Abstract
The cap structure and the poly(A) tail of eukaryotic mRNAs act synergistically to enhance translation. This effect is mediated by a direct interaction of eukaryotic initiation factor 4G and poly(A) binding protein (PABP), which brings about circularization of the mRNA. Of the two recently identified PABP-interacting proteins, one, Paip1, stimulates translation, and the other, Paip2, which competes with Paip1 for binding to PABP, represses translation. Here we studied the Paip2-PABP interaction. Biacore data and far-Western analysis revealed that Paip2 contains two binding sites for PABP, one encompassing a 16-amino-acid stretch located in the C terminus and a second encompassing a larger central region. PABP also contains two binding regions for Paip2, one located in the RNA recognition motif (RRM) region and the other in the carboxy-terminal region. A two-to-one stoichiometry for binding of Paip2 to PABP with two independent Kds of 0.66 and 74 nM was determined. Thus, our data demonstrate that PABP and Paip2 could form a trimeric complex containing one PABP molecule and two Paip2 molecules. Significantly, only the central Paip2 fragment, which binds with high affinity to the PABP RRM region, inhibits PABP binding to poly(A) RNA and translation.
The mRNA 5′ cap structure (termed cap m7GpppN, where N is any nucleotide) and the 3′ poly(A) tail play important roles in translation and its control. The 5′ cap is bound by eukaryotic initiation factor 4F (eIF4F), which consists of three proteins (eIF4E, eIF4A, and eIF4G). eIF4E directly contacts the cap. eIF4A exhibits RNA-dependent ATPase activity and RNA helicase activity (34, 35) and is thought to unwind the mRNA secondary structure in the 5′ untranslated region to promote ribosome binding (for reviews see references 14, 16, and 39). eIF4G functions as a protein scaffold by binding to eIF4E, eIF4A, and eIF3, a factor tightly associated with the 40S ribosomal subunit (16, 18, 24, 26, 38). The mRNA 3′ poly(A) tail is bound by the poly(A) binding protein (PABP). One PABP molecule is bound to every 25 adenosine residues, although 12 adenosines are sufficient for binding (3, 4, 36, 37). PABP contains four RNA recognition motifs (RRMs), followed by a proline-rich C-terminal region (1, 36).
The cap and the poly(A) tail synergistically enhance translation (for reviews see references 13, 19, 38, and 41). The “closed loop” model (19), whereby the mRNA circularizes via protein-protein interactions, is consistent with this synergism. The circularization of the mRNA might promote translation via shunting of terminating ribosomes, or alternatively it may influence initiation factor activity and thereby aid in ribosome recycling (33). A large body of evidence documents the association of PABP with eIF4G (17, 25, 32, 40). Circularization of mRNA was demonstrated in a reconstituted PABP-eIF4G-eIF4E system where the circularized mRNAs were visualized via atomic force microscopy (42).
We reported earlier on a mammalian translational coactivator, Paip1, which binds directly to PABP (7). Paip1 exhibits significant homology to the central portion of eIF4G, which interacts with eIF4A (18), and accordingly, Paip1 also interacts with eIF4A. Paip1 overexpression in COS-7 cells enhances translation of a reporter luciferase gene (7). We recently cloned another PABP-interacting protein, Paip2 (21). Paip2 is an acidic protein (pI = 3.9) with a predicted molecular mass of 14.5 kDa which preferentially represses translation of poly(A)-containing mRNAs. Paip2 competes with Paip1 for PABP binding. Furthermore, Paip2 decreases binding of PABP to oligo(A) RNA (21).
Here we studied the interaction of Paip2 with PABP. We mapped two PABP binding sites in Paip2, a short 16-amino-acid stretch located in the C terminus and a larger central acid-rich region. Paip2 interacts with two independent sites in PABP, one encompassing segments of RRMs 2 and 3 and the other in the C-terminal region. Furthermore, we show that only the interaction of Paip2 with the PABP amino-terminal site results in translational inhibition.
MATERIALS AND METHODS
Vectors.
The constructs pACTAG-2-Paip2 and pcDNA3-Paip2 were designed as described previously (21). For construction of pGEX-GTH-Paip2 fragments 1–42, 1–48, 1–58, 1–75, 1–111, and 76–111, the respective partial Paip2 coding regions were PCR amplified using pcDNA3-Paip2 as a template. The resulting fragments were digested with BamHI/EcoRV and ligated to pGEX-GTH (20) digested with BamHI/SmaI. To construct pGEX-GTH-Paip2 fragments 43–127, 76–127, and 96–127, Paip2 PCR products were digested with XbaI, blunt ended with the Klenow fragment of Escherichia coli DNA polymerase, and digested with BamHI. The resulting fragments were ligated to pGEX-GTH digested with BamHI/SmaI. pGEX-GTH-Paip2 fragments 106–127, 112–127, and 121–127 were constructed by annealing the forward primers (5′-AAT TAT GCT TGT GGT CAA GAG CAA TCT GAA TCC AAA TGC AAA GGA GTT TGT TCC-3′, 5′-AAT TAT GAA TCC AAA TGC AAA GGA GTT TGT TCC TGG GGT GAA GTA CGG AAA TAT-3′, and 5′-AAT TAT GGG GGT GAA GTA CGG AAA TAT TGG-3′) to the reverse primers (5′-AAT TTC AAA TAT TCC CGT ACT TCA CCC CAG GAA CAA ACT CCT TTG CAT TTG GAT-3′, 5′-AAT TTC AAA TAT TTC CGT ACT TCA CCC CAG GAA CAA ACT CCT TTG CAT TTG GAT-3′, and 5′-AAT TTC AAA TAT TTC CGT ACT TCA CCC CCA-3′) (Dalton Chemical Laboratories), respectively. The annealed products were ligated into pGEX-GTH linearized with EcoRI. pGEX-GTH-Paip2(105–120) was constructed by annealing the forward primers (5′-GAT CCA TGC TTG TGG TCA AGA GCA ATC TGA ATC CAA ATG CAA AGG AGT TTG TTC CTT GAG GG-3′) to the reverse primers (5′-CCC TCA AGG AAC AAA CTC CTT TGC ATT TGG ATT CAG ATT GCT CTT GAC CAC AAG CAT G-3′) (Dalton Chemical laboratories); the resulting annealed product was ligated into pGEX-GTH linearized with BamHI/SmaI. To generate pGEX-GTH-Paip2 fragments 12–75, 22–75, 35–75, and 42–75, the Paip2 PCR products were digested with BamHI/EcoRV and ligated into pGEX-GTH digested with BamHI/SmaI.
To generate pcDNA3-GST, the glutathione S-transferase (GST) coding region was PCR amplified from pGEX-2T, digested with BamHI/XbaI, and ligated directionally into pcDNA3 (Invitrogen). To generate pcDNA3-GST-Paip2, the Paip2 coding region was PCR amplified from pBluescriptKS-Paip2 and digested with BamHI/XbaI, blunt ended with the Klenow fragment of Escherichia coli DNA polymerase (MBI Fermentas), and ligated into pcDNA3-GST, cut with XbaI, and blunt ended with the Klenow fragment of E. coli DNA polymerase (MBI Fermentas).
For construction of FLAG-HMK-Paip2, the Paip2 coding region was PCR amplified using pcDNA3-Paip2 as a template. The resulting PCR product was digested with EcoRI and ligated into pARΔr1 (5). To construct pGEX6P3-FLAG-HMK-Paip2, the FLAG-HMK-Paip2 coding region was in turn PCR amplified using the pARΔr1-Paip2 construct as a template. The resulting PCR product was digested with BamHI and ligated into pGEX6P3 (Amersham Pharmacia Biotech [APB]).
PABP constructs were designed as follows: pGexHA was constructed by annealing the forward primer (5′-AAT TCT ACC CAT ACG ATG TTC CTG ACT ATG CGG GC-3′), coding for the cDNA of the hemagglutinin (HA) tag, to the reverse primer (5′-TCG AGC CCG CAT AGT CAG GAA CAT CGT ATG GGT AG-3′) (Sheldon Biotechnology Center, McGill University). The annealed product was ligated into pGEX6P3 between EcoRI and XhoI in the multiple cloning site. To construct pGEX-PABP-RRM1, -RRM2, -RRM3, -RRM4, -C1, -RRM1-2, and -RRM3-4, cDNAs encoding amino acids 1 to 98, 99 to 189, 190 to 289, 290 to 368, 369 to 494, 1 to 189, and 190 to 368 of PABP, respectively, were synthesized by PCR. The resulting fragments were digested with BamHI and EcoRI and ligated into pGEX-HA. To construct pGEX-PABP-C2, cDNAs encoding amino acids 495 to 633 of PABP were synthesized by PCR, digested with BamHI and EcoRI, and ligated into pGEX6P3. pGEX-PABP-RRM2-3 and -RRM1-4 were constructed using cDNA encoding amino acids 99 to 289 and 1 to 368 of PABP, respectively, synthesized by PCR, digested with BamHI, and ligated directionally into the BamHI site of pGEX6P3. pET3B PABP-His was constructed by ligating cDNA encoding PABP-His into pET3B.
Protein expression and purification.
FLAG-HMK-PABP and His-PABP were expressed and purified as described previously (7). For purification of hPABP fragments 1–190, 172–392, 1–97, 83–190, 172–287, and 261–392, E. coli BL21 cells were transformed with the various pGEX6P-hPABP constructs and lysed by sonication, and proteins were purified on glutathione-Sepharose (APB). Proteins were dialyzed into cleavage buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT] [APB]) and digested with PreScission protease (APB) for 4 h at 4°C to cleave the GST tag from the fusion protein. The mixture was incubated with glutathione-Sepharose resin (APB) to remove both the GST tag and the PreScission protease. The remaining proteins were dialyzed against phosphate-buffered saline.
PABP-His was purified as follows: E. coli BL21 λDE3 cells were transformed with pET3B PABP-His. After incubation at 37°C and induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (3 h), cells were centrifuged and resuspended in high-salt buffer (2 M NaCl, 20 mM Tris-HCl [pH 7.5], protease inhibitor cocktail [Boehringer]). The suspension was sonicated and centrifuged at 10,000 rpm for 30 min in a Du Pont Sorvall RC-5B centrifuge (SS-34 rotor). The supernatant was incubated on ice (2 h) and ultracentrifuged at 45,000 rpm for 3 h in a Beckmann Optima L-90K ultracentrifuge (60-T: rotor). The supernatant was incubated with Talon Metal Affinity Resin (Clontech). The resin was washed two times with high-salt buffer and two times with wash buffer (20 mM Tris-HCl [pH 7.5], 300 mM KCl, 10% glycerol, 10 mM imidazole). Protein was eluted with elution buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 10% glycerol, 250 mM imidazole). The protein was dialyzed against phosphate-buffered saline.
GST pull-downs.
Purified proteins (2 μg) were incubated with a 50% slurry of glutathione 4B-Sepharose (25 μl; APB) and incubated end-over-end for 3 h at 4°C. The mixture was centrifuged at 3,000 rpm for 10 s in a Sorvall GLC-1 centrifuge, and the resin was washed four times with 1 ml of buffer A (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, 0.5% Nonidet P-40). Proteins were eluted with 2× Laemmli sample buffer. Samples were boiled at 95°C for 4 min, resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and stained with Coomassie R-250.
Western blotting.
Membranes were incubated overnight at 4°C with one of the following antibodies and dilutions: mouse monoclonal anti-HA, 1:1,000 (Berkeley Antibody Company); anti-FLAG antibody, 1:1,000 (Sigma); rabbit polyclonal anti-Paip2, 1:1,000 (21); rabbit polyclonal anti-PABP, 1:500 (2); and rabbit polyclonal anti-GST, 1:1,000 (8). After a wash with Tris-buffered saline–Tween (TBST), the membranes were incubated with either donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G at 1:5,000 (APB) or sheep anti-mouse horseradish peroxidase-conjugated immunoglobulin G at 1:5,000 (APB) for 30 min. Membranes were washed with TBST four times, and the signals were detected using an ECL kit (APB) and exposure to X-ray film (Du Pont).
Far-Western analysis.
The procedure for far-Western analysis was performed as previously described (7). The membrane was incubated overnight at 4°C in hybridization buffer containing 32P-labeled FLAG-HMK-PABP or FLAG-HMK-Paip2 (250,000 cpm/ml; as described previously [5]). The membrane was washed four times with hybridization buffer and exposed to an X-ray film (Du Pont).
Experimental controls for Biacore experiments.
To obtain quantitative kinetic measurements of the Paip2-PABP interactions, experiments were conducted on an SPR (surface plasmon resonance)-based Biacore biosensor. In a typical Biacore experiment, one of the binding partners (the ligand in Biacore terminology) is immobilized on the sensor chip surface. A solution containing the other binding partner (the analyte in Biacore terminology) is injected over the sensor chip surface. The mass accumulation of the analyte on the surface, as it binds to the ligand, is recorded in arbitrary resonance units (RUs), which are directly proportional to mass. In preliminary experiments, injection of both full-length PABP and the C-terminal portion of PABP (GST-PABP-C2) over a control dextran surface resulted in a significant increase in the SPR signal with time, indicating that full-length PABP and its C terminus bind nonspecifically to the dextran surface. In contrast, no interaction was detectable when Paip2 or the RRM1-4 and RRM2-3 truncated PABP proteins were injected over the control surface (data not shown). Hence, all subsequent experiments were carried out with full-length PABP, GST-PABP-C2, or Paip2 as ligands and with Paip2 or the different RRMs of PABP as analytes. To avoid any avidity artifacts that may be caused by GST-induced dimerization of GST-fused proteins, we cleaved and removed the GST tags from all the species that were used as analytes.
Immobilization of the recombinant proteins on Biacore sensor chips.
Recombinant proteins (PABP-His, FLAG-HMK-Paip2, GST-PABP-C2) were immobilized on different surfaces of CM5 sensor chips using the standard amine coupling procedure (9). During the immobilization step the flow rate was set at 5 μl/min. Reagents were injected in the following order: 0.05 M N-hydroxysuccinimide–0.2 M N-ethyl-N′-(3-dimethylaminopropyl)-carbodiimidehydrochloride mixture (35 μl), recombinant protein solutions (5, 10, and 20 μg/ml for PABP-His, FLAG-HMK-Paip2, and GST-PABP-C2, respectively) in 10 mM sodium acetate (pH 4.5) (or pH 3.5 for FLAG-HMK-Paip2) until the desired amount of protein was coupled (800, 250, and 1,800 RU for PABP-His, FLAG-HMK-Paip2, and GST-PABP-C2, respectively). A solution of 0.1 M ethanolamine–HCl (pH 8.5; 35 μl) was then used to block the remaining activated carboxyl groups. Mock surfaces were also generated using the same procedure by replacing the protein solution with running buffer.
Kinetic assays on the Biacore.
Kinetic experiments were carried out at 25°C at a flow rate of 5 μl/min, except for the mass transport experiments, for which different injections of Paip2 or PABP-RRMs were performed at flow rates ranging from 5 to 50 μl/min. The data collection rate of the apparatus was set to 10 Hz for every kinetic assay. HEPES-buffered saline (20 mM HEPES [pH 7.4], 150 mM NaCl, 3.4 mM EDTA, 0.05% Tween 20) was used as the running buffer and for diluting all the injected proteins.
Different concentrations of Paip2 were injected over each PABP surface (including a mock surface) for 300 s, followed by a 300-s-long buffer injection. Injection times were shortened to 180 s when PABP RRM1-4 and PABP RRM2-3 were injected over Paip2 surfaces. Regeneration of the sensor chip was accomplished by two 5-μl pulse injections of 120 mM HCl solution, followed by an EXTRACLEAN procedure according to the manufacturer's instructions (BIAcore Upgrade Instrument Handbook; Pharmacia).
Biacore data preparation and analysis.
Sensorgrams were prepared and each set was subjected to curve fitting with numerical integration methods using the SPRevolution software package (12, 30). Data preparation was performed as described elsewhere (9, 30). Sensorgrams generated using a mock (blank) surface were subtracted from the corresponding experimental sensorgrams, and the resulting curves were transformed to concentration units using the molecular weight of the injected species. After subtraction, all the curves were reduced to 400 evenly spaced sampling points. For each set of individual curves, corresponding to injections of various concentrations of protein over the same surface, integration was carried out using the different kinetic models available in the SPRevolution software (9 and SPRevolution user manual, available online at http://www.bri.nrc.ca/csrg/equip.htm#biacore).
Mathematical modeling and parameter estimation.
The schematic representation of the models used for the data analysis and their related sets of differential rate equations are listed elsewhere (9; SPRevolution user manual [see above for URL]).
For each model, the kinetic parameters and the active quantity of ligand covalently coupled to the matrix were considered global parameters (i.e., the same value applies to all curves within a set). Moreover, two local parameters were added for each curve to take into account the refractive index changes at the beginning of the wash-on and wash-off phases, respectively. The thermodynamic dissociation constants were calculated from the kinetic constants determined by global fitting.
Evaluation of the quality of the fit for the various kinetic models.
For each set of residuals (difference between the experimental values and the values calculated by numerical integration for each kinetic model), the following three statistical values were calculated.
(i) The standard deviation (SD) of the residuals.
(ii) The “+ or − signs” statistic (Z1) (6): each residual is replaced by its sign value (+ or −), and the following statistic is then calculated on the newly created data set: Z1 = [n × (R1 − 1) − 2 × n1 × n2]/[(2 × n1 × n2)(2 × n1 × n2 − n)/(n − 1)]1/2 where R1 is the number of positive runs, n1 is the “+” number, and n2 is the “−” number.
(iii) the “run up and down” statistic (Z2) (6): using the residuals set x(i), a new set of data, y, is created with y(i) = x(i) − x × (i + 1). As for the above test, each y value is then replaced by its sign value (+ or −). If we call R2 the number of positive y(i) values of the runs, then the statistic Z2 equals {R2 − [(2 × n − 1)/3]}/[(16 × n − 29)/90]1/2.
Assuming that the residuals are independent, the Z1 and Z2 statistics follow a normal law with a mean equal to zero and a variance equal to one.
Filter binding assay.
Filter binding assays were performed essentially as described previously (21) with the following modifications. Reaction mixtures contained A25 RNA (15,000 to 25,000 cpm; final concentration, 0.1 nM) in 50 μl of filter binding buffer (FBB) (20 mM HEPES-KOH [pH 7.5], 100 mM KCl, 2 mM DTT, 2 mM MgCl2). Reaction mixtures were filtered through a 0.45-μm-pore-size nitrocellulose membrane, which was hydrated with FBB, on a Dot Blot apparatus (Bio-Rad) followed by a 500-μl wash with FBB. The membrane was dried and cut for each well, and retained counts per minute (bound) were estimated by liquid scintillation counting.
RESULTS
Characterization of PABP binding sites in Paip2.
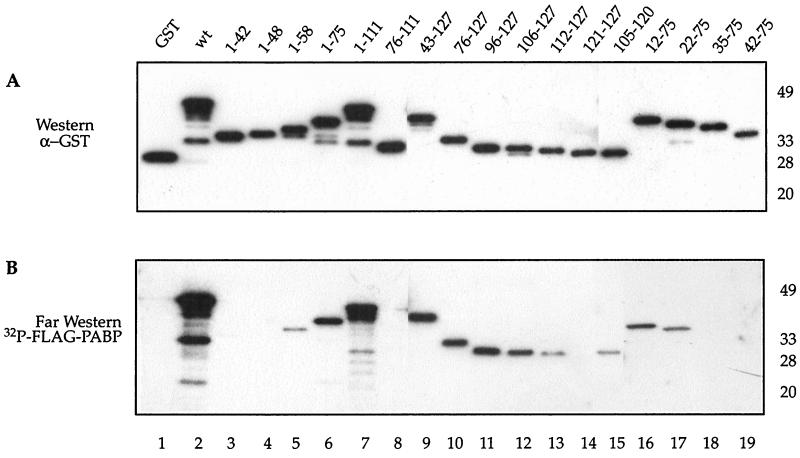
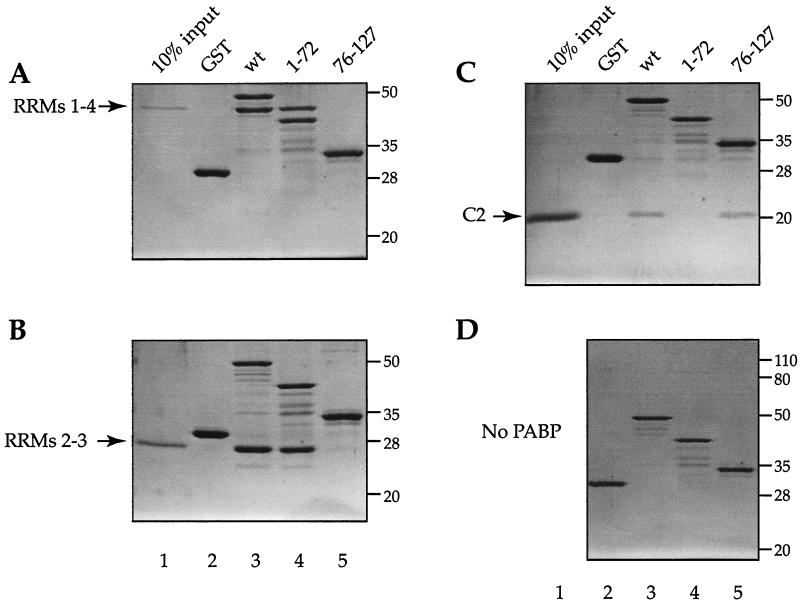
To identify the PABP binding site in Paip2, fragments of Paip2 were generated and expressed in E. coli as GST fusion proteins (Fig. 1A and C). The interaction of the Paip2 fragments with PABP was determined by far-Western analysis (5). Approximately equal amounts of full-length protein were loaded on the gel as determined by a Western blot using an antiserum against GST (Fig. 1A; the presence of additional bands is due to protein degradation). A duplicate membrane was used for far-Western analysis using 32P-labeled FLAG-HMK-PABP as a probe (Fig. 1B). Relative binding was evaluated visually (Fig. 1C). GST-Paip2(wt), but not GST alone, interacted with 32P-labeled FLAG-HMK-PABP (Fig. 1B, compare lanes 1 and 2). Fragments of Paip2 with C-terminal truncations past amino acid 58, i.e., Paip2 fragments 1–42 and 1–48, failed to interact with PABP (lanes 3 and 4), while Paip2(1–58) interacted only weakly with PABP (lane 5). In contrast, Paip2 fragments 1–75 and 1–111 bound strongly to PABP (lanes 6 and 7, respectively). Since Paip2(1–75) interacted with PABP almost as well as Paip2(1–111), the region consisting of amino acids 76 to 111 does not appear to contribute significantly to the interaction of Paip2 with PABP. This is consistent with the failure of Paip2(76–111) to bind to PABP (lane 8). The above results suggest that the central region of Paip2 contains a PABP binding site. To demarcate the N-terminal boundary of the central binding region, N-terminal truncations were created in a fragment having amino acid 75 as the C-terminal border. N-terminal truncations of this region up to amino acid 22 retained some PABP binding, since both Paip2(22–75) and Paip2(12–75) bound to PABP (lanes 17 and 16, respectively). Deletion of residues from the N terminus past amino acid 22, i.e., Paip2(35–75) or Paip2(42–75), abolished PABP binding (lanes 18 and 19, respectively). These results indicate that the entire region encompassing the glutamic-acid-rich residues of Paip2(22–75) is the minimal central domain required for PABP binding (Fig. 1C).
FIG. 1.
Identification of PABP binding sites in Paip2. Purified Paip2 and Paip2 fragments (0.1 μg) were resolved by SDS–15% polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane. The membrane was probed with a rabbit polyclonal anti-GST antibody (A) or processed for far-Western analysis using 32P-labeled FLAG-HMK-PABP as a probe (B), as described in Materials and Methods. Positions of molecular weight markers are shown at right. (C) Schematic diagram of the GST-Paip2 fragments with a summary of the PABP-Paip2 interaction results. Black boxes represent the region rich in glutamic acids, and grey represents the Paip2 C-terminal PABP binding site. The sequences at the top exhibit homology to Paip1.
N-terminal deletions revealed a second PABP binding site. Deletions of Paip2 up to amino acid 106 had no significant effect on PABP binding (lanes 9 to 12), but a further deletion (fragment 112–127) reduced binding dramatically (lane 13). Paip2(105–120) exhibits some binding, albeit weak, to PABP (lane 15). These results indicate that a second PABP-interacting site resides between amino acids 105 and 120. Taken together, the deletion analysis demonstrates that Paip2 possesses two independent sites for PABP binding, a short 16-amino-acid domain in the C-terminal region of the protein and a second central binding domain spanning the entire acid-rich region of Paip2 (Fig. 1C).
Characterization of Paip2 binding sites in PABP.
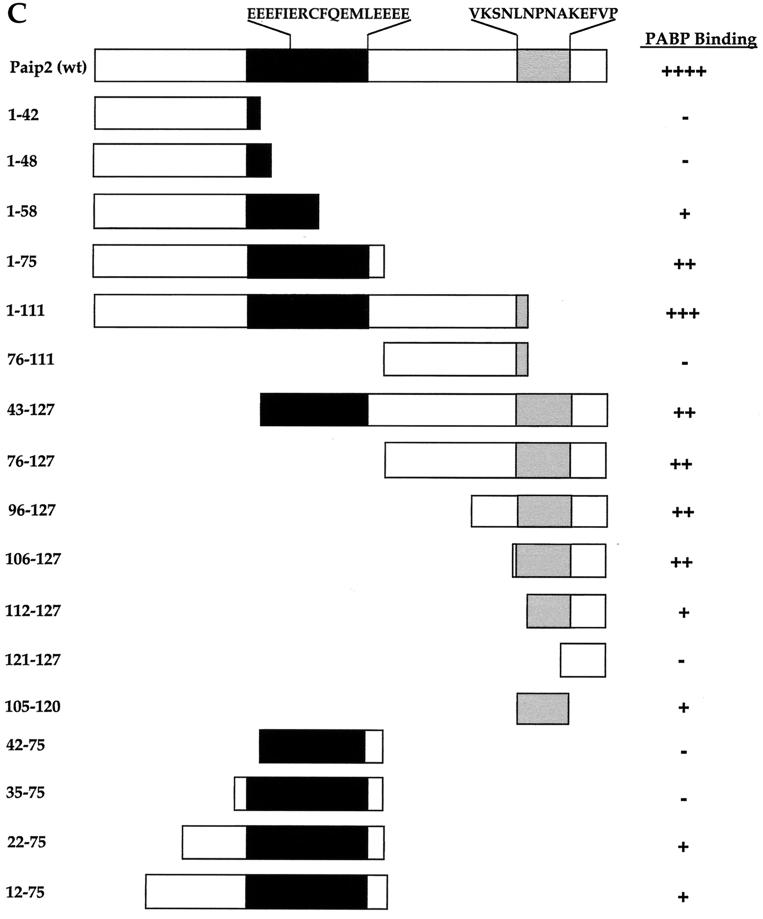
We next wished to study the Paip2 binding sites in PABP. Fragments of PABP were generated as GST fusion proteins and expressed in E. coli (Fig. 2A and B, top panels, and C). Approximately equal amounts of full-length protein were loaded on the gel as determined by a Western blot using an antiserum against GST (Fig. 2A and B, top panels; the presence of additional bands is due to protein degradation). Duplicate membranes were used for far-Western analysis using 32P-labeled FLAG-HMK-Paip2 as a probe on individual PABP RRMs (Fig. 2A, middle and lower panels) or combinations of RRMs (RRM1-2, RRM3-4, RRM2-3, RRM1-4; Fig. 2B, middle and lower panels); relative binding was evaluated visually (Fig. 2C). Paip2 interacts weakly with the individual RRM2 and RRM3 fragments (Fig. 2A, lower panel, lanes 2 and 3), yet it interacts strongly with a fragment containing both RRMs (Fig. 2B, middle panel, lane 4), suggesting that the binding site spans the junction of these two RRMs. Weak interactions are also observed with RRM1-2 and RRM3-4 (Fig. 2B, lower panel, lanes 1 and 2). No interaction could be detected with RRM1 or RRM4 alone (Fig. 2A, middle and lower panels, lanes 1 and 4). Paip2 also significantly interacts with the second half of the C-terminal region of PABP, termed C2 (Fig. 2A, middle panel, lane 6), but not with C1, the first half (Fig. 2A, middle panel, lane 5). Based on the far-Western analysis, Paip2 appears to exhibit a lower affinity for the C-terminal portion of PABP than for its binding site residing between RRMs 2 and 3 (Fig. 2B, middle panel, compare lanes 4 and 5 to lane 7). However, it is possible that other factors, such as differential protein denaturation, affect the interaction. The interactions were studied quantitatively (see below) in the Biacore experiments. Taken together, these data demonstrate the presence of two regions within PABP for Paip2 binding: an apparent high-affinity site within the RRMs and an apparent weaker-affinity site in the C terminus (Fig. 2C).
FIG. 2.
Identification of Paip2 binding sites in PABP. PABP fragments were resolved by SDS–15% polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane. The blots were probed with a rabbit polyclonal anti-GST antibody or processed for far-Western analysis using 32P-labeled HMK-Paip2 as a probe, as described in Materials and Methods. Lanes contain the following amounts of protein. (A) Individual RRMs, 1 μg each; PABP-C1 and -C2, 0.5 μg; GST, 1 μg; and PABP-His, 0.5 μg. (B) Combinations of RRMs, 0.5 μg each; PABP-C2, 0.5 μg; GST, 1 μg; and PABP-His, 0.5 μg. Positions of molecular weight markers are shown at right. (C) Schematic diagram of the GST-PABP fragments with a summary of the Paip2-PABP interaction results. Shaded areas represent domains that contain Paip2 interaction sites in PABP.
PABP interacts specifically with Paip2 as determined by SPR.
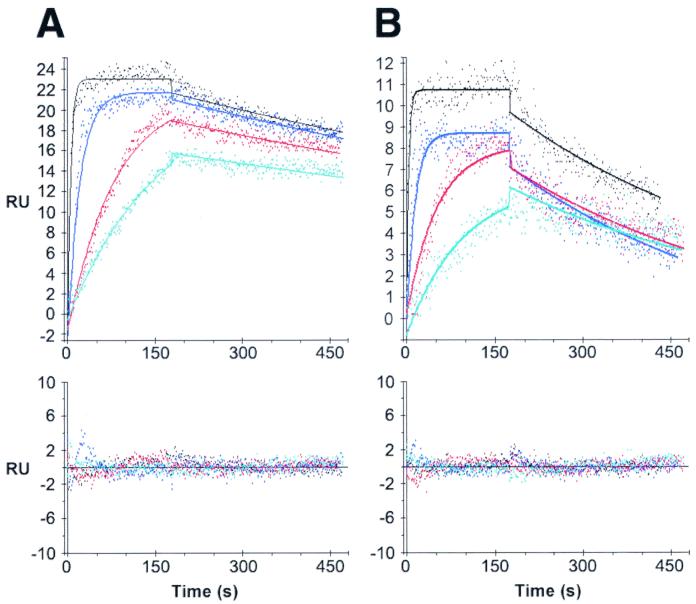
As a preliminary experiment, PABP (1,400 RU) was coupled to a dextran matrix on a CM5 chip and 64 nM Paip2 was injected over this surface or a mock surface. Numerical integration of the resulting curve, after blank subtraction, using a simple kinetic model did not give a good fit as judged by the variance in the residuals (data not shown). This deviation from a simple one-to-one model could result from the existence of a more complex interaction between the two proteins. Alternatively, it may be due to nonoptimized experimental conditions. To minimize artifacts such as mass transport and rebinding effects (15, 28, 29, 43) and steric hindrance or crowding problems (31), we coupled the minimum amount of PABP required to obtain an optimal signal-to-noise ratio when injecting Paip2 (less than 1,000 RU of PABP were coupled). The absence of a mass transport step was verified by injecting the same Paip2 solution (1 nM) at different flow rates ranging from 5 to 50 μl/min over the PABP surface. After data treatment, the curves at the different flow rates were superimposable (data not shown).
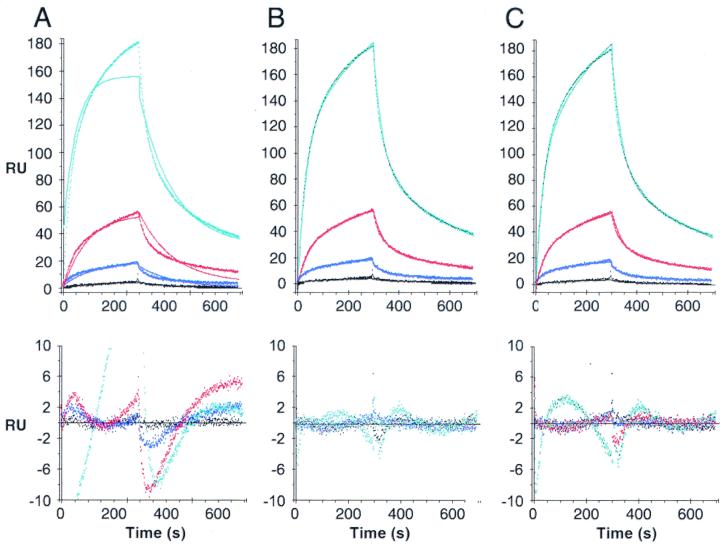
PABP interacts with Paip2 with a 1:2 stoichiometry.
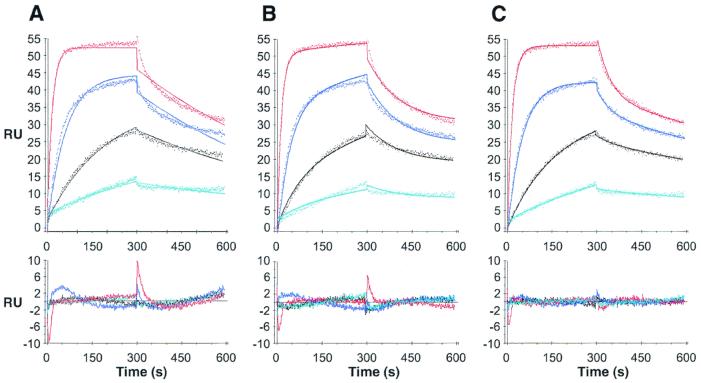
Paip2, at increasing concentrations (from 1 to 64 nM), was injected over the PABP surface, and the resulting sets of curves were analyzed by curve fitting with numerical integration methods. When a kinetic model adequately depicts a molecular interaction, the residuals will be minimal and distributed randomly around a zero value. The analysis of the sensorgrams gave poor fits when a simple one-to-one interaction model was applied (Fig. 3A). Since PABP possesses two binding sites for Paip2 as determined by far-Western blotting, we applied more complex models. The first model includes an initial binding event involving one binding site in each of the PABP and Paip2 molecules, followed by a rearrangement of the complex such that two sites in a PABP molecule are interacting with two sites in a Paip2 molecule (Fig. 3B). The second model assumes that one PABP molecule binds to two Paip2 molecules through two independent and distinct binding sites (2:1 stoichiometry) (Fig. 3C). The latter model fit better than the rearrangement model (evident from the lower values of the SD of the residuals and the Z1 and Z2 statistics; Table 1 and Fig. 3, bottom panels). The kinetic constants from the fittings of the Paip2-PABP interaction are listed in Table 1. The affinity of one Paip2 binding site in PABP is approximately 100-fold higher than that of the other (Kd1 = 0.66 nM; Kd2 = 74 nM). This results from the combination of a 3.6-fold difference in kass (kass1 = 1.14 × 106; kass2 = 3.1 × 105 M−1 s−1) and a 30-fold difference in kdiss (kdiss1 = 7.6 × 10−4; kdiss2 = 2.3 × 10−2 s−1). These results bolster and extend the far-Western results, which demonstrated two binding sites for Paip2 in PABP with apparent different affinities.
FIG. 3.
SPR analysis of the interaction between PABP and Paip2. Paip2 (1, 4, 16, and 64 nM; cyan, black, blue and red lines, respectively) was injected over a PABP surface (800 RU) and over a mock surface (no PABP coupled). Data were treated and integrated using a simple model (A) or models depicting a rearrangement of the protein complex (B) or the existence of two independent binding sites in PABP (C), as described in Materials and Methods. Top panels: experimental sensorgrams (dots) and the calculated fits (solid lines). Bottom panels: corresponding residuals (difference between calculated and experimental data points). Kinetic constants are listed in Table 1.
TABLE 1.
Paip2-PABP full-length kinetic and thermodynamic constants calculated by fitting the experimental data set shown in Fig. 3 to various kinetic models
| Parameter (unit) | Value with kinetic model (mean ± SD)
|
||
|---|---|---|---|
| Simple model | Rearrangement model | 2-site model | |
| kass1 (M−1 s−1) | (0.92 ± 0.02)106 | (1.03 ± 0.03)106 | (1.14 ± 0.02)106 |
| kdiss1 (s−1) | (1.43 ± 0.05)10−3 | (6.9 ± 0.6)10−3 | (7.6 ± 0.4)10−4 |
| kass2 (M−1 s−1) | NAa | (2.9 ± 0.4)10−3b | (3.1 ± 0.2)105 |
| kdiss2 (s−1) | NA | (3.3 ± 27.5)10−5 | (2.3 ± 0.1)10−2 |
| Surface activity (RU) | 223.6 ± 2.5 | 272.8 ± 3.8 | 175.5 ± 2.3 |
| Kd1 (nM)c | 1.6 ± 0.1 | 6.6 ± 0.7 | 0.66 ± 0.05 |
| Kd2 (nM)c | NA | 0.012 ± 0.009d | 74.0 ± 8.2 |
| SD of residuals (RU) | 1.43 | 1.08 | 0.668 |
| Z1 statistic | 26.7 | 24.8 | 13.9 |
| Z2 statistic | 1.2 | 0.6 | 0.4 |
NA, not applicable.
In s−1.
Kdi = kdiss i/kass i.
No unit.
The RRM1-4, RRM2-3, and GST-PABP-C2 truncation mutants interact with Paip2 with a 1:1 stoichiometry.
We next studied Paip2–PABP-RRM1-4, Paip2–PABP-RRM2-3, and Paip2–PABP-C2 interactions to further validate the two-to-one-stoichiometry model and to derive kinetic and thermodynamic values for the individual binding sites. GST-PABP-C2 was used as the ligand with Paip2 as an analyte. Paip2–PABP-RRM1-4 or Paip2–PABP-RRM2-3 interactions were studied by using Paip2 as the ligand and RRM1-4 and RRM2-3 as analytes. For binding of RRM1-4 and RRM2-3 to Paip2, the interactions were consistent with a simple one-to-one model as judged by the curve fits (Fig. 4) and the values of the residual statistics (Table 2). In the case of the GST-PABP-C2 interaction with Paip2, a satisfying fit with a simple model was not obtained. However, a model consistent with a change in the conformation of the Paip2–PABP-C2 complex provided the best fit when more complex kinetic models were applied (Fig. 5 and Table 3). The kinetic and equilibrium constants related to the fittings of the interactions of PABP truncation mutants with Paip2 are listed in Tables 2 and 3. Strikingly, the equilibrium dissociation constants and the kinetic constants for the interactions of the PABP fragments with Paip2 were remarkably similar to those calculated for the two sites in the context of the full-length PABP molecule. Namely, the Kds for RRM1-4 and RRM2-3 are 0.31 and 0.85 nM, respectively, which are comparable to the Kd1 value from the two-site model fitting of full-length PABP (0.66 nM). This results from the kinetic constants for RRM1-4 and RRM2-3 being similar to those of the first site in full-length PABP. Specifically, the kass values for RRM1-4 and RRM2-3 are 1.9 × 106 and 2.7 × 106 M−1 s−1, respectively, which are comparable to the kass1 from the two-site model fitting of full-length PABP (1.14 × 106 M−1 s−1). The kdiss values for RRM1-4 and RRM2-3 are 6 × 10−4 and 2.3 × 10−3 s−1, respectively, which are comparable to the kdiss1 values from the two-site model fitting of full-length PABP (7.6 × 10−4 s−1).
FIG. 4.
SPR analysis of the interaction between PABP RRM1-4 or RRM2-3 and Paip2. RRM1-4 (3.12, 6.25, 25, and 100 nM; cyan, red, blue and black lines, respectively) (A) or RRM2-3 (3.12, 6.25, 25, and 100 nM; green, red, blue and black lines, respectively) (B) were injected over a Paip2 surface (250 RU) and over a mock surface. Data were treated and integrated with a simple model. Top panels: experimental sensorgrams (dots) and the calculated fits (solid lines). Bottom panels: corresponding residuals. Kinetic constants are listed in Table 2.
TABLE 2.
Paip2–RRM1-4 and Paip2–RRM2-3 kinetic and thermodynamic constants calculated by fitting the experimental data set shown in Fig. 5 to a simple one-to-one model
| Parameter (unit) | Value with simple kinetic model (mean ± SD) for:
|
|
|---|---|---|
| RRM1-4 | RRM2-3 | |
| kass (M−1 s−1) | (1.9 ± 0.04)106 | (2.7 ± 0.2)106 |
| kdiss (s−1) | (6 ± 0.03)10−4 | (2.3 ± 0.1)10−3 |
| Surface activity (RU) | 9 ± 0.1 | 7 ± 0.1 |
| Kd (nM) | 0.31 ± 0.01 | 0.85 ± 0.09 |
| SD of residuals (RU) | 0.765 | 0.673 |
| Z1 statistic | 12.0 | 7.2 |
| Z2 statistic | 0.4 | 1.0 |
FIG. 5.
SPR analysis of the interaction between PABP-C2 and Paip2. Paip2 (1, 4, 16, and 64 nM; black, blue, red and cyan lines, respectively) was injected over a PABP-C2 surface (1,800 RU) and over a mock surface. Data were treated and integrated with a simple model (A) or with models depicting a conformational change (rearrangement) of the protein complex (B) or the existence of two independent binding sites in PABP-C2 (C). Top panels: experimental sensorgrams (dots) and the calculated fits (solid lines). Bottom panels: corresponding residuals. Kinetic constants are listed in Table 3.
TABLE 3.
Paip2–PABP-C2 kinetic and thermodynamic constants calculated by fitting the experimental data set shown in Fig. 6 to various kinetic models
| Parameter (unit) | Value with kinetic model (mean ± SD)
|
||
|---|---|---|---|
| Simple model | Rearrangement model | 2-site model | |
| kass1 (M−1 s−1) | (2.2 ± 0.6)104 | (1.1 ± 0.02)105 | (2.0 ± 0.1)105 |
| kdiss1 (s−1) | (1.3 ± 0.03)10−2 | (2.3 ± 0.03)10−2 | (2.5 ± 0.05)10−2 |
| kass2 (M−1 s−1) | NAa | (3.0 ± 0.04)10−3b | (2.6 ± 0.1)104 |
| kdiss2 (s−1) | NA | (2.0 ± 0.03)10−3 | (1.8 ± 0.05)10−3 |
| Surface activity (RU) | 4,162 ± 75 | 1,677 ± 67 | 810 ± 24 |
| Kd1 (nM)c | 573 ± 180 | 210 ± 9 | 125 ± 9 |
| Kd2 (nM)c | NA | 0.67 ± 0.04d | 67 ± 5 |
| Kdapp (nM) | 85 ± 8e | ||
| SD of residuals (RU) | 3.78 | 0.86 | 1.25 |
| Z1 statistic | 30.6 | 19.3 | 21.1 |
| Z2 statistic | 1.2 | 0.1 | 0.2 |
NA, not applicable.
In s−1.
Kd i = kdiss i/kass i.
No unit.
Kdapp = [KA1 × (1 + KA2)]−1 with KA1 = Kd1−1 and KA2 = Kd2−1.
Data from the PABP-C2 experiments also match data obtained with full-length PABP. Indeed, the apparent Kd from the conformational change model for GST-PABP-C2 is 85 nM, which is comparable to the Kd2 from the two-site model fitting of full-length PABP (74 nM). However, a comparison of the kinetic constants for GST-PABP-C2 versus site 2 in full-length PABP is not possible due to the difference in the models that best depict the interactions (a conformational change model versus a simple interaction in the context of two-sited full-length PABP).
The excellent agreement between the kinetic and equilibrium constants when comparing full-length and truncated PABP validates the two-site model for the full-length PABP-Paip2 interaction and, importantly, indicates that binding of Paip2 to the two sites in PABP is noncooperative. Since Paip2 was used as ligand when binding to RRM1-4 and RRM2-3 and as an analyte when binding to full-length PABP, the consistency of the results also suggests that our immobilization strategy did not alter the affinities of the PABP binding sites or introduce any bias in the kinetic analysis due to heterogeneity which might be caused by protein immobilization (22).
Two binding regions on Paip2 interact selectively with defined PABP fragments.
To determine which segments of Paip2 interact with the different PABP fragments, GST pull-down experiments were performed. GST-Paip2(wt) interacted with all the PABP fragments tested: RRM1-4, RRM2-3, and C2 (Fig. 6A to C, lanes 3), while no interaction was observed with GST alone (Fig. 6A to C, lanes 2). Furthermore, the interaction of Paip2(1–75) was strong with PABP RRM2-3 and RRM1-4 (Fig. 6A and B, lanes 4). While no interaction of Paip2(1–75) was observed with the C-terminal fragment of PABP (Fig. 6C, lane 4), this PABP fragment interacted with the C-terminal region of Paip2(76–127), which contains the second PABP binding site (Fig. 6C, lane 5). None of the various Paip2 mutants contained degradation products that comigrated with the PABP fragments, as shown when they were incubated alone with the resin (Fig. 6D, lanes 2 to 5). Taken together, these data demonstrate that the C-terminal region of Paip2 interacts with the C-terminal region of PABP and that the central glutamic acid-rich region of Paip2 interacts with the amino-terminal PABP RRMs. Furthermore, the results are also consistent with the Biacore data, which show that the interaction of Paip2 with the C-terminal portion of PABP is much weaker than that with the RRM region.
FIG. 6.
Binding of recombinant PABP fragments to Paip2 fragments. GST pull-down of PABP RRMs 1 to 4 (A), PABP RRMs 2 and 3 (B), PABP-C2 (C), or no PABP with GST-Paip2 fragments (D). Proteins (2 μg) were incubated with glutathione 4B-Sepharose (25 μl) for 1 h at 4°C and washed four times with 1 ml of buffer A. Bound proteins were eluted by boiling samples in 2× Laemmli sample buffer and resolved by SDS–12.5% or 15 to 20% polyacrylamide gel electrophoresis. The gel was stained with Coomassie R-250. Positions of molecular weight markers are shown at right.
Functional significance of the two PABP binding sites in Paip2.
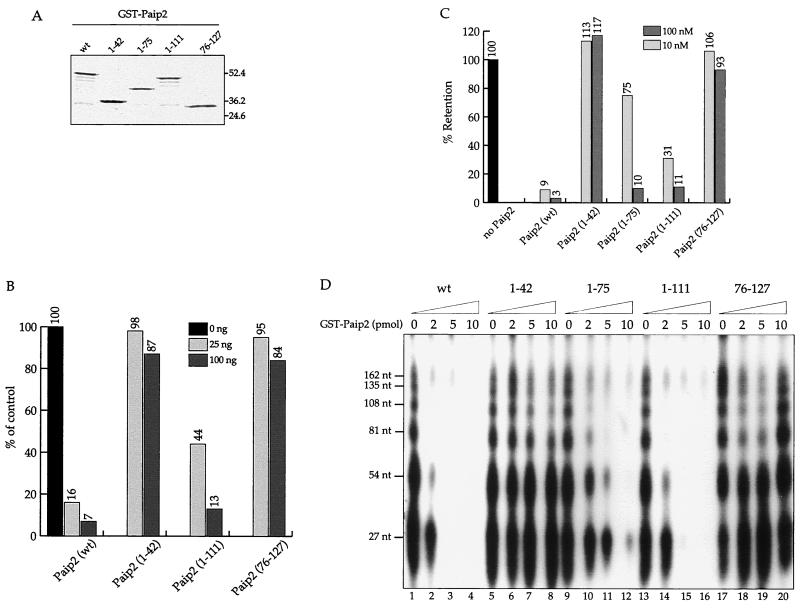
To address the biological significance of the two independent PABP binding sites in Paip2, we first examined their contribution to the inhibitory effect of Paip2 on translation. GST-tagged recombinant full-length Paip2 and three fragments of Paip2 (1–42, 1–111, and 76–127), which contain only one or neither of the two binding sites, were expressed in E. coli and purified (Fig. 7A). A Paip2 fragment, Paip2(1–111), which contains the N-terminal PABP binding domain but lacks the carboxy-terminal domain, was only two times less inhibitory for translation than full-length Paip2 (Fig. 7B). Although equal mass amounts were used in this experiment, the differences in molar amounts are not more than 20%. However, the Paip2 fragment which contains the carboxy-terminal PABP binding domain (76–127) or a fragment which contains neither of the PABP binding sites had only a marginal effect (13 to 16%) on translation even at the highest concentration used (100 ng) (Fig. 7B). It is not clear whether this effect is physiologically significant, since both fragments (76–127 and 1–42) exhibit this small effect at the highest concentration. The results can be readily explained by the 100-fold difference in affinities to PABP between the PABP-binding N-terminal and C-terminal domains of Paip2. Next, we wished to correlate the translational inhibitory activity of the Paip2 fragment with the inhibition of PABP-poly(A) binding. Two assays were employed: a filter binding assay (Fig. 7C) and a PABP-poly(A)-organizing activity assay (Fig. 7D). The latter assay is based on the finding that PABP forms a poly(A) ribonucleoprotein structure, with a repeating pattern of ∼27 nucleotides that is revealed after limited nuclease digestion (3). Consistent with the translation inhibition data, fragments that contained the N-terminal PABP binding site (1–75 and 1–111) strongly inhibited the binding of PABP to A25 RNA (Fig. 7C). Furthermore, they effectively disrupted the repeating structure of the poly(A) ribonucleoprotein (Fig. 7D, lanes 10 to 12 and 14 to 16). In sharp contrast, the Paip2 fragments which contain the C-terminal PABP binding site (76–127) or neither of the PABP binding sites (1–42) had no effect on PABP binding, as determined by the filter binding assay (Fig. 7C) or the poly(A)- organizing activity assay (Fig. 7D, lanes 6 to 8 and 18 to 20).
FIG. 7.
Functional dissection of Paip2. (A) Coomassie R-250 staining of wild-type (wt) and indicated truncated mutants of GST-Paip2. The positions of prestained molecular weight markers are also shown. (B) Effects of Paip2 mutants on translation. Krebs-2 cell-free translation reactions (12.5 μl) were programmed with 25 ng of capped poly(A)+ luciferase mRNA in the absence or presence of GST-Paip2 wild type or the indicated GST-Paip2 mutants at 30°C for 60 min, as described previously (21). Following incubation, 3-μl aliquots were assayed for luciferase activity using the luciferase assay kit (Promega) in a Lumat LB 9507 bioluminometer (EG&G Berthold). Relative luciferase activities (average of two independent determinations) are shown; the value obtained in the absence of added GST-Paip2 was set as 100%. (C) Inhibition of PABP binding to poly(A) by Paip2 mutants. Filter binding assays were performed as described in Materials and Methods. His-PABP (10 nM) and various GST-Paip2 fusion proteins (10 or 100 nM) were incubated with 32P-labeled A25 RNA. Reaction mixtures were then filtered through a nitrocellulose membrane. The radioactivity corresponding to the A25 RNA, which was retained on the membrane in the presence of PABP alone, was set at 100%. The relative levels of retention of the A25 RNA for the different GST-Paip2 proteins are shown. Each result shown is the average of results of at least two independent experiments, which did not differ by more than 10%. (D) Effect of wild-type and mutant GST-Paip2 on the poly(A)-organizing activity of PABP. The poly(A)-organizing activity of PABP was assayed in a total volume of 50 μl with radiolabeled poly(A) (0.5 × 106 cpm) and His-PABP (0.15 μg, 2.1 pmol) essentially as described previously (3, 21). GST-Paip2 wild-type or mutant proteins either were not added (lanes 1, 5, 9, 13, and 17) or were present in the reactions at 2- (lanes 2, 6, 10, 14, and 18), 5- (lanes 3, 7, 11, 15, and 19), and 10-pmol (lanes 4, 8, 12, 16, and 20) amounts. Following PABP-poly(A) complex formation, the mixtures were subjected to limited digestion with micrococcal nuclease and analyzed on a 7 M urea-containing 10% polyacrylamide gel (21).
Taken together, these data show that the strong N-terminal PABP binding site in Paip2 is responsible for the inhibition of binding of poly(A) to PABP and consequently for translational inhibition. The C-terminal PABP binding domain of Paip2, which binds to PABP with a 100-fold-weaker affinity than the N-terminal domain, failed to inhibit the interaction of PABP with poly(A) and consequently failed to inhibit translation. Its possible function is addressed in the Discussion.
DISCUSSION
Paip2 is a translational repressor both in vivo and in vitro. Paip2 inhibits translation by decreasing the affinity of PABP for poly(A) and by competing with Paip1 for PABP binding (21). In this paper, we mapped the mutual binding sites of Paip2 and PABP in each of the proteins. Far-Western analysis revealed that both proteins contain two binding sites (Fig. 1 and 2). Furthermore, GST pull-down experiments showed that the central acidic portion of Paip2 interacts strongly with PABP RRM2-3, whereas the C-terminal binding site of Paip2 exhibits a weaker interaction with the C-terminal region of PABP (Fig. 6).
To better characterize and quantitate Paip2-PABP interactions, we used an SPR-based biosensor (the Biacore) combined with numerical methods to fit the data to various kinetic models (9, 11, 27). Consistent with our far-Western and GST pull-down results, a model depicting the binding of two Paip2 molecules to two independent binding sites in full-length PABP (2:1 stoichiometry) best fits the Biacore data (Fig. 3 and Table 1). Experiments conducted on the Biacore with PABP-RRM1-4, PABP-RRM2-3, and PABP-C2 fragments supported the proposed model for the Paip2-PABP interaction, since for each PABP fragment, which should contain only one of the two binding sites, the interactions were fitted by models describing a 1:1 stoichiometry (Fig. 4 and 5). Moreover, the apparent equilibrium dissociation constants (Kds) calculated for the interactions of the PABP fragments with Paip2 (Tables 2 and 3) were strikingly similar to those calculated for the two Paip2 binding sites in the context of the full-length PABP molecule (Table 1). Not only does this validate the 2:1 stoichiometry model for the Paip2-PABP interaction, but it also indicates that the structures of the two binding sites must be maintained in the isolated fragments and that the two sites are noncooperative in full-length PABP. A comparison of the equilibrium constants indicates that the RRM and C2 binding sites correspond to the high- and low-affinity sites within PABP, respectively. The 2:1 stoichiometry of the Paip2-PABP interaction will have to be confirmed via analytical ultracentrifugation, isothermal calorimetry, or any alternate means of evaluating stoichiometry.
Far-Western analysis showed that RRM2-3 bound Paip2 almost as well as RRM1-4. The Biacore data support these results. For both RRM2-3 and RRM1-4, the interaction was well fitted with a simple model (Fig. 4) and the equilibrium constants were in the same range. A comparison of their kinetic constants (Table 2) reveals a significant difference in that the truncation from RRM1-4 to RRM2-3 caused a fourfold increase in the dissociation rate (from 6 × 10−4 to 23 × 10−4 s−1), suggesting that RRMs 1 and 4 stabilize the interaction between Paip2 and PABP.
A single inconsistency in model fitting exists when comparing full-length PABP to PABP fragments for the Paip2–PABP-C2 interaction. The best fit for the Paip2–PABP-C2 interaction was observed with a model depicting a rearrangement, i.e., a change in conformation (Fig. 5), while for full-length PABP, the binding of each of the two sites was depicted as a simple interaction, albeit within a two-to-one stoichiometry model. This difference in the best-fitting kinetic model for the PABP-C2 binding site may result from limitations in detecting a more complex binding mechanism using model-fitting Biacore data, i.e., a conformational change within the PABP-C2 site may be undetectable when superimposed on a 2:1 stoichiometry model. In any case, the conformational change model depicts the stoichiometry of the Paip2–PABP-C2 interaction as one-to-one, as expected, and the apparent Kds from PABP-C2 and the low-affinity site of full-length PABP are almost identical (Tables 1 and 3). Interestingly, recent nuclear magnetic resonance studies show a dramatic shift in a number of amino acids in the C terminus of PABP upon binding to Paip2, confirming the possibility of a change in conformation at this site (23).
The results of the Paip2-PABP binding study explain the functional properties of Paip2. Paip2 effectively inhibits translation both in vitro and in vivo, by competing with poly(A) and Paip1 for PABP binding (21). Remarkably, all the inhibitory biochemical activities [PABP-poly(A) RNA interaction and translation] of Paip2 are affected by its central PABP binding domain and not by its C-terminal site. This is consistent with the affinity of this domain for PABP binding being much stronger (100-fold) than that of the C-terminal site. Binding of Paip2 to RRMs 2 and 3 of PABP could be responsible for affecting PABP's affinity for poly(A) via direct steric hindrance. Thus, the distinct modes of binding of Paip2 to the RRM and C-terminal regions of PABP may selectively disrupt various PABP activities. What then is the function of Paip2 binding to the C-terminal region of PABP? This domain remains largely uncharacterized. A BLAST sequence similarity search using the C-terminal PABP binding site of Paip2 resulted in a large number of proteins with significant identity (10, 23), suggesting that this region of Paip2 may represent a general PABP binding motif. Whether these proteins with sequence similarity are physiological binding partners of PABP remains to be determined, but competition between Paip2 and other PABP-interacting partners containing this motif is an attractive possibility. One of these proteins is the termination factor eRF3, which has been shown biochemically to interact with PABP (16a). Paip1 also contains this motif and binds to the same two regions in PABP as Paip2 (G. Roy and A. Kahvejian, unpublished observations), indicating that the competition between Paip1 and Paip2 is direct.
In conclusion, we have shown that PABP possesses two distinct Paip2 binding sites, one located within the RRM 2 and 3 regions and the other in the C-terminal domain. Moreover, we demonstrated that Paip2 possesses two PABP binding regions, one within the amino-terminal glutamic acid-rich domain, which binds to the amino-terminal PABP region, and the other within the C-terminal region, which binds to the C-terminal region of PABP. Only one of these interactions, between the N-terminal fragments of PABP and Paip2, is important for the inhibitory activities of Paip2. The newly described complex interactions between PABP and its associated proteins are consistent with the many regulatory roles that PABP plays in the control of gene expression.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work, and their order is arbitrary.
We thank S. Grothe and C. Lister for excellent technical assistance, S. Pyronnet for critical review of the manuscript, and R. C. Deo and S. K. Burley for helpful discussions. This research was supported by grants from the National Institute of Canada and the Howard Hughes Medical Institute International Scholar Program to N.S. N.S. is a Canadian Institute of Health Research Distinguished Scientist and a Howard Hughes Medical Institute International Scholar. K.K., A.K., and G.R. were recipients of doctoral studentships from the Medical Research Council of Canada. G.D.C. is supported by the Protein Engineering Network of Centers of Excellence (PENCE).
REFERENCES
- 1.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina E, Neumann M, Pavlakis G N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 3.Baer B W, Kornberg R D. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer B W, Kornberg R D. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 6.Bradley J V. Distribution-free statistical tests. Englewood Cliffs, N.J: Prentice-Hall Inc.; 1968. [Google Scholar]
- 7.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 8.Craig A W, Svitkin Y V, Lee H S, Belsham G J, Sonenberg N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. doi: 10.1128/mcb.17.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Crescenzo G, Grothe S, Lortie R, Debanne M T, O'Connor-McCourt M. Real-time kinetic studies on the interaction of transforming growth factor alpha with the epidermal growth factor receptor extracellular domain reveal a conformational change model. Biochemistry. 2000;39:9466–9476. doi: 10.1021/bi992987r. [DOI] [PubMed] [Google Scholar]
- 10.Deo R C, Sonenberg N, Burley S K. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4414–4419. doi: 10.1073/pnas.071552198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher R J, Fivash M, Casas-Finet J, Erickson J W, Kondoh A, Bladen S V, Fisher C, Watson D K, Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994;3:257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fivash M, Towler E M, Fisher R J. BIAcore for macromolecular interaction. Curr Opin Biotechnol. 1998;9:97–101. doi: 10.1016/s0958-1669(98)80091-8. [DOI] [PubMed] [Google Scholar]
- 13.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 14.Gingras A C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 15.Glaser R W. Antigen-antibody binding and mass transport by convection and diffusion to a surface: a two-dimensional computer model of binding and dissociation kinetics. Anal Biochem. 1993;213:152–161. doi: 10.1006/abio.1993.1399. [DOI] [PubMed] [Google Scholar]
- 16.Hershey J W B, Merrick W C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- 16a.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 17.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson A. Poly(A) metabolism and translation. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 20.Jensen T H, Jensen A, Kjems J. Tools for the production and purification of full-length, N- or C-terminal 32P-labeled protein, applied to HIV-1 Gag and Rev. Gene. 1995;162:235–237. doi: 10.1016/0378-1119(95)00328-4. [DOI] [PubMed] [Google Scholar]
- 21.Khaleghpour K, Svitkin Y V, Craig A W, DeMaria C T, Deo R C, Burley S K, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein. Mol Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 22.Kortt A A, Oddie G W, Iliades P, Gruen L C, Hudson P J. Nonspecific amine immobilization of ligand can be a potential source of error in BIAcore binding experiments and may reduce binding affinities. Anal Biochem. 1997;253:103–111. doi: 10.1006/abio.1997.2333. [DOI] [PubMed] [Google Scholar]
- 23.Kozlov G, Trempe J F, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 25.Le H, Tanguay R L, Balasta M L, Wei C C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 26.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton T A, Myszka D G, Chaiken I M. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 28.Myszka D G, He X, Dembo M, Morton T A, Goldstein B. Extending the range of rate constants available from BIACORE: interpreting mass transport-influenced binding data. Biophys J. 1998;75:583–594. doi: 10.1016/S0006-3495(98)77549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myszka D G, Morton T A, Doyle M L, Chaiken I M. Kinetic analysis of a protein antigen-antibody interaction limited by mass transport on an optical biosensor. Biophys Chem. 1997;64:127–137. doi: 10.1016/s0301-4622(96)02230-2. [DOI] [PubMed] [Google Scholar]
- 30.O'Connor-McCourt M, De Crescenzo G, Lortie R, Lenferink A, Grothe S. The analysis of surface plasmon resonance-based biosensor data using numerical integration. In: Lundahl P, Lundqvist A, Greijer E, editors. Quantitative analysis of biospecific interactions. Chur, Switzerland: Harwood Academic Press; 1998. pp. 176–190. [Google Scholar]
- 31.O'Shannessy D J, Winzor D J. Interpretation of deviations from pseudo-first-order kinetic behavior in the characterization of ligand binding by biosensor technology. Anal Biochem. 1996;236:275–283. doi: 10.1006/abio.1996.0167. [DOI] [PubMed] [Google Scholar]
- 32.Piron M, Delaunay T, Grosclaude J, Poncet D. Identification of the RNA-binding, dimerization, and eIF4.GI-binding domains of rotavirus nonstructural protein NSP3. J Virol. 1999;73:5411–5421. doi: 10.1128/jvi.73.7.5411-5421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preiss T, Hentze M W. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 34.Rogers G W J, Richter N J, Merrick W C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 35.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs A B, Bond M W, Kornberg R D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 37.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 39.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–270. [Google Scholar]
- 40.Tarun S Z J, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 41.Tarun S Z J, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 42.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 43.Yarmush M L, Patankar D B, Yarmush D M. An analysis of transport resistances in the operation of BIAcore; implications for kinetic studies of biospecific interactions. Mol Immunol. 1996;33:1203–1214. doi: 10.1016/s0161-5890(96)00075-2. [DOI] [PubMed] [Google Scholar]