Abstract
Iron-sulfur clusters are ubiquitous cofactors required for various essential metabolic processes. Conservation of proteins required for their biosynthesis and trafficking allows for simple bacteria to be used as models to aid in exploring these complex pathways in higher organisms. Cyanobacteria are among the most investigated organisms for these processes, as they are unicellular and can survive under photoautotrophic and heterotrophic conditions. Herein, we report the potential role of Synechocystis PCC6803 NifU (now named SyNfu) as the principal scaffold protein required for iron-sulfur cluster biosynthesis in that organism. SyNfu is a well-folded protein with distinct secondary structural elements, as evidenced by circular dichroism and a well-dispersed pattern of 1H-15N HSQC NMR peaks, and readily reconstitutes as a [2Fe-2S] dimeric protein complex. Cluster exchange experiments show that glutathione can extract the cluster from holo-SyNfu, but the transfer is unidirectional. We also confirm the ability of SyNfu to transfer cluster to both human ferredoxin 1 and ferredoxin 2, while also demonstrating the capacity to deliver cluster to both monothiol glutaredoxin 3 and dithiol glutaredoxin 2. This evidence supports the hypothesis that SyNfu indeed serves as the main scaffold protein in Synechocystis, as it has been shown to be the only protein required for viability in the absence of photoautotrophic conditions. Similar to other NFU-type cluster donors and other scaffold and carrier proteins, such as ISCU, SyNfu is shown by DSC to be structurally less stable than regular protein donors, while retaining a relatively well-defined tertiary structure as represented by 1H-15N HSQC NMR experiments.
1. Introduction
Iron-sulfur (Fe-S) clusters are critical cofactors that are required for a myriad of biochemical processes, including electron transport, transcriptional and translation regulation, enzymatic activity, DNA repair, and replication.1, 2 Several distinct cluster species have been found with [2Fe-2S], [3Fe-4S], and [4Fe-4S] forms being most prevalent, and the typical cluster-coordinating environment contains the cysteine thiol-group in addition to other amino acids, such as histidine in the Rieske-type clusters.3, 4 Substitution of amino acid ligands at iron-sulfur cluster binding residues invariably leads to disruption of cluster trafficking and enzymatic function, and natural mutations often lead to lethal disease phenotypes.5–7 Their universal and ubiquitous nature in biochemical systems underscores the importance of Fe-S cluster cofactors, as they are present in all kingdoms of life, and highly conserved pathways exist for their synthesis and delivery to target proteins.8
In higher organisms, pathways involving Fe-S cluster biosynthesis and their concomitant trafficking are much more complex; therefore, cyanobacteria are regarded as excellent model systems for the elucidation of these pathways. This allows for subsequent comparison to more complex systems, as cyanobacteria are also capable of completing oxygenic photosynthesis but have much simpler genomic and cellular structures 9. Furthermore, cyanobacteria, such as Synechocystis PCC6803, have been fully sequenced,10 and databases are readily available to investigate the complex web of protein-protein interactions.11
In particular, Synechocystis contains the ubiquitously conserved gene clusters involved in Fe-S cluster biosynthesis: the suf (sulfur utilization factor) operon, some of the isc (Fe-S cluster) genes, and a single nif (nitrogen fixation) gene.1 These gene clusters are utilized throughout the kingdoms of life for the biosynthesis of Fe-S clusters, and the proteins produced also demonstrate high levels of conservation in both sequence and function.8 Although all three gene clusters are present in Synechocystis, only the gene products from the nif and suf operons are required for survival under photoautotrophic conditions, while only the nif gene is required under heterotrophic conditions.12, 13
In cyanobacteria, the suf pathway for Fe-S cluster biosynthesis has been implicated in the response to oxidative stress and iron starvation, in addition to Fe-S cluster biosynthesis related to photosynthesis.13 The SUF pathway consists of sufABCDE, sufS, and sufR genes.14, 15 SufA has been shown to be involved in remediating oxidative stress, or as an Fe-S cluster carrier protein, and is not required for viability.12, 15 SufE and SufS both act in the liberation of sulfur, as SufS is a cysteine desulfurase and SufE enhances its activity.14–16 The other gene products from the SufBCD operon have been characterized extensively in E. coli, as they have been shown to act as a scaffolding protein, which is able to donate Fe-S clusters to downstream apo proteins.14, 15, 17 Despite this, the role of the SufBCD complex as a scaffold has yet to be investigated at the protein level in Synechocystis.
The only nif gene reported for Synechocystis PCC6803 is ssl2667 and corresponds to the formerly named NifU (SyNfu). The SyNfu protein is composed of 76 amino acids and was initially studied based on its sequence homology with the C-terminal domain of Azotobacter vinelandii NifU.18 Both proteins contain a conserved C-X-X-C motif that is utilized in [2Fe-2S] cluster binding.18, 19 Furthermore, SyNfu was able to transfer its [2Fe-2S] cluster to apo ferredoxin.18 The requirement for SyNfu and its Fe-S cluster binding and transfer capabilities to maintain cellular viability suggest that Nfu could act as an Fe-S cluster scaffold in Synechocystis PCC6803.
SyNfu also demonstrates homology to the NFU family of proteins that are characterized by the same C-X-X-C Fe-S cluster binding motif. However, many of these proteins are more complex than SyNfu, in that they are comprised of additional domains, the functions of which are not completely clear. The similarity between the cluster binding C-terminal domain of human NFU and SyNfu is 46%, and therefore higher than the extent of conservation between SyNfu and the C-terminal domain of A. vinelandii NifU, which is 43% (Fig. 1). This striking degree of similarity and the conservation of cluster binding residues suggest that the Fe-S cluster binding properties may be similar; therefore, we set out to investigate the functional, spectroscopic, and biochemical characteristics of Synechocystis PCC6803 NifU to more completely reveal its role in Fe-S cluster biosynthesis in cyanobacteria and gain additional insight into the function of the NFU domain.
Fig. 1.
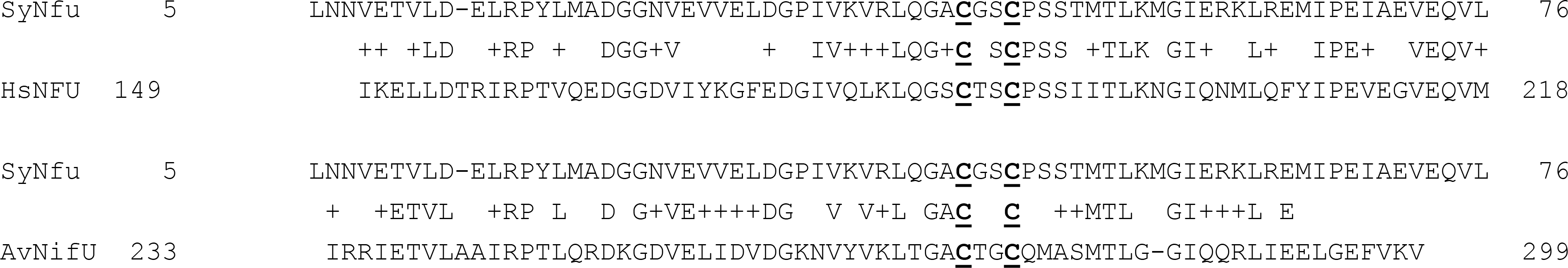
Sequence alignment of Synechocystis PCC6803 NifU and the corresponding C-terminal regions of NifU-like domains of human NFU and A. vinelandii NifU. The show 43% sequence identity with A. vinelandii and 46% sequence identity with human NFU. Sequence similarity is indicated by (+). The conserved C-X-X-C motif shown to ligate [Fe-S] clusters is highlighted by bold underlined red Cys.
2. Material and Methods
2.1. Cloning of SyNfu
Synechocystis PCC6803 gene ssl2667 18, encoding the only NifU-like protein in this cyanobacterium, was synthesized by GenScript. The gene was amplified by PCR to introduce the restriction sites of NdeI and HindIII using the forward primer 5’- CCA ATC ATA TGG AAC TGA CCC T −3’ and the reverse primer 5’- CCA ATA AGC TTT TAC AGC ACT TGT TCC −3’. The PCR product was purified via gel extraction and ligated into the pET28(b+) vector between the above-mentioned restriction sites to generate the expression vector for SyNfu in E. coli BL21 (DE3) cells.
2.2. Expression and Purification of SyNfu
Synechocystis PCC6803 NifU cells were grown overnight in 10 mL of Luria-Bertani (LB) broth media with kanamycin (30 mg/mL) at 37 °C. The overnight cultures were diluted 1:1000 in LB media containing kanamycin until the OD600 reached 0.6. Protein expression was induced with 100 mg/L of isopropyl β-D-1-thiogalactopyranoside (IPTG), and cultures were incubated overnight at 37 °C. Cell pellets were collected by centrifugation at 4,330g for 15 min at 4 °C, and resuspended in 30 mL of 50 mM HEPES, 100 mM NaCl, pH 7.5. Resuspended pellets were incubated with 30 mg lysozyme and 0.6 mg DNase I for 30 min at 4 °C and lysed by use of a dismembranator. Cell lysate was centrifuged at 28,982g for 50 min at 4 °C, and the supernatant was applied to a Ni-NTA column. Protein was eluted with a buffer containing 50 mM HEPES, 100 mM NaCl, 250 mM imidazole, pH 7.5 and concentrated by Amicon ultrafiltration over a 10 kDa membrane. Protein purity was assayed by a 12% SDS-PAGE gel that was visualized with Coomassie Blue staining. Imidazole was removed with dialysis at 4 °C against a buffer containing 50 mM HEPES, 100 mM NaCl, pH 7.5. Purified SyNfu was lyophilized and resuspended in buffer to a known concentration to determine the ε280 = 865.5 M−1 cm−1. The sequence alignment view was generated using Clustal Omega.20
2.3. Protein Expression and Purification of Tm NifS and Additional Cluster Acceptor Proteins
Purification of Thermatoga maritima (Tm NifS) was performed as previously reported.21–23 The expression vector for human ferredoxin-1 (Hs FDX1) was kindly provided by J. Markley and protein was expressed and purified according to literature procedures.24 Purification of human ferredoxin-2 (Hs FDX2) was performed as previously reported.25 The ferredoxins purified as holo proteins and were then subsequently converted to apo forms by treatment with 100 mM EDTA, 5 mM DTT and 8M urea in a buffered solution, pH 7.5.
A construct of human GLRX2 (comprising residues 56–161) with a tobacco etch virus cleavable N-terminal His6 tag in expression vector pNic-Bsa4 was kindly provided by Drs. Kavanagh, Muller-Knapp and Oppermann and protein was expressed and purified as previously reported.26 The yeast Grx3 (Q03835, residues 36–285) open reading frame containing both the Trx-like and Grx-like domains, but lacking the first 35 amino acids (Δ1–35), was purified as previously reported.27
2.4. Thermodynamic Investigation of SyNfu by DSC and VT-CD
Both 0.3 mM and 0.7 mM solutions of apo SyNfu were dialyzed against 50 mM HEPES, 100 mM NaCl, pH 7.5 at 4 °C. The resulting dialysis buffers were used as reference for DSC. Reference buffers and protein samples were thoroughly degassed using a MicroCal Thermovac2 (Malvern Instruments Inc.) prior to analysis on a MicroCal VP-DSC (Malvern Instruments Inc.).28 The data were obtained using a differential mode at a rate of 1.0 °C per min from 10 °C to 100 °C and analyzed using OriginPro software (Origin Labs) and fit to either two-state or non-two-state models. A 0.3 mM solution of apo-SyNfu was buffer exchanged into PBS buffer, pH 7.5. VT-CD experiments were performed on a JASCO J-815 CD spectrometer in a quartz 1 cm cuvette. The wavelength at 222 nm was monitored which is indicative of secondary structure in proteins. The protein was heated at a rate of 1.0 °C per min from 25 °C to 95 °C. Data was analyzed and fit to a two-state model with OriginPro software
2.5. 15N-labeling and 1H-15N HSQC Characterization
Apo SyNfu was uniformly 15N-labeled using conventional techniques. Briefly, cells were grown in M9 minimal media supplemented with 15NH4Cl. Cells were grown, and protein was purified as mentioned above. Apo SyNfu was then buffer-exchanged by a PD-10 desalting column and equilibrated into 40 mM phosphate buffer, pH 6.5, 10% D2O at 298 K. 1H-15N HSQC spectra were acquired on a Bruker 800 MHz spectrometer, equipped with a TXI cryoprobe, and an Avance III HD Console. Standard water suppression and 1H-15N HSQC pulse sequence was performed for collection of data. Apodization, zero-filling and Fourier Transform were performed followed by phase correction. Data was visualized with NMRFAM Sparky version 3.115.29
2.6. Structural Analysis by Circular Dichroism (CD) and Secondary Structure Prediction
Secondary structure prediction was performed using 150 μM apo SyNfu in PBS buffer pH 7.5. Scans from 240–200 nm were performed in a 1 cm cuvette and data processed using JASCO Spectramanager II Analysis software. Secondary structure prediction program K2D330 was then utilized to obtain estimates of % α-helix and β-sheet. All spectra were represented in Origin 7.0.
2.7. Reconstitution of SyNfu
A mixture of approximately 200 μM purified SyNfu, 2 μM Tm NifS, 5 mM DTT, and 2 M urea was alternatively degassed, and argon-purged for 1 h. Degassed solutions of ferric chloride and L-cysteine were added separately and dropwise to the mixture to have final concentrations of 0.6 M for each. The solution was incubated for 45 min with stirring, and an additional 180 μL of 10 mM FeCl3 and L-cysteine were added. After another 45 min of stirring, excess iron and sulfide were removed by separation on a PD-10 column, equilibrated with an argon-purged solution of 50 mM HEPES, 100 mM NaCl, pH 7.5. Reconstituted protein was eluted with 3.5 mL of the equilibration buffer. The protein concentration was determined by absorbance on UV-Vis with ε420 = 10,000 M−1 cm−1, and the success of the reconstitution was determined by absorbance at 330 nm and 415 nm. Protein reconstitution was also completed in the absence of Tm NifS with degassed solutions of ferric chloride and sodium sulfide (final concentrations of 0.6 M).
2.8. EPR Characterization
A 200 μM solution of SyNfu was prepared in 50 mM HEPES buffer. Samples were reduced with 2 mM dithionite and frozen in liquid nitrogen prior to analysis. Samples were analyzed with an X-band EMXPlus EPR with ColdEdge Cryostat. Sample temperature was varied from 10–60 K, microwave frequency of 9.4 GHz and a microwave power of 30 dB. Data were analyzed with OriginPro software and g-values extracted using EasySpin software.
2.9. Oligomeric State Determination
Apo and holo SyNfu were analyzed for the oligomerization state via dynamic light scattering. Samples at 2 mg/mL in 50 mM HEPES, 100 mM NaCl, pH 7.5 were degassed and filtered through a 0.2 μm filter and added to a Hëllma quartz cuvette in a Malvern Zetasizer Nano Zen3600 Dynamic Light Scattering (DLS) instrument. Samples met the criteria of a poly dispersity index (PDI) of less than 0.2 and derived count rate of greater than 2000. Data were converted to natural logarithm to determine the aggregation state.
2.10. [Fe2S2](GS)4] Synthesis
The cluster used was synthesized as previously reported.31 Briefly, ferric chloride (20 mM) and sodium sulfide (20 mM) was added to 10 mL 40 mM glutathione solution, pH 8.6. A volume (40 mL) of ethanol was added to the mixture and mixed by vortexing. The precipitate was collected by centrifugation at 13,000 rpm for 10 min, washed twice with ethanol and dried under vacuum.
2.11. Fe-S Cluster Uptake Monitored by CD
The ability of SyNfu to take up an Fe-S cluster from [2Fe-2S](GS)4 was examined by circular dichroism (CD). CD scans were recorded on a JASCO J-815 CD spectrometer in a quartz 1 cm anaerobic cuvette from 600 – 300 nm at a scan rate of 200 nm/min at 25°C with a 2 min interval between each accumulation. SyNfu (50 μM) in 50 mM HEPES, 100 mM NaCl, pH 7.5, was thoroughly degassed in the presence of 5 mM DTT and transferred to the anaerobic cuvette. Solid [2Fe-2S](GS)4 was resuspended in degassed 50 mM HEPES, 100 mM NaCl pH 7.5 and added to the argon-purged anaerobic cuvette via a gas-tight syringe to a final concentration of 100 μM to initiate the reaction. Spectra were processed using JASCO Spectramanager II Analysis software and were represented in Origin 7.0.
2.12. Kinetics of Glutathione Extraction of the Fe-S Cluster in Holo SyNfu
Glutathione has previously been shown to extract the [2Fe-2S] cluster from holo proteins to form the [2Fe-2S](GS)4 complex by monitoring the change in the charge transfer bands at 330 nm and 415 nm.31 Holo SyNfu demonstrated a larger change in absorbance at 330 nm than at 415 nm, so transfer of the cluster from SyNfu to glutathione was monitored at that wavelength. Degassed, reconstituted holo SyNfu in 50 mM HEPES, 100 mM NaCl, pH 7.5 was incubated with 1 mM GSH in an anaerobic cuvette and the absorbance at 420 nm on a Cary WinUV spectrophotometer was monitored every 2 min over the course of 1 h. The change in absorbance at 330 nm was plotted against time and fit to an exponential decay to obtain the kobs. A control reaction of holo SyNfu in the absence of excess GSH was carried out under the same conditions.
2.13. Cluster Transfer Reactivity
Kinetic studies of cluster transfer experiments were performed based on the experimental design of Johnson and coworkers.32,33 Reactions were performed on a JASCO J-815 CD spectrophotometer in a quartz 1 cm anaerobic cuvette from 600 – 300 nm at a scan rate of 200 nm/min at 25°C with a 2 min interval between each accumulation. Reactions that reached completion within the first 10 min were analyzed over a 10 nm wavelength scale based on the peak of interest, with 10 sec intervals between accumulations. Spectra were processed using JASCO Spectramanager II Analysis software and were represented in Origin 7.0.
Reactions in 50 mM HEPES, 100 mM NaCl, pH 7.5 were prepared by degassing a mixture of 40 μM apo protein, and 5 mM DTT, and were then transferred to an anaerobic cuvette via a gas tight syringe. Degassed holo protein at 20 μM was added to the cuvette to initiate the reaction. The concentration of [2Fe-2S] in the reaction for each holo protein was determined via standard iron quantitation methods.34
The kinetic rate constants for cluster transfer were obtained by monitoring and converting the change in CD signal to the percentage of cluster transferred over time. The plot of time versus percent cluster transfer was fit to an exponential to obtain an observed first-order rate constant kobs that was used to obtain the second-order rate constant.
3. Results
3.1. 1H-15N HSQC NMR Characterization of SyNfu
Sequence alignment of SyNfu to the related C-terminal domains of NFU and NifU proteins shows that NifU from A. vinelandii shares 43% similarity, while human NFU is 46% similar. Given this level of sequence similarity it was of interest to examine the overall structural properties of apo SyNfu to determine if it displayed characteristics of a well-folded protein with defined secondary structural elements. To this end, cells containing the SyNfu expression plasmid were grown, and the protein was 15N-labeled for use in 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectroscopy experiments. The chemical shift is not only sensitive to pH, temperature, and salt, but also to the chemical environment; therefore, a simple 1H-15N HSQC experiment was conducted to yield insight into how well the protein is folded and structurally defined in solution.
The Biological Magnetic Resonance Bank (BMRB) contains a vast database of experimental data concerning chemical shifts and can facilitate comparisons for this purpose.35 If a protein is unfolded or denatured, most of the observable resonances fall within a narrow 1H chemical shift window centered between 8.1–8.4 ppm, while for a well-folded protein with distinct secondary structural features, the observed chemical shifts are resolved and well-dispersed.36–38 The experimental HSQC spectrum (Fig. 2) shows that the expressed apo protein is folded and contains defined secondary structural features, given the chemical shifts observed to span from 6.5 – 9.5 ppm.35 A comparison of these results to those obtained previously for the N- and C-terminal domains of human NFU shows similarity in the degree of peak dispersion,39 and also display resonances in the ranges characteristic of α-helices and β-sheets, which have been noted in the solved C-terminal NMR structure for human NFU.40
Fig. 2.
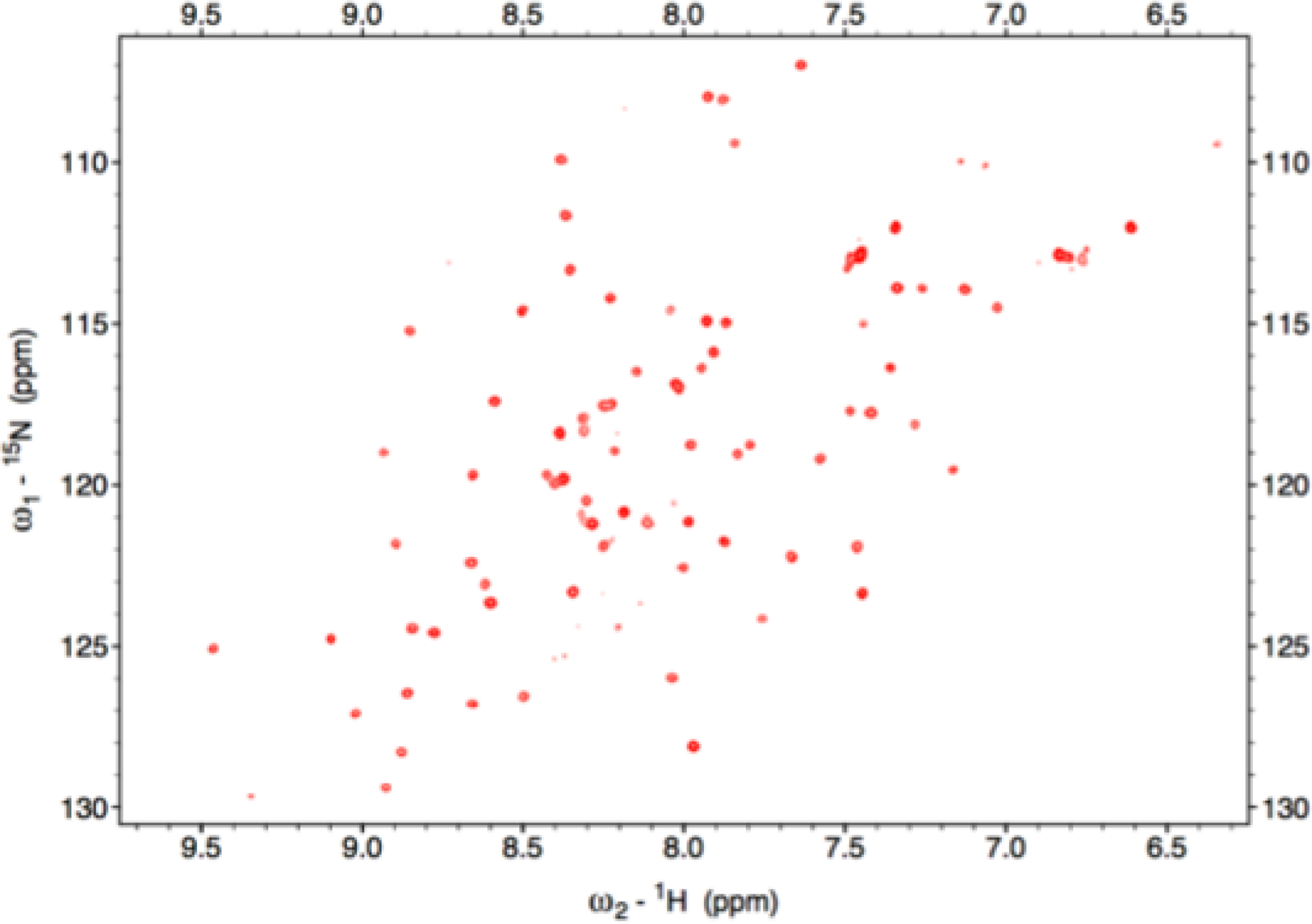
1H-15N HSQC of 15N-labeled SyNfu taken at 800 MHz in 40 mM sodium phosphate buffer, 100 mM NaCl, pH, 7.4 at 298 K.
3.2. Secondary Structure Estimation via Circular Dichroism
The high degree of signal dispersion in the 1H-15N HSQC spectrum (Fig. 2) suggests that the protein contains distinct secondary structural elements, consistent with the C-terminal structure of human NFU, which shows the presence of α-helices and β-sheets,40 and so we further investigated the structural characteristics of SyNfu. Circular dichroism (CD) is an efficient technique for predicting secondary structure, using the chirality of the backbone in the far UV region. Coupled with experimental data and known protein structures from the PDB, algorithms such as K2D3 can yield insight into the secondary structural motifs of proteins with unknown structures.30 To that end, apo SyNfu was buffer exchanged into PBS pH 7.5 to remove the background observed from more traditional buffer components, such as HEPES, and scanned from 240–200 nm. From the K2D3 algorithm, SyNfu is estimated to contain similar amounts of α-helical and β-sheet character with 28.7 and 24.5 percent respectively (Table 1 and Fig. 4a). The significant presence of large amounts of secondary structure, as evidenced by CD and well-dispersed peaks in the 1H-15N HSQC NMR spectrum, shows that despite its small size, SyNfu is well-structured in the apo state.
Table 1.
Percentages of Secondary Structural Features from the K2D3 Algorithm.
| Secondary Structural Element | % Secondary Structure |
|---|---|
| α-helix | 28.7 |
| β-sheet | 24.5 |
Fig. 4.
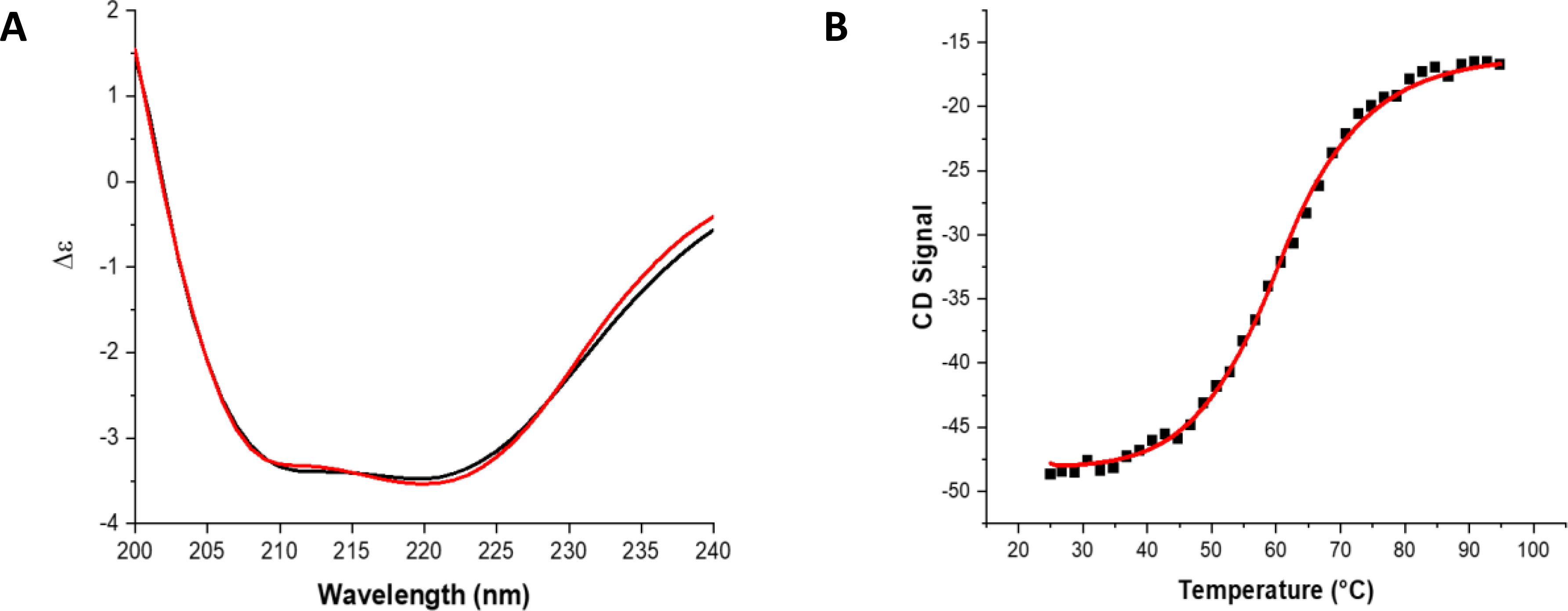
CD spectra and VT-CD melt of apo-SyNfu. A) Overlay of 150 uM Sy-NifU CD spectra (black) and K2D3 prediction (red), B) VT-CD melt of 300 μM apo-SyNfu monitored at 222 nm in PBS buffer, pH 7.5.
3.3. Stability and Thermodynamics of Apo SyNfu
SyNfu consists of a single Nfu domain that has high homology to the C-terminal domain of the human NFU protein. Previous thermodynamic studies on the isolated C-terminal NFU domain have demonstrated that the protein exhibits molten globule character, as shown by both differential scanning calorimetry (DSC) and NMR experiments.28 Given the high level of evolutionary conservation, we decided to further examine the stability and thermodynamic properties of apo SyNfu. DSC thermal melt experiments were performed with apo SyNfu at two concentrations. At 0.3 mM two unfolding events occurred, one at 45.5 °C and another at 66.6 °C, while at 0.7 mM a much broader melting curve was observed (Fig. 3 and Table 2). The thermodynamic properties were also confirmed via VT-CD at 0.3 mM, which obtained a similar melting temperature and enthalpy of unfolding (Fig. 4 and Table 2). At both concentrations, the melting transitions are broad suggesting a molten globule conformation; however, at higher concentrations, the first transition is missing, consistent with protein aggregation. Interestingly, the unfolding event is similar to that observed for the C-terminal construct of human NFU and is supportive of a conformationally dynamic domain.39, 41
Fig. 3.
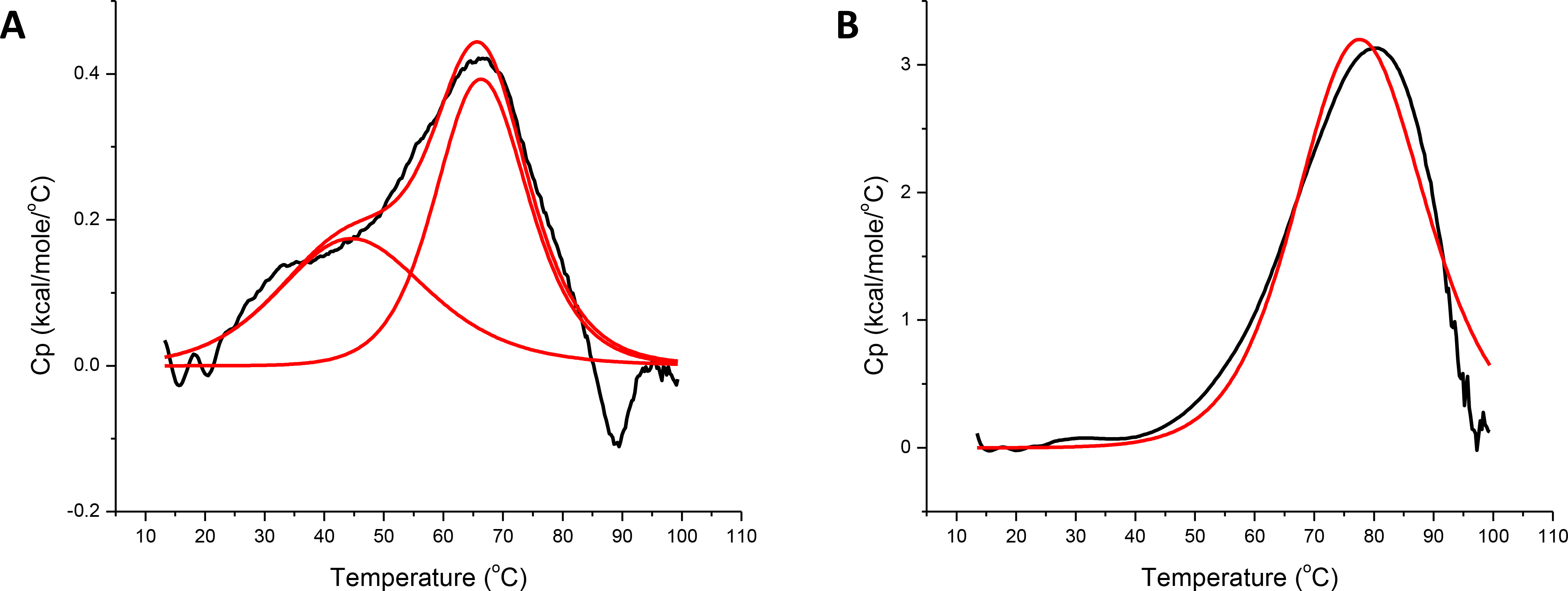
DSC profiles for A, 0.3 mM SyNfu and B, 0.7 mM SyNfu in 50 mM HEPES, 100 mM NaCl, pH 7.5. Origin was used to fit the data to a one-peak non two-state model. Curves shown in A represent both the individual and merged heats of unfolding.
Table 2.
DSC Melting Transitions and Enthalpies of Melting
| [Sy-NifU] (Technique) | Tm1 (°C) | Tm2 (°C) | ΔH1 (kcal/mol) | ΔH2 (kcal/mol) |
|---|---|---|---|---|
| 0.3 mM (DSC) | 66.6 ± 0.3 | 45.5 | 8.2 ± 0.6 | 5.4 ± 0.7 |
| 0.7 mM (DSC) | 78.2 ± 1.0 | N/A | 93.2 ± 1.0 | N/A |
| 0.3 mM (VT-CD) | 60.6 ± 0.2 | N/A | 9.3 ± 0.2 | N/A |
3.4. Spectroscopic Characterization of the [2Fe-2S] Cluster of SyNfu
Previous reports have indicated that SyNfu purifies primarily in the holo state with a bound [2Fe-2S] cluster;18 however, under our purification conditions SyNfu is purified entirely in the apo form. This may be due to the placement of the His-tag as our construct utilizes an N-terminal His-tag, while the previous authors utilized a C-terminal His-tag. Nevertheless, the functional holo protein, with bound cluster, SyNfu was successfully reconstituted, both chemically and enzymatically, with sodium sulfide and Tm NifS/L-cysteine, respectively (Fig. 5). Both reconstitution methods produced the desired holo SyNfu that yielded Fe-S cluster bound UV spectra with characteristic peaks at 330 nm and 415 nm (Fig. 5A), matching previously reported spectra for this protein.18 The holo protein also provided a CD signal, yielding the holo spectrum shown in (Fig. 5B) that resembles that obtained for holo human NFU.34 Reconstitution yields were in the range of 40% as determined from UV-Vis measurements and overall protein measurements. These yields are in the range for the reconstitution of other NFU systems that have been documented previously.
Fig. 5.
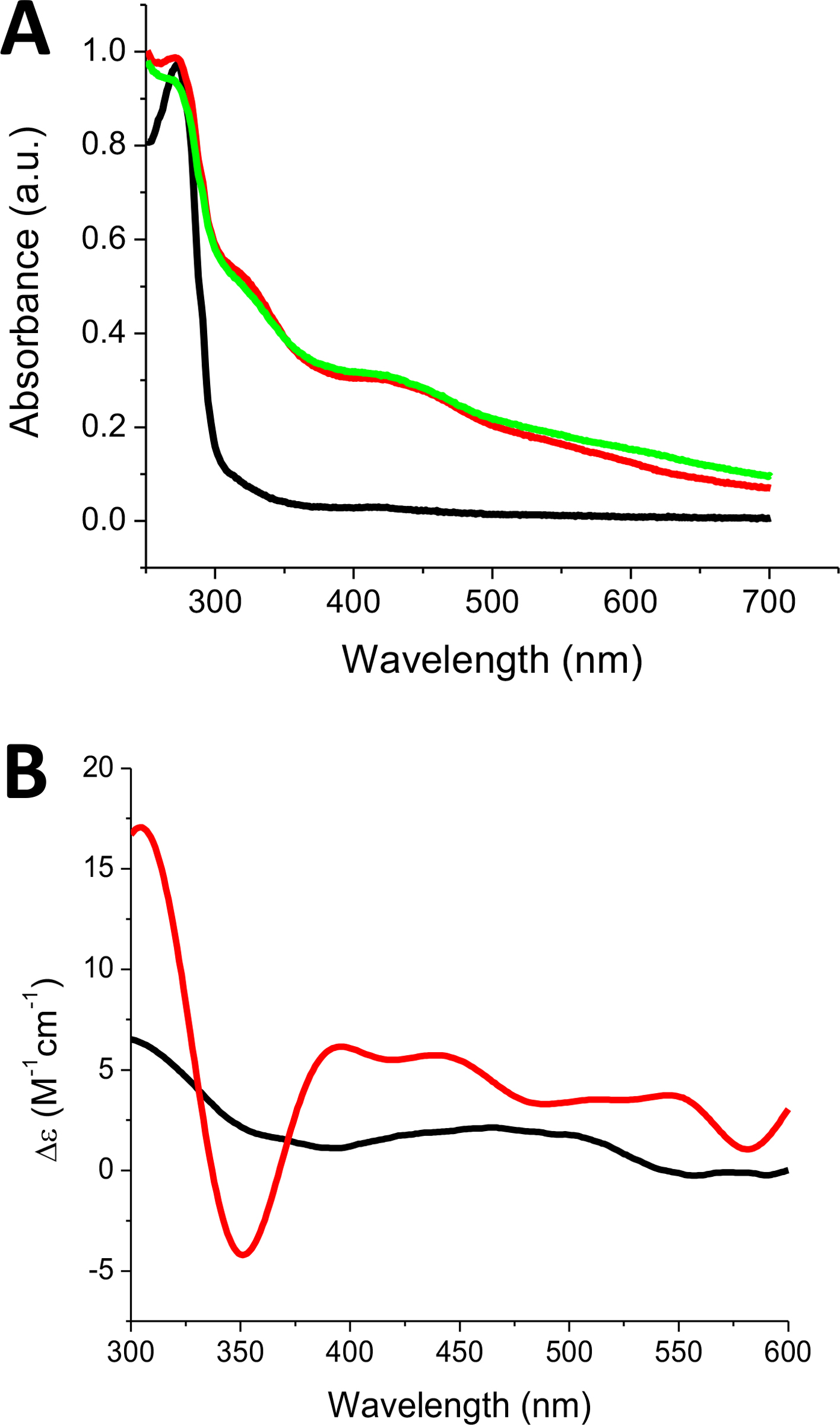
Reconstituted holo SyNfu spectra recorded on (A) UV-Vis and (B) CD. Apo SyNfu is shown in (black). For (A), Holo SyNfu reconstituted with Na2S (green) and holo SyNfu reconstituted with Tm NifS and L-Cys (red). For (B), a representative holo spectrum is shown in (red) and apo spectrum shown in (black).
3.6. EPR Characterization of [2Fe-2S] SyNfu
NifU and NFU proteins can vary in cluster nuclearity, depending on the organism; for example, E. coli NfuA is reported to contain a [4Fe-4S] cluster cofactor, while the C-terminal region of A. vinelandii contains a [2Fe-2S] cluster and human NFU appears to primarily bind a [2Fe-2S] cluster.19, 34, 42, 43 Previous reports of SyNfu have shown that the protein binds one [2Fe-2S] cluster, which was thought to bridge two SyNfu monomers to form a dimer,18 and was able to deliver that cluster and produce an active form of Synechocystis PCC6803 Fdx. To confirm the presence of a [2Fe-2S] cluster bound to SyNfu, X-band EPR spectroscopy was performed. Dithionite-reduced holo-SyNfu showed EPR spectra shown below (Fig. 6a), with g-values similar to those observed for the human holo-BOLA3 homodimer of 2.01 and 2.00. 44 Temperature dependent measurements, showed an inverse linear relationship between temperature and integrated peak area (Fig. 6b), which is indicative of the relaxation properties of a 2Fe-2S cluster. If a 4Fe-4S cluster is expected, a more exponential decay in the integrated peak area would be expected as the temperature is increased. These results further confirm previous reports that SyNfu binds a [2Fe-2S] cluster in its holo form.
Fig. 6.
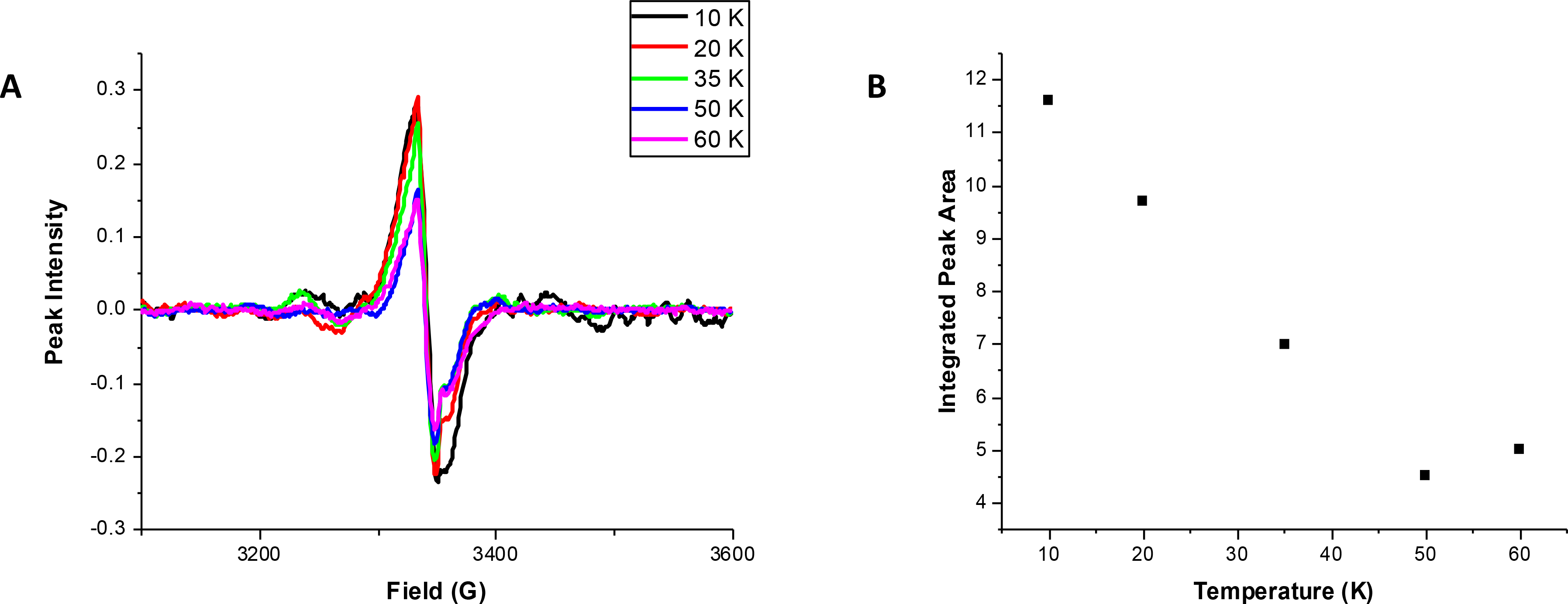
X-band EPR spectra of holo SyNfu. A) Temperature dependence spectra of holo SyNfu reduced with 2 mM dithionite. B) Plot of integrated peak area as a function of temperature.
3.7. Oligomeric State Determination
A previous report on SyNfu speculated that the holo form is a dimer, likely bridged by a [2Fe-2S] cluster, as evidenced by non-denaturing PAGE and iron-staining.18 We were therefore interested in examining both the apo and holo protein with dynamic light scattering (DLS) to confirm the oligomeric states of the apo and holo SyNfu. Aliquots of apo SyNfu, a reconstitution mixture after 1 h, and a reconstitution mixture after 2 h were analyzed. Analysis of the elution profile for apo SyNfu showed that the protein was found mainly in a monomeric form, while the reconstitution mixture after 1 h also showed a similar trace with a small peak forming at approximately twice the size (Fig. 7). After the full reconstitution time of 2 h, a peak corresponding to a complex twice the size of that found for the apo protein was observed, indicating that the holo-form of SyNfu is dimeric.18
Fig. 7.
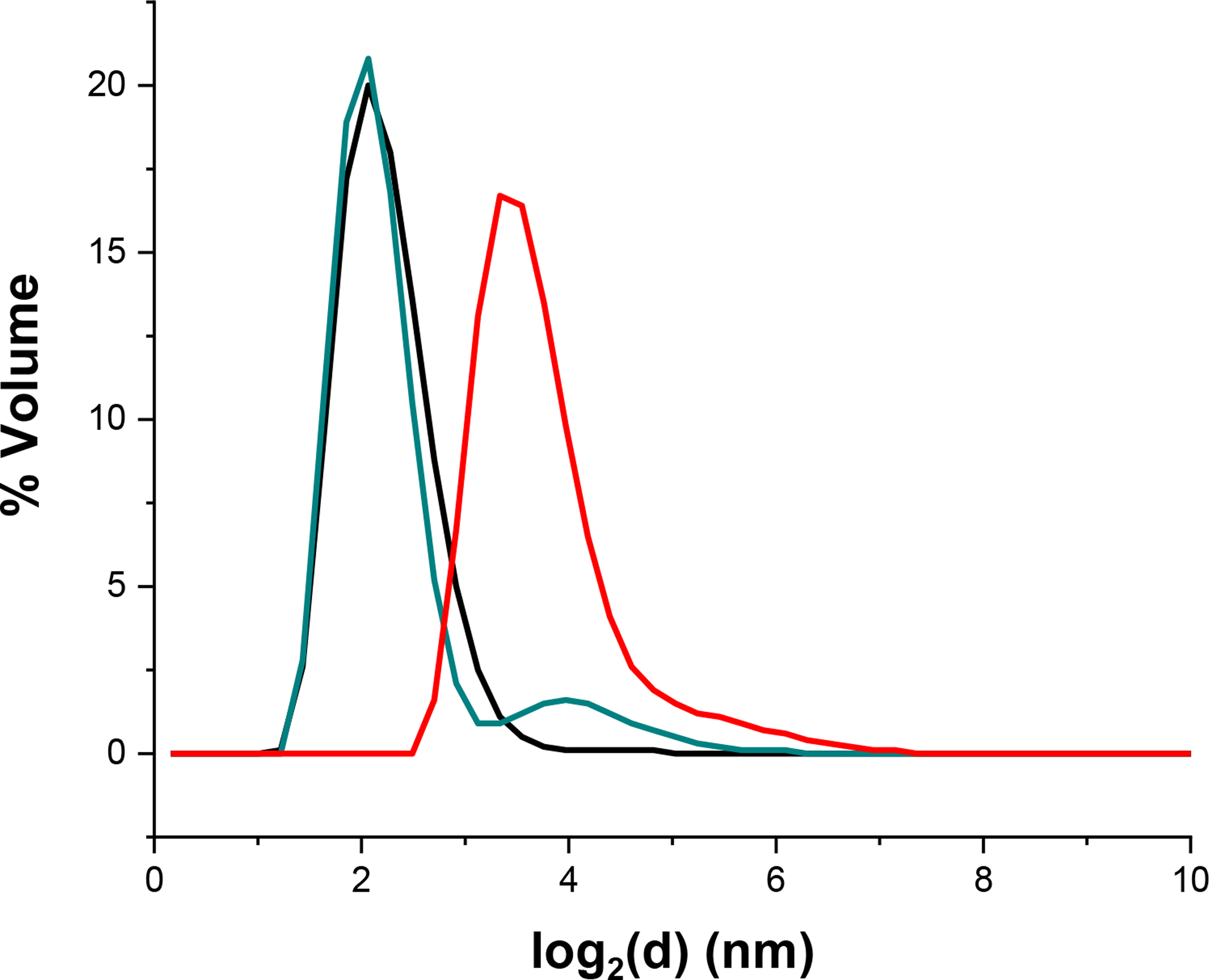
Dynamic light scattering of apo (black), reconstitution after 1 h (cyan), and reconstitution after 2 h (red) of SyNfu.
3.8. Transfer of the [2Fe-2S] Cluster from Holo SyNfu
Our laboratory has previously reported a hydrolytically stable glutathione-complexed Fe-S cluster that is likely of physiological relevance and has been shown to reconstitute apo Fe-S proteins, such as the Fe-S cluster scaffold ISU,31 and serves as a substrate for the mitochondrial ABCB7 exporter.45 Furthermore, we have previously demonstrated that the [2Fe-2S](GS)4 cluster can be formed by the addition of excess GSH to holo proteins, as the cluster is extracted from the protein by the glutathione.
Given its role in organism viability, SyNfu is believed to function as the primary scaffold in Synechocystis, and so we sought to examine the exchangeability of cluster between the apo protein and glutathione ligands, as well as the ability of the holo protein to transfer the bound cluster to apo target proteins. We found that SyNfu cannot be reconstituted by the [2Fe-2S](GS)4 cluster (data not shown). However, GSH is able to extract the cluster from the holo protein at a rate of 158 ± 5.2 M−1min−1 as monitored by the decrease in the absorbance at 330 nm on UV-Vis, resulting from the lower extinction coefficient of the [2Fe-2S](GS)4 complex31 (Fig. 8A). The rate of GSH-induced cluster extraction is linearly dependent on the concentration of GSH, consistent with a mechanism requiring pre-oligomerization of glutathione prior to cluster extraction,76 and/or rate-limiting substitution of the protein Cys – Fe3+ bonds. There is no significant loss of cluster in the absence of GSH, demonstrating that the holo protein is relatively stable and that the observed decrease in absorbance does not derive from cluster degradation (Fig. 8B). Interestingly, the role of glutathione in Synechocystis PCC6803 appears to be more important in the oxidative stress response, while the glutathione precursor gamma-glutamylcysteine appears to be sufficient under optimal growth conditions.46, 47 This suggests that in Synechocystis the role of glutathione may be less important in the trafficking of iron-sulfur clusters given that another small molecule thiol can help mediate cellular functions in this cyanobacterium.
Fig. 8.
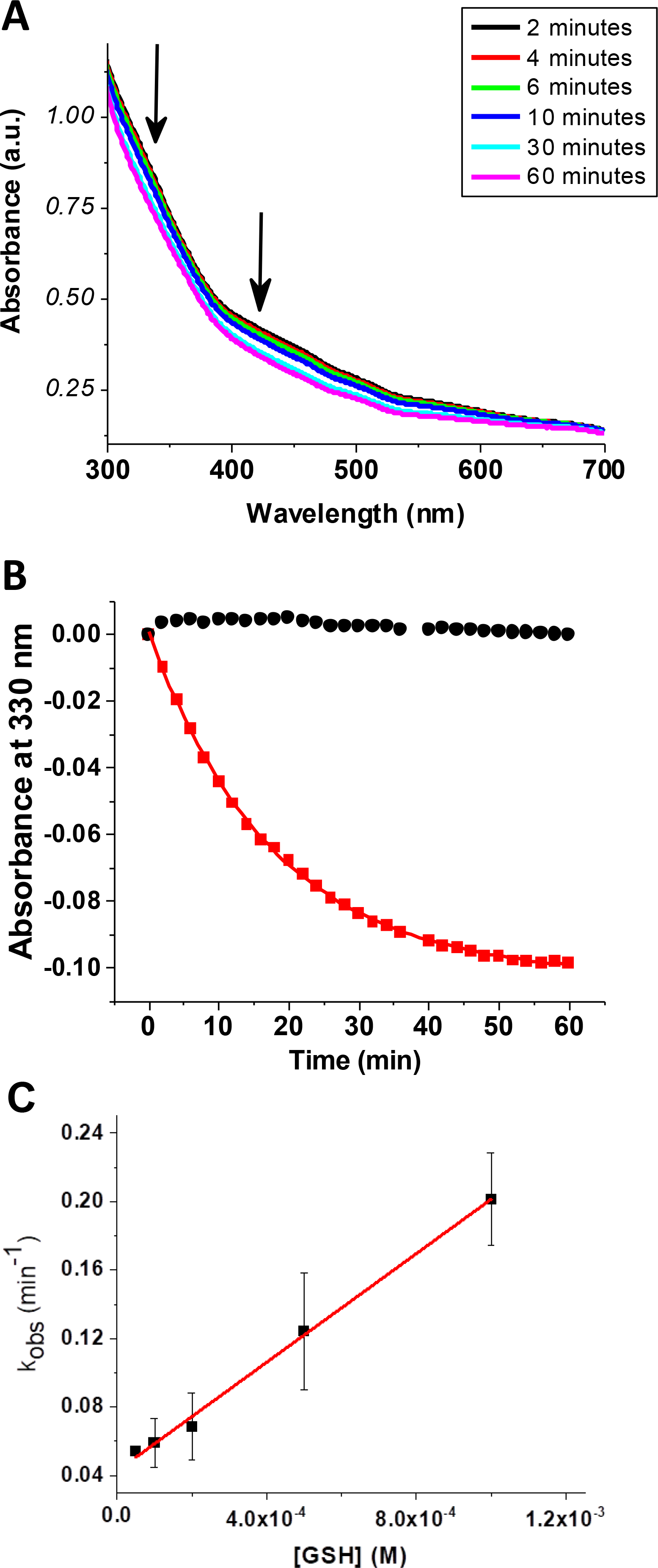
Extraction of the [2Fe-2S] cluster from reconstituted holo SyNfu to form the [2Fe-2S](GS)4 complex monitored by UV-Vis. a) Time course measurement showing a decrease in the characteristic Fe-S cluster bands at 420 and 330 nm. b) Change in absorbance for SyNfu in the absence (black) and presence (red) of 1 mM GSH. c) Linear dependence of observed rate constant with GSH concentration.
Previous work on SyNfu has shown the ability of the holo protein to transfer the [2Fe-2S] cluster to Synechocystis Fdx and to a yeast mitochondrial Fdx18. We were further able to demonstrate transfer from SyNfu to human FDXs 1 and 2 by use of circular dichroism (CD), since each Fe-S cluster bound protein generates a unique CD spectrum particular to their cluster binding environment in the range of 300 – 600 nm; these transfer reactions also reflect the high conservation of these Fe-S cluster proteins across different organisms.48 Following addition of holo SyNfu to an apo FDX1 or FDX2, a peak centered around 440 nm was observed to increase with time as the cluster was transferred (Fig. 9). Based on the initial concentration of the [2Fe-2S] cluster, a second-order rate constant was determined for each reaction.32, 33 For transfer to FDX1 from holo SyNfu, a rate constant of 2015 ± 416 M−1min−1 was calculated, and for the transfer to FDX 2, it was determined as 5642 ± 84 M−1min−1 (Table 3).
Fig. 9.
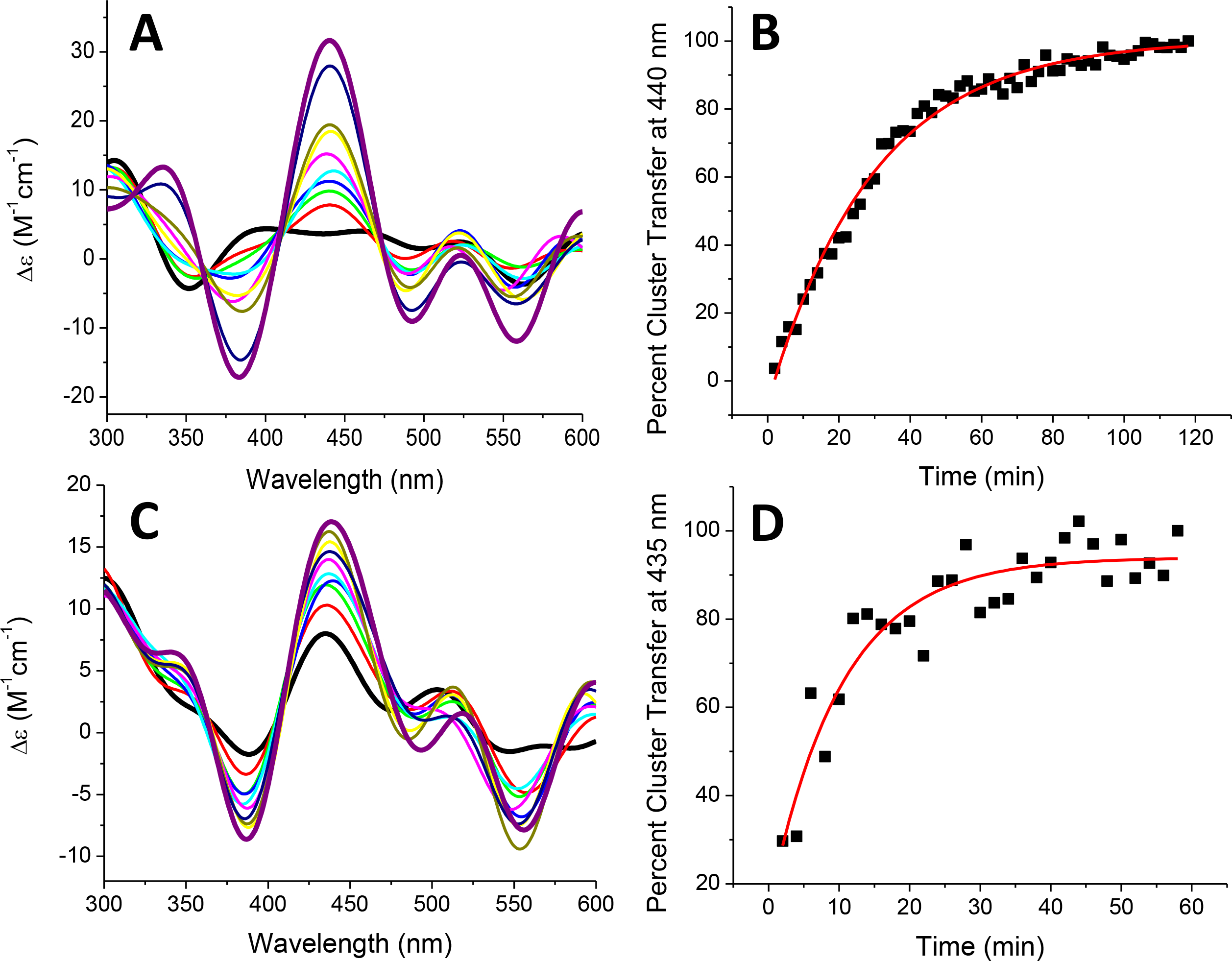
Time-dependent [2Fe-2S] cluster transfer from holo SyNfu to apo human FDX1 (A) and apo human FDX2 (C) recorded by CD in 50 mM HEPES, 100 mM NaCl, pH 7.5 at a 1:1 concentration ratio. Spectra were recorded every 2 min after the addition of holo SyNfu and converted to percent cluster transfer, (B) or (D), to calculate the second-order rate constant based on the concentration of the [2Fe-2S] cluster. The rate constants for cluster transfer to apo FDX1 and FDX2 were calculated to be 2015 M−1min−1 and 5642 M−1min−1, respectively. For both (A) and (C), the starting time (tinitial) is denoted by the (black) trace, and the tfinal is denoted by the (purple) trace.
Table 3.
Second-order rates for the transfer of the [2Fe-2S] cluster from holo SyNfu to various apo target proteins and to GSH for formation of the [2Fe-2S](GS)4.
| cluster transfer reaction | rate constant (M−1min−1) |
|---|---|
| SyNfu to human FDX1 | 2015 ± 416 |
| SyNfu to human FDX2 | 5642 ± 84 |
| SyNfu to human GLRX2 | 1220 ± 320 |
| SyNfu to S. cerevisiae Grx3 | 21821 ± 4736 |
| SyNfu to [2Fe-2S](GS)4 | 158 ± 5 |
Since no other cluster transfers from SyNfu PCC6803 havePCC6803 has been demonstrated, we examined the ability of SyNfu to transfer its Fe-S cluster to another potential target family: the glutaredoxins. Glutaredoxins are also highly conserved proteins and exist in either monothiol or dithiol forms, depending on the number of cysteines in their active site.49 Typically, glutaredoxins function in electron transfer, catalyze disulfide reduction, iron sensing and regulation, and Fe-S cluster transfer and delivery.50 In Synechocystis, they have not been fully characterized, but the glutaredoxins are known to be required for stress adaptation 51 and arsenate reduction.52 Transfer to the glutaredoxins was monitored by CD (Fig. 10), as mentioned above, to obtain an observed rate (kobs) constant for the reaction. The kobs and the initial concentration of [2Fe-2S] were used to calculate the second-order rate. Transfer to glutaredoxin 2 (GLRX2) was somewhat slow, requiring 2 h for the CD signal to begin to level off, which resulted in a second-order rate of 1220 ± 320 M−1min−1. For glutaredoxin 3 (Grx3), the CD spectrum looked like glutaredoxin 3 initially, with no trace of the initial holo SyNfu peaks. Hence, we monitored the reaction on the interval of 10 sec, which resulted in a very fast but measurable transfer to yield a second-order rate of 21821 ± 4736 M−1min−1.
Fig. 10.
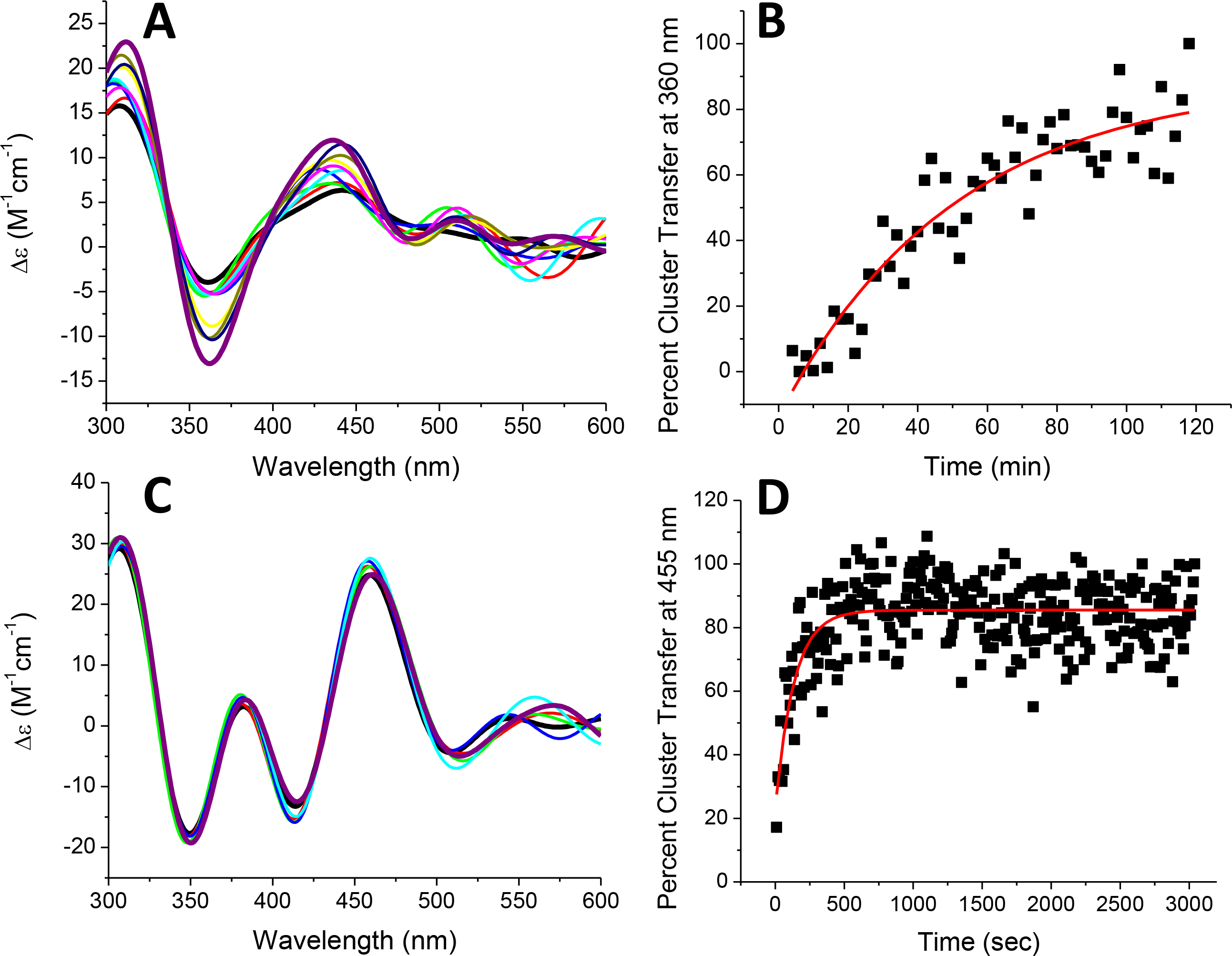
Time-dependent [2Fe-2S] cluster transfer from holo SyNfu to apo human GLRX2 (A) and apo S. cerevisiae Grx3 (C) recorded by CD in 50 mM HEPES, 100 mM NaCl, pH 7.5 at a 1:1 concentration ratio. Spectra were recorded every 2 min (A, B, and C) or 10 sec (D) after the addition of holo SyNfu, and converted to percent cluster transfer, (B) or (D), to calculate the second-order rate constant based on the concentration of the [2Fe-2S] cluster. The rate constants for of cluster transfer to apo GLRX2 and Grx3 were calculated to be 1220 M−1min−1 and 21821 M−1min−1, respectively. For both (A) and (C), the starting time (tinitial) is denoted by the (black) trace, and the tfinal is denoted by the (purple) trace.
4. Discussion
While many Fe-S cluster proteins are highly conserved, some variations in cluster nuclearity and potential function have been observed, depending on the organism under study. This is particularly evident for proteins that contain NifU or NFU domains. As shown in (Fig. 1), the cluster binding motif is highly conserved from cyanobacteria to humans, but these proteins can have additional domains beyond the cluster binding domain, which are thought to confer additional functional specificity and perhaps determine the cluster nuclearity.53 For example, NFU proteins have been shown to bind either [2Fe-2S] or a labile [4Fe-4S] cluster in vitro, depending on the source of the organism, and they are capable of donating to downstream trafficking proteins and other enzymes such as aconitase.34, 40, 42, 43 In contrast, the C-terminal domain of NifU and the NfuA construct in A. vinelandii ligate a [2Fe-2S] and a [4Fe-4S] cluster, respectively.19, 54 Further, NfuA from Synechococcus sp. 7002 has been shown to ligate a [4Fe-4S] cluster, which can donate further to the apo PsaC component in photosystem I, albeit at an elevated pH of 10.0.55 The SyNfu protein from PCC6803 has been previously reported to contain a [2Fe-2S] cluster, which can be subsequently transferred to Fdx1 to create a fully active ferredoxin protein.18, 56 Despite this heterogeneity, the NifU and NFU domains share similar functions in cluster biosynthesis and trafficking and are required for viability, highlighting evolutionary conservation in the role of these Fe-S cluster proteins.
To this extent, cyanobacterial Fe-S cluster proteins serve as models for understanding these pathways, as they are simple unicellular organisms that contain all of the Fe-S cluster genes reported in higher organisms. While the model organism Synechocystis PCC6803 contains all of these genes, only some of the suf genes and the single nif gene are required for viability.12 The typical IscU scaffold found in other organisms is absent in Synechocystis and the IscA gene is not required.12, 15 Further characterization of the suf genes have shown that SufA is dispensable.12, 15 The other suf genes, corresponding to the SufBCD, SufS, and SufR proteins, have been shown to be important for optimal growth under photoautotrophic conditions; however, these same knockdown experiments also showed that they were not as vital under heterotrophic conditions.13 Given that the only other Fe-S protein required for viability corresponds to the product of the nif gene, SyNfu, the preponderance of evidence suggests that this protein may act as the main scaffold in Synechocystis PCC6803.
Accordingly, we were interested in determining the overall structural characteristics of apo SyNfu via 1H-15N HSQC experiments. The as-purified 15N-labeled protein was indeed folded, given the well-resolved and dispersed signals observed (Fig. 2). The protein was shown to have a wide range of resonances from 6.5–9.5 ppm, consistent with secondary structural elements that correspond to α-helices and β-sheet structures 35–37, 57, 58, and they were similar to those shown in the NMR structure of the C-terminal construct of human NFU.40 CD experiments were also performed to estimate the percentage of secondary structural elements via the analysis program K2D3. Results show that the protein contains similar levels of α-helical and β-sheet character (Table 1), showing that this small single domain protein is well structured in the apo state.
Both thermodynamic and 1H-15N HSQC NMR experiments have been performed on the full-length and N- and C-terminal constructs of human NFU,39, 41 and the structures for the N- and C-terminal constructs have also been determined.40 These experiments show that the structural and thermodynamic properties of the specific domain(s) in question vary depending on the particular construct, and specifically whether the isolated domain or complete protein is considered. Apo SyNfu displayed similar behavior to that of the C-terminal construct of human NFU in thermal melt experiments, indicative of a less stable conformation in the apo state, and most likely reflecting conformational isomerism as previously described.59 The transition from a two-state melting curve to a one-state transition suggests aggregation at higher concentrations with stabilization. This would also be consistent with stabilization of the human C-terminal NFU domain following contacts with the N-terminal domain.41 The combination of molten globule-like DSC behavior, but well-defined 1H-15N HSQC spectral signatures is reminiscent not only of the behavior of the human C-terminal NFU domain (particularly in the presence of the N-terminal domain), but also of the behavior of Thermatoga maritima IscU, which exhibited properties consistent with a molten globule, but well-defined 1H-15N HSQC spectra,59 and is a paradigm for dynamic conformational interchange between distinct well-defined conformers, as is human ISCU1.60–63
As-purified apo SyNfu was successfully reconstituted by use of both NifS/L-cysteine and inorganic sulfide as the sulfur source, resulting in the formation of holo SyNfu with characteristic Fe-S cluster charge transfer peaks observed in UV-Vis and CD spectra (Fig. 5) that are consistent with prior reports.18, 56 The CD spectrum for SyNfu closely resembles that of the full-length holo human NFU, further indicating that the cluster environment for these two proteins is very similar.34 EPR characterization of holo SyNfu also suggests a [2Fe-2S] cluster bound form (Fig. 6), as evidenced by the comparison to that of holo BOLA3 homodimer and inverse linear relationship between temperature and integrated peak area, which is indicative of a [2Fe-2S] cluster.44
Previous reports have suggested that the nature of holo SyNfu is that of a dimeric complex based upon native PAGE and iron-staining techniques. To confirm the nature of the interaction, apo SyNfu and reconstitution mixtures at 1 and 2 h were analyzed by DLS. Analysis of these aliquots shows that apo SyNfu is monomeric, while the reconstitution mixture at 1 h shows a slight increase in a peak approximately double the size, suggesting that a dimer complex is beginning to form. At 2 h, the mixture contains a peak at twice the size of that of apo SyNfu, showing that the holo protein forms a dimeric complex. This follows previous reports of NifU and NFU proteins forming dimers, in addition to other Fe-S cluster protein complexes, such as Grx5-Bola3, Grx5-Bola1, and the Bola3 homodimer.34, 42, 44, 64, 65
Our laboratory has previously reported a glutathione-based [2Fe-2S] cluster that most likely exists in the cellular labile iron pool and is readily formed by glutathione extraction of the [2Fe-2S] core from the ISU scaffold protein.31, 66 This provides a biosynthetic pathway for the formation of [2Fe-2S](GS)4 that can then be consequently utilized to reconstitute other downstream Fe-S cluster proteins. SyNfu did not show reconstitution via the glutathione complexed cluster, consistent with the requirement for a structurally well-defined ligand set, where in this case, it would be the required positioning of a pair of Cys residues to affect the rapid chelation of a terminal iron in the cluster. It is also consistent with previous observations for apo ferredoxins and glutaredoxins, reflecting a need for a structured pocket or partial coordination motif 34 to accept cluster from a donor due to the need for rapid consecutive ligand substitutions to prevent the formation of hydrolytically unstable intermediates.31
We also investigated the ability of free GSH to extract the bound cluster from holo reconstituted SyNfu to form the [2Fe-2S](GS)4 complex. GSH was able to slowly remove cluster from SyNfu as evidenced by the observed rate constant, which is significantly lower than any protein-protein transfers examined thus far, but it is also within the same range as that for human NFU 34. This suggests that in this organism, the equilibrium between Fe-S cluster-bound SyNfu and the pool of the [2Fe-2S](GS)4 complex lies toward the cluster-bound protein. This also confirms prior reports showing that glutathione is required under oxidative stress conditions, while the precursor gamma-glutamylcysteine is able to mediate cellular function under normal growth conditions.46, 47
Synechocystis PCC6803 contains nine Fdx proteins in total, which account for a variety of functions within the cell. Fdx1 is most important and is involved in photosynthesis, while Fdx1 also participates in ferredoxin-glutaredoxin-thioredoxin crosstalk pathways, along with Fdx7 and Fdx9. The other FDX proteins are expressed under oxidative stress or heavy metal conditions.67 Given the large number of Fdx proteins within this cyanobacterium, and the conservation of this protein throughout many species, it is encouraging that holo SyNfu can successfully reconstitute both apo human FDX1 and FDX2, in agreement with prior reports18, 56 and further supports SyNfu’s role as a scaffold capable of delivering cluster to ferredoxin proteins with various functions. Cluster delivery to FDX2 was approximately 3-times faster than that of FDX1; however, both of these transfer reactions demonstrated larger rate constants relative to the transfers to A. vinelandii Fdx from either IscU or Grx5.68, 69 The relatively fast transfer from SyNfu supports a role for SyNfu in the delivery of Fe-S clusters to apo protein targets, and differences in the rate constants between FDX1 and FDX2 most likely reflect minor differences in the cluster binding pocket.70, 71
Another important, and conserved group of proteins involved in Fe-S cluster trafficking are the glutaredoxins (GLRXs or Grxs). We examined the ability of holo SyNfu to transfer cluster to both human GLRX2 and yeast Grx3. Cluster transfer to Grx3 was observed to be approximately 20-times faster than the transfer to GLRX2 and most likely again reflects differences in the cluster binding pocket: GLRX2 is a dithiol glutaredoxin and Grx3 is a monothiol glutaredoxin. Previously we have shown that the transfer from holo human NFU to apo Grx3 occurs quite quickly at second-order rate of 36,200 ± 7700 M−1min−1 72, which is in the same range as what was observed for holo SyNfu.72 Synechocystis PCC6803 contains three glutaredoxins: two dithiol glutaredoxins, Grx1 and Grx2, and a single monothiol glutaredoxin, Grx3. Synechocystis PCC6803 Grx1 and Grx2 have been shown to help remediate sodium selenate and oxidative conditions, while also being dispensable under normal growth conditions.73 Synechocystis PCC6803 Grx3, however, is a monothiol glutaredoxin containing the essential CGFS sequence required for iron-sulfur cluster ligation, and it has been shown to rescue yeast deficient in the mitochondrial GLRX5 protein.74, 75 This evidence supports a role for SyNfu as an early scaffold protein upon which clusters are built, so that they can be subsequently transferred to other downstream proteins such as the Grxs and Fdxs.
In summary, we have demonstrated that SyNfu can be both chemically and enzymatically reconstituted as a [2Fe-2S] cluster protein. Further, it can donate its cluster to both human FDX1 and FDX2 as well as monothiol yeast Grx3 and human GLRX2, albeit at an attenuated rate. Accumulating evidence from studies of Synechocystis has shown that SyNfu is indeed the scaffold protein by which clusters are built and subsequently shuttled to other downstream apo proteins, as IscU is not present in Synechocystis and IscA is not required for viability. Our current experimental evidence further supports this hypothesis as SyNfu has been shown to be the only protein from the group of conserved Fe-S cluster genes important for viability under both photoautotrophic and heterotrophic conditions.12, 13 These experiments provide further evidence for that proposal and elaborate on the complex Fe-S cluster biosynthesis pathways present in cyanobacteria.
Finally, these studies further demonstrate the dynamic conformational behavior and flexibility that appear to be common features of iron-sulfur cluster scaffolding and trafficking domains, presumably required for the molecular mechanism of cluster extrusion and delivery. The observations on exchange chemistry with glutathione and glutathione-complexed cluster again reinforce the key requirement for an at least partially structured environment to accept cluster (or not) from the glutathione complex or glutathione ligand, and the likely physiological role of such cluster complexes.
Reactivity of SyNfu determined quantitatively against a variety of cluster acceptors
New insights on structure-function details provided by NMR, DSC and VT-CD experiments
Support for SyNfu as the major scaffold protein in Synechocystis
Acknowledgments
This work was supported by a grant from the National Institutes of Health [AI072443]. Z.T. acknowledges funding from the Pelotonia Fellowship.
Footnotes
Declaration of competing interests
The authors have no conflict of interest to disclose. All authors have approved the final version of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson DC, Dean DR, Smith AD and Johnson MK, Structure, Function, and Formation of Biological Iron-Sulfur Clusters, Annu. Rev. Biochem, 2005, 74, 247–281. [DOI] [PubMed] [Google Scholar]
- 2.Rouault TA, Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease, Disease Models & Mechanisms, 2012, 5, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson DC, Dean DR, Smith AD and Johnson MK, Structure, function, and formation of biological iron-sulfur clusters, Annu Rev Biochem, 2005, 74, 247–281. [DOI] [PubMed] [Google Scholar]
- 4.Qi W and Cowan JA, Structural, Mechanistic and Coordination Chemistry of Relevance to the Biosynthesis of Iron-Sulfur and Related Iron Cofactors, Coord Chem Rev, 2011, 255, 688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouault TA, Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease, Dis Model Mech, 2012, 5, 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stehling O, Wilbrecht C and Lill R, Mitochondrial iron-sulfur protein biogenesis and human disease, Biochimie, 2014, 100, 61–77. [DOI] [PubMed] [Google Scholar]
- 7.Wachnowsky C, Fidai I and Cowan JA, Iron-sulfur cluster biosynthesis and trafficking - impact on human disease conditions, Metallomics, 2018, 10, 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lill R and Mühlenhoff a. U.., Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases, Annual Review of Biochemistry, 2008, 77, 669–700. [DOI] [PubMed] [Google Scholar]
- 9.Kato Shin-ichiro, Mihara Hisaaki, Kurihara Tatsuo, Yoshimura T and Esaki N, Gene cloning, purification, and characterization of two cyanobacterial NifS homologs driving iron-sulfur cluster formation, Bioscience, Biotechnology, and Biochemistry, 2000, 64, 2412–2419. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura Yasukazu, Kaneko Takakazu, Hirosawa Makoto, N. Miyajima and a. S. Tabata, CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803, Nucleic Acids Research, 1998, 26, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim W-Y, Kang S, Kim B-C, Oh J, Cho S, Bhak J and Choi J-S, SynechoNET: integrated protein-protein interaction database of a model cyanobacterium Synechocystis sp. PCC 6803, BMC Bioinformatics, 2008, 9, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balasubramanian R, Shen G, Bryant DA and Golbeck JH, Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002, J Bacteriol, 2006, 188, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang SS, Jiang HB, Song WY, Chen M and Qiu BS, Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions, Planta, 2017, 246, 927–938. [DOI] [PubMed] [Google Scholar]
- 14.Outten FW, Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria, Biochim Biophys Acta, 2015, 1853, 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Iron-Sulfur Cluster Biogenesis and Iron Homeostasis in Cyanobacteria, Front Microbiol, 2020, 11, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tirupati B, Vey JL, Drennan CL and Bollinger JM Jr., Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803, Biochemistry, 2004, 43, 12210–12219. [DOI] [PubMed] [Google Scholar]
- 17.Ayala-Castro C, Saini A and Outten FW, Fe-S cluster assembly pathways in bacteria, Microbiol Mol Biol Rev, 2008, 72, 110–125, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishio K and Nakai M, Transfer of iron-sulfur cluster from NifU to apoferredoxin, J Biol Chem, 2000, 275, 22615–22618. [DOI] [PubMed] [Google Scholar]
- 19.Fu Weiguang, Jack Richard F., Morgan Vance, Dean Dennis R. and Johnson a. M. K., nifU Gene Product from Azotobacter vinelandii Is a Homodimer That Contains Two Identical [2Fe-2S] Clusters, Biochemistry, 1994, 33, 13455–13463 [DOI] [PubMed] [Google Scholar]
- 20.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD and Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Molecular Systems Biology, 2011, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster M, Mansy S, Hwang J, Penner-Hahn J, Surerus K and Cowan J, A mutant human IscU protein contains a stable [2Fe-2S]2+ center of possible functional significance, J. Am. Chem. Soc, 2000, 122, 6805–6806. [Google Scholar]
- 22.Mansy SS, Xiong Y, Hemann C, Hille R, Sundaralingam M and Cowan JA, Crystal structure and stability studies of C77S HiPIP: a serine ligated [4Fe-4S] cluster, Biochemistry, 2002, 41, 1195–1201. [DOI] [PubMed] [Google Scholar]
- 23.Nuth M, Yoon T and Cowan JA, Iron-Sulfur Cluster Biosynthesis: Characterization of Iron Nucleation Sites for Assembly of the [2Fe-2S]2+ Cluster Core in IscU Proteins, J. Am. Chem. Soc, 2002, 124, 8774–8775. [DOI] [PubMed] [Google Scholar]
- 24.Xia B, Cheng H, Bandarian V, Reed GH and Markley JL, Human ferredoxin: overproduction in Escherichia coli, reconstitution in vitro, and spectroscopic studies of iron-sulfur cluster ligand cysteine-to-serine mutants, Biochemistry, 1996, 35, 9488–9495. [DOI] [PubMed] [Google Scholar]
- 25.Qi W, Li J and Cowan JA, Human ferredoxin-2 displays a unique conformational change, Dalton Trans, 2013, 42, 3088–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi W and Cowan JA, Mechanism of glutaredoxin-ISU [2Fe-2S] cluster exchange, Chem. Commun, 2011, 47, 4989–4991. [DOI] [PubMed] [Google Scholar]
- 27.Fidai I, Wachnowsky C and Cowan JA, Mapping cellular Fe-S cluster uptake and exchange reactions - divergent pathways for iron-sulfur cluster delivery to human ferredoxins, Metallomics, 2016, 8, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Ding S and Cowan JA, Thermodynamic and Structural Analysis of Human NFU Conformational Chemistry, Biochemistry, 2013, 52, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee W, Tonelli M and Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics, 2015, 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis-Jeune C, Andrade-Navarro MA and Perez-Iratxeta C, Prediction of protein secondary structure from circular dichroism using theoretically derived spectra, Proteins, 2012, 80, 374–381. [DOI] [PubMed] [Google Scholar]
- 31.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF and Cowan JA, Glutathione complexed Fe-S centers, J Am Chem Soc, 2012, 134, 10745–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mapolelo DT, Zhang B, Randeniya S, Albetel A-N, Li H, Couturier J, Outten CE, Rouhier N and Johnson MK, Monothiol glutaredoxins and A-type proteins: partners in Fe–S cluster trafficking, Dalton Trans, 2013, 42, 3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shakamuri P, Zhang B and Johnson MK, Monothiol Glutaredoxins Function in Storing and Transporting [Fe2S2] Clusters Assembled on IscU Scaffold Proteins, Journal of the American Chemical Society, 2012, 134, 15213–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wachnowsky C, Fidai I and Cowan JA, Iron-sulfur cluster exchange reactions mediated by the human Nfu protein, J Biol Inorg Chem, 2016, 21, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H and Markley JL, BioMagResBank, Nucleic Acids Res, 2008, 36, D402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS and Sykes BD, Chemical-Shifts as a Tool for Structure Determination, Method Enzymol, 1994, 239, 363–392. [DOI] [PubMed] [Google Scholar]
- 37.Peti W, Smith LJ, Redfield C and Schwalbe H, Chemical shifts in denatured proteins: resonance assignments for denatured ubiquitin and comparisons with other denatured proteins, J Biomol NMR, 2001, 19, 153–165. [DOI] [PubMed] [Google Scholar]
- 38.Sibley AB, Cosman M and Krishnan VV, An Empirical Correlation between Secondary Structure Content and Averaged Chemical Shifts in Proteins, Biophysical Journal, 2003, 84, 1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y and Cowan JA, Iron-sulfur cluster biosynthesis: characterization of a molten globule domain in human NFU, Biochemistry, 2009, 48, 7512–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai K, Liu G, Frederick RO, Xiao R, Montelione GT and Markley JL, Structural/Functional Properties of Human NFU1, an Intermediate [4Fe-4S] Carrier in Human Mitochondrial Iron-Sulfur Cluster Biogenesis, Structure, 2016, 24, 2080–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Ding S and Cowan JA, Thermodynamic and structural analysis of human NFU conformational chemistry, Biochemistry, 2013, 52, 4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wachnowsky C, Hendricks AL, Wesley NA, Ferguson C, Fidai I and Cowan JA, Understanding the Mechanism of [4Fe-4S] Cluster Assembly on Eukaryotic Mitochondrial and Cytosolic Aconitase, Inorg Chem, 2019, 58, 13686–13695. [DOI] [PubMed] [Google Scholar]
- 43.Tong WH, Jameson GN, Huynh BH and Rouault TA, Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster, Proc Natl Acad Sci U S A, 2003, 100, 9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wachnowsky C, Rao B, Sen S, Fries B, Howard CJ, Ottesen JJ and Cowan JA, Reconstitution, characterization, and [2Fe-2S] cluster exchange reactivity of a holo human BOLA3 homodimer, J Biol Inorg Chem, 2019, 24, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson SA, Wachnowsky C and Cowan JA, Defining the mechanism of the mitochondrial Atm1p [2Fe-2S] cluster exporter, Metallomics, 2020, DOI: 10.1039/c9mt00286c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cameron JC and Pakrasi HB, Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803, Plant Physiol, 2010, 154, 1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron JC and Pakrasi HB, Glutathione in Synechocystis 6803: a closer look into the physiology of a gshB mutant, Plant Signal Behav, 2011, 6, 89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens PJ, Thomson AJ, Dunn JBR, Keiderling TA, Rawlings J, Rao KK and Hall DO, Circular Dichroism and Magnetic Circular Dichroism of Iron-Sulfur Proteins, Biochemistry, 1978, 22, 4770–4778. [DOI] [PubMed] [Google Scholar]
- 49.Picciocchi Antoine, Saguez Cyril, Boussac Alain, Corinne Cassier-Chauvat and a. F. Chauvat, CGFS-Type Monothiol Glutaredoxins from the Cyanobacterium Synechocystis PCC6803 and Other Evolutionary Distant Model Organisms Possess a Glutathione-Ligated [2Fe-2S] Cluster, Biochemistry, 2007, 46, 15018–15026. [DOI] [PubMed] [Google Scholar]
- 50.Couturier J, Przybyla-Toscano J, Roret T, Didierjean C and Rouhier N, The roles of glutaredoxins ligating Fe–S clusters: Sensing, transfer or repair functions?, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2015, 1853, 1513–1527. [DOI] [PubMed] [Google Scholar]
- 51.Sánchez-Riego AM, López-Maury L and Florencio FJ, Glutaredoxins are essential for stress adaptation in the cyanobacterium Synechocystis sp. PCC 6803, Frontiers in Plant Science, 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Maury L, Sanchez-Riego AM, Reyes JC and Florencio FJ, The Glutathione/Glutaredoxin System Is Essential for Arsenate Reduction in Synechocystis sp. Strain PCC 6803, Journal of Bacteriology, 2009, 191, 3534–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, Talla E, Brochier-Armanet C, Garcia Serres R, Latour JM, Ollagnier-de Choudens S, Fontecave M and Barras F, Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier, Mol Microbiol, 2012, 86, 155–171. [DOI] [PubMed] [Google Scholar]
- 54.Bandyopadhyay S, Naik SG, O’Carroll IP, Huynh BH, Dean DR, Johnson MK and Dos Santos PC, A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier, J Biol Chem, 2008, 283, 14092–14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Z, Heinnickel M, Krebs C, Shen G, Golbeck JH and Bryant DA, Biogenesis of iron-sulfur clusters in photosystem I: holo-NfuA from the cyanobacterium Synechococcus sp. PCC 7002 rapidly and efficiently transfers [4Fe-4S] clusters to apo-PsaC in vitro, J Biol Chem, 2008, 283, 28426–28435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seidler A, Jaschkowitz K and Wollenberg M, Incorporation of iron-sulphur clusters in membrane-bound proteins, Biochem Soc Trans, 2001, 29, 418–421. [DOI] [PubMed] [Google Scholar]
- 57.Asakura T, Taoka K, Demura M and Williamson MP, The relationship between amide proton chemical shifts and secondary structure in proteins, J Biomol NMR, 1995, 6, 227–236. [DOI] [PubMed] [Google Scholar]
- 58.Mielke SP and Krishnan VV, Characterization of protein secondary structure from NMR chemical shifts, Prog Nucl Magn Reson Spectrosc, 2009, 54, 141–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansy SS, Wu SP and Cowan JA, Iron-sulfur cluster biosynthesis: biochemical characterization of the conformational dynamics of Thermotoga maritima IscU and the relevance for cellular cluster assembly, J Biol Chem, 2004, 279, 10469–10475. [DOI] [PubMed] [Google Scholar]
- 60.Kim JH, Fuzery AK, Tonelli M, Ta DT, Westler WM, Vickery LE and Markley JL, Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB, Biochemistry, 2009, 48, 6062–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JH, Tonelli M and Markley JL, Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase, Proc Natl Acad Sci U S A, 2012, 109, 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Markley JL, Kim JH, Dai Z, Bothe JR, Cai K, Frederick RO and Tonelli M, Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery, FEBS Lett, 2013, 587, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai K, Frederick RO, Kim JH, Reinen NM, Tonelli M and Markley JL, Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU), J Biol Chem, 2013, 288, 28755–28770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen S, Rao B, Wachnowsky C and Cowan JA, Cluster exchange reactivity of [2Fe-2S] cluster-bridged complexes of BOLA3 with monothiol glutaredoxins, Metallomics, 2018, 10, 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sen S, Hendricks AL and Cowan JA, Cluster exchange reactivity of [2Fe-2S]-bridged heterodimeric BOLA1-GLRX5, FEBS J, 2020, DOI: 10.1111/febs.15452. [DOI] [PubMed] [Google Scholar]
- 66.Li J and Cowan JA, Glutathione-coordinated [2Fe-2S] cluster: a viable physiological substrate for mitochondrial ABCB7 transport, Chem Commun (Camb), 2015, 51, 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassier-Chauvat C and Chauvat F, Function and Regulation of Ferredoxins in the Cyanobacterium, Synechocystis PCC6803: Recent Advances, Life (Basel), 2014, 4, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chandramouli K and Johnson MK, HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction, Biochemistry, 2006, 45, 11087–11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shakamuri P, Zhang B and Johnson MK, Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins, J Am Chem Soc, 2012, 134, 15213–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheftel AD, Stehling O, Pierik AJ, Elsasser HP, Muhlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R and Lill R, Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis, Proc Natl Acad Sci U S A, 2010, 107, 11775–11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qi W, Li J and Cowan JA, Human ferredoxin-2 displays a unique conformational change, Dalton Trans, 2013, 42, 3088–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wachnowsky C, Fidai I and Cowan JA, Cytosolic iron-sulfur cluster transfer-a proposed kinetic pathway for reconstitution of glutaredoxin 3, FEBS Lett, 2016, 590, 4531–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marteyn B, Domain F, Legrain P, Chauvat F and Cassier-Chauvat C, The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803, Mol Microbiol, 2009, 71, 520–532. [DOI] [PubMed] [Google Scholar]
- 74.Molina-Navarro MM, Casas C, Piedrafita L, Belli G and Herrero E, Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria, FEBS Lett, 2006, 580, 2273–2280. [DOI] [PubMed] [Google Scholar]
- 75.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C and Chauvat F, CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster, Biochemistry, 2007, 46, 15018–15026. [DOI] [PubMed] [Google Scholar]
- 76.Qi W, Li J, Chain CY, Pasquevich GA, Pasquevich AF, Cowan JA, Glutathione-complexed iron-sulfur clusters. Reaction intermediates and evidence for a template effect promoting assembly and stability, Chem. Commun 2013, 49, 6313–6315, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]


