Abstract
Ageing in diverse species ranging from the simple nematode Caenorhabditis elegans to humans is associated with a marked decrease of neuronal function and increased susceptibility to neurodegeneration. Accumulating findings also indicate that alterations in neuronal functionality with age are associated with a decline in mitochondrial integrity and function. The rate at which a mitochondrial population is refreshed is determined by the coordination of mitochondrial biogenesis with mitophagy, a selective type of autophagy targeting damaged or superfluous mitochondria for degradation. Coupling of these opposing processes is crucial for maintaining cellular energy homeostasis, which eventually contributes to health span. Here, we focus on the role of mitophagy in nervous system function in the context of normal physiology and disease. First, we consider the progress that has been made over the last decade in elucidating the mechanisms that govern and regulate mitophagy, placing emphasis on the PINK1/Parkin-mediated mitophagy. We further discuss the contribution of mitophagy to the maintenance of neuronal homeostasis and health as well as recent findings implicating impaired mitophagy in age-related decline of the nervous system function and consequently in the pathogenesis of neurodegenerative diseases.
Keywords: Ageing, Energy homeostasis, Mitophagy, Neurodegeneration, Neuronal health, Neuron
Introduction
Ageing, a natural degradation process, has a dramatic impact on the quality of human life as it is associated with a decline in physical, mental and emotional functionality. When viewed from a biomedical perspective, ageing is a major risk factor for disease onset and progression. The ageing of the nervous system in particular is believed to be linked with neurodegenerative diseases; however, the cellular and molecular pathways that connect these processes are still poorly understood. Neurons are highly polarized post-mitotic cells typically composed of a soma (or a cell body), an axon and a dendritic arborisation. For their survival and function, they depend on dynamic cellular processes such as neuronal growth and maturation, axonal migration, synapse formation and elimination, among others (Nicholls and Budd 2000).
Mitochondria have essential roles in such processes, and interestingly, their proteomes were found to display differences across distinct types of neuronal cells in the mouse brain. Such differences in mitochondrial protein expression reflect specialization for different functions, revealing some diversity in mitochondrial functionality in the central nervous system (Bray 2019). Neurons require a continuous recycling of their mitochondrial pool and a tight regulation of mitochondrial transport to particular regions in order to meet their specific metabolic requirements, further ensuring various neuronal functions. Different quality control mechanisms, including those involved in mitochondrial proteostasis, mitochondrial metabolism, signalling cascades, lipid biogenesis, mitochondrial turnover and mitochondrial dynamics, enable adaptation of mitochondrial function to cellular requirements under physiological and stress conditions (Song et al. 2021).
Alterations in mitochondrial quality and function have been associated with accelerated ageing and various pathological conditions (Sun et al. 2016). Accordingly, coordination of mitochondrial biogenesis with selective mitochondrial autophagy (known as mitophagy) modulates the mitochondrial content and thus contributes to the maintenance of energy homeostasis, at both the cellular and organismal level, ultimately prolonging health span (Palikaras et al. 2015). In this review, we focus on the role of mitophagy in neuronal function and dysfunction. Although multiple mitophagy pathways exist, we largely restrict our consideration to the PINK1/Parkin pathway. Furthermore, we highlight the contribution of mitophagy to neuronal physiology and discuss how defects in mitophagy mechanisms impair cellular and organismal energy metabolism, ultimately accelerating ageing and age-related neurodegeneration.
Molecular pathways of mitophagy
Mitophagy shares many proteins with the autophagy process; however, the selectivity of the autophagosome for mitochondria is governed by proteins that are distinct from known autophagy-related proteins (ATG) and activated in response to specific stimuli. Readers are encouraged to consult recent comprehensive reviews detailing the intricacies of the mitophagy process and the diversity in the underlying mechanisms (Ashrafi and Schwarz 2013; Pickles et al. 2018). Below, we focus on the PTEN-induced putative kinase protein 1 (PINK1)/E3 ubiquitin ligase Parkin pathway because it is the most well-studied mitophagy pathway and it is conserved in diverse species, including Caenorhabditis elegans, Drosophila melanogaster and mammals (there is no homologue in yeast and plants) (Narendra et al. 2008; Onishi et al. 2021; Pickles et al. 2018).
PINK1/Parkin-mediated mitophagy
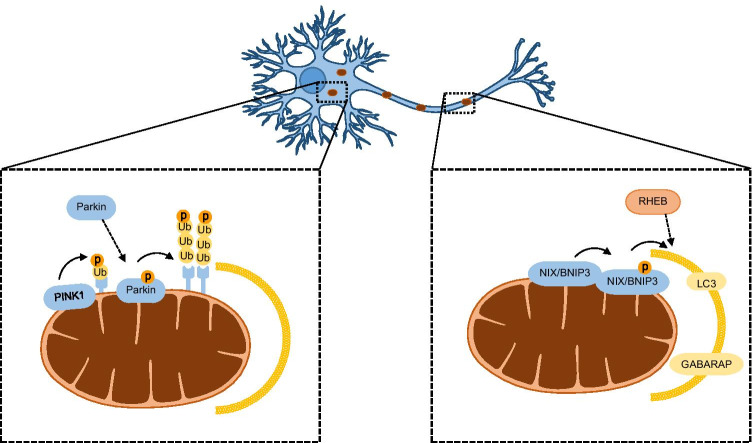
Mutations in PINK1 and Parkin have been associated with autosomal recessive cases of Parkinson’s disease (PD), which is the second most common neurodegenerative disorder. It is a progressive bradykinetic disorder characterized by the loss of dopaminergic neurons in the substantia nigra pars compacta and accumulation of intracellular inclusions composed of α-synuclein aggregates in specific brain stem, spinal cord and cortical regions (Lees et al. 2009). Genetic epistasis analysis in Drosophila demonstrated that PINK1 and Parkin act in the same pathway, with PINK1 functioning upstream of Parkin to modulate mitochondrial function (Clark et al. 2006; Park et al. 2006). Under physiological conditions, PINK1 bearing an amino-terminal mitochondrial targeting sequence is imported into mitochondria through the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIMM23) complexes. When PINK1 reaches the inner mitochondrial membrane, its transmembrane segment is cleaved by the matrix processing peptidase (MPP) and the presenilin-associated rhomboid-like protease (PARL). Cleaved PINK1 retrotranslocates from the mitochondria to the cytosol, where it is degraded through the N-end rule proteasome pathway (Yamano and Youle 2013). Under conditions of mitochondrial damage or stress, however, PINK1 is stabilized on the outer mitochondrial membrane (OMM). Following its activation through autophosphorylation, PINK1 phosphorylates Parkin on Ser65 of its ubiquitin-like domain (Ubl), promoting its recruitment onto depolarised mitochondria. Furthermore, phosphorylated Parkin undergoes conformational changes in its inhibitory domains that stimulate its E3 ubiqutin ligase activity (Trempe et al. 2013). PINK1 also phosphorylates Ser65 of ubiquitin (Ub) attached to OMM-localized proteins, which in turn binds to Parkin leading to its further recruitment and activation (Fig. 1). Once activated, Parkin conjugates ubiquitin chains onto OMM proteins, yielding more substrates for PINK1 to phosphorylate, with consequent Parkin recruitment and activation (Fig. 1). Such a feedforward mechanism drives the selective elimination of defective mitochondria through mitophagy (Harper et al. 2018; Nguyen et al. 2016; Ordureau et al. 2014).
Fig. 1.
Neuronal mitophagy main pathways. The PINK1/Parkin pathway functions mainly in the soma and the receptor-mediated mitophagy mainly in neuronal axons. PINK1 phosphorylates OMM ubiquitin to recruit and phosphorylate Parkin for activation. Parkin ubiquitinates OMM proteins, which are phosphorylated by PINK1, for further Parkin recruitment. In the receptor-mediated mitophagy, NIX/BNIP3 becomes integrated in the OMM upon mitophagy induction. Phosphorylation of BNIP3 and NIX by an unknown kinase promotes interaction with LC3 and GABARAP in the autophagosome. NIX/BNIP3 also promotes interaction of RHEB with depolarized mitochondria, which is physiologically located in the axonal cytoplasm, and is recruited to facilitate retrograde movement and fusion with lysosomes. GABARAP, GABAA receptor-associated protein; LC3, microtubule-associated proteins 1A/1B light chain 3; NIX/ BNIP3, NIP3-like protein X/BCL2/ adenovirus E1B-interacting protein 3; OMM, outer mitochondrial membrane; RHEB, Ras homologue enriched in brain GTP-binding protein; Ub, ubiquitin
Quantitative proteomics and live-cell imaging analysis demonstrated that Parkin forms canonical (K48 and K63) and non-canonical (K6 and K11) ubiquitin chain linkages. The importance of various types of ubiquitin chains to mitophagy was highlighted by studies investigating the role of deubiquitinating enzymes, such as USP15, USP30 and USP35. By removing Parkin-generated ubiquitin chains from the mitochondrial surface, these deubiquitinases compete with mitophagy, to maintain a balance between ubiquitination and deubiquitination events. This delicate balance is important for cellular energy homeostasis (Bingol et al. 2014; Cornelissen et al. 2014; Cunningham et al. 2015; Wang et al. 2015).
S65-phosphorylated ubiquitin (pS65-Ub) chains, including K63-linked, derived from the orchestrated functions of PINK1 and Parkin are considered to act as molecular signals that recruit autophagy receptors such as sequestosome 1(SQSTM1) or p62 (Geisler et al. 2010), optineurin (OPTN), nuclear dot protein 52 (NDP52) (Heo et al. 2015; Lazarou et al. 2015; Sahlender et al. 2005), neighbour of the BRCA1 gene 1 (NBR1) (Chan et al. 2011) and Tax1-binding protein 1 (TAX1BP1) (Lazarou et al. 2015) in depolarised mitochondria. In turn, these receptors bind ubiquitinated cargo through their ubiquitin binding domain, promoting their linkage with microtubule-associated proteins 1A/1B light chain 3 (LC3)/GABAA receptor-associated protein (GABARAP) in the autophagosome. The specific cargo engulfed by an autophagosome is subsequently delivered to a lysosome for degradation (Nguyen et al. 2016).
The PINK1/Parkin pathway interfaces with other mitochondrial quality control mechanisms, including mitochondria-derived vesicles (MDVs) (McLelland et al. 2016, 2014), mitochondrial dynamics (Pryde et al. 2016), the mitochondrial unfolded protein response (UPRmt) (Pickles et al. 2018) and activation of the proteasomal system (Tanaka et al. 2010) to maintain a healthy mitochondrial network, thereby contributing to metabolic homeostasis. It is also worth to note that mitochondrial protein import efficiency has emerged as a central regulator of several of the aforementioned mitochondrial stress response pathways, such as mitophagy, UPRmt and the proteasome (Melber and Haynes 2018).
Mitophagy in physiology
Mitophagy has a housekeeping role in maintaining a functional mitochondrial network under physiological conditions. Healthy mitochondria provide cells with energy, essential cofactors (such as iron-sulphur clusters and heme) and lipids, but also act as intracellular signalling platforms for apoptotic and innate immune pathways (Bahat et al. 2021; Tan and Finkel 2020). Thus, mitophagy is crucial, in particular, for post-mitotic cells such as neurons that need to survive throughout the lifetime of an organism. In these cases, defective or dysregulated mitophagy leads to bioenergetics deficits, with detrimental consequences at both the cellular and whole organism level.
The PINK1/Parkin pathway is the most well characterized mechanism of mitophagy, especially in neuronal cells. Interestingly, mitophagy through the PINK1/Parkin pathway occurs gradually in neurons and much slower than in other cells, as evidenced by comparing cultured hippocampal neurons with HeLa cells. Specifically, autophagosome fusion with lysosome and acidification occur hours after the stimulus of mitophagy has emerged (Evans and Holzbaur 2020b). Recently, it was shown that another E3 ubiquitin ligase, Mdm2, takes part in the PINK1/Parkin pathway. Mdm2 is directly activated by Parkin, without the assistance of other proteins, enhancing protein ubiquitination, and thus mitophagy (Kook et al. 2020). This suggests that the direct protein–protein interaction between two ligases is essential to maintain healthy neurons (Kook et al. 2020). Nevertheless, the importance of PINK1/Parkin pathway in neuronal basal mitophagy has not been clarified yet, and many studies have shown that PINK1 knockout does not influence mitophagy under physiological conditions (Devireddy et al. 2015; McWilliams et al. 2018; Puschmann et al. 2017). This is further verified by a parallel pathway mediated by the mitochondrial ubiquitin ligase 1 (Mul1), which lies on the OMM and appears to compensate for PINK1/Parkin loss (Yun et al. 2014). Furthermore, a recent study on the autophagy receptor SQSTM1 showed that SQSTM1 deletion in cortical neurons inhibits PINK1 accumulation on mitochondria, and this event does not affect mitochondrial degradation (Poon et al. 2021). Altogether, these studies emphasize the existence and importance of Parkin-independent pathways in basal mitophagy.
Protein ubiquitination and deubiquitination by Parkin are crucial for mitophagy. The mitochondrial Rho GTPase Miro is a Parkin ubiquitination substrate and is proven to be essential for the PINK1/Parkin pathway. Miro recruits Parkin and induces its binding on damaged mitochondria, while its degradation arrests mitochondrial motility, isolating mitochondrial damage to a specific region for mitophagy to occur (Lopez-Domenech et al. 2021; Wang et al. 2011; Zheng et al. 2019). PINK1 accumulation in the mitochondria and Miro degradation act as protective mechanisms for neurons as they prevent damaged mitochondria from being transported to the axons and simultaneously promote mitophagy for immediate removal (Liu et al. 2012). Specifically, PINK1 and Parkin are negative regulators of Miro, as loss of PINK1 contributes to enhanced Miro protein levels in Drosophila dopaminergic neurons (Liu et al. 2012). Moreover, poly- and mono-ubiquitination of voltage-dependent anion channel 1 (VDAC1) by Parkin promotes mitophagy and suppresses apoptosis, respectively (Ham et al. 2020). USP30 and USP33 are mitochondrial deubiquitinases that supress PINK1/Parkin-mediated mitophagy by preventing Parkin ubiquitination of proteins (Bingol et al. 2014; Niu et al. 2020). In addition, USP30 inhibits delivery of lysosomes to neuronal mitochondria (Bingol et al. 2014). Deletion of USP30 or USP33 induces translocation of PRKN1/Parkin to depolarised mitochondria to promote mitophagy (Bingol et al. 2014; Niu et al. 2020). Thus, deubiquitination is a mechanism to slow down neuronal mitophagy when required, as discussed above.
Recent studies have discovered important regulators of the PINK1/Parkin pathway in neuronal mitophagy. Sigma 1 receptor (Sig 1R) is an endoplasmic reticulum (ER) chaperone protein that is widely expressed in neurons. Its importance in mitophagy was highlighted by the fact that Sig 1R deletion impairs mitochondrial degradation in mouse retinal explants and cultured cells treated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), as evidenced by the accumulation of mitophagic markers VDAC1 and TIMM23 (Yang et al. 2019). Notably, PINK1/Parkin signalling was not affected, suggesting that Sig 1R depletion prevents only the autophagosome–lysosome fusion event and not the initiation of mitophagy (Yang et al. 2019). It is also possible that mitophagy inhibition and accumulation of VDAC1 and TIMM23 result from the impairment of Parkin-independent pathways. By contrast, it was recently shown that knockdown of Sig 1R inhibits PINK1 stabilization and Parkin recruitment on depolarized mitochondria of dopaminergic neurons in culture and eventually impairs mitophagy (Wang et al. 2021). Finally, mitochondrial expression of DJ-1, a protein involved in the oxidative stress response, was increased in primary cortical neurons following CCCP treatment. DJ-1 overexpression induced Parkin translocation, suggesting it regulates Parkin, and this regulation occurs vice versa, as enhanced DJ-1 in the mitochondria is mediated by Parkin (Joselin et al. 2012).
Mitochondria are probably shaped in the soma and then translocated and anchored at the axons and dendrites of neurons, which raises the question whether mitophagy is differentially regulated in each region (Fig. 2). Under physiological conditions, Parkin levels vary among different brain regions, indicating that its processing is also different (McWilliams et al. 2018). Initially, it was shown that Parkin-mediated mitophagy occurs solely in the soma of neurons (Cai et al. 2012), but it was later proved that it also takes place in the axons (Ashrafi et al. 2014; Wang et al. 2011). The theory that mitochondria are transferred back to the soma for degradation is compelling, as motility and degradation of neuronal mitochondria are tightly regulated processes (Cardanho-Ramos et al. 2020). Parkin translocation in neurons reduces the anterograde mitochondrial motility, probably to prevent damaged mitochondria from entering the axons, or arrests mitochondrial motility completely (Cai et al. 2012; McWilliams et al. 2018; Wang et al. 2011). Overexpression of PINK1 enhances retrograde mitochondrial movement, but not anterograde, while PINK1 or Parkin knockdown promotes anterograde mitochondrial motility (Devireddy et al. 2015; Wang et al. 2011). Under mild stress, the majority of mitochondria are unable to recruit Parkin and thus are transported retrogradely (Lin et al. 2017). Thus, mitophagy in the soma is probably required to sustain the healthy mitochondrial population in the axons, because, upon stress, mitochondria are transported from the axons to the soma for degradation (Lin et al. 2017).
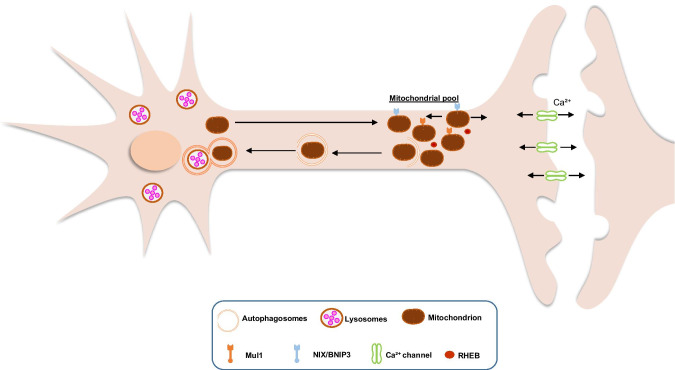
Fig. 2.
Basal neuronal mitophagy. In vivo studies show that PINK1 and Parkin are not activated neither required for mitophagy under physiological conditions. Mitochondria are generated in the soma and transferred in the axons and synapses where there is high ATP and Ca2+ buffering demand. Mitochondria are anchored there but can move anterogradely and retrogradely. PINK1/Parkin-independent mitophagy of depolarized or superfluous mitochondria will initiate until the formation of autophagosomes. Then the autophagosomes will be transported to the soma for fusion with lysosomes and degradation with the help of RHEB. Since PINK1/Parkin signalling is not essential for basal mitophagy, probably Mul1 and NIX/BNIP3 are involved. Mul1, mitochondrial ubiquitin ligase 1; NIX/BNIP3, NIP3-like protein X/BCL2/ adenovirus E1B- interacting protein 3; RHEB, Ras homologue enriched in brain GTP-binding protein
A recent study showed that treatment with CCCP induces translocation of Ras homologue enriched in brain GTP-binding protein (RHEB) from the axonal cytoplasm to damaged mitochondria that are subsequently moved in a retrograde direction, for degradation in the soma. RHEB is not involved in the PINK1/Parkin somal basal mitophagy but in the axonal receptor-mediated mitophagy. Indeed, the outer mitochondrial membrane protein NIP3-like protein X (NIX; also known as BCL2/adenovirus E1B-interacting protein 3-like, BNIP3L) has been shown to facilitate the association of RHEB with damaged mitochondria, thereby targeting them to autophagosomes in axons (Fig. 1) (Han et al. 2020). There is also the case that the PINK1/Parkin pathway is specific to the soma, while in the axons, mitophagy is mainly Parkin-independent (Cai et al. 2012). This is further supported by the fact that Parkin or PINK1 deletion in Drosophila affects the somal mitochondria but not the ones located in the axons or junctions (Devireddy et al. 2015; Sung et al. 2016). Collectively, mitophagy initiation can occur in the axons, in physiological conditions or under mild stress; once autophagosomes are formed, they are probably transported to the soma where lysosomes are located for degradation (Fig. 2) (Maday et al. 2012; McWilliams et al. 2018).
Mitophagy is well-studied during ageing and neurodegeneration, but under physiological conditions, the research is limited. This can be attributed to the fact that no standard approaches exist to stimulate the extent of damage that induces basal mitophagy in neurons. CCCP is a nonspecific ionophore that is commonly used to induce global damage in mitochondria, as it uncouples mitochondrial oxidative phosphorylation (Cai et al. 2012; Han et al. 2020). On the other hand, antimycin A is a milder depolarizer, which inhibits respiratory complex III and affects neuronal mitochondrial motility, but still induces partial neuronal loss (Ashrafi et al. 2014; Wang et al. 2011). Valinomycin is even milder than antimycin A, because it does not alter the pH gradient and induces Parkin recruitment without critical neuronal loss (Puschmann et al. 2017). Thus, artificially induced mitophagy in cells cannot mimic basal mitophagy, and only in vivo studies can shed light into this process. However, Evans and Holzbaur (2020a, b) managed to simulate low levels of mitochondrial damage in vitro, by simply removing antioxidants from the media of hippocampal neurons, resulting in enhanced ROS production, without affecting the mitochondrial morphology and dynamics. Overall, these results suggest that basal and mildly induced mitophagy occurs also in the axons, but when mitophagy is accompanied by neuronal loss, mitochondria are degraded in the soma or transferred there for degradation, probably to prevent neuronal apoptosis (Fig. 3). Finally, mitochondrial damage may not be the sole initiator of neuronal basal mitophagy, as it is a crucial process for cell differentiation and fate, and thus, it would be interesting to identify other types of stimuli for mitophagy induction, besides depolarization and damage.
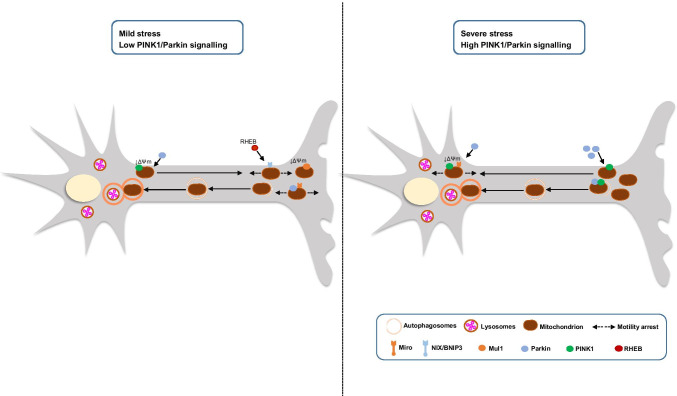
Fig. 3.
Mitophagy is differentially regulated upon mild and toxic stress. Under mild stress, mitochondria become depolarized, but only a few mitochondria recruit Parkin. Low levels of Parkin induce anterograde motility. The motility of axonal mitochondria is arrested, either by Parkin recruitment and Miro degradation or translocation of RHEB, and they undergo mitophagy immediately to isolate damage. After initiation of mitophagy and formation of autophagosomes in the axons, autophagosomes are transferred to the soma for fusion with lysosomes and degradation. Under mild stress, the entire or majority of mitochondrial population undergo Parkin-independent mitophagy. Under severe stress, the PINK1/Parkin pathway is mainly activated. Parkin recruitment and degradation of Miro arrest mitochondrial motility to prevent damage from spreading. High levels of Parkin increase retrograde motility of the axonal mitochondria. Thus, stress-induced mitophagy occurs mainly in the soma, like during neurodegeneration. ΔΨm, mitochondrial membrane potential; Mul1, mitochondrial ubiquitin ligase 1; NIX/BNIP3, NIP3-like protein X/BCL2/ adenovirus E1B- interacting protein 3; RHEB, Ras homologue enriched in brain GTP-binding protein
Mitophagy in ageing
Accumulating findings indicate that mitophagy declines with age. Indeed, assessment of in vivo mitophagy using a transgenic mouse model that expresses a mitochondrial-targeted form of the fluorescent reporter Keima (mt-Keima) revealed that basal levels of mitophagy are reduced by approximately 70% in the dentate gyrus region of old mt-Keima mice compared to young animals. This area of the brain is enriched for neural stem cells and has a key role in hippocampal memory formation and learning (Sun et al. 2015). The age-dependent decline in mitophagy is further supported by a recent study where Western blot analysis from 28- and 48-week-old mice with dysfunctional mitochondria showed a gradual increase in LC3II/LC3I ratio and p62 levels, which indicates impaired mitochondrial autophagy (Galizzi et al. 2021).
A growing body of evidence now suggests that age-related reduction of mitophagy is not simply a corollary of the ageing process, but rather, it has a causative role in senescence decline. Consistent with this, a recent study demonstrated that p53-deficient mice, which have elevated Parkin-dependent mitophagy, exhibit a better cardiac performance with increasing age compared to their wild-type counterparts. Furthermore, overexpression of Parkin mitigated cardiac functional decline in aged mice, corroborating the beneficial effects of mitophagy against cardiovascular ageing (Hoshino et al. 2013). In addition, emerging observations suggest that impaired mitophagy contributes to a human premature ageing disorder known as Werner syndrome (WS). In fact, it has been shown that stimulation of mitophagy in WS human samples and WS invertebrate models through NAD+ repletion ameliorates disease phenotypes (Fang et al. 2019a).
Studies in Drosophila uncovered a direct link between Parkin and ageing. Flies carrying null alleles of parkin displayed locomotor defects as a consequence of apoptotic muscle degeneration and reduced longevity (Greene et al. 2003). On the other hand, ubiquitous or neuron-specific overexpression of Parkin was shown to extend adult Drosophila lifespan without affecting reproduction, physical activity or feeding behaviour. Long-lived Parkin overexpressing flies displayed a reduction in both the number and size of protein aggregates, reduced Mitofusin levels and increased mitochondrial activity during ageing. These findings demonstrated that Parkin overexpression improves the capacity of the proteostasis network and alters mitochondrial dynamics, thereby promoting health span and longevity (Rana et al. 2013).
An intriguing perspective on the roles of PINK1 and Parkin in modulating healthy ageing was provided by using a transgenic Drosophila model that expresses the mitophagy reporter mt-Keima. It was shown that mitophagy occurs in Drosophila flight muscle and dopaminergic neurons in vivo even under physiological conditions. More importantly, mitophagy in these cell types increases during ageing, and this age-associated increase is abrogated when PINK1 or Parkin is depleted. Furthermore, knockdown of the Drosophila homologues of the deubiquitinases USP15 and USP30 rescues, at least partially, the mitophagy defects in parkin-deficient flies. These findings provide experimental evidence that PINK1 and Parkin are crucial components of age-dependent mitophagy in Drosophila in vivo (Cornelissen et al. 2018).
In C. elegans, mitophagy was shown to be required for longevity under conditions of reduced insulin signalling, impaired mitochondrial function or under caloric restriction, a condition known to extend lifespan from yeast to primates. Knockdown of either pdr-1 (Parkinson’s disease-related-1 gene encoding the nematode Parkin orthologue), pink-1 or dct-1 (DAF-16/FOXO-controlled germline-tumour affecting-1), which encodes a putative homologue to the mammalian NIX/BNIP3L and BNIP3 (BCL2/ adenovirus E1B- interacting protein 3), shortens the lifespan of insulin signalling-defective daf-2(e1370) animals and calorie-restricted eat-2(ad465) mutants. These findings establish that different longevity-promoting processes, including caloric restriction, are associated with mitophagy. Moreover, mitophagy deficiency impairs mitochondrial function and compromises the response to stress. These detrimental effects trigger a mitochondrial retrograde signalling cascade through SKN-1 (SKiNhead-1), the nematode homologue of NRF2 transcription factor, which induces expression of both mitochondrial biogenesis genes and mitophagy by upregulating dct-1 (Palikaras et al. 2015). Collectively, these findings provide critical insights into the mechanisms by which mitophagy modulates neuronal and whole-body ageing.
Mitophagy in age-related pathologies: focus on neurodegeneration
Beyond ageing, multiple lines of evidence suggest an intriguing link between mitophagy and age-associated pathologies, including neurodegenerative diseases. As discussed above, genetic and biochemical studies established that mutations in PINK1 and Parkin genes are associated with autosomal recessive cases of PD, supporting early observations that implicate mitochondrial damage in PD pathogenesis (Pickrell and Youle 2015). Interestingly, genetic deficiency of the mitochondrial protein phosphoglycerate mutase family member 5 (PGAM5), which is involved in PINK1 stabilization on damaged mitochondria, leads to PD-like movement abnormalities and dysfunctional dopaminergic neurons in mice (Lu et al. 2014). Moreover, mutations in leucine-rich repeat kinase 2 (LRRK2), which affect mitochondrial function and mitophagy, have been associated with autosomal dominant PD. Such mutations impair the degradation of Miro, thereby delaying the arrest of mitochondria upon depolarization and the subsequent axonal mitophagy. Indeed, in neurons derived from induced pluripotent stem cells from patients with inherited or sporadic PD, degradation of Miro is reduced, and consequently, the initiation of mitophagy is delayed. Notably, lowering of Miro levels in LRRK2G2019S human neurons and Drosophila PD models rescued neurodegenerative phenotypes (Hsieh et al. 2016).
Along similar lines, LRRK2 was shown to affect mitochondrial function by impairing PINK1/Parkin-dependent mitophagy in cell lines and primary fibroblasts from PD patients. This adverse effect is exacerbated by the most frequent G2019S mutation that increases LRRK2 kinase activity. Moreover, LRRK2 impaired the interactions between Parkin and Drp1 and their targets on the outer mitochondrial membrane early in mitophagy when damaged mitochondria tend to aggregate. Consistently, impaired protein–protein interactions and the PINK1/Parkin-dependent mitophagy defect can be rescued by the inhibition of LRRK2 kinase activity (Bonello et al. 2019). Taken together, these findings support the notion that defective mitophagy, leading to mitochondrial failure and subsequently reduced energy supply, contributes to PD pathogenesis.
Accumulation of damaged mitochondria in neurons is also a hallmark of Alzheimer’s disease (AD), the most common form of dementia. Extracellular deposits of amyloid beta (Aβ) peptide and intraneuronal tangles of hyper-phosphorylated forms of microtubule-associated protein tau (p-tau) are the histopathological characteristics of the disease (Braak and Braak 1991). Neurons affected in AD experience mitochondrial abnormalities and decreased mitochondrial bioenergetics that occur early in AD pathogenesis and may contribute to AD pathologies (Yao et al. 2009). However, the role of mitophagy in AD progression is still elusive. Recently, it was shown that Parkin-mediated mitophagy is induced in cultured primary neurons derived from mutant hAPP Tg mice and AD patient brains. In this context, mitophagosomes were aberrantly accumulated, most likely because of increased autophagic flux and defective degradation of autophagic substrates due to reduced lysosomal efficiency (Ye et al. 2015). In addition, electron microscopy of the hippocampal mitochondria from normal and transgenic amyloid precursor protein/pesenilin-1 (APP/PS1) AD mice showed decreased mitophagy events, along with elevated ROS production in AD mice (Zhang et al. 2021).
There is compelling evidence from studies in animal and cellular models of AD and also in patients with sporadic late-onset AD suggesting that mitophagy perturbations result in synaptic failure and decreased cognitive function. This occurs by promoting Aβ and tau accumulation through increased oxidative stress and impaired cellular bioenergetics. These abnormalities, in turn, may cause mitophagy alterations (Kerr et al. 2017). In a recent study, defects in mitophagy were detected in post-mortem hippocampal samples of AD patients, in induced pluripotent stem cell-derived human AD neurons and in mouse and nematode AD models. Importantly, the restoration of mitophagy ameliorated cognitive deficits in nematode and mouse models of AD by inhibiting Aβ plaques and tau hyper-phosphorylation. In Aβ and tau C. elegans models of AD, memory improvement was mediated by the PINK-1-, PDR-1- or DCT-1-dependent pathways (Fang et al. 2019b). Taken together, these findings support the view that mitophagy defects and eventually perturbation of mitochondrial homeostasis are intimately linked to AD pathology. Accordingly, genetic or pharmacological modulation of mitophagy would be an attractive therapeutic intervention for AD.
Similarly, recent evidence supports the involvement of mitochondrial dysfunction and defective mitophagy in the pathogenesis of Huntington’s disease (HD). This is one of a number of dominant inherited late-onset neurodegenerative disorders caused by aberrant polyglutamine expansion in the gene encoding the huntingtin protein (Orr 2001). Recent work on cellular and mouse models of HD and cells from HD patients suggested that cytosolic and mitochondrial cargo are inefficiently engulfed by autophagosomes under these diseased states, and consequently, their removal is perturbed (Martinez-Vicente et al. 2010). Along similar lines, mitophagy was significantly reduced in a mouse model of HD expressing human Huntingtin’s transgene (HTT) along with the mt-Keima reporter compared to its control counterpart (Sun et al. 2015). In agreement, neuronal expression of mutant huntingtin (mHtt) was shown to affect mitochondrial morphology in a Drosophila model of HD. PINK1 overexpression rescued mitochondrial abnormalities, ameliorated neuronal integrity and increased survival in HD flies. These neuroprotective effects were dependent on Parkin and required mitofusin and VDAC1/Porin. In line with this, PINK1 overexpression, at least partially, rescued mitophagy defects in HD striatal cells derived from a HdhQ111 knock-in mouse model of HD. Collectively, these findings indicate that mitophagy is impaired in mHtt-linked diseases and its restoration may ameliorate HD pathology (Khalil et al. 2015).
Concluding remarks and future perspectives
Multiple quality control pathways help mitochondria maintain proper function under physiological conditions and in response to stress. Perturbations in mitochondrial function and defects in mitochondrial quality control systems are hallmarks of ageing and age-related pathologies. Over the last decade, several studies highlight the roles of mitophagy in distinct physiological and pathological contexts. Monitoring in vivo mitophagy using animal disease models helped uncover the involvement of impaired mitophagy in many human pathologies, including neurodegenerative disorders. Therefore, modulation of mitophagy by pharmacological agents seems to hold great promise for therapeutic applications. Indeed, many natural and/or synthetic compounds, including NAD+ precursors (Fang et al. 2019a), resveratrol (Varghese et al. 2020), spermidine (Schroeder et al. 2021) and urolithin A (Ryu et al. 2016) have been shown to alleviate various disease phenotypes by modulating mitophagy in patient-derived samples and in model organisms. Importantly, oral consumption of urolithin A has been found to improve mitochondrial and cellular health in a human clinical trial (Andreux et al. 2019). Although the role of mitophagy in both physiological and pathological contexts is being increasingly appreciated, issues related to tissue- and disease- specificity are still poorly understood. Moreover, important questions remain unanswered regarding the complex interplay between different mitophagy pathways and the interface of mitophagy with other mitochondrial quality control systems under physiological conditions. A better understanding of such crosstalk will provide critical insights into the roles of mitophagy in neuronal function and dysfunction and will eventually contribute towards developing emergent intervention strategies to fight numerous pathological conditions in humans. As an initial step to address these topics, future studies should involve the development of novel genetically encoded reporters to monitor neuronal mitophagy in vivo as well as genetic and pharmacological interventions to modulate mitophagy in cell type- and tissue-specific contexts.
Acknowledgements
We apologize to those colleagues whose work could not be referenced due to space limitations.
Funding
D. T. is supported by HealthAge (a joint training and research program on Lifespan Regulation Mechanisms in Health and Disease – GA-812830). Work in the authors’ laboratory is funded by grants from the European Research Council (ERC-GA695190-MANNA) and the General Secretariat for Research and Innovation of the Greek Ministry of Development and Investments.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Andreux PA, Blanco-Bose W, Ryu D, Burdet F, Ibberson M, Aebischer P, Auwerx J, Singh A, Rinsch C (2019) The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat Metab 1:595–603. 10.1038/s42255-019-0073-4 [DOI] [PubMed]
- Ashrafi G, Schwarz TL (2013) The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 20:31–42. 10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed]
- Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL (2014) Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol 206:655–670. 10.1083/jcb.201401070 [DOI] [PMC free article] [PubMed]
- Bahat A, MacVicar T, Langer T (2021) Metabolism and innate immunity meet at the mitochondria. Front Cell Dev Biol 9:720490. 10.3389/fcell.2021.720490 [DOI] [PMC free article] [PubMed]
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510:370–375. 10.1038/nature13418 [DOI] [PubMed]
- Bonello F, Hassoun SM, Mouton-Liger F, Shin YS, Muscat A, Tesson C, Lesage S, Beart PM, Brice A, Krupp J, Corvol JC, Corti O (2019) LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson’s disease. Hum Mol Genet 28:1645–1660. 10.1093/hmg/ddz004 [DOI] [PubMed]
- Braak H, Braak E (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82:239–259. 10.1007/BF00308809 [DOI] [PubMed]
- Bray N (2019) Many makes of mitochondria. Nat Rev Neurosci 20:645. 10.1038/s41583-019-0229-y [DOI] [PubMed]
- Cai Q, Zakaria HM, Simone A, Sheng ZH (2012) Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol 22:545–552. 10.1016/j.cub.2012.02.005 [DOI] [PMC free article] [PubMed]
- Cardanho-Ramos C, Faria-Pereira A, Morais VA (2020) Orchestrating mitochondria in neurons: cytoskeleton as the conductor. Cytoskeleton (hoboken) 77:65–75. 10.1002/cm.21585 [DOI] [PMC free article] [PubMed]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441:1162–1166. 10.1038/nature04779 [DOI] [PubMed]
- Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, De Strooper B, Verstreken P, Vandenberghe W. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen T, Vilain S, Vints K, Gounko N, Verstreken P, Vandenberghe W (2018) Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. eLife 7:e35878. 10.7554/eLife.35878 [DOI] [PMC free article] [PubMed]
- Cunningham CN, Baughman JM, Phu L, Tea JS, Yu C, Coons M, Kirkpatrick DS, Bingol B, Corn JE. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat Cell Biol. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- Devireddy S, Liu A, Lampe T, Hollenbeck PJ (2015) The organization of mitochondrial quality control and life cycle in the nervous system in vivo in the absence of PINK1. J Neurosci 35:9391–9401. 10.1523/JNEUROSCI.1198-15.2015 [DOI] [PMC free article] [PubMed]
- Evans CS, Holzbaur EL (2020a) Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. eLife 9: e50260. 10.7554/eLife.50260 [DOI] [PMC free article] [PubMed]
- Evans CS, Holzbaur ELF (2020b) Quality control in neurons: mitophagy and other selective autophagy mechanisms. J Mol Biol 432:240–260. 10.1016/j.jmb.2019.06.031 [DOI] [PMC free article] [PubMed]
- Fang EF, Hou Y, Lautrup S, Jensen MB, Yang B, SenGupta T, Caponio D, Khezri R, Demarest TG, Aman Y, Figueroa D, Morevati M, Lee HJ, Kato H, Kassahun H, Lee JH, Filippelli D, Okur MN, Mangerich A, Croteau DL, Maezawa Y, Lyssiotis CA, Tao J, Yokote K, Rusten TE, Mattson MP, Jasper H, Nilsen H, Bohr VA (2019a) NAD(+) augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat Commun 10:5284. 10.1038/s41467-019-13172-8 [DOI] [PMC free article] [PubMed]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, Rocktaschel P, Croteau DL, Akbari M, Greig NH, Fladby T, Nilsen H, Cader MZ, Mattson MP, Tavernarakis N, Bohr VA (2019b) Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci 22:401–412. 10.1038/s41593-018-0332-9 [DOI] [PMC free article] [PubMed]
- Galizzi G, Palumbo L, Amato A, Conigliaro A, Nuzzo D, Terzo S, Caruana L, Picone P, Alessandro R, Mule F, Di Carlo M (2021) Altered insulin pathway compromises mitochondrial function and quality control both in in vitro and in vivo model systems. Mitochondrion 60:178–188. 10.1016/j.mito.2021.08.014 [DOI] [PubMed]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131. 10.1038/ncb2012 [DOI] [PubMed]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A 100:4078–4083. 10.1073/pnas.0737556100 [DOI] [PMC free article] [PubMed]
- Ham SJ, Lee D, Yoo H, Jun K, Shin H, Chung J (2020) Decision between mitophagy and apoptosis by Parkin via VDAC1 ubiquitination. Proc Natl Acad Sci U S A 117:4281–4291. 10.1073/pnas.1909814117 [DOI] [PMC free article] [PubMed]
- Han S, Jeong YY, Sheshadri P, Cai Q (2020) Mitophagy coordination with retrograde transport ensures the integrity of synaptic mitochondria. Autophagy 16:1925–1927. 10.1080/15548627.2020.1810919 [DOI] [PMC free article] [PubMed]
- Harper JW, Ordureau A, Heo JM (2018) Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol 19:93–108. 10.1038/nrm.2017.129 [DOI] [PubMed]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW (2015) The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell 60:7–20. 10.1016/j.molcel.2015.08.016 [DOI] [PMC free article] [PubMed]
- Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S (2013) Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 4:2308. 10.1038/ncomms3308 [DOI] [PubMed]
- Hsieh CH, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St Lawrence E, Schule B, Krainc D, Palmer TD, Wang X (2016) Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell 19:709–724. 10.1016/j.stem.2016.08.002 [DOI] [PMC free article] [PubMed]
- Joselin AP, Hewitt SJ, Callaghan SM, Kim RH, Chung YH, Mak TW, Shen J, Slack RS, Park DS (2012) ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet 21:4888–4903. 10.1093/hmg/dds325 [DOI] [PubMed]
- Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, Fang EF. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151–166. doi: 10.1016/j.tins.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil B, El Fissi N, Aouane A, Cabirol-Pol MJ, Rival T, Lievens JC. PINK1-induced mitophagy promotes neuroprotection in Huntington's disease. Cell Death Dis. 2015;6:e1617. doi: 10.1038/cddis.2014.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook S, Zhan X, Thibeault K, Ahmed MR, Gurevich VV, Gurevich EV. Mdm2 enhances ligase activity of parkin and facilitates mitophagy. Sci Rep. 2020;10:5028. doi: 10.1038/s41598-020-61796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees AJ, Hardy J, Revesz T. Parkinson's disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- Lin MY, Cheng XT, Tammineni P, Xie Y, Zhou B, Cai Q, Sheng ZH. Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron. 2017;94:595-610 e6. doi: 10.1016/j.neuron.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, Takahashi R, Hattori N, Imai Y, Lu B. Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Domenech G, Howden JH, Covill-Cooke C, Morfill C, Patel JV, Burli R, Crowther D, Birsa N, Brandon NJ, Kittler JT. Loss of neuronal Miro1 disrupts mitophagy and induces hyperactivation of the integrated stress response. EMBO J. 2021;40:e100715. doi: 10.15252/embj.2018100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, Zhang Y, Dawson VL, Dawson TM, Lenardo M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson's-like movement disorder. Nat Commun. 2014;5:4930. doi: 10.1038/ncomms5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Lee SA, McBride HM, Fon EA. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214:275–291. doi: 10.1083/jcb.201603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 2018;27:439-449 e5. doi: 10.1016/j.cmet.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, Haynes CM. UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Padman BS, Lazarou M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- Niu K, Fang H, Chen Z, Zhu Y, Tan Q, Wei D, Li Y, Balajee AS, Zhao Y. USP33 deubiquitinates PRKN/parkin and antagonizes its role in mitophagy. Autophagy. 2020;16:724–734. doi: 10.1080/15548627.2019.1656957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40:e104705. doi: 10.15252/embj.2020104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, Wells JA, Gygi SP, Schulman BA, Harper JW. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT. Beyond the Qs in the polyglutamine diseases. Genes Dev. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pickles S, Vigie P, Youle RJ (2018) Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A, Saini H, Sethi S, O'Sullivan GA, Plun-Favreau H, Wray S, Dawson LA, McCarthy JM. The role of SQSTM1 (p62) in mitochondrial function and clearance in human cortical neurons. Stem Cell Reports. 2021;16:1276–1289. doi: 10.1016/j.stemcr.2021.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde KR, Smith HL, Chau KY, Schapira AH. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. J Cell Biol. 2016;213:163–171. doi: 10.1083/jcb.201509003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschmann A, Fiesel FC, Caulfield TR, Hudec R, Ando M, Truban D, Hou X, Ogaki K, Heckman MG, James ED, Swanberg M, Jimenez-Ferrer I, Hansson O, Opala G, Siuda J, Boczarska-Jedynak M, Friedman A, Koziorowski D, Rudzinska-Bar M, Aasly JO, Lynch T, Mellick GD, Mohan M, Silburn PA, Sanotsky Y, Vilarino-Guell C, Farrer MJ, Chen L, Dawson VL, Dawson TM, Wszolek ZK, Ross OA, Springer W. Heterozygous PINK1 p. G411S increases risk of Parkinson's disease via a dominant-negative mechanism. Brain. 2017;140:98–117. doi: 10.1093/brain/aww261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Felix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22:879–888. doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S, Hofer SJ, Zimmermann A, Pechlaner R, Dammbrueck C, Pendl T, Marcello GM, Pogatschnigg V, Bergmann M, Muller M, Gschiel V, Ristic S, Tadic J, Iwata K, Richter G, Farzi A, Ucal M, Schafer U, Poglitsch M, Royer P, Mekis R, Agreiter M, Tolle RC, Sotonyi P, Willeit J, Mairhofer B, Niederkofler H, Pallhuber I, Rungger G, Tilg H, Defrancesco M, Marksteiner J, Sinner F, Magnes C, Pieber TR, Holzer P, Kroemer G, Carmona-Gutierrez D, Scorrano L, Dengjel J, Madl T, Sedej S, Sigrist SJ, Racz B, Kiechl S, Eisenberg T, Madeo F. Dietary spermidine improves cognitive function. Cell Rep. 2021;35:108985. doi: 10.1016/j.celrep.2021.108985. [DOI] [PubMed] [Google Scholar]
- Song J, Herrmann JM, Becker T. Quality control of the mitochondrial proteome. Nat Rev Mol Cell Biol. 2021;22:54–70. doi: 10.1038/s41580-020-00300-2. [DOI] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T (2015) Measuring In Vivo Mitophagy. Mol Cell 60:685–696. 10.1016/j.molcel.2015.10.009 [DOI] [PMC free article] [PubMed]
- Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Tandarich LC, Nguyen K, Hollenbeck PJ. Compartmentalized regulation of parkin-mediated mitochondrial quality control in the Drosophila nervous system in vivo. J Neurosci. 2016;36:7375–7391. doi: 10.1523/JNEUROSCI.0633-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JX, Finkel T (2020) Mitochondria as intracellular signaling platforms in health and disease. J Cell Biol 219(5):e202002179. 10.1083/jcb.202002179 [DOI] [PMC free article] [PubMed]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- Varghese N, Werner S, Grimm A, Eckert A (2020) Dietary mitophagy enhancer: a strategy for healthy brain aging? Antioxidants 9(10):932. 10.3390/antiox9100932 [DOI] [PMC free article] [PubMed]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Serricchio M, Jauregui M, Shanbhag R, Stoltz T, Di Paolo CT, Kim PK, McQuibban GA. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wan C, He T, Han C, Zhu K, Waddington JL, Zhen X (2021) Sigma-1 receptor regulates mitophagy in dopaminergic neurons and contributes to dopaminergic protection. Neuropharmacology 196:108360. 10.1016/j.neuropharm.2020.108360 [DOI] [PubMed]
- Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Shen H, Li J, Guo LW. SIGMAR1/Sigma-1 receptor ablation impairs autophagosome clearance. Autophagy. 2019;15:1539–1557. doi: 10.1080/15548627.2019.1586248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Sun X, Starovoytov V, Cai Q. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer's disease patient brains. Hum Mol Genet. 2015;24:2938–2951. doi: 10.1093/hmg/ddv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Puri R, Yang H, Lizzio MA, Wu C, Sheng ZH, Guo M. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife. 2014;3:e01958. doi: 10.7554/eLife.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fang Y, Zhao X, Zheng Y, Ma Y, Li S, Huang Z, Li L. miR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol Ther Nucleic Acids. 2021;24:822–831. doi: 10.1016/j.omtn.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YR, Zhang XN, Chen Z. Mitochondrial transport serves as a mitochondrial quality control strategy in axons: implications for central nervous system disorders. CNS Neurosci Ther. 2019;25:876–886. doi: 10.1111/cns.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]