Abstract
Regulation of gene expression by small non-coding RNAs, such as miRNAs and siRNAs, is an inherent part of complex biological processes that allow function and survival of eukaryotic cells. The type III ribonuclease DICER is widely recognized as a key step in the production of miRNAs and siRNAs, although it also has non-canonical functions such as DNA repair and induction of apoptosis. DICER is at the cornerstone of most biological processes; hence, its mRNA and protein levels are subject to multiple layers of regulation. Accordingly, DICER derangement is related to disease and leads to accelerated aging. Interventions that boost DICER function hold great potential as strategies to promote health and longevity. In this review, we will summarize the structural features of DICER, describe its canonical and non-canonical roles, and discuss the most common regulatory mechanisms having an impact on DICER abundance and function. We will also touch upon the current literature demonstrating that DICER deregulation can lead to diseases, thus highlighting the importance of DICER in warranting the beneficial effects of health-promoting interventions.
Keywords: DICER, miRNAs, RNAi, Aging
Introduction
Organisms must regulate their biological processes in order to survive and succeed as species. This includes fine-tuning gene expression, which is partially accomplished by post-transcriptional regulation by microRNAs (miRNAs) and other types of small non-coding RNAs (ncRNAs) (Ebert and Sharp 2012). miRNAs guide post-transcriptional silencing of mRNA targets by translational repression and/or mRNA destabilization (Eichhorn et al. 2014). First discovered in the nematode Caenorhabditis elegans (C. elegans), they have now been identified in a wide variety of eukaryotes. Based on the most stringent definition criteria, there are at least 500 human miRNAs, many of them conserved in other animals. Given that each miRNA can target hundreds of mRNAs, miRNAs are predicted to have a wide impact over the transcriptome/translatome (Bartel 2018).
In humans, the canonical miRNA processing pathway initiates with the transcription of miRNA genes by RNA polymerase II giving rise to “pri-miRNAs”, which fold back and form a hairpin structure that is processed by the microprocessor complex containing the double-stranded RNA-binding protein (dsRBP) DGCR8 and the RNase III DROSHA. The product, a ~ 70-nt stem-loop called “pre-miRNA”, is exported to cytoplasm by Exportin-5, and there, the DICER complex recognizes and cleaves the pre-miRNA loop hairpin, generating a miRNA duplex of ~ 22-nt with a 2-nt overhang at the 3′-end. DICER and the auxiliary proteins will load and deliver the miRNA duplex onto argonaute (AGO) proteins, thus forming the RNA-induced silencing complex (RISC). One strand of the duplex, called the “passenger” strand, is removed, while the other, the “guide” strand, remains in the complex to direct the silencing of target transcripts (Ha and Kim 2014; Bartel 2018). Although a large number of miRNAs requires DROSHA and DICER for their biogenesis, some miRNAs bypass the cleavage activity of these enzymes. The miRtrons are miRNAs generated from intronic regions and do not depend upon DROSHA-mediated cleavage; instead, they undergo a lariat-debranching processing by the debranching enzyme 1 (DBR1) (Ruby et al. 2007). Similarly, miRNAs derived from small nucleolar RNAs (snoRNAs) and from transfer RNAs (tRNAs) are alternatively processed and do not require DROSHA (Scott et al. 2009; Li et al. 2018). To our knowledge, miR-451 is the only miRNA described to date to be processed independently of DICER (Cheloufi et al. 2010; Stavast and Erkeland 2019).
At the same time DICER is a key enzyme in the miRNA pathway, it is also a central player in the RNA interference (RNAi) pathway, a more ancient gene-silencing pathway involved in a myriad of eukaryotic processes such as viral defense, transposon silencing, and development (MacRae et al. 2006; Shabalina and Koonin 2008; Bartel 2018). In the RNAi pathway, DICER cleaves long double-stranded RNAs (dsRNAs) to generate small interfering RNAs (siRNAs) (Shabalina and Koonin 2008; Kurzynska-Kokorniak et al. 2015).
While DICER has been recognized to process various ncRNAs other than just miRNAs and siRNAs, accumulating evidence also shows that DICER operates beyond its function in small RNA biogenesis (Rybak-Wolf et al. 2014; Burger and Gullerova 2015; Kurzynska-Kokorniak et al. 2015; Song and Rossi 2017). Some of these non-canonical DICER functions are independent of its ribonuclease activity, including the association with P-bodies/RNA granules by passive binding to target RNAs (Rybak-Wolf et al. 2014) and promotion of apoptosis after acquisition of deoxyribonuclease activity (Nakagawa et al. 2010).
DICER was first discovered 20 years ago in Drosophila melanogaster (Bernstein et al. 2001) and soon shown to be conserved in many species, although certain differences in DICER domains, isoforms, catalytic activity, and number of family members exist across phyla (Kurzynska-Kokorniak et al. 2015; Song and Rossi 2017). DICER abundance and function are tightly controlled by several layers of regulatory mechanisms, which go from transcriptional (Jafarnejad et al. 2013) to post-translational regulation (Rigbolt et al. 2011; Gross et al. 2014). These layers of regulation have to be fine-tuned to ensure dynamic changes in DICER function to occur in response to environmental challenges without disrupting homeostasis. Accordingly, disbalance in DICER levels and activity can accelerate aging and cause disease, including cancer and metabolic disease (Hill et al. 2009; Mori et al. 2012; Emde et al. 2015; Aryal et al. 2019), while promotion of DICER function may be beneficial (Mori et al. 2012; Pinto et al. 2018; Guerra et al. 2019; Rocha et al. 2020).
In this review, we will cover the roles of each DICER domain as well as the canonical and non-canonical functions of DICER. Then, we will provide an overview on how DICER is regulated. Finally, we will discuss physiological implications of DICER dysregulation on disease. Due to space constraints, we will focus mostly on C. elegans and mammalian data.
DICER structure
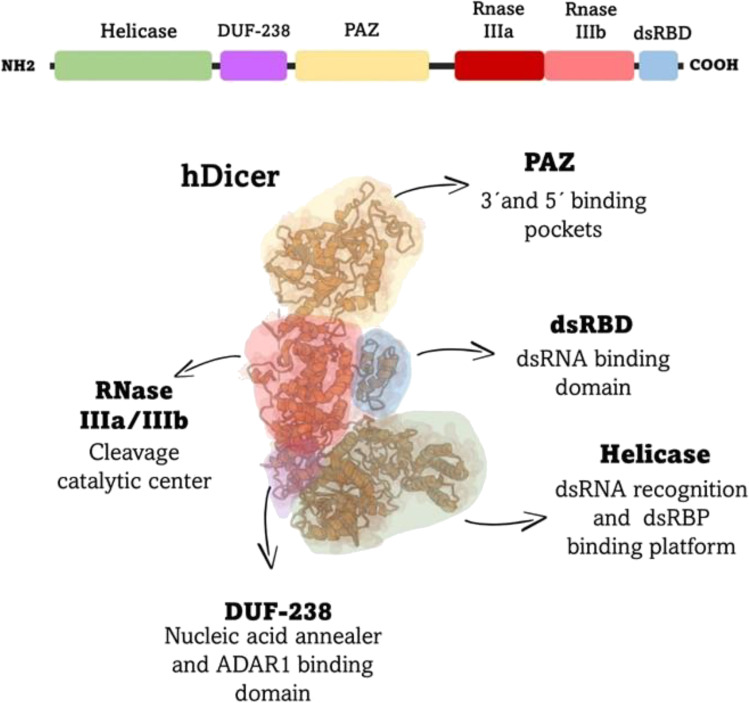
DICER is a multidomain endonuclease that belongs to the fourth class of type III ribonuclease (RNase III) enzymes. DICER is encoded by a single gene in vertebrates and C. elegans, although it can be present in more paralogs in other species. For instance, Drosophila melanogaster has two DICER paralogs, while Arabidopsis thaliana presents four (Paturi and Deshmukh 2021). In humans, DICER is encoded by the DICER1 gene located at chromosome 14, comprising a region of about 72 kbp containing 29 exons. Human DICER (hDICER) consists of 1922 amino acids (~ 220 kDa) which represents the largest and most complex member of the RNase III family (Zhang et al. 2004). Recently, a study by Liu and colleagues reconstructed the 3D structure of hDICER using cryo-electron microscopy, revealing its architecture as an L-shaped molecule which consists of a N-terminal helicase, a DUF283 domain, a Platform-PAZ-connector helix domain, two RNase III domains (RNase IIIa and RNase IIIb), and a C-terminal double-stranded RNA-binding domain (dsRBD) (Liu et al. 2018) (Fig. 1). Below a more detailed description of the DICER domains and their function:
Fig. 1.
hDICER domains and their respective roles. Schematic illustration of domain arrangement of hDICER and the 3D cartoon representation of hDICER domains colored, and their main functions. The 3D cartoon representation was obtained from RCSB Protein Data Bank (Burley et al. 2021), ID: 5ZAL (Liu et al. 2018) using Mol*viewer (Sehnal et al. 2021). The RNA, present in the cryo-EM map, has been omitted.
Helicase
DICER’s helicase domain is structurally organized into three lobes consisted of HEL1, HEL2i, and HEL2 subdomains (Liu et al. 2018). These subdomains are conserved in DExD/H and RIG-I-like helicase families. While DExD/H-box helicases unwind RNA or DNA duplexes, RIG-l-like helicases translocate along these nucleic acids (Byrd and Raney 2012). Regarding spatial organization, the HEL1 subdomain links the long and short arms of the “L” via interaction with the DUF283 and RNase IIIb domains (Liu et al. 2018). HEL2 is located between the other two helicase subdomains, on a C-shaped arrangement, whereas HEL2i rests at the end of the short arm of the “L” (Liu et al. 2018). The helicase domain discriminates pre-siRNAs and pre-miRNAs depending on the dsRNA termini, in a process that can be fueled or not by ATP. For example, C. elegans and D. melanogaster DICER cleave dsRNAs with blunt termini in an ATP-dependent manner, while processing of pre-miRNAs, which carry a 3’-overhang, does not require ATP (Welker et al. 2011; Sinha et al. 2018). The helicase domains from vertebrate DICER are not ATP-dependent (Zhang et al. 2002), and their discrimination mechanism relies on the interaction with the apical loop of pre-miRNA substrates. Besides dsRNA discrimination, the helicase domain serves as a platform for binding double-strand RNA-binding proteins (dsRBPs) (Lee et al. 2006).
DUF238
Although the DUF283 (domain of unknown function) of hDICER has structural similarities with dsRBPs, it does not bind dsRNAs, but instead, binds single-stranded nucleic acids and acts as a nucleic acid annealer, facilitating the hybridization between RNA or DNA complementary strands (Kurzynska-Kokorniak et al. 2016). This domain is also important for the binding of adenosine deaminase acting on RNA 1 (ADAR1), a protein that partners with DICER to increase efficiency of pri-miRNA cleavage and miRNA transfer to AGO proteins (Ota et al. 2013). Interestingly, hDICER with deleted DUF238 has increased binding but reduced cleavage efficiency of dsRNA, whereas it can bind and cut pre-miRNA substrates as efficient as the wild type hDICER (Ma et al. 2008).
PAZ domain
The Piwi/Ago/Zwille (PAZ) is a highly conserved domain found in DICER and AGO proteins. This domain possesses a variant of the OB (oligonucleotide/oligosaccharide-binding) fold, a structure involved in single-stranded nucleic acid binding (Song et al. 2003). The PAZ domain has adjacent elements, the structured linker platform and the connector helix (Tian et al. 2014). The PAZ domain has a 3’ binding pocket where the 3’ overhang of the RNA substrate is connected by hydrogen bonds, while the platform has a phosphate binding pocket that accommodates the 5’-phosphate end (Song et al. 2003; Tian et al. 2014). A truncated version of hDICER lacking the PAZ domain is not able to process pre-miRNA or dsRNA substrates, but still retains both RNase and DNase activity over short single-stranded RNA and DNAs, suggesting that in the absence of the PAZ domain, non-canonical products can be generated (Wojnicka et al. 2020).
RNase domains
DICER has two RNase domains: RNase IIIa and IIIb. These domains form a dimer that constitutes the single dsRNA cleavage center, the catalytic core of DICER, which processes dsRNAs generating shorter RNA fragments of 18–24 bp with an 2-nt 3´ overhang (Zhang et al. 2004; MacRae et al. 2006). DICER RNase III domains, in the presence of divalent metal ions (e.g., Mg2+), catalyze the hydrolysis of a phosphodiester bond within each strand of the dsRNA, in a way that the RNase IIIa domain cleaves the 3´hydroxyl-bearing strand, whereas the RNase IIIb domain cleaves the 5′ phosphate-bearing RNA strand (Zhang et al. 2004; MacRae et al. 2006).
dsRBD
The dsRBD is a binding domain that comprises 65–70 amino acids folding into a αβββα conformation (Masliah et al. 2013). The main function of the dsRBD is to bind to dsRNA. Even though dsRBD can bind to specific dsRNA sequences, binding of this domain to substrates appears to be shape-dependent (Masliah et al. 2013). The dsRBD was proven necessary for RNA substrate binding in the absence of the PAZ domain (Ma et al. 2012), and its deletion reduces processing of blunt dsRNA, making hDICER processing more dependent on the presence of the 3′-overhang (Zhang et al. 2004). Interestingly, a nuclear localization signal is present in the dsRBD, which supports evidence of noncanonical functions of DICER in the nucleus (Pong and Gullerova 2018).
DICER-associated proteins
DICER can bind to other proteins to form a complex that enhances small RNA processing. DICER-interacting dsRBPs are present in different species, such as R2D2 and Loqs-PD in D. melanogaster, RDE-4 in C. elegans, and PACT and TRBP in humans (Liu et al. 2003; Masliah et al. 2013). In humans, the binding of PACT and TRBP to DICER seems to be mutually exclusive, which leads to distinctive effects on substrate selection and cleavage (Masliah et al. 2013). In association with PACT, hDICER has a cleavage preference for pre-miRNAs rather than pre-siRNAs, whereas this is not evident when hDICER is bound to TRBP (Lee et al. 2013). TRBP increases substrate recognition by DICER and enhances the stability of DICER/dsRNA complexes, leading to a better cleavage efficiency (Chakravarthy et al. 2010). TRBP also participates in assembling the RISC and is one of the components of this complex (Chendrimada et al. 2005; Ma et al. 2008; Masliah et al. 2013). Moreover, evidence suggest that PACT and TRBP may affect the cleavage site of dsRNAs, thus allowing generation of different sized miRNAs, also called iso-miRs (Masliah et al. 2013). Interestingly, as it is the case for some miRNAs, TRBP interferes with the position where hDICER cleaves the precursor and switches guide strand selection (Masliah et al. 2013). In addition to PACT and TRBP, ADAR1 has also been shown to bind to hDICER. The complex ADAR1/hDICER enhances pre-miRNA cleavage reaction and promotes loading of miRNA onto the RISC (Ota et al. 2013).
DICER function
dsRNA and pre-miRNA processing
DICER was firstly recognized for its role in siRNA processing. In a seminal work, Bernstein and colleagues showed that D. melanogaster DICER cleaves dsRNA precursors into ~ 22-nt small dsRNAs revealing a fundamental role for DICER in the RNAi pathway (Bernstein et al. 2001). Shortly after, DICER was also shown to cleave hairpin RNA structures and be essential for miRNA maturation (Hutvágner et al. 2001).
In some species, including humans, DICER recognizes different dsRNA substrates by its PAZ and helicase domains. Studies using recombinant D. melanogaster Dcr-2 show that dsRNA containing a 2-nt 3’ overhang interacts with the PAZ domain in an ATP-independent manner, whereas dsRNA with blunt termini is threaded through the helicase domain, cleaved and then the 2-nt 3’ overhang of the dsRNA is recognized by the PAZ domain (Sinha et al. 2018). Regarding the cleavage position, different models have been proposed, including the 3’ counting rule, the 5’ counting rule, and the loop counting rule, which consider the distance from either the 3’-end, the 5’-end, or the loop of the pre-miRNA (MacRae et al. 2007; Park et al. 2011; Gu et al. 2012). Recently, it was confirmed that several pre-miRNAs are indeed cleaved by hDICER using at least one of these counting rules (Luo et al. 2021). Regardless of the model, the physical distance between the PAZ (3’-end and 5’-end binding pockets) and the RNase IIIa catalytic site, which is about 58 Å, is determinant for the cleavage site (Liu et al. 2018). The catalysis mechanism of the RNase III domain is shared with all the enzymes in the RNase III family, which use metal ion-mediated hydrolysis (Nowotny and Yang 2006). Each RNase III domain catalyzes the hydrolytic cleavage of one strand of the dsRNA substrate, a process facilitated by the presence of divalent cations in the catalytic sites (MacRae et al. 2006).
Non-canonical functions
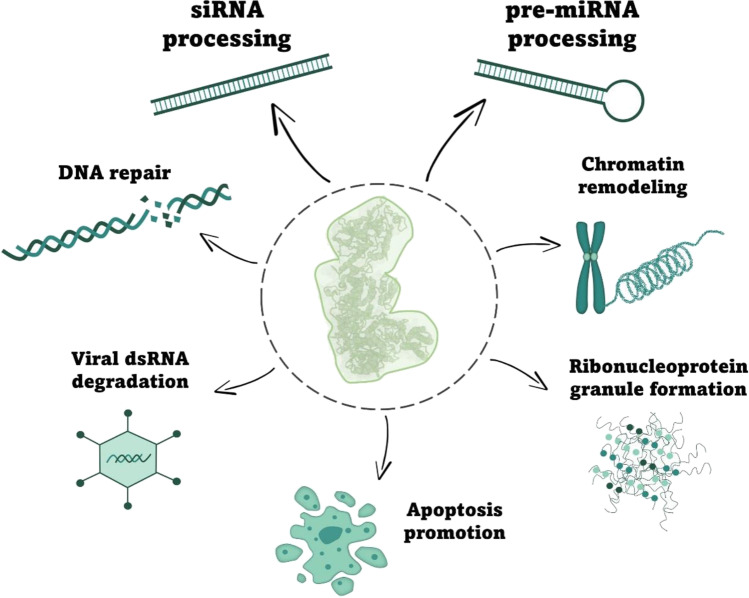
In addition to dsRNA and pre-miRNA processing, DICER has alternative functions, some of which are related to its nuclear localization (Fig. 2). DICER has been reported to have a role in DNA repair; this ribonuclease is highly expressed in the developing cerebellum in mice, and its deficiency increases DNA damage in that region (Swahari et al. 2016). Moreover, in vitro evidence shows that DNA double-strand breaks (DSBs) induce hDICER phosphorylation leading to accumulation of DICER in the nucleus, where it is then recruited to DSB sites to facilitate DNA damage response (Burger et al. 2017). In C. elegans cells, DICER (DCR-1) was implicated with chromatin remodeling in response to UV radiation, a phenomenon shown to be independent of DICER catalytic activity (Chitale and Richly 2017). In addition, DICER mediates the recruitment of the histone methyltransferase MMSET to the DNA damage site, which leads to efficient nucleotide excision repair (Chitale and Richly 2018). Conversely, C. elegans CED-3 caspase cleaves DCR-1 to generate a truncated version with deoxyribonuclease activity, which produces 3' hydroxyl DNA breaks, promoting apoptosis (Nakagawa et al. 2010). In addition, C. elegans DCR-1 is required for ribonucleoprotein granule formation, suggesting a role in RNA localization and assembly of RNA–protein complexes (Beshore et al. 2011). Indeed, later evidence has also shown that hDICER and C. elegans DCR-1 were functionally linked to RNA granules. hDICER and DCR-1 interact with mRNAs carrying “passive” sites that sequestrate DICER endonuclease activity (Rybak-Wolf et al. 2014). Many of these mRNAs with “passive” sites were shown to encode RNA granule-associated genes, and their translated proteins were able to interact with DICER (Rybak-Wolf et al. 2014). Recent data has also identified the role of an alternatively spliced hDICER isoform capable of processing viral dsRNA into siRNA to protect stem cells from viral infection (Poirier et al. 2021). These exciting findings defy the previous assumption that, by having the dsRNA processing inhibited by its own helicase domain, mammalian DICER would not use RNAi to fight viral infection as plants and invertebrates do.
Fig. 2.
Functions of DICER. Illustrative scheme showing the canonical (siRNA and pre-miRNA processing) and non-canonical functions (chromatin remodeling, DNA repair, ribonucleoprotein granule formation, apoptosis promotion, and viral dsRNA degradation) of DICER
DICER regulation
Our knowledge about the molecular mechanisms controlling DICER expression and function under basal conditions is limited, although it is clear that different stress conditions such as autophagy, apoptosis, and hypoxia culminate in DICER regulation (Matskevich and Moelung 2008; Gibbings et al. 2012; Van Den Beucken et al. 2014). It is important to elucidate the regulatory mechanisms affecting DICER levels and/or function given the association between DICER and disease pathogenesis (Mori et al. 2012; Boon et al. 2013; Theotoki et al. 2020). Below are examples of the current knowledge regarding these mechanisms:
Transcriptional regulation
Transcriptional regulation of the DICER gene is complex considering the wide variety of mRNA isoforms generated by a combination of alternative promoters and alternative splicing. Four DICER mRNA variants encode full-length hDICER (containing 1922-amino acid residues) only diverging in their 5’- and 3’-untranslated regions (UTRs) (https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000100697;r=14:95086228-95158010). Other variants, however, may result in alternative open reading frames or change mRNA stability or translational efficiency. For example, in mice, an intronic MT-C retrotransposon promoter gives rise to an oocyte-specific Dicer mRNA variant which is translated into a DICER isoform that lacks the N-terminal helicase domain and confers more efficient production of small RNAs from long dsRNA, thus boosting the endogenous RNAi pathway in mouse oocytes (Flemr et al. 2013).
Overall, the transcriptional regulation of the DICER gene is operated by selective transcriptional factors and controlled by epigenetic mechanisms. In breast cancer cells, hypoxia reduces DICER expression by silencing the DICER1 promoter through oxygen-dependent inhibition of H3K27me3 demethylases KDM6A/B (Van Den Beucken et al. 2014). There are several transcription factors shown to control DICER expression on a cell-, tissue- or developmental stage-dependent manner. SOX4 plays a role in the regulation of embryonic development and determination of cell fate, and its function partially relies on its ability to trigger DICER1 transcription (Jafarnejad et al. 2013). Microphthalmia-associated transcription factor (MITF) triggers the expression of the DICER1 gene in melanocytes by binding to a conserved regulatory sequence (Levy et al. 2010). The DICER1 promoter can also be targeted by p53, p63, or TAp63 (Boominathan 2010; Su et al. 2010).
Post-transcriptional regulation
The level of DICER transcripts does not always mirror the protein levels, which illustrates the complexity of DICER regulation and highlights the importance of post-transcriptional mechanisms (Kurzynska-Kokorniak et al. 2015). The notion that DICER abundance is determined by post-transcriptional mechanisms is further supported by evidence showing that pharmacological interventions that are expected to interfere with transcription, such as the use of histone deacetylase inhibitors, slightly change DICER1 mRNA levels while markedly decrease DICER protein levels in human cells (Wiesen and Tomasi 2009). How exactly DICER protein abundance is controlled remains elusive, although many post-transcriptional mechanisms have been described.
Several alternative splicing variants have been identified for the DICER transcript. Some of these splicing variants encode shorter isoforms of the DICER protein, while others do not encode any protein (Kurzynska-Kokorniak et al. 2015). Alternative splicing or promoter selection may also create alternative open reading frames and change the 5’-UTR, thus impacting the stability of the DICER transcript and its translation (Irvin-Wilson and Chaudhuri 2005; Singh et al. 2005; Potenza et al. 2010). Indeed, DICER mRNA translation efficiency is highly dependent on the nature of the 5’-UTR (Irvin-Wilson and Chaudhuri 2005; Singh et al. 2005). The 5’-UTRs within the DICER transcript may be comprised by a combination of three unique leader exons (1A, 1B and 1C) and three alternatively spliced exons (AS1, AS2, and AS3). DICER transcripts with 5’-UTRs containing any of these alternatively spliced exons are less efficiently translated compared to the transcript variants without them (Singh et al. 2005). Moreover, DICER transcripts containing the leader exon 1B have the lowest translational efficiency (Singh et al. 2005).
In differentiated human epithelial cells and numerous cancer cell lines, shorter DICER mRNA variants are produced through alternative processing of the pre-mRNA (Grelier et al. 2009; Hinkal et al. 2011). Moreover, a splicing variant found in neuroblastoma cells lacks one of the protein coding exons, causing the translation of an alternative open reading frame with a premature stop codon that gives rise to a protein lacking the dsRBD and defective in one of the two RNase III domains (Potenza et al. 2010).
The 3’-UTR also plays a key role in post-transcriptional regulation of the DICER mRNA. DICER 3’-UTR is targeted by various miRNAs, including miR-103/107 (Grelier et al. 2009; Martello et al. 2010; Hinkal et al. 2011), miR-192 (Feinberg-Gorenshtein et al. 2013), and members of the let-7 miRNA family (Forman et al. 2008; Hinkal et al. 2011). DICER mRNA variants may also have shorter 3’-UTR lacking miRNA-binding sites and thus escaping from miRNA-mediated repression (Stavast and Erkeland 2019). These shorter variants are polyadenylated at certain sites of the transcript and are more abundant in cancer cell lines (Hamaya et al. 2012; Stavast and Erkeland 2019).
Another factor regulating the expression of DICER is the export of DICER mRNA to the cytoplasm. It has been demonstrated the saturation of Exportin-5 with other RNAs such as pre-miRNAs, viral RNAs, and some tRNAs decreases DICER protein abundance by reducing mRNA export (Bennasser et al. 2011).
Post-translational regulation
DICER protein undergoes post-translational modifications such as phosphorylation (Rigbolt et al. 2011) and SUMOylation (Gross et al. 2014). These modifications may affect DICER abundance and/or activity. In C. elegans, during oogenesis, DCR-1 becomes phosphorylated within the RNase IIIb and dsRBD domains by extracellular signal-kinase (ERK), and this is an essential step for DCR-1 nuclear translocation (Drake et al. 2014). In addition, phosphorylation of DCR-1 inhibits its function, and it has been shown that DCR-1 gets rapidly dephosphorylated prior to fertilization (Drake et al. 2014). DICER has 10 lysine moieties which are described to change DICER activity upon SUMOylation (Gross et al. 2014). Interestingly, it has been shown that alveolar macrophages from smokers undergo downregulation of several miRNAs due to SUMOylation-related decrease in DICER activity, indicating environmental factors can affect DICER function through post-translational modification (Gross et al. 2014). Indirectly, phosphorylation of TRBP by Ras signaling may also stabilize DICER and increase miRNA biogenesis in human cells (Paroo et al. 2009).
The physiological and pathophysiological roles of DICER
Given the role of miRNAs in regulating crucial cellular and organismal processes, disturbances in their production are largely connected to disease (Mori et al. 2014; Foulkes et al. 2014; Kurzynska-Kokorniak et al. 2015; Emde et al. 2015). Disruption of DICER, being a key enzyme in the miRNA biogenesis pathway, is pervasive for life and health span. For example, whole-body Dicer knockout mice die early during development (Bernstein et al. 2003), and various models of tissue-specific deletions in Dicer exhibit premature lethality (Chen et al. 2008; Mori et al. 2014). Indeed, a growing body of evidence places DICER at the cornerstone of pathways related to stress resistance, metabolism, and longevity. In mice, DICER protein levels are downregulated in adipose tissue during obesity or normal aging, which culminates in reduced miRNA levels (Mori et al. 2012; Oliverio et al. 2016). Consistently, adipose-specific removal of Dicer causes lipodystrophy, impairs metabolism, and accelerates aging (Mori et al. 2014; Reis et al. 2016). Age-related DICER downregulation can be prevented by dietary restriction (Mori et al. 2012), an intervention that prolongs lifespan. Furthermore, in C. elegans and mice, several dietary restriction protocols not only upregulate DCR-1/DICER but also depend on dcr-1/Dicer to drive their beneficial effects on metabolism and lifespan (Reis et al. 2016; Guerra et al. 2019), suggesting a positive association between DICER levels/activity and health span. Consistently, overexpression of dcr-1 in C. elegans increases lifespan and stress resistance (Mori et al. 2012). Furthermore, treatment with enoxacin—a broad spectrum fluoroquinolone that binds to TRBP and facilitates pre-miRNA processing by DICER—increases lifespan in C. elegans (Pinto et al. 2018) and ameliorates metabolism and counteracts obesity in mice (Rocha et al. 2020).
Altered post-transcriptional regulation of DICER may also impinge on health. Mice harboring constitutively phosphorylated DICER that accumulates in the nucleus display abnormal metabolism, increased cellular stress and accelerated aging (Aryal et al. 2019). This can be explained in part by decreased levels of some miRNAs, but promotion of DICER nuclear functions may also be involved (Aryal et al. 2019).
Cellular stress appears to be the main driver for DICER dysregulation. Stress resulted from pathogenic ALS-causing mutations leads to stress granule formation and reorganization of the DICER complex, which in turn impairs miRNA biogenesis (Emde et al. 2015). DICER expression is also downregulated by different pro-oxidative agents (Mori et al. 2012).
In humans, mutations in the DICER1 gene are linked to cancer. Heterozygous germline mutations in the DICER1 gene are the underlying cause of a modestly penetrant, autosomal dominant syndrome known as DICER1 syndrome (Foulkes et al. 2014). The syndrome involves increased risk for several types of cancer, including the pediatric cancer pleuropulmonary blastoma, thyroid gland neoplasia, and ovarian tumors (Hill et al. 2009; Foulkes et al. 2014; Schultz et al. 2020), as well as other developmental abnormalities (Hill et al. 2009). Most of the tumors affect individuals before the age of 40 years.
Conclusion
DICER is an evolutionarily conserved enzyme playing a central role in small RNA biogenesis and involved in a wide range of biological processes, from antiviral responses to regulation of gene expression (Fig. 2). DICER abundance and activity are tightly regulated, and defects in such regulation lead to disease. Future research assessing the landscape of DICER regulatory mechanisms, modifications, and isoforms across different conditions will be instrumental for the development of new pharmacological, genetic, and/or dietary strategies aimed at safeguarding DICER function to prevent disease and increase health and lifespan.
Author contribution
CAVS, GTS, MDI, and MAM wrote the paper. CAVS, GTS, and MDI contributed equally.
Funding
Authors were supported by grants of FAPESP (2019/01587–1 to C.A.V-J and 2019/25958–9 to M.A.M), CNPq (310287/2018–9 to M.A.M.) and CAPES (“This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001” to G.T-S and M.D.I).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Carlos A. Vergani-Junior, Guilherme Tonon-da-Silva and Mehmet Dinçer Inan have contributed equally.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aryal NK, Pant V, Wasylishen AR, et al. Constitutive dicer1 phosphorylation accelerates metabolism and aging in vivo. Proc Natl Acad Sci U S A. 2019;116:960–969. doi: 10.1073/pnas.1814377116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Chable-Bessia C, Triboulet R, et al. Competition for XPO5 binding between dicer mRNA, pre-miRNA and viral RNA regulates human dicer levels. Nat Struct Mol Biol. 2011;18:323–327. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001 doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Beshore EL, McEwen TJ, Jud MC, et al. C. elegans dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev Biol. 2011;350:370–381. doi: 10.1016/j.ydbio.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29:613–639. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- Burger K, Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and dicer. Nat Rev Mol Cell Biol. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- Burger K, Schlackow M, Potts M, et al. Nuclear phosphorylated dicer processes doublestranded RNA in response to DNA damage. J Cell Biol. 2017;216:2373–2389. doi: 10.1083/jcb.201612131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci. 2012;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, et al. Targeted deletion of dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S, Richly H. DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 2017;45:5901–5912. doi: 10.1093/nar/gkx261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S, Richly H. DIC ER- and MMS ET-catalyzed H4K20me2 recruits the nucleotide excision repair factor XPA to DNA damage sites. J Cell Biol. 2018;217:527–540. doi: 10.1083/jcb.201704028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M, Furuta T, Suen KM, et al. A requirement for ERK-dependent dicer phosphorylation in coordinating oocyte-to-embryo transition in C.elegans. Dev Cell. 2014;31:614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, et al. MRNA Destabilization Is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde A, Eitan C, Liou L-L, et al. Dysregulated mi RNA biogenesis downstream of cellular stress and ALS -causing mutations: a new mechanism for ALS. EMBO J. 2015;34:2633–2651. doi: 10.15252/embj.201490493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg-Gorenshtein G, Guedj A, Shichrur K, et al. miR-192 directly binds and regulates Dicer1 expression in neuroblastoma. PLoS One. 2013;8:e78713. doi: 10.1371/journal.pone.0078713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Priest JR, Duchaine TF. DICER1: Mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelier G, Voirin N, Ay AS, et al. Prognostic value of dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross TJ, Powers LS, Boudreau RL, et al. A Microrna processing defect in smokers’ macrophages is linked to sumoylation of the endonuclease DICER. J Biol Chem. 2014;289:12823–12834. doi: 10.1074/jbc.M114.565473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra BA, Brandão BB, Pinto SS, et al. Dietary sulfur amino acid restriction upregulates DICER to confer beneficial effects. Mol Metab. 2019;29:124–135. doi: 10.1016/j.molmet.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hamaya Y, Kuriyama S, Takai T, et al (2012) A distinct expression pattern of the long 3′-untranslated region dicer mRNA and its implications for posttranscriptional regulation in colorectal cancer. Clin Transl Gastroenterol 3. 10.1038/ctg.2012.12 [DOI] [PMC free article] [PubMed]
- Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325(80):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkal GW, Grelier G, Puisieux A, Moyret-Lalle C. Complexity in the regulation of dicer expression: dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br J Cancer. 2011;104:387–388. doi: 10.1038/sj.bjc.6606022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(80):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Irvin-Wilson CV, Chaudhuri G. Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res. 2005;7:1–7. doi: 10.1186/bcr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarnejad SM, Ardekani GS, Ghaffari M, et al. Sox4-mediated dicer expression is critical for suppression of melanoma cell invasion. Oncogene. 2013;32:2131–2139. doi: 10.1038/onc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Koralewska N, Pokornowska M, et al. The many faces of dicer: the complexity of the mechanisms regulating dicer gene expression and enzyme activities. Nucleic Acids Res. 2015;43:4365–4380. doi: 10.1093/nar/gkv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Pokornowska M, Koralewska N, et al. Revealing a new activity of the human dicer DUF283 domain in vitro. Sci Rep. 2016;6:1–13. doi: 10.1038/srep23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, et al. Differential roles of human dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–6576. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Robinson KC, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/J.CELL.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu Z, Sheng J. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (basel) 2018;9:246. doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301(80):1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang J, Cheng H, et al. Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell. 2018;173:1191–1203.e12. doi: 10.1016/j.cell.2018.03.080. [DOI] [PubMed] [Google Scholar]
- Luo QJ, Zhang J, Li P, et al. RNA structure probing reveals the structural basis of dicer binding and cleavage. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-23607-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol. 2012;422:466–476. doi: 10.1016/j.jmb.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by dicer. Science. 2006;311(80):195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, et al. A microRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Masliah G, Barraud P, Allain FHTH-T. RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Cell Mol Life Sci. 2013;70:1875–1895. doi: 10.1007/s00018-012-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matskevich AA, Moelung K. Stimuli-dependent cleavage of dicer during apoptosis. Biochem J. 2008;412:527–534. doi: 10.1042/BJ20071461. [DOI] [PubMed] [Google Scholar]
- Mori MA, Raghavan P, Thomou T, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Thomou T, Boucher J, et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J Clin Invest. 2014;124:3339–3351. doi: 10.1172/JCI73468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, et al. Caspase-dependent conversion of dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328(80):327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio M, Schmidt E, Mauer J et al (2016) Dicer1–miR-328–Bace1 signalling controls brown adipose tissue differentiation and function. Nat Cell Biol 18(3):328–336. 10.1038/ncb3316 [DOI] [PubMed]
- Ota H, Sakurai M, Gupta R, et al. ADAR1 forms a complex with dicer to promote MicroRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, et al. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paturi S, Deshmukh MV. A glimpse of “dicer biology” through the structural and functional perspective. Front Mol Biosci. 2021;8:304. doi: 10.3389/fmolb.2021.643657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S, Sato VN, De-Souza EA, et al. Enoxacin extends lifespan of C. elegans by inhibiting miR-34-5p and promoting mitohormesis. Redox Biol. 2018;18:84–92. doi: 10.1016/j.redox.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier EZ, Buck MD, Chakravarty P, et al. An isoform of dicer protects mammalian stem cells against multiple RNA viruses. Science. 2021;373(80):231–236. doi: 10.1126/science.abg2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pong SK, Gullerova M. Noncanonical functions of microRNA pathway enzymes – Drosha, DGCR8. Dicer and Ago Proteins FEBS Lett. 2018;592:2973–2986. doi: 10.1002/1873-3468.13196. [DOI] [PubMed] [Google Scholar]
- Potenza N, Papa U, Scaruffi P, et al. A novel splice variant of the human dicer gene is expressed in neuroblastoma cells. FEBS Lett. 2010;584:3452–3457. doi: 10.1016/j.febslet.2010.06.045. [DOI] [PubMed] [Google Scholar]
- Reis FCG, Branquinho JLO, Brandão BB, et al. Fat-specific dicer deficiency accelerates aging and mitigates several effects of dietary restriction in mice. Aging (Albany NY) 2016;8:1201–1222. doi: 10.18632/aging.100970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KTG, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3–rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Rocha AL, de Lima TI, de Souza GP, et al. Enoxacin induces oxidative metabolism and mitigates obesity by regulating adipose tissue miRNA expression. Sci Adv. 2020;6:eabc6250. doi: 10.1126/SCIADV.ABC6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Jens M, Murakawa Y, et al. A variety of dicer substrates in human and C. elegans. Cell. 2014;159:1153–1167. doi: 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- Schultz KAP, Stewart DR, Kamihara J, et al (2020) DICER1 Tumor Predisposition. GeneReviews®
- Scott MS, Avolio F, Ono M, et al. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D, Bittrich S, Deshpande M, et al. Mol∗Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431–W437. doi: 10.1093/nar/gkab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Bevan SC, Patil K, et al. Extensive variation in the 5′-UTR of dicer mRNAs influences translational efficiency. Biochem Biophys Res Commun. 2005;335:643–650. doi: 10.1016/j.bbrc.2005.07.138. [DOI] [PubMed] [Google Scholar]
- Sinha NK, Iwasa J, Shen PS, Bass BL. Dicer uses distinct modules for recognizing dsRNA termini. Science. 2018;359(80):329–334. doi: 10.1126/science.aaq0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song MS, Rossi JJ. Molecular mechanisms of dicer: endonuclease and enzymatic activity. Biochem J. 2017;474:1603–1618. doi: 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavast CJ, Erkeland SJ (2019) The non-canonical aspects of MicroRNAs: many roads to gene regulation. Cells 8:1465. 10.3390/cells8111465 [DOI] [PMC free article] [PubMed]
- Su X, Chakravarti D, Cho MS, et al. TAp63 suppresses metastasis through coordinate regulation of dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swahari V, Nakamura A, Baran-Gale J, et al. Essential function of dicer in resolving DNA damage in the rapidly dividing cells of the developing and malignant cerebellum. Cell Rep. 2016;14:216–224. doi: 10.1016/j.celrep.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theotoki EI, Pantazopoulou VI, Georgiou S, et al. Dicing the disease with dicer: the implications of dicer ribonuclease in human pathologies. Int J Mol Sci. 2020;21:7223. doi: 10.3390/ijms21197223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, et al. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human dicer. Mol Cell. 2014;53:606–616. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Beucken T, Koch E, Chu K, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:1–13. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, et al. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnicka M, Szczepanska A, Kurzynska-Kokorniak A. Unknown areas of activity of human ribonuclease dicer: a putative deoxyribonuclease activity. Molecules. 2020;25:1414. doi: 10.3390/molecules25061414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, et al. Human dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, et al. Single processing center models for human dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]