Abstract
RNA-binding proteins (RBPs) play an important role in RNA metabolism, regulating the stability, localization, and functional dynamics of RNAs. Alternation in the RBP-RNA network has profound implications in cellular physiology, and is related to the development and spread of cancer in certain cases. To regulate the expression of specific genes and their biological activities, various strategies have been applied to target RBPs for cancer treatments, including small-molecule inhibitors, small-interfering RNA, peptides, and aptamers. Recently, the deployment of the CRISPR-Cas9 technology has provided a new platform for RBP screening and regulation. This review summarizes the delivery systems of the CRISPR-Cas9 system and their role in RBP-based cancer therapeutics, including identification of novel RBPs and regulation of cancer-associated RBPs. The efficient delivery of the CRISPR-Cas9 system is important to the profound understanding and clinical transition of RBPs as cancer therapeutic targets.
Keywords: RNA-binding protein, CRISPR-Cas9, cancer, delivery strategy, RNA, CRISPR screening, RBP editing
Graphic abstract
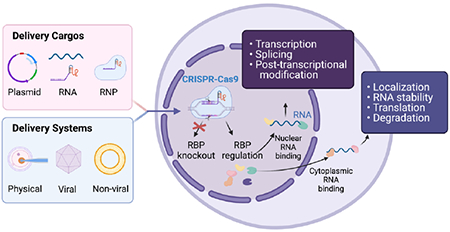
Delivery of the CRISPR-Cas9 system for RBP editing and RNA regulation.
1. Introduction
RNA binding proteins (RBPs) control intrinsic networks of RNA metabolism, modulating various post-transcriptional RNA processes, including splicing, polyadenylation, transport, translation, and stability [1]. The modular structure and multiple repeats of RNA-binding domains (RBDs) of RBP allow them to coordinately bind to coding or non-coding RNA with different sequence specificities and affinities, thereby forming a functional ribonucleoprotein (RNP) complex [2]. Among over 1,500 validated human RBPs, about 600 structurally distinct RBDs were identified [3]. The classical RBDs include RNA recognition motif (RRM), double-stranded RNA-binding domain (dsRBD), K-homology domain (KH), zinc-finger domain (ZnF), Piwi domain and others [2]. The RBDs are arranged in a variety of ways to enable RNA binding specificity and regulate expansive biological activities. For example, the arrangement of various RBD modules creates different macromolecular binding surfaces to define the RNA-binding specificity. In addition, the length of the linker between two RBDs affects the applicable RNA range. Longer linkers allow the two domains to recognize various target sets while short linkers recognize contiguous nucleic acids sequences [2].
Given the importance of RBP functions in RNA processing and the subsequent cellular activities, it is not surprising that RBP malfunction may cause carcinogenesis through chromosomal rearrangements, mutations, and gene amplification [4–7]. Although the changes in RBP mRNA levels might be small across multiple cancer cell lines, these changes may result in large-scale disruption in the downstream regulatory networks [8]. Therefore, tampering RBP functions holds great potential in cancer therapeutics as RBPs coordinate complex RNA-protein and protein-protein interactions. Various strategies have been applied to target RBP for cancer therapeutics, including small-molecule inhibitors, peptides, aptamers, antisense oligonucleotides, and siRNA [9]. In the last decade, the breakthrough of the RNA-guided DNA endonuclease CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated proteins) system has led to dramatic advances in the biomedical field, which provides a unique strategy to modulate RBP activities and related RNA expressions [10]. Originated in prokaryotes as a protective mechanism against viral infections, the programmable CRISPR-Streptococcus pyogenes (SpCas9) is extensively utilized across multiple disciplines [11,12]. The single-guide RNA (sgRNA) guides the Cas9 endonuclease to the desired site to create a double-stranded break (DSB), which is then repaired by non-homologous end-joining (NHEJ) or homology-directed repair (HDR) [13]. The flexible design of sgRNAs greatly simplifies the process of knocking out or correcting mutations in the targeted RBPs for cancer treatment (Fig. 1).
Fig. 1.
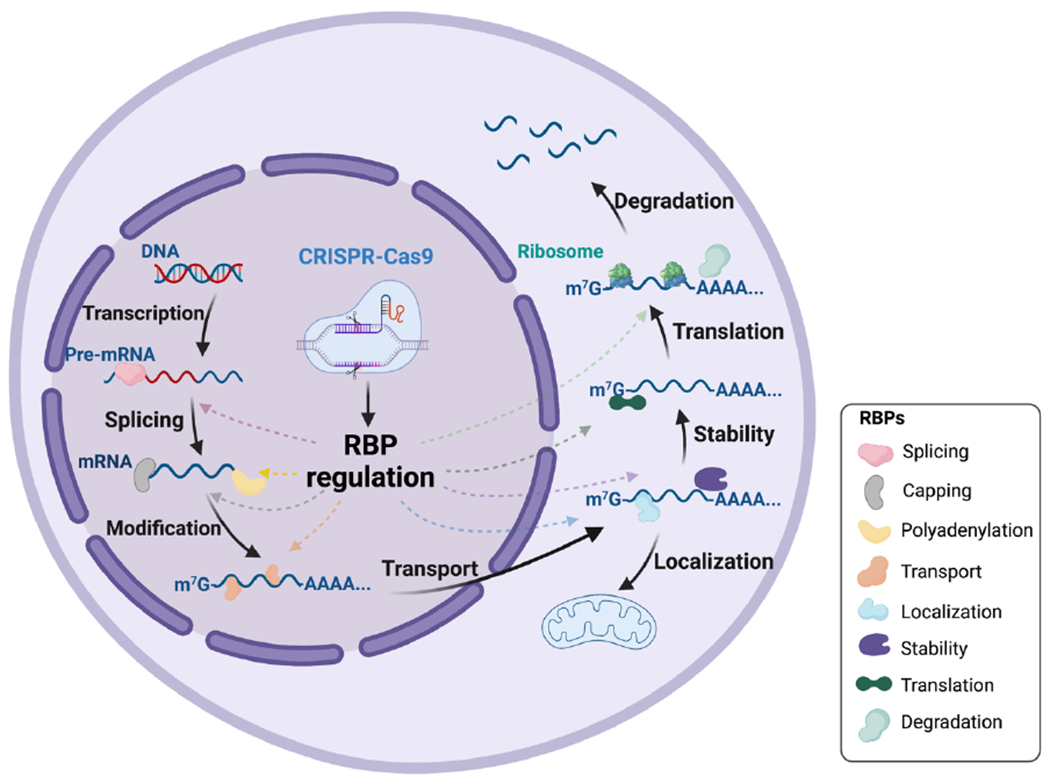
CRISPR-Cas9 system for RBP editing and RNA regulation. Directed by the sgRNA, CRISPR-Cas9 endonuclease could either knockout or edit the targeted RBP. RBP regulation post CRISPR delivery can affect several post-transcriptional RNA processes that occur in the nucleus (splicing, capping, polyadenylation and other post-transcriptional modifications) and cytoplasm (transport, localization, stability, translation, and degradation). Figure created using BioRender.com.
The CRISPR system has been applied in a variety of ways to explore RBP-based cancer therapies. First, CRISPR facilitates the identification of new RBP targets for cancer treatment. For example, CRISPR-Cas9 mediated genome knockout validates functional studies of RBPs through loss-of-function screens, and further cross-linking immunoprecipitation (CLIP) and RNA-sequencing can be used to recognize upregulated and downregulated genes following RBP knockdown [14]. Second, CRISPR can be used to reverse the cancer-causing situations by targeting cancer-associated RBPs, including knockout of oncogenic RBPs, upregulation of tumor-suppressive RBPs, and correction of tumor-specific RBP mutations [9]. However, several challenges remain for the delivery of CRISPR in diseased cells for RBP regulation. One challenge is the selection of delivery platforms and CRISPR components according to their physiological properties and biological functions, as the CRISPR system necessitates all parts of the system to be delivered into the same cell for effect. Furthermore, the selective targeting of tumor cells and identification of key RBPs are essential for the CRISPR system to exert its anti-tumor functions. This article overviews recent advances in the delivery and application of CRISPR-Cas9 in RBP-based cancer therapeutics.
2. Delivery platforms of the CRISPR-Cas9 system
CRISPR-Cas9 system has been applied for genome editing in different forms [15,16]. The CRISPR system has been delivered in three forms of cargoes: DNA plasmids that encode both Cas9 protein and the sgRNA, mRNAs that encode Cas9 protein and separate sgRNAs, and ribonucleoprotein (RNP) complexes that contain Cas9 protein and sgRNA [17,18]. Different components of the CRISPR system can be relatively easily delivered into cellular models. However, CRISPR components must be protected from multiple degradation mechanisms while circulating in vivo. Therefore, the selection and optimization of a CRISPR delivery system can greatly affect gene editing efficiency. Currently, several clinical trials utilize CRISPR-mediated genome editing for various therapeutic areas. Table 1 highlights CRISPR-mediated cancer therapeutics in clinical trials. CRISPR delivery can be broadly grouped into three categories: physical, viral, and non-viral delivery. There are advantages and disadvantages to each delivery method, which are described in this section.
Table 1.
Current Ex vivo CRISPR-mediated cancer therapeutics in clinical trials
| Phase | Cancer Type | Delivery Mode | Editing Target | ID |
|---|---|---|---|---|
| I/II | B-cell leukemia | Electroporation of CRISPR sgRNA to disrupt endogenous TCR and B2M. | Allogeneic CD19-CAR modified T cells with βTCRα, TCRβ, β-2 microglobulin (B2M) knockout. | NCT03166878 |
| I/II | B cell leukemia, B cell lymphoma | Undefined | Allogeneic CD19 and CD20 or CD22 CAR-T cells. | NCT03398967 |
| I | Metastatic non-small cell lung cancer | Electroporation of Cas9 and sgRNA plasmids to target exon 2 of PD-1. | Modified autologous T cells with programmed cell death protein 1 (PD-1) knockout. | NCT02793856 |
| I | Esophageal cancer | Electroporation of CRISPR/Cas9 DNA to disrupt PD-1 | PD-1 knockout engineered T cells. | NCT03081715 |
| I | Advanced refractory cancer | Electroporation of Cas9 RNP with three sgRNA for gene editing of TRAC, TRBC, and PDCD1. | Knockdown of endogenous T cell receptor (TCR) and PD-1 and knock-in of cancer-specific TCR transgene in autologous isolated T-cells. | NCT03399448 |
| I | Solid tumors | Electroporation of Cas9 RNP with sgRNAs targeting PD-1 and TCR. | PD-1 and TCR gene knockout CAR-T Cells. | NCT03545815 |
| I | B-cell lymphoma, non-Hodgkin lymphoma | Undefined | Allogeneic CD19-directed CAR T cell immunotherapy (CTX110). | NCT04035434 |
| I | Relapsed or refractory acute leukemia lymphocytic | Electroporation of CRISPR sgRNA to disrupt endogenous HPK1 in Autologous T cells. | Autologous T Cells with endogenous hematopoietic progenitor kinase 1 (HPK1) knockout. | NCT04037566 |
| I | Multiple myeloma | Undefined | Anti-BCMA allogeneic engineered T cells (CTX120). | NCT04244656 |
| I | Gastro-intestinal (GI) cancer | Undefined | CISH (Cytokine-induced SH2 protein) modified tumor-infiltrating lymphocytes (TIL). | NCT04426669 |
| I | Relapsed or refractory T or B Cell lymphoma, renal cell carcinoma | Undefined | Anti-CD70 allogeneic engineered T cells (CTX130). |
NCT04438083
NCT04502446 |
| I | B acute lymphoblastic leukemia | Lentiviral vector CRISPR guides for genome editing of CD52 and TRAC and transient Cas9 protein. | Allogeneic engineered human T cells to disrupt CD52 and T cell receptor α (TRAC.) | NCT04557436 |
| I | Relapsed or refractory B cell non-Hodgkin lymphoma, various lymphoma | AAV vector with Cas9 chRDNA (CRISPR hybrid RNA-DNA) technology to insert CD19-specific CAR into T cell, knockout TRAC and PD-1. | Allogeneic anti-CD19 CAR-T cells (CB-010). | NCT04637763 |
2.1. Physical Delivery
Physical delivery methods utilize external forces to disrupt the cellular membrane and deliver the cargo into the cells. These methods include microinjection, electroporation, microfluidic constriction, and hydrodynamic delivery [19]. Although the physical delivery of the CRISPR system is currently deemed too invasive for clinical applications in humans, advances have been made towards the in vivo delivery in mice [20,21]. Due to the high delivery efficiency even in difficult-to-transfect cells, physical delivery is commonly used for initiating CRISPR delivery for ex vivo applications.
2.1.1. Microinjection
Microinjection is a commonly used type of mechanical delivery, which utilizes mechanical force to pierce the cell membrane and deposit the cargo into the cell. The advantage of microinjection is its high efficiency, specificity, and controlled dosage of all components. However, the microinjection process is laborious, technically challenging, and limited to a single injection per cell [22]. Microinjection is commonly used for single-cell applications and has been used to create various transgenic animals [23]. CRISPR-Cas9 system was utilized to generate multiple gene mutant mice using single microinjection in zygote by co-injecting Cas9 mRNA with multiple sgRNAs targeting different genes which resulted in a biallelic mutation at 80% efficiency [22]. Additionally, microinjection of the CRISPR-Cas9 system has been utilized in other mammalian animals as well, including sheep and cynomolgus monkeys [24,25]. Microinjection of CRISPR components is a viable and standardized method for genome editing in various animal models, but is limited to single-cell applications.
2.1.2. Electroporation
Electroporation uses high voltage electrical current pulses to temporarily permeabilize the cellular membrane, allowing larger components, such as nucleic acids, to enter the cell [26]. Since the electric pulse can be applied to multiple cells simultaneously, electroporation foregoes the need to individually inject single cells as in microinjection and is a useful technique for generating a population of mutant mammalian cells. A side-by-side comparison of mutagenesis between microinjection and electroporation found simple knock-in allele and indel mutagenesis was similar between the two methods [27]. However, a comparison between the two methods of manipulation found that the electroporation method was statistically more efficient at creating knock-in alleles and embryos show higher rates of survival as compared to microinjection methods [27].
Electroporation is well suited for in vitro or ex vivo applications because the required large voltage is often not suitable for in vivo applications. However, there have been some reported instances of electroporation to deliver CRISPR components in vivo in mice utilizing in utero electroporation of the developing brain [28,29]. Organ-specific gene knockout in the brain can be possible by delivering the CRISPR-Cas9 system with electroporation in vivo. In another study, a single plasmid encoding for CRISPR-Cas9/sgRNA targeting the human Rhodopsin (RHO) gene was injected via electroporation to the subretinal area of RHO mutant mice which significantly reduced mutant RHO protein expression [20]. By utilizing the specificity of the CRISPR-Cas9 system, specific locus within the genome can be manipulated in vitro to study and identify cancer-related human chromosomal translocations and the role of specific genes on cell proliferation [30,31].
Nucleofection is a type of specialized electroporation which is designed to deliver cargoes directly to the nuclei of cells. The nucleofection method does not rely on nucleus disassociation, usually during the cell division cycle, for the nucleic acid to enter the nucleus. Therefore, this method of delivery is especially useful for delivering DNA cargo for non-dividing cells, such as neurons [32]. DNA plasmids encoding CRISPR-Cas9/sgPD-1 and CD133-CAR piggyBac transposon were co-nucleofected into primary T cells, which generated CD133-specific chimeric antigen receptor (CAR) T cells with PD-1 deficiency [33]. Compared to non-transfected T cells or traditional CAR T cells, injection of these engineered CAR T cells to the tumor sites showed improved survival in mouse orthotopic glioma model. The improved efficacy of engineered CAR T cells for cancer immunotherapy shows how the nucleofection-delivered CRISPR system can be utilized as an ex vivo therapeutic option for cancer treatments [33]. In a report, allogeneic CD34+ hematopoietic stem and progenitor cells (HSPCs) were nucleofected with a CRISPR-Cas9 RNP system to create a mutant CCR5 gene [34]. These engineered HSPCs were transplanted into a patient who was diagnosed with both HIV-1 infection and acute lymphoblastic leukemia. Although engraftment of CRISPR edited HSPCs did not cure HIV-1 infection at 19 months, the patient showed up to 8.28% of bone marrow cells that displayed CCR5 disruption, and no associated off-target side effects which exhibited that allogeneic CRISPR modified cell transplantation can be a valuable therapeutic vector. Cas9 RNP nucleofection avoids the integration of exogenous DNA and reduces the possibility of off-target editing because of the transient nature of RNPs. Because nucleofection is highly specific and efficacious at delivering large cargoes such as RNPs, it can be a useful tool for engineering specific cells for ex vivo applications.
2.1.3. Microfluidic Constriction/Microfluidic Squeezing
Microfluidic constriction or microfluidic squeezing is a process of passing cells through a microtechnology-enabled device to perforate the cell membrane temporarily. Through the transiently disrupted cellular membrane, various macromolecule cargoes such as DNA, RNA, protein, or RNP can be passively diffused into the cytoplasm without the need for other forms of delivery vectors [35]. Han et al. optimized the microfluidic constriction device concerning fluid flow rates, cell constriction pore size and shape, and duration of the cell passage through the constriction pore. Taken together, the optimized microfluidic microchip device showed increased delivery of CRISPR/Cas9 encoding DNA plasmids across various cell types while simultaneously enhancing the survivability of these cells [36]. Microfluidics constriction has also been shown to efficiently deliver CRISPR RNP complexes to primary CD4+ T cells [37]. In addition, a comparison between electroporation and microfluidic squeezing methods showed that cells manipulated through microfluidic squeezing displayed less up-regulated cytokine secretion when adoptively transferred into mice models in vivo [38]. Taken together, the microfluidic constricted method of delivery of the CRISPR system can be an appealing delivery strategy for in vitro and ex vivo applications.
2.1.4. Hydrodynamic injection
Unlike previously discussed methods of physical delivery which were more applicable for in vitro or ex vivo delivery, hydrodynamic injection is an in vivo physical delivery technique. By injecting a relatively large amount of fluid carrying genetic materials into the bloodstream, the sudden increase in pressure will temporarily increase the permeability of capillary endothelium and parenchymal cells to promote the passage of large molecules into the cytoplasm [39]. A highly practiced example of hydrodynamic injection is gene delivery to hepatocytes through tail vein injection in rodents. The rapid increase in blood pressure through intravenous injection causes a sharp increase in pressure in the liver sinusoids, which temporarily increases the permeability of hepatocytes to uptake large cargoes. Typically, physiological functions of hydrodynamically treated mice return to baseline levels within 72 hours post-injection, which suggests that hydrodynamic injection is a viable method of gene delivery in vivo [40]. Different tissues have been specifically targeted by modifying injection sites which also demonstrates the safety and applicability of hydrodynamic injection for clinical applications [39]. Hydrodynamic injection of CRISPR plasmid DNA that encodes for Cas9 and sgRNA that targets genes Pten and p53 caused direct mutation of both genes in the liver of mice in vivo which subsequently resulted in the generation of liver tumors [41]. An advantage of hydrodynamic injection is that a naked nucleic acid cargo can be effectively delivered, reducing potential adverse side effects related to delivery vectors [17].
2.2. Viral Vectors
Viral vectors can efficiently infect a wide variety of mammalian cells, which is an advantageous feature for delivering CRISPR components. However, because the immune system has been designed to identify and fight off any potential viral pathogens, decreased efficiency of delivery can occur due to host immune system-mediated response. The viral vectors do not cause serious illnesses in humans, and most often, are engineered to reduce host immune responses but can still trigger adverse immune responses [42]. Another advantage of viral vectors is the ability to design the vector to incorporate the CRISPR DNA into the host genome for stable expression. However, this feature may cause adverse side effects, such as off-target effects and insertional mutagenesis [43]. Therefore, careful administration in response to potential side effects should be considered before utilizing viral vector-mediated delivery. Currently, recombinant Adeno-associated Virus (AAV) is the leading delivery system for CRISPR delivery in vivo [44]. Other forms of viral vectors include adenoviral vectors (AdVs) and lentiviral vectors (LV) and are also discussed in this section.
2.2.1. Adeno-associated Virus (AAV) vectors
Adeno-associated Viruses (AAV) are a member of the Parvoviridae family and are commonly found in humans. The virus is small, measuring ~26 nm in diameter and the genome is a single-stranded DNA around 4.7 kb in length. Because of the small physical size and the genome length, AAV can only carry smaller genetic cargo, limited to less than 5 kb of genomic material [44]. AAVs’ non-pathogenic nature, high transfection efficiency, low immunogenicity and cytotoxicity make them one of the leading delivery systems for in vivo studies[45]. However, AAV can still trigger host immune response which can lower the efficacy and increase related toxicity. The capsid protein coating of AAV can be engineered to create new AAV vectors for higher transfection efficiency and confer transgene expression at lower doses [46]. In addition, the capsid protein can be optimized to deliver AAV to specific cells or tissues and to reduce binding affinity to AAV antibodies to reduce host immune response [47,48]. The flexibility and diversity of AAV capsid structure is a key advantage to AAV-mediated CRISPR delivery in vivo.
To bypass the limitations in cargo size, a different version of the Cas9 protein from S. aureus rather than Streptococcus pyrogenes can be used. This Cas9 protein (SaCas9) is equally potent, yet smaller in size, which reduces the overall length of the cargo and creates the possibility to include other tags or markers within one AAV particle [49]. Another strategy to utilize AAV for CRISPR delivery despite its size limitation is to employ different populations of AAVs that deliver separate CRISPR system components. Although this method is more complicated because both components of the CRISPR system must be transduced into the same cell for efficacy, this methodology has been utilized in various ways. Separate AAVs encapsulating DNA coding for SaCas9 and sgRNA targeting MeCP2 knocked down MeCP2 protein in the mouse brain in vivo, which resulted in approximately 80% co-transduction efficiency and reduction of MeCP2 protein levels by 60% [50]. In another study, Yang et al. delivered two separate AAVs, one containing DNA plasmid encoding for SaCas9 and the other expressing sgRNA and donor DNA into newborn mice with partial deficiency to ornithine transcarbamylase to correct disease phenotype in treated mice through HDR-mediated repairs [51]. In addition, an intein-mediated split-Cas9 system, which consists of two separate AAV vectors carrying Cas9 C-terminal and Cas9 N-terminal respectively, showed comparable nuclease activity as native Cas9 [52].
AAV-mediated CRISPR delivery is a valuable tool for exploring the complex cancer genetic network in vivo. Platt et al. developed knock-in mice models to constitutively express Credependent Cas9 and injected with AAV encoding for a single sgRNA to generate mutations in the Kras gene while knocking out both p53 and Lkb1 genes [53]. Mice injected with the AAV-sgRNA encoding for the three genes developed lung tumor, which showcased how the CRISPR genome editing tool can be applied to study the role of multiple genes simultaneously in the complex disease of cancer in vivo [53]. One group reported utilizing AAV-mediated in vivo CRISPR screen to identify functional suppressors in glioblastoma (GBM) by generating a sgRNA library of a mouse-homolog tumor suppressor gene (mTSG) library [54]. Four months post-injection, half of the mice injected with AAV-mTSG library displayed brain tumors, as compared to empty AAV-vector or PBS injected mice. Combining deep targeted-captured sequencing, the drivers for GBM generation could be identified in an endemic mouse model, which is a valuable methodology to analyze cancer genetics in vivo directly [54].
2.2.3. Adenovirus Vectors (AdVs)
Adenovirus (AdV) is a commonly occurring virus that causes mild illnesses in humans. It measures between 80-100 nm in diameter and its double-stranded DNA genome is around 40 kb long, which can package around 8 kb of foreign DNA [55]. Due to its capacity to carry large genetic cargo, AdV vector-mediated delivery can be optimized by including a nuclear localization signal to the CRISPR-Cas9 components [56]. Advances in AdV engineering have created AdV vectors that lack viral genome which allows for up to 37 kb of target DNA delivery [57]. Like AAVs, it can infect many mammalian cells including both dividing and non-dividing cells. An advantage of AdVs is that their genome does not integrate into the host cells, which reduces off-target effects and insertional mutagenesis [58]. Nonetheless, because AdVs can be pathogenic, the introduction of AdV vectors can elicit innate immune responses [59]. An emphasis on increasing the safety profile of AdV vectors in relation to host immune response will greatly improve the therapeutic application of AdV vectors for CRISPR-mediated treatments. Therefore, AdV vectors have been optimized and engineered through various methods, including copolymer encapsulation of AdV vectors and utilization of non-human AdV vectors to limit cross-reactive immunity [60,61].
Ehrke-Schulz et al. utilized high-capacity adenoviral vectors (HCAdVs) which lack viral coding regions to deliver a complete CRISPR/Cas9 system with sgRNA to disrupt HPV16 or HPV18 oncogene E6. In HPV-positive cervical cancer cell lines, transduction of CRISPR-HCAdVs exhibited signs of cell apoptosis [62]. In a separate study, separate AdV vectors carrying DNA encoding for Cas9 or sgRNA targeting the oncogenic mutation-specific EGFR triggered accurate interruption at the oncogenic mutation and diminished tumor volume in the xenograft mouse model of human lung cancer [63]. Furthermore, recombinant AdVs have been reported to efficiently transduce over 90% of hepatocytes in vivo [64]. AdVs encapsulating plasmid encoding for Cas9 with sgRNA targeting Pten were injected into mice. Although AdV vector-associated immunotoxicity and Cas9 specific cellular immune response were found in the liver, the treated mice showed Nonalcoholic Steatohepatitis (NASH) like phenotype consistent with a Pten mutation [65].
2.2.4. Lentivirus Vectors (LV)
Lentiviruses, one type of retroviruses, are ~100 nm in diameter, which are capable of packaging around 8kb of genetic material [66]. Similar to AAVs and AdVs, LVs are capable of infecting a wide variety of mammalian cells. Because retroviruses integrate into the host genome, LV can be disadvantageous for delivering CRISPR/Cas systems [67]. To circumvent gene integration, non-integrating LV vectors have been engineered [68]. A non-integrating LV vector was used to establish an immunocompetent metastatic renal cell carcinoma in mice which is useful for creating therapeutic gene editing models in vivo [69]. However, the integrative nature of LV can be harnessed for creating gene libraries for genetic screenings [70]. For example, Chen et al. generated more than 67,000 sgRNA to screen genes that play a role in lung metastasis in mice in vivo. Mouse non-small-cell lung cancer (NSCLC) line was transduced with a lentivirus expressing Cas9 fused to GFP to generate a transduced cell line that constitutively expresses Cas9-GFP. The genome-wide mouse sgRNA library was then transduced to the Cas9-GFP expressing cells. The transduced cells were grown in vitro then transplanted into immunocompromised mice which identified loss-of-function mutations that promote cancer metastasis. This study exhibits CRISPR/Cas9 mediated in vivo genome screening of genes in cancer proliferation [71].
Due to its high infectivity and ability to transduce non-dividing cells, such as dendritic cells, lentivirus vectors have been used to generate engineered CAR-T cells for acute lymphoblastic leukemia (ALL) treatment [72]. Lentiviral vectors are currently utilized in numerous clinical trials for ex vivo CRISPR mediated therapeutics [73].
2.3. Non-viral delivery
An emerging field in the delivery of CRISPR systems is non-viral forms of delivery. This term broadly encompasses all forms of organic and inorganic delivery methods including lipid and lipid-derived nanoparticles, polymer-based particles, cell-penetrating peptides, nucleic acids nanoparticles, and inorganic nanoparticles [17]. Nonviral vectors offer attractive features such as low immunogenicity, flexibility in cargo size and complexity, and large-scale production capacity [74]. Recent advances in material science and novel biomaterial engineering have created biocompatible compounds that can efficiently and safely deliver CRISPR components in vivo. Additionally, chemical modifications of these synthetic vectors display enhanced delivery efficiency, targeting specificity, and reduction of adverse immune responses. Modifications to the CRISPR components by intracellular trafficking modules guide the components to the desired intracellular locations. Selected non-viral systems mediated CRISPR delivery in vivo are described in this section.
2.3.1. Lipid and lipid-derived nanoparticles
Lipid and lipid-derived nanoparticles (LNPs) are frequently used for CRISPR system delivery [18]. LNPs are amphiphilic systems that are composed of multiple different hydrophobic and hydrophilic components, such as cationic or ionizable lipids, neutral lipids such as phospholipids or cholesterol, and polyethylene glycol (PEG)-lipid [75]. LNPs offer many benefits to CRISPR delivery, including high encapsulation efficiency, biocompatibility, and delivery efficiency. Since the discovery of lipid-mediated gene transfer, cationic lipids or ionizable lipids have been applied to nucleic acids delivery in cells [76–79]. These lipids form tight structures with anionic nucleic acids at low pH, which helps to encapsulate and protect the nucleic acids to be delivered into cells. Due to its versatility in structure and manipulation, LNPs have been used to deliver all three forms of CRISPR cargoes. An amino-ionizable lipid was utilized to form a lipid nanoparticle with Cas9 encoding mRNA and sgRNA targeting PLK1. The formulated CRISPR-LNPs were injected into mice model of aggressive orthotopic glioblastoma, leading to improved survival by 30% [80]. In addition, the LNPs were engineered for antibody-directed delivery by coating the LNP surface with cell-targeting antibodies, specifically anti-EGFR, to reduce offsite toxicities and reach disseminated tumors [80]. In a separate study, CRISPR/Cas9 plasmid expressing sgRNA targeting a region of the BCR-ABL gene was encapsulated in poly(ethylene glycol)-b-poly(lactic acid-co-glycolic acid) (PEG-PLGA) based cationic lipid-assisted polymeric nanoparticles (CLANs) to specifically target the disrupted BCR-ABL gene in chronic myeloid leukemia in mice. Intravenous injection of CLANs disrupted the BCR-ABL gene and improved survival in treated mice [81]. The PEG-PLGA based CLAN formulation was optimized for plasmid DNA encapsulation and protection from DNases during circulation [81]. Because RNP complexes denature at low pH, Wei et al. established a lipid nanoparticle formulation that can be positively charged at neutral pH by including a permanently cationic lipid DOTAP to allow for encapsulation of RNP complex at neutral pH [82]. This lipid nanoparticle formulation was delivered in vivo encapsulating the Cas9/sgDMD RNP complex in a mouse model of Duchenne muscular dystrophy (DMD), which corrected disease-associated mutation and restored disease phenotype [82]. Another strategy to deliver RNP complexes is through the design of virus-like nanoparticles (VLN). This nanoplatform features a mesoporous silica nanoparticle (MSN) based core that encapsulates the CRISPR system, which is then entirely encapsulated in a lipid membrane exterior. This system exhibits some advantages because the MSN core features a large cargo capacity that can be specifically designed to specified size and charge. In addition, the lipid coating can be specifically tailored for cell-specific targeting and increased circulation [83]. Liu et al. utilized this adaptability of VLN to co-deliver CRISPR RNP with sgRNA targeting PD-L1 and axitinib in tumor-bearing mice in vivo [84].
2.3.2. Polymer-based nanoparticles (polyplexes)
Polymer-based nanoparticles can form tight nanocomplexes with nucleic acids or proteins. Similar to LNPs, polymer-based nanoparticles demonstrate high encapsulation efficiency, versatility in structure, and ease of modification and manufacturing [85]. A common polymeric vector used is polyethenimine (PEI) which can be branched or linear. Branched PEI has high DNA packaging and good endosomal escape, while the linear PEI displays significantly reduced cytotoxicity compared to the branched PEI [86]. Therefore, assessment of the structure and concentration of PEI should be evaluated for efficient, effective, but most importantly, safe delivery. By optimizing the concentration of PEI in relation to other lipids and polymers, a functional yet nontoxic delivery vehicle can be achieved. For example, a lipopolymer complex made up of PEG-PEI-Cholesterol (PPC) encapsulating a plasmid that encodes for CRISPR-Cas9 and sgRNA targeting VEGFA fused with osteosarcoma cell-specific aptamer (LC09) demonstrated selective distribution of the CRISPR components to orthotopic and metastatic osteosarcoma. Thereby decreasing VEGFA expression in osteosarcoma cells and inhibited further metastasis [87]. In addition, a chain-shattering Pt(IV)-backboned polymeric particle was developed to more efficiently release the encapsulated DNA plasmid encoding for Cas9 and sgRNA targeting EZH2 during the endo/lysosomal escape [88]. In vivo application of this Pt(IV) polyplex nanoparticle in prostate cancer tumor-bearing mice showed a decrease in tumor burden compared to cisplatin treatment which showcases how this polyplex delivery of CRISPR components can be utilized as an effective anti-tumor treatment [88]. Another study developed a poly(amide-amine)-poly( β-amino ester) hyperbranched copolymer (hPPC) to deliver CRISPR-Cas9 encoding DNA to target HPV E7 oncogene in HPV-positive cervical cancer cells [89]. hPPC molecule exhibited strong plasmid condensation and high transfection efficiency while maintaining low cytotoxicity. In vivo biodistribution of the hPPC polyplex particle showed uptake in tumor tissue and inhibition of tumor growth in HPV positive cervical cancer tumor-bearing mice [89].
2.3.3. Inorganic nanoparticles
Inorganic nanoparticle is a broad term that is used for a wide variety of metallic and nonmetallic synthetic compounds [90]. In particular, gold nanoparticles have been used for CRISPR delivery in vivo. Lee et al. synthesized a vehicle named CRISPR-Gold which is comprised of a gold nanoparticle core conjugated with Cas9 RNP and donor DNA, which is then encapsulated with cationic polymer Pasp(DET) to deliver CRISPR RNP complexes in vivo and repair genes via homology-directed repair [91]. Administration of CRISPR-Gold nanoparticles not only conferred gene editing in vivo but also could be well tolerated without toxicity [91].
3. CRISPR-assisted RBP screening in cancer
Using large-scale quantitative methods, Gerstberger and co-workers revealed that the human genome may contain 1,542 or more RBPs. The large repertoire of RBPs underlies the complexity of RNA metabolism and post-transcriptional regulation [3]. Recently, Van Nostrand et al. built an elaborate RBP-RNA regulatory network to study the functions of 356 human RBPs using integrative approaches [14]. Common RBP analysis starts with cross-linking immunoprecipitation (CLIP) binding assays followed by sequencing, which help identify a set of RNA elements that directly bind to each RBPs. RBP functions are then determined through CRISPR- or short hairpin RNA (shRNA)-induced knockdown followed by sequencing (KD-RNA-Seq) [14]. Compared with RNA interference (RNAi)-induced gene knockdown, the recent implementation of CRISPR-Cas9 system-based knockout greatly promoted high-throughput loss-of-function screens, with fewer off-target effects and a more thorough depletion of the target gene [92–95]. Despite the variety of CRISPR delivery platforms described in the previous section, lentivirus-based delivery is still most commonly used in both in vitro (Fig. 2A) and in vivo (Fig. 2B) studies for RBP screening.
Fig. 2.
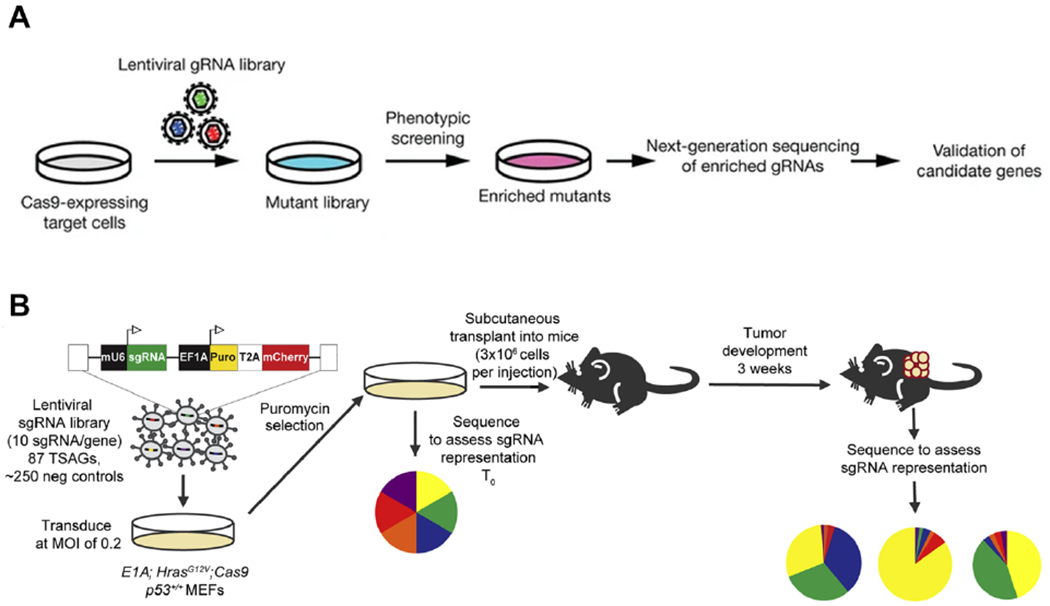
Pooled CRISPR-Cas9 screens can identify functional RBPs in tumor progression. (A) Schematic of in vitro genetic screening with genome-wide lentiviral sgRNA libraries (Reprint from Koike-Yusa et al. [93]). (B) Schematic of in vivo genetic screening with genome-wide lentiviral sgRNA libraries to assess sgRNA representation (Reprint from Bieging-Rolett et al. [96]).
RBPs interact with RNAs to form ribonucleoprotein complexes that coordinate RNA processing and post-transcriptional gene regulation (PTGR). Some RBPs are key molecular determinants of alternative splicing and play an essential role in gene regulation. Perturbations in alternative splicing are causally associated with the occurrence of cancer [97]. A genome-wide CRISPR-Cas9 knockout screen identified RBP heterogeneous nuclear ribonucleoprotein L (HNRNPL) as an essential factor for prostate cancer cell growth by regulating the alternative splicing of RNAs encoding the androgen receptor [98]. More recently, Wang et al. uncovered an interactive RBPs network in acute myeloid leukemia (AML) using CRISPR-Cas9 screens. The sgRNAs were cloned into GFP-tagged lentivirus vector and transduced into a Cas9-expressing AML cell line, MOLM-13. Through the loss-of-function pooled screening, transcriptome analysis of AML patients, and cell survival assays, RBM39 was identified as a top targeting candidate for AML maintenance and survival [99]. Gene ontology (GO) analysis revealed RBM39’s role in alternative splicing as evidenced by the up-regulation of splicing factors SRSF10 and HNRNPH1 in AML patients. Using the CRISPR-Cas9 technology, the authors also identified essential RNA binding domains of RBM39, RRM1 and RRM2, which are critical for pre-mRNA splicing and AML survival [99].
Post-transcriptional modifications in RNA metabolism, such as polyadenylation and methylation, are also tightly regulated by RBPs during fundamental cellular processes and have been investigated in oncogenesis. Davis et al. identified proper post-transcriptional polyadenylation of RUNX1, a key hematopoietic transcription factor, is important in maintaining the balance of hematopoietic stem cells (HSCs) division and differentiation [100]. Alternative polyadenylation of RUNX1 produces functional antagonistic protein isoforms RUNX1a and RUNX1b/c. RUNX1b/c is the dominant isoform in the healthy hematopoietic system, whereas RUNX1a hinders HSC differentiation and is overexpressed in AML patients. To accurately detect changes in post-transcriptional processing of RUNX1, the authors developed a split GFP minigene reporter by exclusively targeting RBPs. GFP reporter accurately recapitulates the two isoforms and monitors their formation. Lentiviral delivery of CRISPR-Cas9 sgRNAs alters the expressions of fluorescent proteins and the calculated βscores indicate the level of sgRNA enrichment. HNRNPA1 and KNERBS1 were identified as mutually antagonistic RBPs during the formation of RUNX1a isoforms, with HNRNPA1 inhibiting and KNERBS1 activating the RUNX1a forming process. In another study, Wang et al. identified ZFP36L2 as a critical AML differentiation regulator through surface-antigen guided CRISPR-Cas9 screens [101]. ZFP36L2 preferentially interacts with the 3’UTR of myeloid differentiation mRNAs, mediating mRNA deadenylation and degradation to maintain the undifferentiated state of leukemia. Genetic inhibition of ZFP36L2 was performed using a CRISPR interference (CRISPRi) system, in which the sgRNA is designed to target the downstream enhancer cluster of ZFP36L2, thereby increasing the stability of myeloid differentiation mRNAs and triggering myeloid differentiation in AML cells [101]. More recently, an N6-methyladenosine (m6A) writer complex, Methyltransferase like 3 (METTL3), stood out as the top candidate regulating LPS-induced macrophage activation using TNF-α readout for pooled CRISPR-Cas9 screens [102]. METTL3-deficient macrophages exhibited decreased overall RNA m6A methylation level and reduced TNF-α expression upon LPS stimulation. Since macrophage activation is involved in multiple innate immune responses, Mettl3 conditional knockout mice demonstrated increased susceptibility towards bacterial infections and accelerated tumor growth.
In addition to the above-mentioned cellular screening, CRISPR screening has also been conducted in mouse tumor models. To identify new dependencies in myeloid leukemia, Bajaj et al. conducted a genome-wide CRISPR-Cas9 screen in a blast crisis chronic myeloid leukemia (bcCML) mouse model for leukemia stem cells (LSCs) regulators [103]. In the BCR-ABL/NUP98-HOXA9-driven mouse model of bcCML, the undifferentiated bcCML stem cells were isolated from the spleen and then transduced with the Brie library (AddGene 73633)-encapsulated lentiviral vectors. An aliquot of cells was collected before and after puromycin selection for sequencing, and 35 million postselection cells were transplanted in sub-lethally irradiated B6 mice for 7 days before collecting leukemic cells. While few sgRNAs were depleted in the in vitro selection process, around 3500 genes were depleted over 3-fold in vivo. After the Enrichr analysis on genes depleted over 3-fold, gene ontology (GO) selection of RBP genes known to bind mRNA, and exclusion of genes with generalized functions, RBP Staufen2 (Stau2) was found to be enriched in immature LSCs [103].
Integrative analysis revealed Stau2 regulates a network of chromatin-binding factors and affects global histone methylation. CRISPR-mediated Stau2 knockout not only reduces the colony-forming ability of bcCML in vitro, but also reduces bcCML establishment in vivo [103]. More recently, Bieging-Rolett et al. identified ZMAT3 as a key RNA splicing factor downstream of p53 (Fig. 2B). The lentiviral sgRNA libraries targeting p53 were used to transduce neoplastic mouse embryonic fibroblasts (MEFs) that express Cas9 and two oncogenes, E1A and HrasG12V. The Cas9-expressing MEFs were collected after puromycin selection and were then subcutaneously transplanted into recipient mice. The authors measured the enrichment of individual sgRNAs in tumors three weeks after transplantation, and found sgRNA targeting Zmat3 is dominant in every tumor, indicating strong tumor-suppressive activity of ZMAT3 [96].
CRISPR-based screening coupled with other integrative analyses provides a promising method for identifying functional RBPs and relevant downstream effectors in the RBP-RNA network, which could serve as important tools to identify novel therapeutic targets in cancers.
4. CRISPR for preclinical therapeutics of RBP editing in cancer
Post-transcriptional control of gene expression closely regulates the normal and pathological phenotypes. RBP plays a key role in regulating gene expressions by binding to the exons, introns or untranslated regions (UTR) of the regulated mRNA [104]. Altered RBP expressions and activities are often observed in cancer, thereby influencing critical pathways in tumor initiation and survival (Fig. 3). Targeting cancer-associated RBPs and their regulatory networks using the CRISPR system provide new opportunities for cancer therapy. Common approaches include targeting oncogenic RBPs, promoting tumor-suppressive RBPs, and correcting cancer-associated RBP mutations.
Fig. 3.
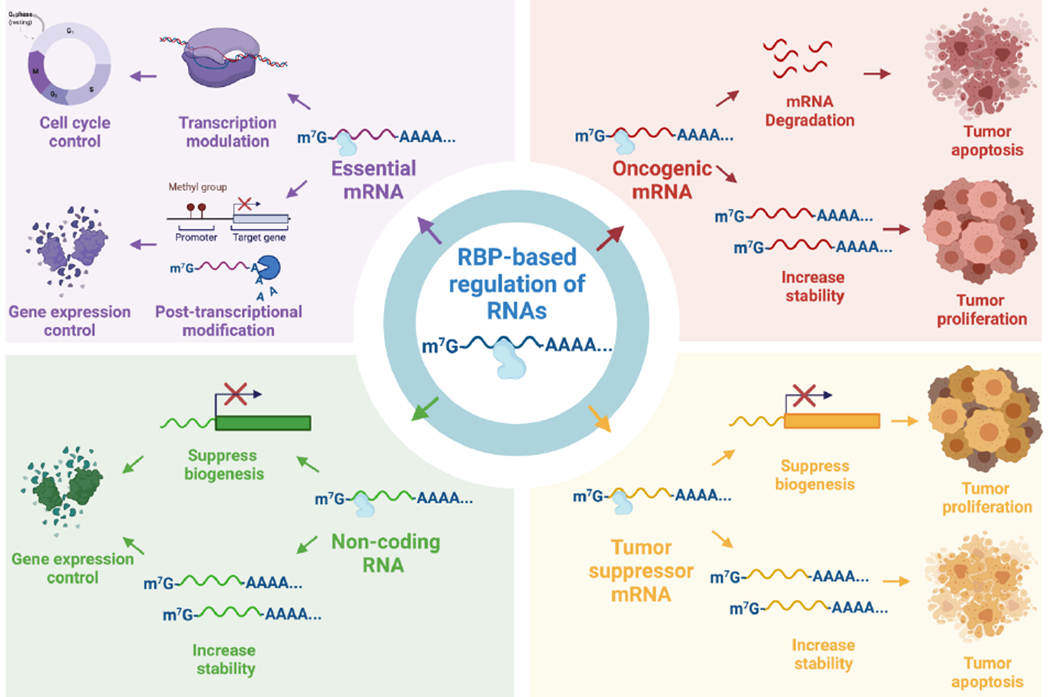
RBP-based regulation of RNAs for cancer therapeutics. RBPs binding to oncogenic or tumor suppressor mRNAs results in an altered expression of the encoded cancer-related proteins, thereby affecting cellular metabolisms such as tumor proliferation or apoptosis. The binding of RBPs with essential mRNAs, which encodes for transcription factors or post-transcriptional modulators, has profound implications in cellular physiology as it can control cell cycle progression and gene expression levels. RBPs can also regulate the expression of non-coding RNAs (ncRNA), which in turn affect the expression of the genes regulated by the ncRNA. Figure created using BioRender.com.
4.1. Selective targeting of oncogenic RBPs
4.1.1. Targeting oncogene transcripts through RBP regulation
The gene regulatory function of RBPs is sometimes hijacked by cancer cells, therefore promoting the expression and stability of oncogenic mRNA transcripts. The RBP insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) is overexpressed in B cell acute lymphoblastic leukemia (B-ALL) with mixed lineage leukemia (MLL) arrangement [105]. The overexpression of IGF2BP3 provides murine bone marrow (BM) cells with a competitive survival advantage and tilts the hematopoietic development toward the B cell/myeloid lineages. CLIP followed by high-throughput sequencing confirmed that IGF2BP3 binds to functional motifs within 3’UTRs of oncogenic target CDK6 and MYC and enhances their expression in the BM. To knock out IGF2BP3 in a human acute leukemia cell line, RS4;11, Palanichamy et al. delivered the LentiCRISPR system with two different sgRNAs, Cr1 and Cr2, targeting the IGF2BP3 locus. Compared to Cr1, Cr2-mediated IGF2BF3 knockout completely abrogated IGR2BP3 protein expression and resulted in significantly reduced RS4;11 cell proliferation [105]. Similarly, Cifdaloz and co-workers identified CUGBP Elav-like family member 1 (CELF1) as a key factor in stabilizing the oncogenic DEK mRNA, thereby controlling other modulators of melanoma cell division and proliferation [106]. shRNA-mediated CELF1 depletion significantly reduced cell proliferation in two human melanoma cell lines, SK-Mel-103 and UACC-62 [106]. More recently, Zhang et al. found that Argonaute 2 (AGO2) mediates pro-oncogenic transcriptomic changes that benefit the survival of hepatocellular carcinoma (HCC) by stabilizing oncogenic MYC transcripts [107]. HCC patients with elevated AGO2 expression had a worse prognosis compared to patients with low AGO2 levels. To determine the role of AGO2 in HCC progression, Zhang et al. cloned sgRNAs targeting the AGO2 into LentiCRISPR v2 plasmid to generate AGO2 HCC cell lines, which significantly reduced the oncogenic MYC transcripts and expression [107]. To confirm the findings in vivo, the authors used shRNA to knock down AGO2 in two HCC cell lines, HuH1 and MHCC97H, and injected the cells into athymic/nude mice. Compared with the control mice, knockdown of AGO2 significantly delayed the growth of both tumors [107].
In addition to lentivirus vectors, non-viral vectors have also been used to deliver CRISPR-Cas9 targeting oncogenic RBPs. The HuR (ELAVL1) is a ubiquitously expressed RBP that can recognize and bind to the AU-rich RNA elements (ARE) within the 3’UTR, thereby regulating mRNA stability and translation [108]. HuR promotes cancer cell survival by binding to pro-oncogenic mRNA transcripts, including WEE1 and IDH1, and is often overexpressed in pancreatic ductal adenocarcinoma (PDA) and colorectal cancer cells [109]. Lal et al. deleted the HuR gene in both PDA and colon cell lines by transfecting the cells with CRISPR-Cas9 plasmid using Lipofectamine 2000 [109]. The HuR-knockout plasmid contained Cas9 protein and sgRNAs targeting the exon 2 or exon 3 in the HuR locus. Among the tested sgRNAs, only the sgRNA targeting exon 3 succeeded in producing a stable HuR-knockout cell line. HuR-deficient PDA cells (MIA Paca-2 and Hs776T) showed increased apoptosis in vitro and were unable to engraft tumor in vivo, suggesting HuR knockout causes a xenograft lethal phenotype [109]. Similarly, HuR-knockout colorectal carcinoma HCT116 cells also demonstrated increased apoptosis in vitro, but were able to produce a colon cancer xenograft in vivo, albeit tumor growth was dramatically reduced compared to the control group [109]. HuR also stabilizes pro-oncogenic long non-coding RNA (lncRNA), NEAT1 and lncRNA-HGBC, and contributes to the development of ovarian and gallbladder carcinogenesis, respectively [110,111].
4.1.2. Targeting cancer cell metabolism through RBP regulation
Cell metabolisms are tightly regulated by transcription factors and epigenetic modifications. In the previous CRISPR-assisted screening section, we discussed the various roles of RBPs in transcriptional regulation that may lead to large-scale disruption of downstream metabolic pathways. For example, RBM39 mediates alternative splicing of HOXA9 transcripts, an essential transcription factor that regulates distinct gene expression and hematopoietic stem cell expansions, thereby promoting the progression and maintenance of AML [99]. In this study, the authors cloned sgRNAs targeting the RRM1 and RRM2 domains of RBM39 into lentiviral vectors and delivered them into Cas9-expressing RN2 cells. The RBM39-knockout RN2 cells were then intravenously injected into sub-lethally irradiated mice, which resulted in a remarkable delay in leukemic progression compared to the control mice [99]. More recently, Musashi1(Msi1) was identified as a key contributor to glioblastoma (GBM) growth by regulating the expression of transcription factors, E2F2 and E2F8, therefore promoting DNA replication and cell cycle progression [112]. CRISPR-mediated Msi1 knockout in two GBM cell lines, U251 and U343, increased the GBM’s sensitivity to cell cycle and DNA replication inhibitors, which provides potential combinational treatment of Msi1 inhibition and cell cycle/DNA replication inhibitors for GBM patients.
In addition to the modulation of transcription factors, RBPs also regulate post-transcriptional modifications for cancer metabolism reprogramming. We discussed in section 3 that Stau2 regulates chromatin-binding factors, which affects global histone methylation and ultimately the growth and maintenance of chronic myeloid leukemia (CML) cells [103]. In this study, the authors generated Stau2 knockout mice through microinjection of Cas9 nuclease and sgRNA targeting Stau2 exon 4 into fertilized C57BL6/N mice zygotes, which resulted in frame-shift mutations that created multiple stop codons and lost multiple RBDs. The establishment of bcCML in vivo was reduced 2-fold in Stau2−/− mice compared to wild type, which increased the likelihood of survival by 13-fold. In addition, the leukemia stem cells (LSCs) from Stau2−/− established leukemia were functionally depleted with reduced colony-forming and self-renewal abilities [103]. More recently, Kosti and co-workers identified SERBP1 as a central regulator of metabolic pathways in glioblastoma (GBM), including methylation and serine biosynthesis [113]. Although CRISPR-Cas9 failed to produce SERBP1 knockout GBM cell lines due to their life-sustaining activities, SERBP1 knockdown via siRNA delayed GBM growth and altered GBM-relevant phenotypes. By disrupting methionine production, SERBP1 knockdown reduced H3K27me3-mediated histone methylation and affected the expression of genes implicated in cancer metabolic pathways [113]. SERBP1 knockdown also upregulated neurogenesis- and neuronal differentiation-associated genes, which are usually downregulated in GBM due to H3K27me3-induced epigenetic silencing. By bringing together the functions in both cancer metabolism and epigenetic regulation, SERBP1 enhances GBM phenotype and promotes a poorly differentiated state of glioma stem cells.
4.2. Restoring anti-tumor activities by targeting RBPs
4.2.1. Restoring p53 tumor-suppressive activities
p53 is a nuclear protein that functions as a transcription factor under diverse stress signals, including hypoxia, DNA damage, oxidative stress, telomere shortening and oncogene activation. p53 activation induces multiple cellular responses that prevent the growth and survival of tumor cells. Among them, induction of cell cycle arrest and programmed cell death is considered most relevant to tumor inhibition [114]. Around half of human cancers observed mutations in p53 tumor suppressors, which mainly occur in the core domains that regulate sequence-specific DNA binding activities of the p53 protein [115]. Restoration of p53 activities through targeted RBP regulations presents a novel anti-tumor strategy. Previously, small molecule Nutlin was often used to activate p53 tumor-suppressive activities by inhibiting the interaction between p53 with MDM2, a negative regulator of p53 [116]. However, the pharmacological outcomes of the treatment, whether it is cell cycle arrest, senescence, or apoptosis, were difficult to predict. Recently, Rizzotto and co-workers revealed that Nutlin-dependent apoptosis associates with the enhanced translation of mRNA containing CG-rich motif mediating p53-dependent death (CGPD-motif) in the 3’UTR. RBPs PCBP2 and DHX30 repress CGPD-motif expression through PCBP2-dependent binding of DHX30 to the 3’UTR of the motif genes, thereby inhibiting p53-dependent apoptosis. DHX30 depletion using shRNA increased CGPD-motif translation and enhanced Nutlin-dependent apoptosis [117].
To identify critical downstream components in p53-mediated tumor suppression, Bieging-Rolett and co-workers transduced mouse embryonic fibroblasts (MEFs) expressing Cas9 and two oncogenes, E1A and HrasG12V, with lentiviral sgRNA libraries targeting p53 tumor-suppressor genes [96]. The transfected cells were then injected into recipient mice and allowed three weeks to grow before sequencing. Results showed that sgRNA targeting Zmat3 dominated every tumor [96]. Zmat3 encodes for RBP ZMAT3, which modulates exon inclusion in a variety of transcripts, including p53 inhibitor MDM4 and MDM2. To determine the role of ZMAT3 in vivo, the authors delivered lentiviral vectors expressing Cre recombinase and sgRNA targeting p53 or zmat3 to the lungs of KrasG12 driven lung tumor mouse model by intratracheal injection. CRISPR-mediated inactivation of p53 and ZMAT3 demonstrated accelerated lung tumorigenesis in KT;H11LSL–Cas9 mice compared to that of control sgRNAs [96]. In a hepatocellular carcinoma mouse model, the authors delivered recombinant transposon vectors expressing oncogene KrasG12D, vectors expressing Sleeping Beauty transposase, and CRISPR vectors expressing Cas9/sgRNAs targeting p53 or Zmat3 in a mixture of 0.9% NaCl solution, which is then delivered to hepatocytes via hydrodynamic injection into mouse lateral tail vein. Similarly, Cas9-mediated p53 and ZMAT3 inactivation both led to the development of hepatocellular carcinoma. In both tumor models, the tumor sizes of Cas9/sgp53 mice were larger than those of Cas9/sgZmat3 mice, suggesting ZMAT3 as an RNA splicing regulator downstream of the p53 tumor suppression pathways [96].
Some RBPs also function as p53 suppressors. For example, Lucchesi and co-workers identified Rbm38 can suppress the translation of p53 by preventing the binding of eukaryotic translation initiation factor 4E (eIF4E) to the p53 mRNA [118]. The authors knocked out Rbm38 in RKO and MCF7 cells using CRISPR-Cas9, and both cell lines showed increased p53 expression. To inhibit the formation of Rbm38-eIF4E complex, they identified an 8 amino acid peptide (Pep8) from Rbm38 that can alleviate Rbm38-mediated p53 suppression and enhance p53 expression [118]. Furthermore, Rbm24, which shares a homogeneous family as Rbm38, also interacts with eIF4E and suppresses p53 translation. Targeting Rbm24 represents a potential strategy to ensure the proper expression of p53 [119].
4.2.2. Increasing the expression of tumor suppressors
p21 is another recognized tumor suppressor downstream of p53 that mediates cell cycle arrest and senescence via both p53-dependent and -independent pathways, and is sometimes considered as an indicator of p53 activities. Liu and co-workers found that CELF6 modulates p21 expression by binding to the 3’UTR of p21 mRNA and increase its stability [120]. LentiCRISPR vector carrying sgCELF6 was used to knock out CELF6 in colorectal carcinoma HCT116 cells. CELF6 depletion not only dramatically reduced p21 expression, but also promoted the proliferation and colony forming-abilities in HCT116 cells, suggesting CELF6 as a putative tumor-suppressive RBP.
Tumor suppressors also exist in the form of microRNA (miRNA). Let-7, a highly conserved tumor suppressor family of miRNAs, represses the expression of multiple oncogenic mRNA targets, including MYC, RAS and HMGA2 [121–123]. Let-7 is downregulated in various tumors, thereby restoring the let-7 functions provides a novel therapeutic pathway. RBP LIN28B has been reported to suppress the biogenesis of let-7 both in vitro and in vivo [124]. Lentivirus-mediated CRISPR-Cas9 silencing of LIN28B has de-repressed let-7, decreased MYC expression and reduced proliferation in a human multiple myeloid cell line, MOLP-8 [125].
Due to the important roles of the innate immunity in tumor immune surveillance and anti-tumor immune response, tumor suppression can be achieved by activating the innate immune pathways. IL-1 receptor-associated kinase 3 (IRAKM) has been identified to negatively regulate TLR4 signaling pathway. N6-methyladenosine (m6A) writer complex METTL3 installs m6A modification on Irakm mRNA and promotes its degradation [102]. CRISPR-mediated knock out of METTL3 decreased overall m6A modification in RAW264.7 cells, slowed down Irakm mRNA degradation, and increased IRAKM expression, which resulted in suppressed TLR4-signaling and macrophage activation upon LPS stimulation [102]. To further confirm the role of m6A modification in the anti-tumor immunity, Mettl3 conditional KO mice (Mettl3flox/flox;Lyzm-Cre mice) was generated and subcutaneously implanted with MC38 murine colon adenocarcinoma cells. As a result, METTL3-deficient mice demonstrated faster tumor growth compared to their wild-type littermates (Mettl3flox/flox mice). Tumor-associated macrophages (TAMs) from METTL3-deficient mice exhibited reduced M1-like markers and increased M2-like markers, which indicates the immunosuppressive nature of the TAMs [102]. Therefore, targeting the m6A modification for macrophage activation represents a potential therapeutic approach in cancer.
4.3. Correcting tumor-specific mutations
Cancer genomic analysis identified recurrent mutations in genes coding the RNA splicing factors SF3B1, U2AF1, and SRSF2 are commonly found in cancer patients [126–129]. While SF3B1 mutation induces uncanny 3’ splice site selection through a different branchpoint [130], U2AF1 and SRSF2 mutations alter their sequence-specific RNA-binding preferences and affinities [131,132]. AML cells containing these spliceosome-mutations are sensitive to small molecule modulators of RNA splicing, such as sulfonamides drugs E7820 and indisulam [99]. In addition to the small molecules that are used for targeting RBPs, the emerging CRISPR-mediated genome editing offers great potential for correcting RBP mutations. To identify the genes affected by the SF3B1 mutation-induced mis-splicing, Inoue and co-workers analyzed pan-cancer mis-spliced events that trigger target mRNA degradation with the CRISPR-Cas9 screen [133]. They found SF3B1 mutation leads to Bromodomain Containing 9 (BRD9) mRNA degradation by inducing inclusion of poison exon, which resulted in a loss of the non-canonical BAF and promoted melanoma tumorigenesis. To correct the mis-splicing of BRD9 caused by SF3B1 mutation, the authors used CRISPR-Cas9 to induce mutagenesis of the poison exon, which significantly slowed the growth of SF3B1-mutant melanoma cells both in vitro and in vivo. Since no donor sequence was provided, the BRD9 mis-splicing mutations were corrected through the NHEJ pathway [133]. In another study, SRSF2 P95 mutation was found to cause 20-30% of the myelodysplastic syndromes (MDS). Chang and co-workers nucleofected a plasmid carrying CRISPR-Cas9/sgRNA targeting the first intron of SRSF2 gene together with a donor plasmid carrying normal SRSF2 allele, which corrected SRSF2 P95L mutations in MDS patient-derived induced pluripotent stem cells (iPSCs) through the HDR repair pathway [134].
Modulating RBP expressions also provide an attractive treatment for cancer-associated mutations. For example, about 85% of Ewing sarcoma (EwS) is caused by a chromosomal translocation of the EWS gene fused with transcription factor FLI1. The EWS-FLI1 fusion underlines EwS pathogenesis and exhibits oncogenic properties by inducing alternative splicing and transcriptional changes [135]. Using a whole-genome CRISPR screen, the oncofetal RBP LIN28B was identified as one of the top candidates that influence EwS evolution and prognosis [136]. LIN28B binds to EWS-FLI1 fusion mRNA and regulates its stability in approximately 10% of Ewing sarcomas. CRISPR-Cas9 mediated depletion of LIN28B in EwS demonstrated decreased expression of EWS-FLI1 fusion protein, thereby abrogating the self-renewal abilities of EwS cells and preventing the occurrence of tumors.
5. Conclusion
RBPs regulate intricate networks of RNA biogenesis and have profound implications for subsequent cellular activities. Alternation in RBP expressions can affect many genes and pathways, which often leads to dysregulation in cell growth and tumorigenesis. Previously, researchers used small molecules, aptamers and peptides to target specific RBPs for cancer treatment. The recently emerging CRISPR-Cas9 technology offers great potential for RBP targeting with simple sgRNA designs and versatile targeting sites. However, several challenges remain in delivering the CRISPR components for RBP-based cancer therapeutics in clinical applications, including low cellular uptake, immunogenicity and selective cell targeting. Multiple platforms have been developed for CRISPR delivery, and each of them has both advantages and disadvantages. The majority of clinical studies nowadays use viral vectors to deliver CRISPR-Cas9/sgRNA plasmid for gene editing, but challenges like immunogenicity, cellular toxicity, potential mutagenesis and carcinogenesis are still yet to be addressed [73]. Concurrently, innovations in nonviral vectors have enabled targeted delivery with reduced immunogenicity and cellular toxicity [18]. In addition, a number of approaches have been developed for screening the delivery platform, such as physical transduction-based high-throughput screening [137–139] and barcoded delivery platforms for accelerated in vivo screening [140,141]. Another hindrance to clinical adoption of the CRISPR-Cas system is potential off-target effects. Off-target effects can be moderated by more proficient and precise engineering of the CRISPR-Cas system to increase the specificity of gene editing. Development of new delivery vehicles with high delivery specificity may limit the potential toxicity and off-target effects of the CRISPR-Cas system, and promote the adoption of the CRISPR system for clinical applications.
In this article, we summarize recent advances in CRISPR-delivery and CRISPR-mediated RBP targeting for cancer treatment. Common applications of CRISPR in RBP studies include: (1) Identifying new RBPs in cancer and understanding their cancer-related functions; (2) Modulating therapeutically relevant RBPs for cancer treatment. With the development of RBP-targeting therapies, we envision the emergence of many new delivery platforms, which in turn promote the translation of RBP-based cancer therapeutics into broad clinical applications.
Acknowledgements
Y.D. acknowledges the support from the NIH through the National Heart, Lung, and Blood Institute (R01HL136652), as well as the start-up fund from the College of Pharmacy at The Ohio State University. J.Y. acknowledges the support from the Professor Sylvan G. Frank Graduate Fellowship. Authors acknowledge that figures were created with biorender.com.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
Y.D. is a scientific advisory board member of Oncorus Inc and serves as a consultant of Rubius Therapeutics. The authors have no competing interests to declare.
Reference
- [1].Glisovic T, Bachorik JL, Yong J, Dreyfuss G, RNA-binding proteins and post-transcriptional gene regulation, FEBS Lett. 582 (2008) 1977–1986. 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lunde BM, Moore C, Varani G, RNA-binding proteins: modular design for efficient function, Nat. Rev. Mol. Cell Biol 8 (2007) 479–490. 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gerstberger S, Hafner M, Tuschl T, A census of human RNA-binding proteins, Nat. Rev. Genet 15 (2014) 829–845. 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kechavarzi B, Janga SC, Dissecting the expression landscape of RNA-binding proteins in human cancers, Genome Biol. 15 (2014) R14. 10.1186/gb-2014-15-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pereira B, Billaud M, Almeida R, RNA-Binding Proteins in Cancer: Old Players and New Actors, Trends Cancer. 3 (2017) 506–528. 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- [6].Gebauer F, Schwarzl T, Valcárcel J, Hentze MW, RNA-binding proteins in human genetic disease, Nat. Rev. Genet 22 (2021) 185–198. 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- [7].Croce CM, Oncogenes and Cancer, N. Engl. J. Med 358 (2008) 502–511. 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- [8].Wang J, Liu Q, Shyr Y, Dysregulated transcription across diverse cancer types reveals the importance of RNA-binding protein in carcinogenesis, BMC Genomics. 16 (2015) S5. 10.1186/1471-2164-16-S7-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mohibi S, Chen X, Zhang J, Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer, Pharmacol. Ther 203 (2019) 107390. 10.1016/j.pharmthera.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity, Science. 337 (2012) 816–821. 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gasiunas G, Barrangou R, Horvath P, Siksnys V, Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria, Proc. Natl. Acad. Sci 109 (2012) E2579–E2586. 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Koonin EV, Makarova KS, Zhang F, Diversity, classification and evolution of CRISPR-Cas systems, Curr. Opin. Microbiol 37 (2017) 67–78. 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ, Modulating DNA Repair Pathways to Improve Precision Genome Engineering, ACS Chem. Biol 13 (2018) 389–396. 10.1021/acschembio.7b00777. [DOI] [PubMed] [Google Scholar]
- [14].Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J-Y, Cody NAL, Dominguez D, Olson S, Sundararaman B, Zhan L, Bazile C, Bouvrette LPB, Bergalet J, Duff MO, Garcia KE, Gelboin-Burkhart C, Hochman M, Lambert NJ, Li H, McGurk MP, Nguyen TB, Palden T, Rabano I, Sathe S, Stanton R, Su A, Wang R, Yee BA, Zhou B, Louie AL, Aigner S, Fu X-D, Lécuyer E, Burge CB, Graveley BR, Yeo GW, A large-scale binding and functional map of human RNA-binding proteins, Nature. 583 (2020) 711–719. 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS, Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases, Genome Res. 24 (2014) 132–41. 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Han HA, Pang JKS, Soh B-S, Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing, J. Mol. Med 98 (2020) 615–632. 10.1007/s00109-020-01893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lino CA, Harper JC, Carney JP, Timlin JA, Delivering CRISPR: a review of the challenges and approaches, Drug Deliv. 25 (2018) 1234–1257. 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yan J, Kang DD, Dong Y, Harnessing lipid nanoparticles for efficient CRISPR delivery, Biomater. Sci (2021). 10.1039/D1BM00537E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fajrial AK, He QQ, Wirusanti NI, Slansky JE, Ding X, A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing, Theranostics. 10 (2020) 5532–5549. 10.7150/thno.43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, Marigo V, Recchia A, In vivo Editing of the Human Mutant Rhodopsin Gene by Electroporation of Plasmid-based CRISPR/Cas9 in the Mouse Retina, Mol Ther Nucleic Acids. 5 (2016) e389 doi: 10 1038/mtna 2016 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu W, Lu Z, Li F, Wang W, Qian N, Duan J, Zhang Y, Wang F, Chen T, Efficient in vivo gene editing using ribonucleoproteins in skin stem cells of recessive dystrophic epidermolysis bullosa mouse model, Proc Natl Acad Sci U A. 114 (2017) 1660–1665. 10.1073/pnas.1614775114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering, Cell. 153 (2013) 910–8. 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hammer RE, Pursel VG, Rexroad CE, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL, Production of transgenic rabbits, sheep and pigs by microinjection, Nature. 315 (1985) 680–3. 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- [24].Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Crénéguy A, Brusselle L, Anegón I, Menchaca A, Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes, PLoS One. 10 (2015) e0136690. 10.1371/journal.pone.0136690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J, Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos, Cell. 156 (2014) 836–43. 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- [26].Weaver JC, Chizmadzhev YA, Theory of electroporation: A review, Bioelectrochem. Bioenerg 41 (1996) 135–160. 10.1016/S0302-4598(96)05062-3. [DOI] [Google Scholar]
- [27].Alghadban S, Bouchareb A, Hinch R, Hernandez-Pliego P, Biggs D, Preece C, Davies B, Electroporation and genetic supply of Cas9 increase the generation efficiency of CRISPR/Cas9 knock-in alleles in C57BL/6J mouse zygotes, Sci Rep. 10 (2020) 17912. 10.1038/s41598-020-74960-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J, Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling, Nat Commun. 6 (2015) 7391. 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H, CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation, Sci Rep 6 (2016) 20611. 10.1038/srep20611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S, Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system, Nat Commun. 5 (2014) 3964. 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- [31].Feng Y, Sassi S, Shen JK, Yang X, Gao Y, Osaka E, Zhang J, Yang S, Yang C, Mankin HJ, Hornicek FJ, Duan Z, Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system, J Orthop Res. 33 (2015) 199–207. 10.1002/jor.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Leclere PG, Panjwani A, Docherty R, Berry M, Pizzey J, Tonge DA, Effective gene delivery to adult neurons by a modified form of electroporation, J Neurosci Methods. 142 (2005) 137–43. 10.1016/j.jneumeth.2004.08.012. [DOI] [PubMed] [Google Scholar]
- [33].Hu B, Zou Y, Zhang L, Tang J, Niedermann G, Firat E, Huang X, Zhu X, Nucleofection with Plasmid DNA for CRISPR/Cas9-Mediated Inactivation of Programmed Cell Death Protein 1 in CD133-Specific CAR T Cells, Hum Gene Ther. 30 (4) 446–458. 10.1089/hum.2017.234. [DOI] [PubMed] [Google Scholar]
- [34].Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, Liu T, Wang X, Zhang B, Zhao L, Hu L, Ning H, Zhang Y, Deng K, Liu L, Lu X, Zhang T, Xu J, Li C, Wu H, Deng H, Chen H, CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia, N Engl J Med. 381 (2019) 1240–1247. 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- [35].Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han M-J, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim K-S, Anderson DG, Langer R, Jensen KF, A vector-free microfluidic platform for intracellular delivery, Proc. Natl. Acad. Sci 110 (2013) 2082–2087. 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L, CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation, Sci. Adv 1 (2015) e1500454. 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Han X, Liu Z, Ma Y, Zhang K, Qin L, Cas9 Ribonucleoprotein Delivery via Microfluidic Cell-Deformation Chip for Human T-Cell Genome Editing and Immunotherapy, Adv. Biosyst 1 (2017) 1600007. 10.1002/adbi.201600007. [DOI] [PubMed] [Google Scholar]
- [38].DiTommaso T, Cole JM, Cassereau L, Buggé JA, Hanson JLS, Bridgen DT, Stokes BD, Loughhead SM, Beutel BA, Gilbert JB, Nussbaum K, Sorrentino A, Toggweiler J, Schmidt T, Gyuelveszi G, Bernstein H, Sharei A, Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo, Proc. Natl. Acad. Sci 115 (2018) E10907–E10914. 10.1073/pnas.1809671115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Suda T, Liu D, Hydrodynamic gene delivery: its principles and applications, Mol Ther. 15 (2007) 2063–9. 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- [40].Kobayashi N, Nishikawa M, Hirata K, Takakura Y, Hydrodynamics-based procedure involves transient hyperpermeability in the hepatic cellular membrane: implication of a nonspecific process in efficient intracellular gene delivery, J Gene Med. 6 (2004) 584–92. 10.1002/jgm.541. [DOI] [PubMed] [Google Scholar]
- [41].Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, Zhang F, Anderson DG, Sharp PA, Jacks T, CRISPR-mediated direct mutation of cancer genes in the mouse liver, Nature. 514 (2014) 380–4. 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nayak S, Herzog RW, Progress and prospects: immune responses to viral vectors, Gene Ther. 17 (2010) 295–304. 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nault J-C, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouzé E, Pilati C, Verret B, Blanc J-F, Balabaud C, Calderaro J, Laurent A, Letexier M, Bioulac-Sage P, Calvo F, Zucman-Rossi J, Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas, Nat. Genet 47 (2015) 1187–1193. 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- [44].Wang D, Tai PWL, Gao G, Adeno-associated virus vector as a platform for gene therapy delivery, Nat Rev Drug Discov. 18 (5) 358–378. 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].He X, Urip BA, Zhang Z, Ngan CC, Feng B, Evolving AAV-delivered therapeutics towards ultimate cures, J. Mol. Med 99 (2021) 593–617. 10.1007/s00109-020-02034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ling C, Li B, Ma W, Srivastava A, Development of Optimized AAV Serotype Vectors for High-Efficiency Transduction at Further Reduced Doses, Hum Gene Ther Methods. 27 (8) 143–9. 10.1089/hgtb.2016.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D, CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox, Biotechnol J. 9 (2014) 1402–12. 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- [48].Huttner NA, Girod A, Perabo L, Edbauer D, Kleinschmidt JA, Büning H, Hallek M, Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies, Gene Ther. 10 (2003) 2139–47. 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- [49].Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, In vivo genome editing using Staphylococcus aureus Cas9, Nature. 520 (2015) 186–191. 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F, In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9, Nat Biotechnol. 33 (2015) 102–6. 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM, A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice, Nat Biotechnol. 34 (2016) 334–8. 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Truong DJ, Kühner K, Kühn R, Werfel S, Engelhardt S, Wurst W, Ortiz O, Development of an intein-mediated split-Cas9 system for gene therapy, Nucleic Acids Res. 43 (2015) 6450–8. 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling, Cell. 159 (2014) 440–55. 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, Renauer P, Bilguvar K, Gunel M, Sharp PA, Zhang F, Platt RJ, Chen S, AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma, Nat. Neurosci 20 (2017) 1329–1341. 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Song X, Liu C, Wang N, Huang H, He S, Gong C, Wei Y, Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy, Adv Drug Deliv Rev. 168 (1) 158–180. 10.1016/j.addr.2020.04.010. [DOI] [PubMed] [Google Scholar]
- [56].Maggio I, Zittersteijn HA, Wang Q, Liu J, Janssen JM, Ojeda IT, van der Maarel SM, Lankester AC, Hoeben RC, Gongalves MAFV, Integrating gene delivery and gene-editing technologies by adenoviral vector transfer of optimized CRISPR-Cas9 components, Gene Ther. 27 (5) 209–225. 10.1038/s41434-019-0119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Palmer D, Ng P, Improved system for helper-dependent adenoviral vector production, Mol Ther. 8 (2003) 846–52. 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- [58].Tatsis N, Ertl HC, Adenoviruses as vaccine vectors, Mol Ther. 10 (2004) 616–29. 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hartman ZC, Appledorn DM, Amalfitano A, Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications, Virus Res. 132 (2008) 1–14. 10.1016/j.virusres.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Beer SJ, Matthews CB, Stein CS, Ross BD, Hilfinger JM, Davidson BL, Poly (lacticglycolic) acid copolymer encapsulation of recombinant adenovirus reduces immunogenicity in vivo, Gene Ther. 5 (1998) 740–746. 10.1038/sj.gt.3300647. [DOI] [PubMed] [Google Scholar]
- [61].Tatsis N, Tesema L, Robinson ER, Giles-Davis W, McCoy K, Gao GP, Wilson JM, Ertl HCJ, Chimpanzee-origin adenovirus vectors as vaccine carriers, Gene Ther. 13 (2006) 421–429. 10.1038/sj.gt.3302675. [DOI] [PubMed] [Google Scholar]
- [62].Ehrke-Schulz E, Heinemann S, Schulte L, Schiwon M, Ehrhardt A, Adenoviral Vectors Armed with PAPILLOMAVIRUs Oncogene Specific CRISPR/Cas9 Kill Human-Papillomavirus-Induced Cervical Cancer Cells, Cancers Basel. 12 (2020). 10.3390/cancers12071934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Koo T, Yoon AR, Cho HY, Bae S, Yun CO, Kim JS, Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression, Nucleic Acids Res. 45 (2017) 7897–7908. 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li Q, Kay MA, Finegold M, Stratford-Perricaudet LD, Woo SL, Assessment of recombinant adenoviral vectors for hepatic gene therapy, Hum. Gene Ther 4 (1993) 403–409. 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- [65].Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, Weng Z, Gao G, Xue W, Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses, Hum Gene Ther. 26 (2015) 432–42. 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Xu CL, Ruan MZC, Mahajan VB, Tsang SH, Viral Delivery Systems for CRISPR, Viruses. 11,1 (2019). 10.3390/v11010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kotterman MA, Chalberg TW, Schaffer DV, Viral Vectors for Gene Therapy: Translational and Clinical Outlook, Annu Rev Biomed Eng. 17 (2015) 63–89. 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- [68].Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, Buch P, MacLaren RE, Anderson PN, Barker SE, Duran Y, Bartholomae C, von Kalle C, Heckenlively JR, Kinnon C, Ali RR, Thrasher AJ, Effective gene therapy with nonintegrating lentiviral vectors, Nat Med. 12 (2006) 348–53. 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- [69].Hu J, Schokrpur S, Archang M, Hermann K, Sharrow AC, Khanna P, Novak J, Signoretti S, Bhatt RS, Knudsen BS, Xu H, Wu L, A Non-integrating Lentiviral Approach Overcomes Cas9-Induced Immune Rejection to Establish an Immunocompetent Metastatic Renal Cancer Model, Mol Ther Methods Clin Dev. 9 (2018) 203–210. 10.1016/j.omtm.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening, Nat Methods. 11 (2014) 783–784. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA, Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis, Cell. 160 (2015) 1246–60. 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kochenderfer JN, Feldman SA, Zhao Y, Xu H, Black MA, Morgan RA, Wilson WH, Rosenberg SA, Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor, J Immunother. 32 (2009) 689–702. 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bulcha JT, Wang Y, Ma H, Tai PWL, Gao G, Viral vector platforms within the gene therapy landscape, Signal Transduct Target Ther. 6 (2021) 53. 10.1038/s41392-021-00487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li L, Hu S, Chen X, Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities, Biomaterials. 171 (2018) 207–218. 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hou X, Zaks T, Langer R, Dong Y, Lipid nanoparticles for mRNA delivery, Nat. Rev. Mater (2021) 1–17. 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fraley R, Subramani S, Berg P, Papahadjopoulos D, Introduction of liposome-encapsulated SV40 DNA into cells, J Biol Chem. 255 (1980) 10431–5. [PubMed] [Google Scholar]
- [77].Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M, Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure, Proc. Natl. Acad. Sci. U. S. A 84 (1987) 7413–7417. 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang Y, Sun C, Wang C, Jankovic KE, Dong Y, Lipids and Lipid Derivatives for RNA Delivery, Chem. Rev (2021). 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang X, Zhao W, Nguyen GN, Zhang C, Zeng C, Yan J, Du S, Hou X, Li W, Jiang J, Deng B, McComb DW, Dorkin R, Shah A, Barrera L, Gregoire F, Singh M, Chen D, Sabatino DE, Dong Y, Functionalized lipid-like nanoparticles for in vivo mRNA delivery and base editing, Sci. Adv 6 (2020) eabc2315. 10.1126/sciadv.abc2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rosenblum D, Gutkin A, Kedmi R, Ramishetti S, Veiga N, Jacobi AM, Schubert MS, Friedmann-Morvinski D, Cohen ZR, Behlke MA, Lieberman J, Peer D, CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy, Sci. Adv 6 (2020) eabc9450. 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liu Y, Zhao G, Xu C-F, Luo Y-L, Lu Z-D, Wang J, Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy, Biomater. Sci 6 (2018) 1592–1603. 10.1039/c8bm00263k. [DOI] [PubMed] [Google Scholar]
- [82].Wei T, Cheng Q, Min Y-L, Olson EN, Siegwart DJ, Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing, Nat. Commun 11 (2020) 3232. 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Tarn D, Ashley CE, Xue M, Carnes EC, Zink J.l., Brinker CJ, Mesoporous silica nanoparticle nanocarriers: biofunctionality and biocompatibility, Acc Chem Res. 46 (2013) 792–801. 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Liu Q, Wang C, Zheng Y, Zhao Y, Wang Y, Hao J, Zhao X, Yi K, Shi L, Kang C, Liu Y, Virus-like nanoparticle as a co-delivery system to enhance efficacy of CRISPR/Cas9-based cancer immunotherapy, Biomaterials. 258 (2020) 120275. 10.1016/j.biomaterials.2020.120275. [DOI] [PubMed] [Google Scholar]
- [85].Chen K, Jiang S, Hong Y, Li Z, Wu Y-L, Wu C, Cationic polymeric nanoformulation: Recent advances in material design for CRISPR/Cas9 gene therapy, Prog. Nat. Sci. Mater. Int 29 (2019) 617–627. 10.1016/j.pnsc.2019.10.003. [DOI] [Google Scholar]
- [86].Ryu N, Kim MA, Park D, Lee B, Kim YR, Kim KH, Baek J.l., Kim WJ, Lee KY, Kim UK, Effective PEI-mediated delivery of CRISPR-Cas9 complex for targeted gene therapy, Nanomed. 14 (10) 2095–2102. 10.1016/j.nano.2018.06.009. [DOI] [PubMed] [Google Scholar]
- [87].Liang C, Li F, Wang L, Zhang ZK, Wang C, He B, Li J, Chen Z, Shaikh AB, Liu J, Wu X, Peng S, Dang L, Guo B, He X, Au DWT, Lu C, Zhu H, Zhang BT, Lu A, Zhang G, Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma, Biomaterials. 147 (2017) 68–85. 10.1016/j.biomaterials.2017.09.015. [DOI] [PubMed] [Google Scholar]
- [88].Zhang Q, Kuang G, He S, Liu S, Lu H, Li X, Zhou D, Huang Y, Chain-shattering Pt(IV)-backboned polymeric nanoplatform for efficient CRISPR/Cas9 gene editing to enhance synergistic cancer therapy, Nano Res. 14 (2021) 601–610. 10.1007/s12274-020-3066-4. [DOI] [Google Scholar]
- [89].Gao X, Jin Z, Tan X, Zhang C, Zou C, Zhang W, Ding J, Das BC, Severinov K, Hitzeroth l.l., Debata PR, He D, Ma X, Tian X, Gao Q, Wu J, Tian R, Cui Z, Fan W, Huang Z, Cao C, Bao Y, Tan S, Hu Z, Hyperbranched poly(β-amino ester) based polyplex nanopaticles for delivery of CRISPR/Cas9 system and treatment of HPV infection associated cervical cancer, J Control Release. 321 (05 10) 654–668. 10.1016/j.jconrel.2020.02.045. [DOI] [PubMed] [Google Scholar]
- [90].Luther DC, Huang R, Jeon T, Zhang X, Lee YW, Nagaraj H, Rotello VM, Delivery of drugs, proteins, and nucleic acids using inorganic nanoparticles, Adv Drug Deliv Rev. 156 (2020) 188–213. 10.1016/j.addr.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang W, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair, Nat. Biomed. Eng 1 (2017) 889–901. 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Glaß M, Michl P, Hüttelmaier S, RNA Binding Proteins as Drivers and Therapeutic Target Candidates in Pancreatic Ductal Adenocarcinoma, Int. J. Mol. Sci 21 (2020) 4190. 10.3390/ijms21114190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K, Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library, Nat. Biotechnol 32 (2014) 267–273. 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- [94].Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, Genome-scale CRISPR-Cas9 knockout screening in human cells, Science. 343 (2014) 84–87. 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F, Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex, Nature. 517 (2015) 583–588. 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bieging-Rolett KT, Kaiser AM, Morgens DW, Boutelle AM, Seoane JA, Van Nostrand EL, Zhu C, Houlihan SL, Mello SS, Yee BA, McClendon J, Pierce SE, Winters IP, Wang M, Connolly AJ, Lowe SW, Curtis C, Yeo GW, Winslow MM, Bassik MC, Attardi LD, Zmat3 Is a Key Splicing Regulator in the p53 Tumor Suppression Program, Mol. Cell 80 (2020) 452–469.e9. 10.1016/j.molcel.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lee SC-W, Abdel-Wahab O, Therapeutic targeting of splicing in cancer, Nat. Med 22 (2016) 976–986. 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG, Wu Q, Feng FY, Loda M, He HH, Liu XS, Brown M, Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E5207–E5215. 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, Hockemeyer K, Ghebrechristos YE, Yoshimi A, Inoue D, Ki M, Cho H, Bitner L, Kloetgen A, Lin K-T, Uehara T, Owa T, Tibes R, Krainer AR, Abdel-Wahab O, Aifantis I, Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia, Cancer Cell. 35 (2019) 369–384.e7. 10.1016/j.ccell.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Davis AG, Einstein JM, Zheng D, Jayne ND, Fu X-D, Tian B, Yeo GW, Zhang D-E, A CRISPR RNA-binding protein screen reveals regulators of RUNX1 isoform generation, Blood Adv. 5 (2021) 1310–1323. 10.1182/bloodadvances.2020002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang E, Zhou H, Nadorp B, Cayanan G, Chen X, Yeaton AH, Nomikou S, Witkowski MT, Narang S, Kloetgen A, Thandapani P, Ravn-Boess N, Tsirigos A, Aifantis I, Surface antigen-guided CRISPR screens identify regulators of myeloid leukemia differentiation, Cell Stem Cell. 28 (2021) 718–731.e6. 10.1016/j.stem.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, Yao J, Mao K, Liu T, Meng F-L, Pan W, Zou Q, Liu J, Zhou Y, Xia Q, Flavell RA, Zhu S, Li H-B, Pooled CRISPR screening identifies m6A as a positive regulator of macrophage activation, Sci. Adv 7 (2021) eabd4742. 10.1126/sciadv.abd4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Bajaj J, Hamilton M, Shima Y, Chambers K, Spinler K, Van Nostrand EL, Yee BA, Blue SM, Chen M, Rizzeri D, Chuah C, Oehler VG, Broome HE, Sasik R, Scott-Browne J, Rao A, Yeo GW, Reya T, An in vivo genome-wide CRISPR screen identifies the RNA-binding protein Staufen2 as a key regulator of myeloid leukemia, Nat Cancer. 1 (2020) 410–422. 10.1038/s43018-020-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Hentze MW, Castello A, Schwarzl T, Preiss T, A brave new world of RNA-binding proteins, Nat. Rev. Mol. Cell Biol 19 (2018) 327–341. 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- [105].Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, Katzman S, Toloue M, Yan W, Basso G, Pigazzi M, Sanford JR, Rao DS, RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation, J. Clin. Invest 126 (2016) 1495–1511. 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cifdaloz M, Osterloh L, Graña O, Riveiro-Falkenbach E, Ximénez-Embún P, Muñoz J, Tejedo C, Calvo TG, Karras P, Olmeda D, Miñana B, Gómez-López G, Cañon E, Eyras E, Guo H, Kappes F, Ortiz-Romero PL, Rodríguez-Peralto JL, Megías D, Valcárcel J, Soengas MS, Systems analysis identifies melanoma-enriched pro-oncogenic networks controlled by the RNA binding protein CELF1, Nat. Commun 8 (2017) 2249. 10.1038/s41467-017-02353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhang K, Pomyen Y, Barry AE, Martin SP, Khatib S, Knight L, Forgues M, Dominguez DA, Parhar R, Shah AP, Bodzin AS, Wang XW, Dang H, AGO2 Mediates MYC mRNA Stability in Hepatocellular Carcinoma, Mol. Cancer Res. MCR. 18 (2020) 612–622. 10.1158/1541-7786.MCR-19-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B, Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis, Int. J. Mol. Sci 14 (2013) 10015–10041. 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, Romeo C, Stout MC, Londin E, Goetz A, Lowder CY, Nevler A, Yeo CJ, Campbell PM, Winter JM, Dixon DA, Brody JR, CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype, Mol. Cancer Res. MCR. 15 (2017) 696–707. 10.1158/1541-7786.MCR-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chai Y, Liu J, Zhang Z, Liu L, HuR-regulated IncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer, Cancer Med. 5 (2016) 1588–1598. 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hu Y-P, Jin Y-P, Wu X-S, Yang Y, Li Y-S, Li H-F, Xiang S-S, Song X-L, Jiang L, Zhang Y-J, Huang W, Chen S-L, Liu F-T, Chen C, Zhu Q, Chen H-Z, Shao R, Liu Y-B, LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis, Mol. Cancer. 18 (2019) 167. 10.1186/s12943-019-1097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Baroni M, Yi C, Choudhary S, Lei X, Kosti A, Grieshober D, Velasco M, Qiao M, Burns SS, Araujo PR, DeLambre T, Son MY, Plateroti M, Ferreira MAR, Hasty P, Penalva LOF, Musashi1 Contribution to Glioblastoma Development via Regulation of a Network of DNA Replication, Cell Cycle and Division Genes, Cancers. 13 (2021) 1494. 10.3390/cancers13071494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kosti A, de Araujo PR, Li W-Q, Guardia GDA, Chiou J, Yi C, Ray D, Meliso F, Li Y-M, Delambre T, Qiao M, Burns SS, Lorbeer FK, Georgi F, Flosbach M, Klinnert S, Jenseit A, Lei X, Sandoval CR, Ha K, Zheng H, Pandey R, Gruslova A, Gupta YK, Brenner A, Kokovay E, Hughes TR, Morris QD, Galante PAF, Tiziani S, Penalva LOF, The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation, Genome Biol. 21 (2020) 195. 10.1186/s13059-020-02115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ryan KM, Phillips AC, Vousden KH, Regulation and function of the p53 tumor suppressor protein, Curr. Opin. Cell Biol 13 (2001) 332–337. 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- [115].Cho Y, Gorina S, Jeffrey PD, Pavletich NP, Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations, Science. 265 (1994) 346–355. 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- [116].Shen H, Maki CG, Pharmacologic activation of p53 by small-molecule MDM2 antagonists, Curr. Pharm. Des 17 (2011) 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Rizzotto D, Zaccara S, Rossi A, Galbraith MD, Andrysik Z, Pandey A, Sullivan KD, Quattrone A, Espinosa JM, Dassi E, Inga A, Nutlin-Induced Apoptosis Is Specified by a Translation Program Regulated by PCBP2 and DHX30, Cell Rep. 30 (2020) 4355–4369.e6. 10.1016/j.celrep.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lucchesi CA, Zhang J, Ma B, Chen M, Chen X, Disruption of the Rbm38-eIF4E Complex with a Synthetic Peptide Pep8 Increases p53 Expression, Cancer Res. 79 (2019) 807–818. 10.1158/0008-5472.CAN-18-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zhang M, Zhang Y, Xu E, Mohibi S, de Anda DM, Jiang Y, Zhang J, Chen X, Rbm24, a target of p53, is necessary for proper expression of p53 and heart development, Cell Death Differ. 25 (2018) 1118–1130. 10.1038/s41418-017-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu G, Zhang Q, Xia L, Shi M, Cai J, Zhang H, Li J, Lin G, Xie W, Zhang Y, Xu N, RNA-binding protein CELF6 is cell cycle regulated and controls cancer cell proliferation by stabilizing p21, Cell Death Dis. 10 (2019) 688. 10.1038/s41419-019-1927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ, MicroRNA Let-7a Down-regulates MYC and Reverts MYC-Induced Growth in Burkitt Lymphoma Cells, Cancer Res. 67 (2007) 9762–9770. 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- [122].Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ, RAS is regulated by the let-7 microRNA family, Cell. 120 (2005) 635–647. 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- [123].Mayr C, Hemann MT, Bartel DP, Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation, Science. 315 (2007) 1576–1579. 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Viswanathan SR, Daley GQ, Gregory RI, Selective blockade of microRNA processing by Lin28, Science. 320 (2008) 97–100. 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Manier S, Powers JT, Sacco A, Glavey SV, Huynh D, Reagan MR, Salem KZ, Moschetta M, Shi J, Mishima Y, Roche-Lestienne C, Leleu X, Roccaro AM, Daley GQ, Ghobrial IM, The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma, Leukemia. 31 (2017) 853–860. 10.1038/leu.2016.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann W-K, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih L-Y, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S, Frequent pathway mutations of splicing machinery in myelodysplasia, Nature. 478 (2011) 64–69. 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- [127].Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ, SF3B1 and Other Novel Cancer Genes in Chronic Lymphocytic Leukemia, N. Engl. J. Med 365 (2011) 2497–2506. 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Jänne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M, Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing, Cell. 150 (2012) 1107–1120. 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, Miller DK, Christ AN, Bruxner TJC, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey U-MH, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM, Genomic analyses identify molecular subtypes of pancreatic cancer, Nature. 531 (2016) 47–52. 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- [130].Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, Corson L, Feala J, Fekkes P, Ichikawa K, Keaney GF, Lee L, Kumar P, Kunii K, MacKenzie C, Matijevic M, Mizui Y, Myint K, Park ES, Puyang X, Selvaraj A, Thomas MP, Tsai J, Wang JY, Warmuth M, Yang H, Zhu P, Garcia-Manero G, Furman RR, Yu L, Smith PG, Buonamici S, Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point, Cell Rep. 13 (2015) 1033–1045. 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- [131].Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, Kielkopf CL, Ley TJ, Walter MJ, Graubert TA, U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing, Leukemia. 29 (2015) 909–917. 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, Li Y, Chung YR, Micol J-B, Murphy ME, Cho H, Kim M-K, Zebari AS, Aumann S, Park CY, Buonamici S, Smith PG, Deeg HJ, Lobry C, Aifantis I, Modis Y, Allain FH-T, Halene S, Bradley RK, Abdel-Wahab O, SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition, Cancer Cell. 27 (2015) 617–630. 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Inoue D, Chew G-L, Liu B, Michel BC, Pangallo J, D’Avino AR, Hitchman T, North K, Lee SC-W, Bitner L, Block A, Moore AR, Yoshimi A, Escobar-Hoyos L, Cho H, Penson A, Lu SX, Taylor J, Chen Y, Kadoch C, Abdel-Wahab O, Bradley RK, Spliceosomal disruption of the non-canonical BAF complex in cancer, Nature. 574 (2019) 432–436. 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Chang C-J, Kotini AG, Olszewska M, Georgomanoli M, Teruya-Feldstein J, Sperber H, Sanchez R, DeVita R, Martins TJ, Abdel-Wahab O, Bradley RK, Papapetrou EP, Dissecting the Contributions of Cooperating Gene Mutations to Cancer Phenotypes and Drug Responses with Patient-Derived iPSCs, Stem Cell Rep. 10 (2018) 1610–1624. 10.1016/j.stemcr.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Selvanathan SP, Graham GT, Erkizan HV, Dirksen U, Natarajan TG, Dakic A, Yu S, Liu X, Paulsen MT, Ljungman ME, Wu CH, Lawlor ER, Üren A, Toretsky JA, Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing, Proc. Natl. Acad. Sci 112 (2015) E1307–E1316. 10.1073/pnas.1500536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Keskin T, Bakaric A, Waszyk P, Boulay G, Torsello M, Cornaz-Buros S, Chevalier N, Geiser T, Martin P, Volorio A, Iyer S, Kulkarni A, Letovanec I, Cherix S, Cote GM, Choy E, Digklia A, Montemurro M, Chebib I, Nielsen PG, Carcaboso AM, Mora J, Renella R, Suva ML, Fusco C, Provero P, Rivera MN, Riggi N, Stamenkovic I, LIN28B Underlies the Pathogenesis of a Subclass of Ewing Sarcoma, Cell Rep. 30 (2020) 4567–4583.e5. 10.1016/j.celrep.2019.12.053. [DOI] [PubMed] [Google Scholar]
- [137].Fan Y, Yen C-W, Lin H-C, Hou W, Estevez A, Sarode A, Goyon A, Bian J, Lin J, Koenig SG, Leung D, Nagapudi K, Zhang K, Automated high-throughput preparation and characterization of oligonucleotide-loaded lipid nanoparticles, Int. J. Pharm 599 (2021) 120392. 10.1016/j.ijpharm.2021.120392. [DOI] [PubMed] [Google Scholar]
- [138].Tomé I, Francisco V, Fernandes H, Ferreira L, High-throughput screening of nanoparticles in drug delivery, APL Bioeng. 5 (2021) 031511. 10.1063/5.0057204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Westhaus A, Cabanes-Creus M, Rybicki A, Baltazar G, Navarro RG, Zhu E, Drouyer M, Knight M, Albu RF, Ng BH, Kalajdzic P, Kwiatek M, Hsu K, Santilli G, Gold W, Kramer B, Gonzalez-Cordero A, Thrasher AJ, Alexander IE, Lisowski L, High-Throughput In Vitro, Ex Vivo, and In Vivo Screen of Adeno-Associated Virus Vectors Based on Physical and Functional Transduction, Hum. Gene Ther 31 (2020) 575–589. 10.1089/hum.2019.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, Castro MG, Anderson SE, Rudoltz TG, Lando GN, Tiwari PM, Kirschman JL, Willett N, Jang YC, Santangelo PJ, Bryksin AV, Dahlman JE, High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing, Proc. Natl. Acad. Sci 115 (2018) E9944–E9952. 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Kremer LPM, Cerrizuela S, Dehler S, Stiehl T, Weinmann J, Abendroth H, Kleber S, Laure A, Andari JE, Anders S, Marciniak-Czochra A, Grimm D, Martin-Villalba A, High throughput screening of novel AAV capsids identifies variants for transduction of adult NSCs within the subventricular zone, Mol. Ther. - Methods Clin. Dev 23 (2021) 33–50. 10.1016/j.omtm.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


