10.1093/schbul/sbn005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raemdonck GAAV, Tjalma WAA, Coen EP, et al. Identification of protein biomarkers for cervical cancer using human cervicovaginal fluid. PLoS ONE. 2014;9:e106488. doi: 10.1371/journal.pone.0106488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazi S, Zahiri J (2021) Post-translational modifications in proteins: resources, tools and prediction methods. Database 2021. 10.1093/database/baab012 [DOI] [PMC free article] [PubMed]
- Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Brown NJ, Vaughan DE, et al. Established and Emerging Plasma Biomarkers in the Prediction of First Atherothrombotic Events. Circulation. 2004;109:IV–6. doi: 10.1161/01.CIR.0000133444.17867.56. [DOI] [PubMed] [Google Scholar]
- Ryo M, Nakamura T, Kihara S, et al. Adiponectin as a biomarker of the metabolic syndrome. Circ J. 2004;68:975–981. doi: 10.1253/circj.68.975. [DOI] [PubMed] [Google Scholar]
- Santos AL, Lindner AB (2017) Protein posttranslational modifications: roles in aging and age-related disease. Oxidative Medicine and Cellular Longevity 2017:e5716409. 10.1155/2017/5716409 [DOI] [PMC free article] [PubMed]
- Schmid AW, Fauvet B, Moniatte M, Lashuel HA. Alpha-synuclein post-translational modifications as potential biomarkers for Parkinson disease and other synucleinopathies. Mol Cell Proteomics. 2013;12:3543–3558. doi: 10.1074/mcp.R113.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzam S, Coleman JRI, Caspi A, et al. A polygenic p factor for major psychiatric disorders. Transl Psychiatry. 2018;8:205. doi: 10.1038/s41398-018-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugart LR. DNA damage as a biomarker of exposure. Ecotoxicology. 2000;9:329–340. doi: 10.1023/A:1026513009527. [DOI] [Google Scholar]
- Shuwen H, Xi Y, Qing Z, et al. Predicting biomarkers from classifier for liver metastasis of colorectal adenocarcinomas using machine learning models. Cancer Med. 2020;9:6667–6678. doi: 10.1002/cam4.3289. [DOI] [Google Scholar]
- Singh A, Shannon CP, Gautier B, et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit S, van Breemen MJ, Hoefsloot HCJ, et al. Assessing the statistical validity of proteomics based biomarkers. Anal Chim Acta. 2007;592:210–217. doi: 10.1016/j.aca.2007.04.043. [DOI] [PubMed] [Google Scholar]
- Soler-Botija C, Gálvez-Montón C, Bayés-Genís A (2019) Epigenetic biomarkers in cardiovascular diseases. Front Genet 0: 10.3389/fgene.2019.00950 [DOI] [PMC free article] [PubMed]
- Song Y, Wang J, Cheng Z, et al. Quantitative global proteome and lysine succinylome analyses provide insights into metabolic regulation and lymph node metastasis in gastric cancer. Sci Rep. 2017;7:42053. doi: 10.1038/srep42053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Wang H, Yin X, et al. Application of NMR metabolomics to search for human disease biomarkers in blood. Clin Chem Lab Med. 2019;57:417–441. doi: 10.1515/cclm-2018-0380. [DOI] [PubMed] [Google Scholar]
- Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szász AM, Lánczky A, Nagy Á, et al (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7:49322–49333. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed]
- Tabák AG, Jokela M, Akbaraly TN, et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam ZY, Ng SP, Tan LQ, et al. Metabolite profiling in identifying metabolic biomarkers in older people with late-onset type 2 diabetes mellitus. Sci Rep. 2017;7:4392. doi: 10.1038/s41598-017-01735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin EP, Zeiler FA, Ercole A, et al (2017) Serial sampling of serum protein biomarkers for monitoring human traumatic brain injury dynamics: a systematic review. Front Neurol 0: 10.3389/fneur.2017.00300 [DOI] [PMC free article] [PubMed]
- Theodorescu D, Wittke S, Ross MM, et al. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Howard AG, Durazo-Arvizu R, et al. Analytical and biological variability in biomarker measurement in the Hispanic Community Health Study/Study of Latinos. Clin Chim Acta. 2016;463:129–137. doi: 10.1016/j.cca.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Zhao Y, Zheng J, et al. Circular RNA: a potential diagnostic, prognostic, and therapeutic biomarker for human triple-negative breast cancer. Molecular Therapy - Nucleic Acids. 2021;26:63–80. doi: 10.1016/j.omtn.2021.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstikov V, Moser AJ, Sarangarajan R, et al. Current status of metabolomic biomarker discovery: impact of study design and demographic characteristics. Metabolites. 2020;10:224. doi: 10.3390/metabo10060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossberg JT, Crooke PS, Henderson MA, et al. Using biomarkers to predict progression from clinically isolated syndrome to multiple sclerosis. J Clin Bioinform. 2013;3:18. doi: 10.1186/2043-9113-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Schiffmann H, Hube-Magg C, et al. Random forest-based modelling to detect biomarkers for prostate cancer progression. Clin Epigenetics. 2019;11:148. doi: 10.1186/s13148-019-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food & Drug Administration (2021) List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools). FDA
- van Rensburg IC, Loxton AG. Transcriptomics: the key to biomarker discovery during tuberculosis? Biomark Med. 2015;9:483–495. doi: 10.2217/bmm.15.16. [DOI] [PubMed] [Google Scholar]
- Venigalla H, Mekala H, Hassan M, et al (2017) An update on biomarkers in psychiatric disorders-are we aware , do we use in our clinical practice ? https://www.semanticscholar.org/paper/An-Update-on-Biomarkers-in-Psychiatric-we-aware-%2C-Venigalla-Mekala/68318bb2083bba0551ca0cb6c068e17abda99616. Accessed 12 Aug 2021
- Verstockt S, Verstockt B, Vermeire S. Oncostatin M as a new diagnostic, prognostic and therapeutic target in inflammatory bowel disease (IBD) Expert Opin Ther Targets. 2019;23:943–954. doi: 10.1080/14728222.2019.1677608. [DOI] [PubMed] [Google Scholar]
- Wahlbeck K, Tuunainen A, Ahokas A, Leucht S. Dropout rates in randomised antipsychotic drug trials. Psychopharmacology. 2001;155:230–233. doi: 10.1007/s002130100711. [DOI] [PubMed] [Google Scholar]
- Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun Y, Teng S, Li K. Prediction of sepsis mortality using metabolite biomarkers in the blood: a meta-analysis of death-related pathways and prospective validation. BMC Med. 2020;18:83. doi: 10.1186/s12916-020-01546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi X, Li T, Huang Y, et al. RNA biomarkers: frontier of precision medicine for cancer. Noncoding RNA. 2017;3:9. doi: 10.3390/ncrna3010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Wen Z, Wang R, et al. Identification of the lipid biomarkers from plasma in idiopathic pulmonary fibrosis by Lipidomics. BMC Pulm Med. 2017;17:174. doi: 10.1186/s12890-017-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Zhao H, Zeng Y. Lipidomics: a promising cancer biomarker. Clin Transl Med. 2018;7:21. doi: 10.1186/s40169-018-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xia B-R, Jin W-L, Lou G. Circulating tumor cells in precision oncology: clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 2019;19:341. doi: 10.1186/s12935-019-1067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu Z, Xiao TS. Post-translational regulation of inflammasomes. Cell Mol Immunol. 2017;14:65–79. doi: 10.1038/cmi.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Sun H, Yan G, et al. Metabolomics for biomarker discovery: moving to the clinic. Biomed Res Int. 2015;2015:e354671. doi: 10.1155/2015/354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun X-F, Shen B, Zhang H. Potential applications of DNA, RNA and protein biomarkers in diagnosis, therapy and prognosis for colorectal cancer: a study from databases to AI-assisted verification. Cancers. 2019;11:172. doi: 10.3390/cancers11020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Hu Y, Zang T, Wang Y (2020) Identifying protein biomarkers in blood for Alzheimer’s disease. Front Cell Dev Biol. 10.3389/fcell.2020.00472 [DOI] [PMC free article] [PubMed]
- Zhu CS, Pinsky PF, Cramer DW, et al. A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prev Res. 2011;4:375–383. doi: 10.1158/1940-6207.CAPR-10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Koch A, Krockenberger K, Großhennig A. Personalized medicine using DNA biomarkers: a review. Hum Genet. 2012;131:1627–1638. doi: 10.1007/s00439-012-1188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2009) Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. Nature 460:748–752. 10.1038/nature08185 [DOI] [PMC free article] [PubMed]