Abstract
Complex protein glycosylation occurs through biosynthetic steps in the secretory pathway that create macro- and microheterogeneity of structure and function. Required for all life forms, glycosylation diversifies and adapts protein interactions with binding partners that underpin interactions at cell surfaces and pericellular and extracellular environments. Because these biological effects arise from heterogeneity of structure and function, it is necessary to measure their changes as part of the quest to understand nature. Quite often, however, the assumption behind proteomics that posttranslational modifications are discrete additions that can be modeled using the genome as a template does not apply to protein glycosylation. Rather, it is necessary to quantify the glycosylation distribution at each glycosite and to aggregate this information into a population of mature glycoproteins that exist in a given biological system. To date, mass spectrometric methods for assigning singly glycosylated peptides are well-established. But it is necessary to quantify glycosylation heterogeneity accurately in order to gauge the alterations that occur during biological processes. The task is to quantify the glycosylated peptide forms as accurately as possible and then apply appropriate bioinformatics algorithms to the calculation of micro- and macro-similarities. In this review, we summarize current approaches for protein quantification as they apply to this glycoprotein similarity problem.
Keywords: Glycoprotein, glycopeptide, similarity, glycosylation, glycoproteomics, bioinformatics
Abbreviations: AGP, α1-acid glycoprotein; CCS, collision cross section; DIA, data-independent analysis; ETD, electron transfer dissociation; EThcD, electron transfer higher-energy collisional dissociation; FDR, false discovery rate; HCD, higher-energy collisional dissociation; MRM, multiple reaction monitoring; PRM, parallel reaction monitoring; PTM, posttranslational modification; QTOF, quadrupole time-of-flight; TDA, target decoy analysis
Graphical Abstract
Highlights
-
•
Singly glycosylated peptides can be identified unambiguously using HCD LC-MS.
-
•
For calculation of glycoprotein similarity, quantify all glycopeptide glycoforms.
-
•
Glycoprotein similarities can be calculated using the Tanimoto coefficient.
-
•
Similarity calculations require high reproducibility of glycoproteomics LC-MS data.
In Brief
To understand the roles of glycoproteins in biological processes, it is necessary to quantify the changes that occur to glycosylation at individual sites and to the whole molecule. That glycoprotein glycosylation is inherently heterogeneous means that the distribution of glycoforms at each glycosite must be quantified in order to inform calculation of molecular similarities. We review analytical and statistical methods for determining glycoprotein molecular similarities from glycoproteomics data.
Biological Roles of Protein Glycosylation
Complex cotranslational protein glycosylation in the endoplasmic reticulum serves as a handle for protein folding quality control and sorting in the secretory pathway. The initial glycan cores become elaborated in the Golgi apparatus by biosynthetic enzymes, the functions of which reflect complex balances of substrate concentrations, kinetic, and transport effects. The resulting mature proteins show macro- and micro-heterogeneity of glycosylation that gives rise to populations of mature molecule proteoforms with a distribution of structures and functions (1). Such glycosylation heterogeneity is an evolutionary mechanism whereby protein function with respect to binding to lectin domains is elaborated, giving rise to organized networks at the cell surface and in extracellular matrices through which cells interact with their surroundings, other cells, and pathogens (2, 3).
If the goal of proteomics is to define the flux of protein expression, then that of glycoproteomics is to define the changes to protein site-specific glycosylation that occur in biological processes. That the expression levels of the biosynthetic enzymes, substrate transporters, and core proteins of the secretory pathway are regulated according to cell type, location, development, and disease is well-established. We therefore expect that glycoprotein structure varies spatially and temporally. This is supported by numerous antibody staining and lectin binding studies (4, 5). But there remains a dearth of information regarding how the glycoprotein site-specific glycosylation varies according to the functional requirements of disparate biological systems.
This review describes progress toward the goal of quantifying changes in glycoprotein site-specific glycosylation from bottom-up mass spectral experiments on intact glycopeptides. We place emphasis on analysis of singly glycosylated peptides using label-free collisional dissociation of glycoproteins from biological samples. Recent advances in approaches for glycopeptide quantification, including metabolic labeling approaches, have been reviewed elsewhere (6) and are not described here.
Assigning Glycopeptides Using Proteomics Methods
Algorithmic assignment of glycopeptides from tandem mass spectra has been covered in recent reviews (7, 8). Briefly, glycopeptides dissociate to produce diagnostic saccharide oxonium ions, neutral losses of monosaccharides from the precursor ion, and peptide backbone product ions. Glycopeptide tandem mass spectra that contain all three types of product ions usually receive the most confident assignments.
With regard to instrumentation, there is a need to distinguish between beam-type collisional dissociation and ion trap resonant collisional dissociation. Beam-type dissociation occurs in triple quadrupole, quadrupole time-of-flight (QTOF), and quadrupole Orbitrap instruments. Multiple collision events between precursor ions and collision gas result in vibrational excitation and dissociation. The extent of dissociation of the precursor ion can be controlled by adjusting the collision energy. Product ions may undergo subsequent dissociation. While beam-type collisional dissociation was used well before the term “higher-energy collisional dissociation (HCD)” was coined, HCD will be used in this review. Resonant collisional dissociation occurs in ion trap instruments whereby a designated precursor ion m/z window is collisionally excited. The resulting product ions, having different m/z values from the precursor ion, are cooled rapidly by the ion trap bath gas, resulting in a lower extent of dissociation than observed with beam-type dissociation. Ion traps have low mass accuracy relative to QTOF and Orbitrap analyzers. Note that ion trap-Orbitrap instruments can be used to generate either beam-type dissociation (HCD) with high mass accuracy Orbitrap detection or resonant collisional dissociation of precursors with low mass accuracy ion trap detection.
At the present time, high-resolution beam-type collisional dissociation tandem mass spectrometry methods (referred to here as HCD) have been established for assigning the composition of glycosylation existing on a glycopeptide (9, 10, 11, 12). Dissociation of glycosidic bonds occurs more readily than that of the peptide backbone, resulting in a situation where dissociation of the glycan posttranslational modification (PTM) alters the overall product ion pattern. Because traditional peptide database search algorithms and quantification tools do not recognize such glycosidic bond cleavage product ions, specialized tools are required for glycopeptides. In the case of peptides with a single N- or O-glycan, HCD tandem mass spectra can define the peptide sequence, glycan composition, and site of glycosylation. Often cited practices for producing high-quality HCD tandem mass spectra include enrichment of glycopeptides prior to tandem MS (13, 14, 15) and use of stepped collision energies (10, 11, 16). By contrast, others have used a single high HCD dissociation energy and long LC-gradients to identify coronavirus S-protein glycosylation (17). A number of commercial (18, 19) academic (20, 21, 22) software programs have been cited in recent glycoproteomics publications.
In glycopeptides where more than one N- or O-glycan is present, glycan dissociation observed in HCD tandem MS often prevents assigning the glycan compositions present at individual glycosites. ETD-based approaches have been used for multiply glycosylated O-glycopeptides (23). In addition, β-O-GlcNAc modification of Ser and Thr residues is particularly labile and requires special methodological consideration, as described in recent reviews (24). For the purpose of the present discussion, the β-O-GlcNAc group dissociates readily during collisional dissociation, making it difficult to assign glycosylated sites (25), and ETD-based methods are therefore preferred (26). Ultraviolet photodissociation also shows great promise for analysis of β-O-GlycNAcylated peptides (27).
Electron-activated dissociation methods produce preferential dissociation of glycopeptide peptide backbone bonds (28). Electron transfer dissociation (ETD) is available on many commercial mass spectrometers and, in principle, produces detailed peptide backbone dissociation with much lower extent of glycan dissociation than observed for collisional dissociation. For best results, supplementary activation of ions resulting from ETD is necessary to separate charge loss ions. Available methods include use of supplemental collisional activation (29, 30, 31), often referred to as electron transfer higher-energy collisional dissociation (EThcD), and activated ion ETD (32, 33). Due to the fragmentation to the glycopeptide glycan, collisional dissociation methods determine the overall glycan composition on the peptide. In favorable cases, the site of glycosylation can be assigned from glycosylated peptide backbone product ions. If more than one glycan is present, however, collisional dissociation typically defines the overall glycan composition but cannot differentiate the compositions at individual peptide sites (34). ETD-based methods produce preferential cleavage of the peptide backbone and can be used to assign multiply glycosylated peptides (35). The duty cycle for ETD methods is lower than for HCD due to the ion–ion reaction and supplemental activation times. For this reason, triggering of ETD spectra based on the presence of oxonium ions from HCD spectra is used to conserve analyzer time (29, 36). Because most of the publically available glycoproteomics data sets use HCD methods and define singly glycosylated peptides, we will focus on this aspect in this review.
Glycopeptide Identification Approaches
The interpretation of glycopeptide tandem mass spectra depends on the completeness of dissociation of the peptide and glycan portions. For collisional dissociation, this depends on the size of the glycan, the peptide sequence, and precursor ion charge state (11). It is important to control the false discovery rate (FDR) for both the peptide and glycan portions of the glycopeptide (16). For this it is necessary to model the extent to which the pattern of peptide+Y ions for a glycopeptide compares with an empirical null model. In order to dissociate as many different glycopeptides as efficiently as possible, researchers have used stepped collision energies (11, 16) at the expense of decreased number of precursor ions selected for tandem MS. As shown in Table 1, there are a number of monosaccharide combinations that can lead to ambiguous tandem MS assignments for glycopeptides.
Table 1.
Monosaccharide combinations that can lead to ambiguous tandem MS assignments
| Saccharide 1 | Mass (Da) | Saccharide 2 | Mass (Da) | Error (Da) |
|---|---|---|---|---|
| NeuAc | 291.0954 | Fuc2 | 292.115 | 1.02 |
| NeuAc, NH3 (adduct) | 308.121 | Hex, Fuc | 308.110 | 0.011 |
| HexNAc2, SO3 (substitution) | 486.115 | Hex3 | 486.158 | 0.0429 |
| HexNAc2, Fuc, NeuAc2 | 1134.407 | Hex7 | 1134.369 | 0.037 |
| NeuAc, Hex | 453.148 | NeuGc, Fuc | 453.148 | 0.0 |
In order to augment the evidence in support for a given glycan attached to a peptide, researchers have used chromatographic retention time modeling (20, 37). Using reversed-phase chromatography, it is known that addition of monosaccharide residues to a glycopeptide glycan induces a consistent shift in retention time (29, 30, 31). The retention time shifts have been related to alteration of hydrophobicity of the glycopeptide relative to the bare peptide in the same chromatography system (38). We developed a linear modeling approach for glycopeptides and applied it to two published studies on N-glycopeptides (37). We showed that the model was able to correct common errors in glycan assignments due to either insufficient dissociation or ambiguous monosaccharide and adduct combinations. We also determined that glycopeptides produce linear trendlines of collision cross section (CCS) versus mass-to-charge ratio (39). These observations support the conclusion that modeling of glycopeptide CCS can be used to augment the assignment of glycopeptides.
To apply target decoy analysis (TDA) (40) to glycopeptides requires generation of an appropriate decoy. A key assumption of TDA is that the scoring pattern of targets and decoys are largely equivalent until the threshold of something being considered a positive or real is reached. If the two sets diverge in scoring well before this threshold, then the assumption is broken and TDA underestimates the FDR. An appropriate decoy for a glycopeptide must produce false scores up to the threshold of reality. The decoy glycopeptides employed in TDA can take several forms. Some softwares, such as GlycReSoft (20), use reversed peptides with identical glycan masses, and no change in glycosite composition; this has the consequence of relying on peptide ions for FDR detection. The pGlyco software (16, 21) creates decoys using mass shifts after generating theoretical fragmentation of reversed peptides and unmodified glycans; its successor, pGlyco2, goes a step further and uses the union of the peptide and glycan FDRs with the glycopeptide FDR subtracted as overlap; this is comprehensive but, like GlycReSoft, runs the risk of removing false glycopeptides since the analysis is composition-agnostic when calculating the overlapping FDR. More complex implementations can help adjust for this, such as permutation to generate new glycans of similar mass; the permuted and nonpermuted glycan decoys can be combined with reversed peptides and unreversed peptides to generate a more comprehensive set of decoys that can be used combinatorially to ensure a more full accounting of false positives is made. This is available in the GlycReSoft multipart search option. GlycoPep Evaluator (41) generates glycopeptides that are isobaric in mass to the target glycopeptides by generating peptides and glycans under a set of rules to roughly match the mass of the targets. This can create a successful set of decoys, but it runs the risk of creating decoys that are in fact true positives instead of false positives. Some softwares (18, 42) fully reverse proteins and find new glycosites and tryptic digests. The challenge is that these can stray too far in mass and general composition to resemble any glycopeptides in the target set. GlycoFragWork (43) combines CID and ETD scoring systems using linear discriminant analysis to generate a single TDA-like evaluation score.
Glycopeptide Quantification
As described in (44), exploratory proteomics studies using label-free or isobaric labeling result in low to moderate levels of quantification precision. Targeted quantification using multiple reaction monitoring (MRM) or parallel reaction monitoring (PRM) increases the quantitative precision. The most precise relative quantification assays employ a stable isotope-labeled standard for every peptide target. In proteomics, targeted quantification tools including Skyline (45) and PeptideAtlas (46) support the development of methods whereby peptides are quantified using tandem MS product ions. These tools allow PTMs that are chemically defined, including phosphorylation, acetylation, and methylation. As mentioned earlier, the glycan PTM itself undergoes dissociation during tandem MS. Such glycan dissociation, combined with glycosylation heterogeneity, is not presently allowed for quantitative proteomics tools.
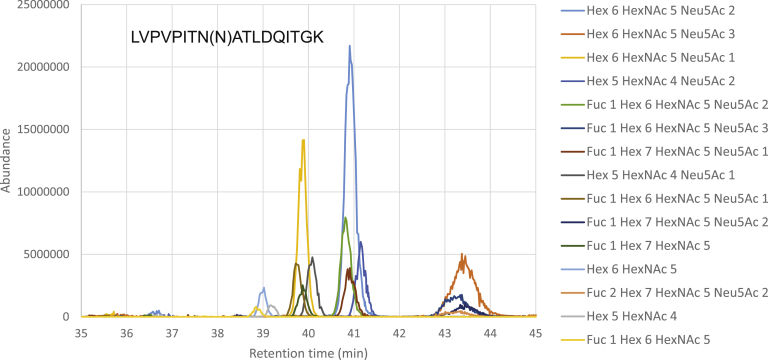
Glycopeptide quantification using MS reaction monitoring has been reviewed in detail elsewhere (6). The following discussion is oriented toward the use of MS reaction monitoring for protein similarity measurement. A number of groups have published MRM/PRM methods for quantification of glycopeptides (47, 48, 49, 50). Because peptide backbone dissociation results in low-abundance product ions, the tandem MS transitions often employ neutral saccharide losses from the precursor and oxonium ions. The number of peptides that can be quantified in a targeted MS experiment is limited by analyzer speed. As shown in Figure 1, the heterogeneous glycoforms for a typical glycopeptide modified with complex glycosylation elute from a reversed-phase chromatography column over a narrow retention time window. In order to define glycoprotein similarity, each of the glycoforms observed must be quantified. Such quantification requires 6 to 8 points across the EIC peak. As a result, data-dependent analysis, which selects precursors based on abundance, resulting in stochastic patterns of precursor ion selection, is poorly suited for complete sampling of the heterogeneous glycopeptides. Thus, for many N-glycopeptides, there is insufficient analyzer speed to quantify all glycoforms using tandem MS transitions. This pattern of coeluting glycopeptides also complicates the task of building a targeted MS method due to the overlapping EICs, a problem that grows in magnitude as the sample complexity increases.
Fig. 1.
Example of overlapping AGP glycopeptide 25 to 42 LVPVPITN(N)ATLDQITGK extracted ion chromatograms from published data (77). The glycosite is given in parenthesis.
Data-independent analysis (DIA) methods have been applied widely in proteomics (51). While the advantage is that all precursor ions are dissociated, the challenge to interpretation of DIA proteomics data is to maintain sufficient selectivity of identification of coeluting peptides. Thus, by reducing the size of the quadrupole window used to step through the mass range, the selectivity increases but the scan rate and sensitivity decrease. To interpret DIA tandem mass spectral data, peptide-centric and spectrum-centric approaches have been described (52). Spectrum-centric methods, including DIA-Umpire (53) and OpenSWATH (54), seek to identify a single peptide for each DIA tandem mass spectrum and use a reference library to assign peptides from the DIA data. Peptide-centric methods such as PECAN (55) query the DIA data for the presence of designated peptides using peptide-specific product ions. As such, these methods tolerate the presence of coisolated peptides better than do spectrum-centric approaches.
The presence of many coeluting glycopeptide glycoforms drives the need to design DIA methods with appropriate high selectivity. SWATH-type DIA was used for the analysis of IgG glycoforms, for which heterogeneity is limited, from unfractionated human plasma (56). A DIA method for analysis of biosynthetically truncated mucin-type O-glycopeptides has been described in which in silico augmented Glyco-DIA spectral libraries were used to assign glycopeptides from unenriched serum (57). A targeted DIA method was used to detect 59 N-glycosites from 41 glycoproteins from HILIC-enriched serum for which glycopeptide Y ions were calculated manually and imported into Skyline to generate transitions for quantification (58). Khoo et al. noted that while the optimal collision energy for glycopeptide depends on the peptide sequence, the dependence on the glycan composition is small by comparison (59). Thus, they showed that setting collision energy to optimize the abundance of the peptide+HexNAc Y1 ion allows the definition of tandem MS transitions that are specific to the peptide sequence but allow detection of any glycan composition. This approach allows for unanticipated glycoforms to be identified using DIA.
Although there remains no easy way to use existing proteomics DIA analysis software for glycoproteomics, due to the previously mentioned glycosylation heterogeneity and glycosidic bond dissociation, a DIA approach based on profiling the abundances of HexNAc and sialic acid oxonium ions was developed for the purpose of comparing the similarities of multiply glycosylated biotherapeutic glycoproteins. Termed broadband collision-induced dissociation (bbCID), this method was used to assign glycopeptides from standard proteins (60). For this, oxonium ions were used to indicate MS1 scans from which precursors were assigned manually and glycopeptides assigned from the corresponding tandem mass spectra.
Statistical Analysis Methods for Calculation of Glycoprotein Similarity
In order to assess the roles of glycoprotein glycosylation in biological mechanisms, it is necessary to determine whether a mutated form of a glycoprotein is similar to the wildtype version. This entails combining the abundances of the glycoforms at each site and using a statistical metric to assess the similarity since more traditional statistical analyses are hamstrung by statistical power. In the field of chemoinformatics, molecular similarity refers to similarity of structural or functional molecular properties (61). It is used in drug design studies and in screening chemical structural databases for available compounds with similar chemical properties (62). Molecular similarity is akin to the inverse distance between a pair of compounds in descriptor space. To enable similarity screening of large compound databases, molecules are represented using molecular screens or molecular fingerprints. A number of commercial (63) and public (64) fingerprint databases have been used to screen orphan drug candidates based on similarity. Similarity measures are then used to compare the molecular structures through the fingerprint information. The Tanimoto coefficient (65) is the most often used similarity measure for comparing chemical structures using molecular fingerprints (66). Originally developed to classify plants, the Tanimoto coefficient uses binary presence/absence data to evaluate co-occurrences that reveal relationships among biological or chemical species. The Tanimoto coefficient corresponds to the ratio of the intersection to the union for a species pair. A hypothesis test for organism similarity based on presence/absence data has been developed using the Tanimoto coefficient with bootstrap and measurement concentration algorithms (67).
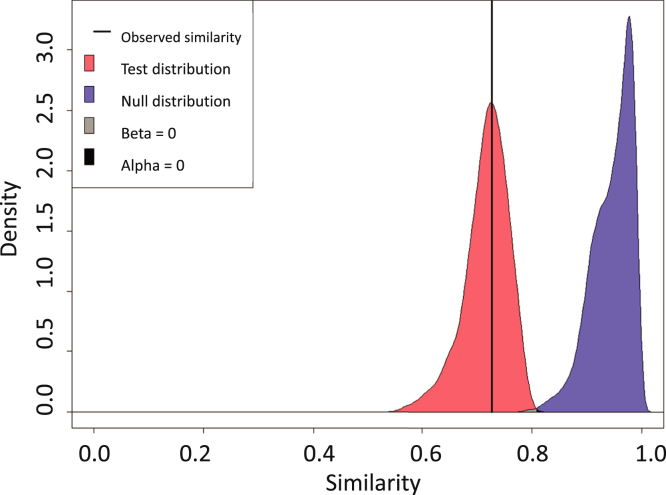
A glycoprotein contains a set of glycosites, each with a distribution of biosynthetically related glycoforms. A glycoproteomics experiment determines the monosaccharide composition and an abundance for each observed glycopeptide glycoform. The complexities of glycoprotein glycosylation patterns require appropriate statistical metrics for measuring the degree of molecular similarity. We recently developed a modified form of the Tanimoto coefficient to determine statistical similarity between pairs of glycoprotein preparations based on the presences and abundances of glycopeptide glycoforms and applied it to compare wildtype and mutant influenza A virus hemagglutinin glycosylation (68). For this purpose, we used a modified form of the Tanimoto coefficient, shown below:
A and B correspond to the glycopeptide abundance vectors for two glycoprotein samples. These abundances are log-scaled and standardized to yield values in the continuous range of 0 to 1. The glycopeptide abundances are measured in technical repeats that are averaged. The PA and PB vectors contain the proportion of observed values to total values across technical replicates, thereby accounting for missing values. K is a distance scaling term , and d(A,B) is the Manhattan distance between A and B. The resulting plot shows two distributions of similarity coefficients, a null hypothesis distribution, and a test hypothesis distribution. The null hypothesis Tanimoto coefficient distribution is calculated from random combinations with replacement of all replicates for samples A and B in order to simulate a joint distribution. The test hypothesis Tanimoto coefficient is calculated from random combinations of replicates of A to those of B. As shown for the idealized case in Figure 2, the degree of overlap of null and test distributions determines the confidence with which we can quantify the glycosylation similarity between the sample groups. The x-axis shows the Tanimoto similarity and the y-axis the distribution density. The null and test distributions are analogous to the null and alternative hypotheses used in statistical inference. If the null and test distributions do not overlap significantly, then the null hypothesis is rejected and the samples are dissimilar. If the null hypothesis is not rejected, then the sample pair is not dissimilar. The area to the right of the point of intersection is analogous to the Type I error (α), corresponding to false-positive rate as results in this region could be from the test comparison but have a higher likelihood of being from the null and therefore a negative comparison. The area to the left of the point of intersection is analogous to Type II error (β), corresponding to the false-negative rate. While results in this region could be from the Null comparison, they have a higher likelihood of being from the test comparison and therefore a positive comparison. The plot shows narrow null and test distributions and acceptable levels of Type I and Type II errors.
Fig. 2.
This example of a similarity comparison shows two glycosites that are differentiable from one another as determined by the low degree of distribution overlap. These glycosites are the same site on the same protein, but one has worse-quality data than the other due to other glycoproteins injected and processed along with it confounding the signal.
Glycopeptide Quantification and Mixture Complexity
Quantitative glycopeptidomics faces challenges that quantitative proteomics does not; focusing on glycoproteoforms intrinsically multiplies ion diversity and reduces quantitation points. Proteomics benefits from the ability to reference multiple peptides when trying to gauge the quantitation of a single proteoform; in contrast, a glycoproteoform can only be evaluated with missed cleavages of the same glycosite. Not only must a complete quantitative assay of a glycoproteome contend with more possibilities than the unglycosylated proteome, but it also must face a decrease in the ability to sample completely the glycopeptide glycoforms to be quantified. To better explain the scale, consider a proteome of 1000 proteoforms. Assuming eight points along the LC elution curve and five peptides per proteoform, this proteome could be quantified by 40,000 successful peptide spectrum matches, 40,000 successful acquisitions. Next, consider the related glycoproteome: based on 20 glycoforms per glycosite, an average of 50% of proteins glycosylated, and just two glycosites per glycosylated protein, there are an expected 20,000 glycoproteoforms. In order to uphold a similar standard of eight points along an elution curve for full quantitation, there needs to be 160,000 successful glycopeptide spectrum matches, 160,000 successful acquisitions. This estimation of a fourfold increase in the sampling power necessary to quantify the glycoproteome is likely to underestimate the multiplicative power of glycosylation because many glycoproteins have more than two glycosites.
The likelihood of precursor ion coisolation must also be taken into consideration in both DDA and DIA experiments. Coisolation of two glycopeptide precursor ions is a more demanding problem than coisolation of two unmodified peptides. In many cases, the peptides can still be identified, assuming that their sequences are not related, but even that is not a certainty. Glycopeptides with a common peptide sequence, however, show a narrow range of LC retention times for the set of glycoforms for a given peptide sequence in m/z space; when these glycopeptides are coisolated and dissociated, many of the same peptide+Yn ions and oxonium ions are observed for both precursors. Precursor ion cation adducts, if present, increase the computational work needed to identify the glycopeptides from one another. As shown in Table 1 (37), there are several common combinations of monosaccharides and adducts that lead to precursor ions masses that may lead to false identifications. In the case when two glycopeptides with unrelated peptide sequences coelute, the fact that product ions from peptide backbone dissociation are often low in abundances can result in peak misidentification.
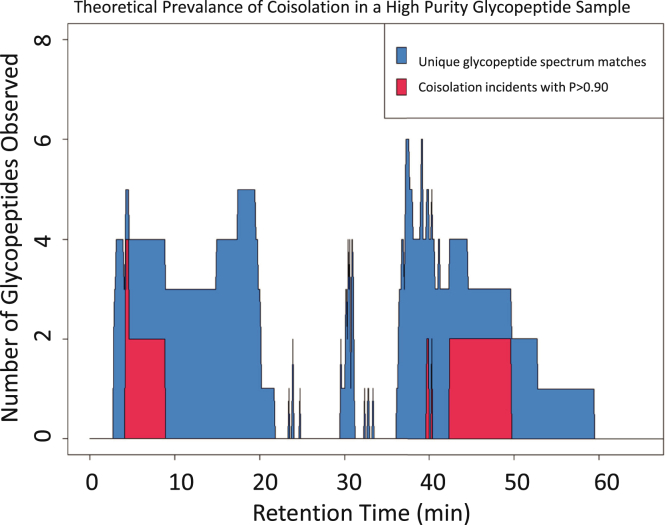
Given that the ability to sample glycopeptide glycoforms using is limited by analyzer speed, DIA methods are of interest for quantifying glycoprotein glycosites. For such DIA studies, it is important to estimate the probability of multiple precursor ions in the scan window. As a conservative example of this prevalence, we examined a previously published data set examining a purified α1-acid glycoprotein (AGP) sample (69). We created a simple model to estimate the degree of coisolation expected from an experiment with 10 u windows. This model is based on 23 total glycan compositions at five different glycosites in AGP. Based on the analytical data, some sites had as few as three glycan compositions and some with as many as 20, for a total of 60 glycopeptides observed over a 65 min gradient. Figure 3 shows the number of glycopeptides observed at a given time and the number of glycopeptides that had a probability of coisolating greater than 90%. The probability was determined based on window size, mass error of 10 ppm, positive charge states from 2 to 5, and observed time windows of glycopeptides; it was assumed that if a glycopeptide was above a signal threshold, it would produce a tandem mass spectrum. The glycopeptide spectrum matches used to generate these data come from a GlycReSoft search from published data (69), and they should bias away from glycopeptides that coisolate as the search is not set up to handle such instances, so any glycopeptides that do present as likely to coisolate are likely to be edge cases. Overall 12 glycopeptides out of 60 would be expected to be present in the same 10 u window or 20% of identified glycopeptides. This degree of susceptibility indicates the need to differentiate glycopeptides present in the same DIA window. As the glycopeptide mixture complexity increases, the search space expands, and the need to differentiate glycopeptides in the same DIA window increases.
Fig. 3.
This shows the number of glycopeptides observed in a purified glycoprotein sample (blue) and the number of coisolation events (red) that would be likely (p > 0.90) given a 10 u window size. The plots are likely skewed downward due to coisolation events reducing the number of glycopeptide observations.
Quality Standards and Reproducibility
The assessment of changes in glycoprotein glycosylation in biological processes requires careful attention to LC-MS reproducibility. While various quality assessment tools exist, none are standard. Many of the tools that glycoproteomic researchers use are repurposed proteomic tools or expansions on general mass spectrometry tools. While these tools are well-designed and have their own merits, they all lack the complete view needed to assess a glycoproteomic experiment; they are not designed to do so. Glycoproteomics requires an increase in standards from proteomics due to the potential for error and requires specific considerations to handle the idiosyncrasies of its identification processes and quantifications. In regard to FDR, TDA (40) is ill-suited to glycopeptide identification as it relies on the assumption that Decoys mimic targets closely enough to genuinely be confused for Targets by the scoring algorithm. As our ability to score and identify glycopeptides improves, the reliability of our FDR estimation via TDA decreases. Various different software have attempted to solve the glycopeptide decoy problem, as discussed above, but none have found a perfect solution as of yet. And while Posterior Error Probability has advanced in recent years (70, 71, 72, 73), it has yet to be implemented, tested, and standardized for glycoproteomics, and many approaches rely on system knowledge that is currently infeasible.
QuaMeter is a quality metric that provides a vast array of metrics regarding the underlying spectra in an experiment and can be applied to any compound class (74). QuaMeter’s assessment of tandem MS coverage, intensity mapping, and noise assessment are useful for performing consistent experiments and adjusting for batch effects (74). The metrics it produces can be compared with benchmarks for the purpose of gauging experimental consistency but do not give insight to identification and quantitation quality.
The Skyline (45) quality tools focus on peptide quantitation. While using Skyline for glycopeptide quantification is possible, it must be carefully implemented to prevent signal contamination between separate glycoproteoforms. At present, glycopeptides and their fragments must be individually added and identified, which makes analysis of a glycopeptide data set time-consuming, especially if an experiment is examining more complex mixtures such as whole tissue. Skyline has an intuitive quantitation assessment tool that uses visualizations to allow the user to ensure that peak boundaries are correct and visual displays to ensure within sample group consistency (29). Glycopeptides need to be treated as ordinary PTMs in MaxQuant, and MaxQuant does not allow for fragment PTMs to contribute to one single identification and therefore quantitation (75, 76). This leads to false-positive identifications as mass shifts are left unaccounted for. MaxQuant can ensure high-quality identifications in the proteomics data used to generate a glycoproteomic search space, but at present cannot be applied directly to glycoproteomics data, meaning that its probabilistic FDR system cannot be applied to glycopeptide identifications.
Conclusions
Rigorous and complete quantification of glycopeptide data is necessary for making the biomedical discoveries required to meet emerging human health problems. Researchers need tools to properly assess the confidence of glycoproteomics results. This will require development of specific tools and standardization to increase interoperability between experiments. Spectral quality tools, while important, do not paint the whole picture. They lack the perspective of how variation in spectral quality influences interpretation of identification and quantification of glycopeptides. A degradation of signal at a specific time in the elution could lead to a vast shift in results leading to spurious conclusions. To address this problem will require development of holistic tools that inform the user of glycopeptides that are likely candidates for error based on spectral information, search space data, and related sample information. While it is possible for researchers to investigate all of these avenues on their own, the complexity and scope of this task are bound to cause error and are not conducive to generating a set of standards.
Looking to the future, there is a golden opportunity to increase standardization for glycoproteomics. All glycoproteomic experiments must have a known search space. Between this known search space and the ability to predict the expected retention time shifts caused by glycosylation, it should be possible to create a comprehensive data quality analysis. This information can be used to create a glycopeptide coisolation likelihood model as well as predict likely candidates for mass-shifting adducts that closely mimic glycan mass shifts. These two models, in conjunction with spectral data, will determine glycopeptides more likely to produce quantification error. The previously described statistical similarity methods allow for comprehensive examination within an experimental group and can identify problematic glycopeptides and samples as a whole. By using all of these methods together, a researcher can build a consensus on the reliability of their glycopeptide quantification data.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
Funding and additional information
The authors were supported by NIH grants U01CA221234 and R01GM133963. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
W. H. performed bioinformatics calculations and wrote the article; J. Z. supervised this work, wrote and edited the article.
References
- 1.Dennis J.W. Genetic code asymmetry supports diversity through experimentation with posttranslational modifications. Curr. Opin. Chem. Biol. 2017;41:1–11. doi: 10.1016/j.cbpa.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Nabi I.R., Shankar J., Dennis J.W. The galectin lattice at a glance. J. Cell Sci. 2015;128:2213–2219. doi: 10.1242/jcs.151159. [DOI] [PubMed] [Google Scholar]
- 3.Cummings R.D. Stuck on sugars - how carbohydrates regulate cell adhesion, recognition, and signaling. Glycoconj. J. 2019;36:241–257. doi: 10.1007/s10719-019-09876-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant O.C., Tessier M.B., Meche L., Mahal L.K., Foley B.L., Woods R.J. Combining 3D structure with glycan array data provides insight into the origin of glycan specificity. Glycobiology. 2016;26:772–783. doi: 10.1093/glycob/cww020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelamegham S., Mahal L.K. Multi-level regulation of cellular glycosylation: From genes to transcript to enzyme to structure. Curr. Opin. Struct. Biol. 2016;40:145–152. doi: 10.1016/j.sbi.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delafield D.G., Li L. Recent advances in analytical approaches for glycan and glycopeptide quantitation. Mol. Cell. Proteomics. 2021;20 doi: 10.1074/mcp.R120.002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu H., Khatri K., Zaia J. Algorithms and design strategies towards automated glycoproteomics analysis. Mass Spectrom. Rev. 2017;36:475–498. doi: 10.1002/mas.21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein J.A., Zaia J. A perspective on the confident comparison of glycoprotein site-specific glycosylation in sample cohorts. Biochemistry. 2020;59:3089–3097. doi: 10.1021/acs.biochem.9b00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebecchi K.R., Wenke J.L., Go E.P., Desaire H. Label-free quantitation: A new glycoproteomics approach. J. Am. Soc. Mass Spectrom. 2009;20:1048–1059. doi: 10.1016/j.jasms.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Khatri K., Staples G.O., Leymarie N., Leon D.R., Turiák L., Huang Y., Yip S., Hu H., Heckendorf C.F., Zaia J. Confident assignment of site-specific glycosylation in complex glycoproteins in a single step. J. Proteome Res. 2014;13:4347–4355. doi: 10.1021/pr500506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinneburg H., Stavenhagen K., Schweiger-Hufnagel U., Pengelley S., Jabs W., Seeberger P.H., Silva D.V., Wuhrer M., Kolarich D. The art of destruction: Optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J. Am. Soc. Mass Spectrom. 2016;27:507–519. doi: 10.1007/s13361-015-1308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thaysen-Andersen M., Packer N.H., Schulz B.L. Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Mol. Cell. Proteomics. 2016;15:1773–1790. doi: 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zacharias L.G., Hartmann A.K., Song E., Zhao J., Zhu R., Mirzaei P., Mechref Y. HILIC and ERLIC enrichment of glycopeptides derived from breast and brain cancer cells. J. Proteome Res. 2016;15:3624–3634. doi: 10.1021/acs.jproteome.6b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu R., Zacharias L., Wooding K.M., Peng W., Mechref Y. Glycoprotein enrichment analytical techniques: Advantages and disadvantages. Methods Enzymol. 2017;585:397–429. doi: 10.1016/bs.mie.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Y., Xie J., Fang P., Yao J., Yan G., Shen H., Yang P. Study on behaviors and performances of universal N-glycopeptide enrichment methods. Analyst. 2018;143:1870–1880. doi: 10.1039/C7AN02062G. [DOI] [PubMed] [Google Scholar]
- 16.Liu M.Q., Zeng W.F., Fang P., Cao W.Q., Liu C., Yan G.Q., Zhang Y., Peng C., Wu J.Q., Zhang X.J., Tu H.J., Chi H., Sun R.X., Cao Y., Dong M.Q., et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017;8:438. doi: 10.1038/s41467-017-00535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bern M., Kil Y.J., Becker C. Byonic: Advanced peptide and protein identification software. Curr. Protoc. Bioinformatics. 2012 doi: 10.1002/0471250953.bi1320s40. Chapter 13:Unit13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diedrich J.K., Pinto A.F.M., Yates J.R. Energy dependence of HCD on peptide fragmentation: Stepped collisional energy finds the sweet spot. J. Am. Soc. Mass Spectrom. 2013;24:1690–1699. doi: 10.1007/s13361-013-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein J., Carvalho L., Zaia J. Application of network smoothing to glycan LC-MS profiling. Bioinformatics. 2018;34:3511–3518. doi: 10.1093/bioinformatics/bty397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng W.F., Liu M.Q., Zhang Y., Wu J.Q., Fang P., Peng C., Nie A., Yan G., Cao W., Liu C., Chi H., Sun R.X., Wong C.C., He S.M., Yang P. pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep. 2016;6:25102. doi: 10.1038/srep25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasir W., Toledo A.G., Noborn F., Nilsson J., Wang M., Bandeira N., Larson G. SweetNET: A bioinformatics workflow for glycopeptide MS/MS spectral analysis. J. Proteome Res. 2016;15:2826–2840. doi: 10.1021/acs.jproteome.6b00417. [DOI] [PubMed] [Google Scholar]
- 23.Pap A., Klement E., Hunyadi-Gulyas E., Darula Z., Medzihradszky K.F. Status report on the high-throughput characterization of complex intact O-glycopeptide mixtures. J. Am. Soc. Mass Spectrom. 2018;29:1210–1220. doi: 10.1007/s13361-018-1945-7. [DOI] [PubMed] [Google Scholar]
- 24.Ma J., Hart G.W. Analysis of protein O-GlcNAcylation by mass spectrometry. Curr. Protoc. Protein Sci. 2017;87:24 10 1–24 10 16. doi: 10.1002/cpps.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Udeshi N.D., O'Malley M., Shabanowitz J., Hunt D.F., Hart G.W. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Wang W.H., Li Z., Shabanowitz J., Hunt D.F., Hart G.W. O-GlcNAc site mapping by using a combination of chemoenzymatic labeling, copper-free click chemistry, reductive cleavage, and electron-transfer dissociation mass spectrometry. Anal. Chem. 2019;91:2620–2625. doi: 10.1021/acs.analchem.8b05688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escobar E.E., King D.T., Serrano-Negrón J.E., Alteen M.G., Vocadlo D.J., Brodbelt J.S. Precision mapping of O-linked N-acetylglucosamine sites in proteins using ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc. 2020;142:11569–11577. doi: 10.1021/jacs.0c04710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Håkansson K., Cooper H.J., Emmett M.R., Costello C.E., Marshall A.G., Nilsson C.L. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 29.Saba J., Dutta S., Hemenway E., Viner R. Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int. J. Proteomics. 2012;2012:560391. doi: 10.1155/2012/560391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Q., Wang B., Chen Z., Urabe G., Glover M.S., Shi X., Guo L.W., Kent K.C., Li L. Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J. Am. Soc. Mass Spectrom. 2017;28:1751–1764. doi: 10.1007/s13361-017-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatri K., Pu Y., Klein J.A., Wei J., Costello C.E., Lin C., Zaia J. Comparison of collisional and electron-based dissociation modes for middle-down analysis of multiply glycosylated peptides. J. Am. Soc. Mass Spectrom. 2018;29:1075–1085. doi: 10.1007/s13361-018-1909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley N.M., Westphall M.S., Hebert A.S., Coon J.J. Implementation of activated ion electron transfer dissociation on a quadrupole-orbitrap-linear ion trap hybrid mass spectrometer. Anal. Chem. 2017;89:6358–6366. doi: 10.1021/acs.analchem.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley N.M., Hebert A.S., Westphall M.S., Coon J.J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 2019;10:1311. doi: 10.1038/s41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein J.A., Meng L., Zaia J. Deep sequencing of complex proteoglycans: A novel strategy for high coverage and site-specific identification of glycosaminoglycan-linked peptides. Mol. Cell. Proteomics. 2018;17:1578–1590. doi: 10.1074/mcp.RA118.000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley N.M., Malaker S.A., Driessen M.D., Bertozzi C.R. Optimal dissociation methods differ for N- and O-glycopeptides. J. Proteome Res. 2020;19:3286–3301. doi: 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh C., Zampronio C.G., Creese A.J., Cooper H.J. Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. J. Proteome Res. 2012;11:4517–4525. doi: 10.1021/pr300257c. [DOI] [PubMed] [Google Scholar]
- 37.Klein J., Zaia J. Relative retention time estimation improves N-glycopeptide identifications by LC-MS/MS. J. Proteome Res. 2020;19:2113–2121. doi: 10.1021/acs.jproteome.0c00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley N.M., Westphall M.S., Coon J.J. Activated ion-electron transfer dissociation enables comprehensive top-down protein fragmentation. J. Proteome Res. 2017;16:2653–2659. doi: 10.1021/acs.jproteome.7b00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glaskin R.S., Khatri K., Wang Q., Zaia J., Costello C.E. Construction of a database of collision cross section values for glycopeptides, glycans, and peptides determined by IM-MS. Anal. Chem. 2017;89:4452–4460. doi: 10.1021/acs.analchem.6b04146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Z., Su X., Go E.P., Desaire H. New glycoproteomics software, GlycoPep evaluator, generates decoy glycopeptides de novo and enables accurate false discovery rate analysis for small data sets. Anal. Chem. 2014;86:9212–9219. doi: 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polasky D.A., Yu F., Teo G.C., Nesvizhskii A.I. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-glyco. bioRxiv. 2020 doi: 10.1101/2020.05.18.102665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayampurath A., Yu C.Y., Song E., Balan J., Mechref Y., Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal. Chem. 2014;86:453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- 44.Carr S.A., Abbatiello S.E., Ackermann B.L., Borchers C., Domon B., Deutsch E.W., Grant R.P., Hoofnagle A.N., R H.U., Koomen J.M., Liebler D.C., Liu T., Maclean B., Mani D.R., Mansfield E., et al. Targeted peptide measurements in biology and medicine: Best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deutsch E.W., Lam H., Aebersold R. PeptideAtlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan W., Sanda M., Wu J., Koomen J., Goldman R. Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC-MS-MRM in liver disease. J. Proteomics. 2015;116:24–33. doi: 10.1016/j.jprot.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darebna P., Novak P., Kucera R., Topolcan O., Sanda M., Goldman R., Pompach P. Changes in the expression of N- and O-glycopeptides in patients with colorectal cancer and hepatocellular carcinoma quantified by full-MS scan FT-ICR and multiple reaction monitoring. J. Proteomics. 2017;153:44–52. doi: 10.1016/j.jprot.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song E., Pyreddy S., Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2012;26:1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong Q., Ruhaak L.R., Stroble C., Parker E., Huang J., Maverakis E., Lebrilla C.B. A method for comprehensive glycosite-mapping and direct quantitation of serum glycoproteins. J. Proteome Res. 2015;14:5179–5192. doi: 10.1021/acs.jproteome.5b00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman J.D., Goodlett D.R., Masselon C.D. Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 2014;33:452–470. doi: 10.1002/mas.21400. [DOI] [PubMed] [Google Scholar]
- 52.Ting Y.S., Egertson J.D., Payne S.H., Kim S., MacLean B., Kall L., Aebersold R., Smith R.D., Noble W.S., MacCoss M.J. Peptide-centric proteome analysis: An alternative strategy for the analysis of tandem mass spectrometry data. Mol. Cell. Proteomics. 2015;14:2301–2307. doi: 10.1074/mcp.O114.047035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsou C.-C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A.-C., Nesvizhskii A.I. DIA-umpire: Comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rost H.L., Rosenberger G., Navarro P., Gillet L., Miladinovic S.M., Schubert O.T., Wolski W., Collins B.C., Malmstrom J., Malmstrom L., Aebersold R. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 55.Ting Y.S., Egertson J.D., Bollinger J.G., Searle B.C., Payne S.H., Noble W.S., MacCoss M.J. PECAN: Library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat. Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanda M., Goldman R. Data independent analysis of IgG glycoforms in samples of unfractionated human plasma. Anal. Chem. 2016;88:10118–10125. doi: 10.1021/acs.analchem.6b02554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye Z., Mao Y., Clausen H., Vakhrushev S.Y. Glyco-DIA: A method for quantitative O-glycoproteomics with in silico-boosted glycopeptide libraries. Nat. Methods. 2019;16:902–910. doi: 10.1038/s41592-019-0504-x. [DOI] [PubMed] [Google Scholar]
- 58.Lin C.-H., Krisp C., Packer N.H., Molloy M.P. Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. J. Proteomics. 2018;172:68–75. doi: 10.1016/j.jprot.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 59.Pan K.T., Chen C.C., Urlaub H., Khoo K.H. Adapting data-independent acquisition for mass spectrometry-based protein site-specific N-glycosylation analysis. Anal. Chem. 2017;89:4532–4539. doi: 10.1021/acs.analchem.6b04996. [DOI] [PubMed] [Google Scholar]
- 60.Couto N., Davlyatova L., Evans C.A., Wright P.C. Application of the broadband collision-induced dissociation (bbCID) mass spectrometry approach for protein glycosylation and phosphorylation analysis. Rapid Commun. Mass Spectrom. 2018;32:75–85. doi: 10.1002/rcm.8016. [DOI] [PubMed] [Google Scholar]
- 61.Willett P. Chemoinformatics - similarity and diversity in chemical libraries. Curr. Opin. Biotechnol. 2000;11:85–88. doi: 10.1016/s0958-1669(99)00059-2. [DOI] [PubMed] [Google Scholar]
- 62.Haranczyk M., Holliday J. Comparison of similarity coefficients for clustering and compound selection. J. Chem. Inf. Model. 2008;48:498–508. doi: 10.1021/ci700413a. [DOI] [PubMed] [Google Scholar]
- 63.Franco P., Porta N., Holliday J.D., Willett P. The use of 2D fingerprint methods to support the assessment of structural similarity in orphan drug legislation. J. Cheminform. 2014;6:5. doi: 10.1186/1758-2946-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franco P., Porta N., Holliday J.D., Willett P. Molecular similarity considerations in the licensing of orphan drugs. Drug Discov. Today. 2017;22:377–381. doi: 10.1016/j.drudis.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 65.Rogers D.J., Tanimoto T.T. A computer program for classifying plants. Science. 1960;132:1115–1118. doi: 10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- 66.Bajusz D., Rácz A., Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 2015;7:20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung N.C., Miasojedow B., Startek M., Gambin A. Jaccard/Tanimoto similarity test and estimation methods for biological presence-absence data. BMC Bioinformatics. 2019;20:644. doi: 10.1186/s12859-019-3118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang D., Hackett W.E., Zhong L., Wan X.F., Zaia J. Measuring site-specific glycosylation similarity between influenza a virus variants with statistical certainty. Mol. Cell Proteomics. 2020;19:1533–1545. doi: 10.1074/mcp.RA120.002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khatri K., Klein J.A., Zaia J. Use of an informed search space maximizes confidence of site-specific assignment of glycoprotein glycosylation. Anal. Bioanal. Chem. 2017;409:607–618. doi: 10.1007/s00216-016-9970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kall L., Storey J.D., MacCoss M.J., Noble W.S. Posterior error probabilities and false discovery rates: Two sides of the same coin. J. Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 71.Kall L., Storey J.D., Noble W.S. Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics. 2008;24:i42–i48. doi: 10.1093/bioinformatics/btn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kall L., Storey J.D., Noble W.S. QVALITY: Non-parametric estimation of q-values and posterior error probabilities. Bioinformatics. 2009;25:964–966. doi: 10.1093/bioinformatics/btp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yi X., Gong F., Fu Y. Transfer posterior error probability estimation for peptide identification. BMC Bioinformatics. 2020;21:173. doi: 10.1186/s12859-020-3485-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma Z.Q., Polzin K.O., Dasari S., Chambers M.C., Schilling B., Gibson B.W., Tran B.Q., Vega-Montoto L., Liebler D.C., Tabb D.L. QuaMeter: Multivendor performance metrics for LC-MS/MS proteomics instrumentation. Anal. Chem. 2012;84:5845–5850. doi: 10.1021/ac300629p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 76.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 77.Khatri K., Klein J.A., White M.R., Grant O.C., Leymarie N., Woods R.J., Hartshorn K.L., Zaia J. Integrated omics and computational glycobiology reveal structural basis for influenza A virus glycan microheterogeneity and host interactions. Mol. Cell. Proteomics. 2016;15:1895–1912. doi: 10.1074/mcp.M116.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]