Abstract
Mammalian splicing factor 1 (SF1; also mammalian branch point binding protein [mBBP]; hereafter SF1/mBBP) specifically recognizes the seven-nucleotide branch point sequence (BPS) located at 3′ splice sites and participates in the assembly of early spliceosomal complexes. SF1/mBBP utilizes a “maxi-K homology” (maxi-KH) domain for recognition of the single-stranded BPS and requires a cooperative interaction with splicing factor U2AF65 bound to an adjacent polypyrimidine tract (PPT) for high-affinity binding. To investigate how the KH domain of SF1/mBBP recognizes the BPS in conjunction with U2AF and possibly other proteins, we constructed a transcriptional reporter system utilizing human immunodeficiency virus type 1 Tat fusion proteins and examined the RNA-binding specificity of the complex using KH domain and RNA-binding site mutants. We first established that SF1/mBBP and U2AF cooperatively assemble in our reporter system at RNA sites composed of the BPS, PPT, and AG dinucleotide found at 3′ splice sites, with endogenous proteins assembled along with the Tat fusions. We next found that the activities of the Tat fusion proteins on different BPS variants correlated well with the known splicing efficiencies of the variants, supporting a model in which the SF1/mBBP-BPS interaction helps determine splicing efficiency prior to the U2 snRNP-BPS interaction. Finally, the likely RNA-binding surface of the maxi-KH domain was identified by mutagenesis and appears similar to that used by “simple” KH domains, involving residues from two putative α helices, a highly conserved loop, and parts of a β sheet. Using a homology model constructed from the cocrystal structure of a Nova KH domain-RNA complex (Lewis et al., Cell 100:323–332, 2000), we propose a plausible arrangement for SF1/mBBP-U2AF complexes assembled at 3′ splice sites.
RNA-binding proteins participate in many pathways of gene expression and often function as part of large protein and RNA assemblies, such as the ribosome or spliceosome. To understand how such assemblies are formed and regulated, it is necessary to examine how individual RNA binding domains recognize their specific RNA sites and how their binding specificity may be modulated when placed in the context of a larger complex. Two of the most common types of eukaryotic RNA-binding domains are the RNP (or RRM) domain and the K homology (KH) domain (46). Structural studies of isolated RNP and KH domains and their complexes with RNA, as well as of other RNA-protein complexes, have elucidated many features important for specific RNA recognition, such as hydrogen bonding interactions and insertion of bases into hydrophobic protein pockets (21, 53). Recent studies with tethered RNP domains and multiprotein-RNA complexes have begun to define how the spatial organization of domains can contribute to RNA-binding specificity (2, 20, 26, 43, 57). Here we examine how a KH domain recognizes its RNA site in conjunction with other proteins that assemble at 3′ splice sites.
KH domains are ∼70 residues in length and, like RNP domains, often are arranged as tandem repeats (13, 49, 58). A subset of KH proteins contain only a single, larger (∼100-amino-acid) domain, a “maxi-KH” domain, which contains additional amino acids within two loop regions. Maxi-KH domains also are flanked by two other conserved regions, QUA1 and QUA2, of unknown function (see reference 58 for a review). In addition, maxi-KH domain proteins contain sequences likely to be bound by SH3 or WW domains, suggesting potential roles in signal transduction, and therefore have been named STAR proteins (signal transduction and activation of RNA) (58). The structures of several isolated KH domains all display similar three-stranded antiparallel β sheets packed against three α helices but have relatively low sequence homology (5, 34, 39; G. Musco, A. Kharrat, G. Stier, F. Faternali, T. J. Gibson, M. Nilges, and A. Pastore, Letter, Nat. Struct. Biol. 4:712–716). Several largely conserved hydrophobic residues are interspersed throughout the domain, and all contain a GXXG motif in the loop connecting helices α1 and α2.
Relatively few physiological RNA sites have been identified for KH domains, but it already is clear that a wide range of RNA structures can be recognized by these domains. Currently known RNA targets range in size from 7 to 75 nucleotides, binding affinities range from 10−6 to 10−9 M, and in some cases one domain is used for recognition while in other cases multiple domains are used (9, 12, 27, 29). Recently, the cocrystal structure of a KH domain from the Nova-2 protein bound to an RNA hairpin was reported (35). This domain primarily recognizes four unpaired nucleotides, which bind in a hydrophobic pocket formed by the α1 and α2 helices and an edge of the β2 strand, with additional contacts made by the flanking GXXG loop, characteristic of all KH domains, as well as by a variable loop between β2 and β3. Given the wide range of RNA sites recognized by KH domains, it will be interesting to determine whether all KH domains, including the larger maxi-KH domains found in STAR proteins, use a similar binding mode.
Mammalian splicing factor 1 (SF1), also known as mammalian branch point binding protein (mBBP) and hereafter designated SF1/mBBP, is a member of the STAR family and participates in the assembly of the spliceosomal E complex (the early mammalian U1 snRNP complex or commitment complex) by binding to the seven-nucleotide branch point sequence (BPS) found at 3′ splice sites (3, 9, 30). SF1/mBBP, together with splicing factor U2AF (which is composed of 65- and 35-kDa subunits), facilitates U2 snRNP binding to 3′ splice sites (32). As SF1/mBBP and U2 snRNP both bind to the BPS, it has been postulated that SF1/mBBP is displaced upon U2 snRNP binding (19, 37, 47, 52). In one case, SF1/mBBP also has been shown to participate in exon definition by binding to a set of seven-nucleotide repeats located in a splicing enhancer of a microexon (15). In its canonical setting, the maxi-KH domain of SF1/mBBP specifically recognizes the BPS while an additional zinc knuckle domain interacts nonspecifically with RNA and raises the overall binding affinity (9, 10). SF1/mBBP also forms a cooperative complex with U2AF65, which binds to the polypyrimidine tract (PPT) just 3′ to the BPS (8, 9, 44). In addition, U2AF65 forms a heterodimer with U2AF35, which recognizes the AG dinucleotide found at intron-exon boundaries 3′ to the PPT (24, 38, 60, 61, 64). Mammalian BPSs show substantial variation (YNCURAY is the consensus sequence, where Y is pyrimidine, R is purine, and N is any nucleotide), and cooperative interactions with U2AF are believed to help SF1/mBBP recognize these diverse sequences. In contrast, the yeast BPS is highly conserved (UACUAAC) and yeast BBP (yBBP) binding appears less dependent on interactions with adjacent protein-RNA complexes (8, 9, 44). Additional protein-protein interactions between SF1/mBBP and the WW motifs of formin-binding proteins 11 and 21 and the SH3 domain of Abl also have been observed (6, 7), but their functional importance remains to be determined.
Like SF1/mBBP, other KH domain proteins bind RNA as part of larger complexes, and in some cases there is evidence that their RNA-binding properties are modulated by interactions with auxiliary proteins or protein-RNA complexes. The hnRNP E1 and E2 proteins contain three KH domains and have been implicated in stabilizing α-globin mRNA and the translational silencing of 15-lipoxygenase mRNA by binding to 3′ untranslated region elements in conjunction with different proteins (41). In the life cycle of poliovirus, these proteins also bind to a viral 5′ cloverleaf structure important for replication and to an internal ribosomal entry site element, switching between the two sites based on the availability of the viral 3CD protein (23). Finally, the RNA-binding affinity of Sam68, a STAR protein that interacts with several cell signaling proteins, appears to be regulated by tyrosine phosphorylation (58).
To further examine how KH domains recognize RNA in the context of a larger assembly and to better characterize the binding properties of a maxi-KH domain, we have used a mammalian cell reporter system to examine the RNA-binding properties of SF1/mBBP. We demonstrate that the U2AF heterodimer is recruited along with SF1/mBBP to 3′ splice site reporters, composed of the BPS, PPT, and AG dinucleotide, in mammalian cell nuclei and that binding to BPS variants correlates with previously measured splicing efficiencies, supporting a role for the SF1/mBBP complex in determining splice site usage. Using mutagenesis, we have defined an RNA-binding surface of SF1/mBBP that agrees well with the one used by Nova-2 (35), suggesting that the maxi-KH domains of STAR proteins and “simple” KH proteins utilize related binding mechanisms. We have generated a structural model of the SF1/mBBP KH domain and, based on the Nova-2 binding arrangement, propose an arrangement of the SF1/mBBP-U2AF complex assembled at 3′ splice sites.
MATERIALS AND METHODS
Plasmid construction.
BPS reporter plasmids were constructed by cloning synthetic oligonucleotide cassettes encoding the sequences shown in Fig. 1 into the AflII and HindIII restriction sites of an human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR)-chloramphenicol acetyltransferase (CAT) reporter plasmid in place of the transcription activation response (TAR) site. This plasmid contains a previously modified HIV-1 LTR (51) in which additional restriction sites have been engineered between the start of transcription and the TAR element. The inserted sequence of the wild-type BPS reporter, beginning at the 5′ end of the transcript and encompassing the BPS, PPT, and AG dinucleotide (shown in boldface) is 5′-GGTCTCTCTGGCTTAAGTTCGTACTAACCCTGTCCCTTTTTTTTCCACAGCAAGCTT, with the AflII and HindIII sites underlined. Plasmids encoding the Tat fusion proteins were constructed by cloning PCR amplification products into the SalI and SpeI restriction sites of a pSV2Tat expressor plasmid (33), creating C-terminal fusions following amino acid 72 of HIV-1 Tat and a linker of three glycines. Tat-fused SF1/mBBP was generated by amplifying a fragment encoding amino acids 2 to 307 from pGEX6P-SF1 (8), and a truncated version, Tat-fused SF1/mBBPΔN, was generated by amplifying a fragment encoding amino acids 69 to 307. Tat-fused U2AF65 was generated by amplifying a fragment encoding full-length U2AF65 (amino acids 1 to 475) from pGEX6P-U2AF65 (8). A variant with an internal deletion, Tat-fused U2AF65Δ95–138, also was constructed. Alanine mutations were introduced into Tat-fused SF1 by PCR mutagenesis.
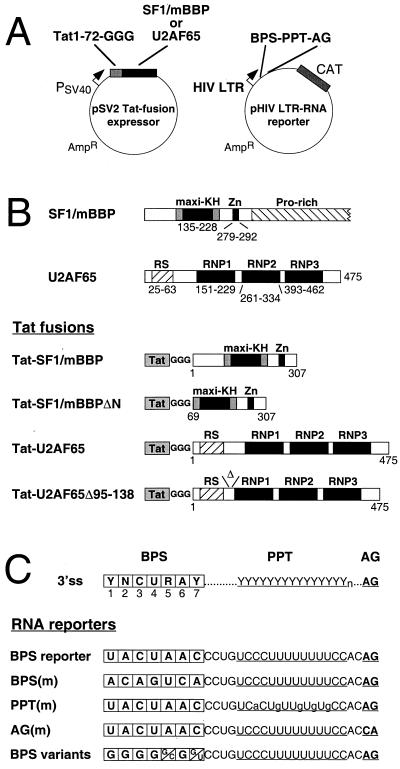
FIG. 1.
(A) Schematic diagrams of the Tat fusion expressor and RNA reporter plasmids. The expressor plasmid encodes HIV-1 Tat residues 1 to 72, followed by a linker of three glycines and the relevant fusion, expressed from a simian virus 40 early promoter (PSV40). The reporter plasmid utilizes a modified HIV-1 LTR to drive CAT expression, with the BPS-PPT-AG sequence replacing the TAR site at the 5′ end of the mRNA. (B) Domain organization of wild-type SF1/mBBP and U2AF65 (top) and the Tat fusion proteins (bottom). The maxi-KH domain with flanking QUA1 and QUA2 regions characteristic of STAR proteins, the Zn knuckle, and the proline-rich region of SF1/mBBP and the RS domain and three RNP domains of U2AF65 are indicated. Numbers refer to amino acid positions in the proteins. The Tat fusions contain amino acids 1 to 72 of Tat followed by the glycine linker. (C) The configuration of the BPS, PPT, and AG dinucleotide at canonical 3′ splice sites (ss) (top) and the sequences of the BPS reporter and mutants (bottom) are shown. BPS variants indicate the single nucleotide substitutions used at each position.
Transient transfection and CAT assays.
Levels of CAT activation were measured by cotransfecting 100 ng of BPS reporter plasmids or 25 ng of the TAR reporter plasmid and 5 ng of the Tat fusion expressor plasmid (except where indicated in the figure legends) into HeLa cells using 5 μl of Lipofectin (Life Technologies) in 3.8-cm2 wells for 4 h. Total plasmid DNA was adjusted to 2 μg with pBluescript DNA. CAT activities were assayed after 44 h by using an appropriate amount of cell extract, as described previously (55). Activities were quantitated using a Molecular Dynamics phosphorimager, and fold activation relative to that for the reporter plasmid alone was calculated. For each experiment, CAT assays were performed in duplicate and percentages of activation for the different reporters or protein mutants relative to those for the wild-type combination were calculated. Percentages of activation were then averaged over three or four separate transfection experiments, and standard deviations of the means (shown in Fig. 3 to 5) were calculated. To control for possible differences in expression levels of the various fusion proteins, CAT activities also were measured on an HIV-1 LTR reporter containing the wild-type TAR element. All fusion proteins contained full-length Tat, including its own RNA-binding domain, and therefore could activate the TAR-containing reporter. Activation levels of the BPS reporters were normalized to the values with the TAR reporter, which showed less-than-twofold differences in activation for all fusion proteins. HeLa cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum.
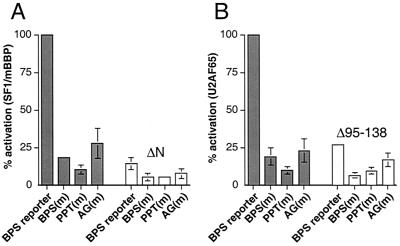
FIG. 3.
Activation by the Tat fusion proteins requires RNA-protein interactions at the BPS, PPT, and AG dinucleotide. (A) Activation of the BPS reporter and mutants by Tat-fused SF1/mBBP (gray bars) and Tat-fused SF1/mBBPΔN (white bars). Tat-expressing (5 ng) and BPS reporter (100 ng) plasmids were cotransfected into HeLa cells, and CAT activities were measured after 44 h. Percent activation was calculated by normalizing to the activation level of the Tat-fused SF1/mBBP-BPS reporter interaction, and standard deviations (bars) were calculated as described in Materials and Methods. (B) Activation of the BPS reporter and mutants by Tat-fused U2AF65 (gray bars) and Tat-fused U2AF65Δ95-138 (white bars). Activities were normalized to the activation level of the Tat-fused U2AF65-BPS reporter interaction.
FIG. 5.
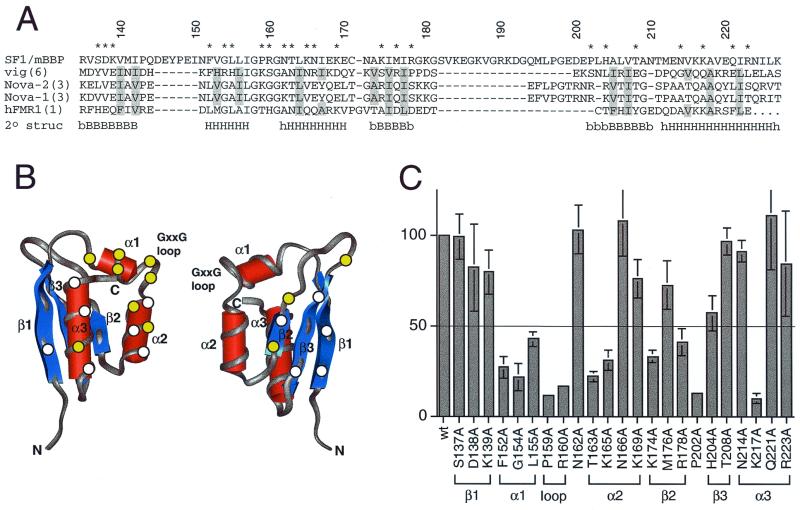
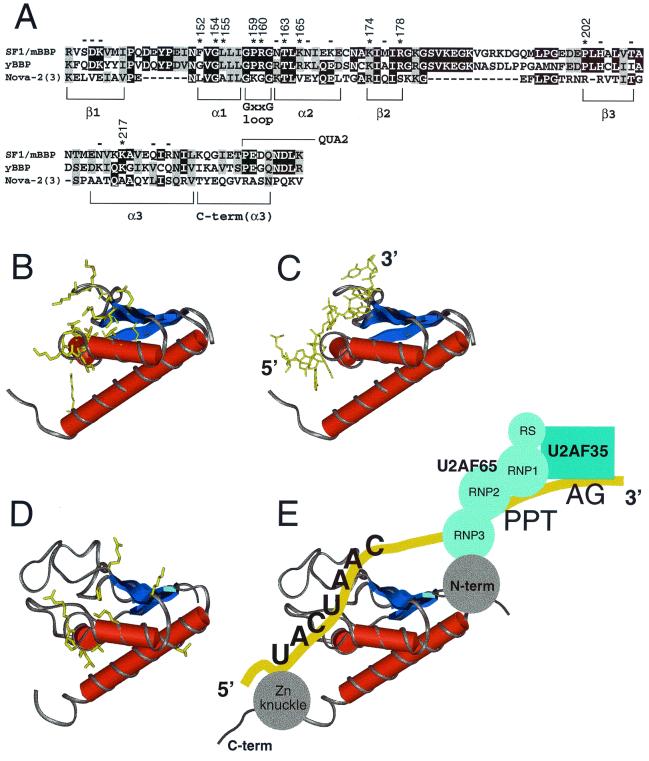
Activation of the BPS reporter by Tat-fused SF1/mBBP mutants. (A) Sequence alignment of the maxi-KH domain of SF1/mBBP and selected KH domains from vigilin (vig; domain 6), Nova-2 (domain 3), Nova-1 (domain 3), and hFMR-1 (domain 1). Secondary structure elements were assigned based on the X-ray and NMR structures of these individual domains (34, 39; Musco et al., letter). B and H, β-sheet and α-helical residues, respectively, which are defined clearly in all structures; lowercase letters, residues that can be assigned to a secondary structure in only one or two structures. Residues considered to be part of the hydrophobic core and therefore not chosen for mutation are shaded. Numbers refer to the SF1/mBBP sequence, and residues marked with an asterisk were mutated to alanines. (B) Averaged NMR structure of the sixth KH domain from vigilin (PDB file 1VIH [39]), shown from two different faces. Circles, approximate locations of amino acids chosen for mutagenesis, assuming that SF1/mBBP adopts a similar fold; yellow circles, positions that decrease activity by at least twofold (see panel C); white circles, positions that have little or no effect. (C) Activities of the Tat-fused SF1/mBBP mutants normalized to the activity of the wild-type (wt) protein with standard deviations (bars) calculated as for Fig. 3. The line corresponds to a twofold decrease in activity. The predicted corresponding units of secondary structures are indicated.
Molecular modeling.
The LOOK algorithm (Molecular Applications Group, Palo Alto, Calif.) was used to construct a three-dimensional model of the SF1/mBBP KH domain based on the crystal structure of the Nova-2 KH domain bound to RNA (35) (PDB file 1EC6; chain A). The amino acid sequence alignment was based on those of Musco et al. (39) and Lewis et al. (35) but was modified to better align three residues within the variable loop between β2 and β3.
RESULTS
Reporter system to monitor SF1/mBBP-RNA interactions in vivo.
We are interested in understanding how the maxi-KH domain of SF1/mBBP recognizes the seven-nucleotide BPS in the context of the complex it forms with other proteins that also assemble at 3′ splice sites. We chose to test whether the Tat hybrid system (33, 54) might be suitable for studying these interactions in vivo. The HIV-1 Tat protein enhances the efficiency of transcriptional elongation from the HIV-1 LTR and binds to an RNA hairpin, known as TAR, located at the 5′ end of the nascent transcript. Heterologous RNA-protein interactions can be used to deliver the Tat activation domain to the LTR (33), and therefore we asked whether SF1/mBBP fused to Tat could activate transcription from an LTR-CAT reporter containing the BPS in place of TAR (Fig. 1A). A fusion protein containing a fragment of SF1/mBBP (residues 1 to 307) linked to the C terminus of full-length Tat (residues 1 to 72) was constructed. This SF1/mBBP fragment includes the maxi-KH domain, surrounding QUA1 and QUA2 regions, and the zinc knuckle (Fig. 1B) and is sufficient for BPS recognition, its interaction with U2AF65, and splicing activity (8, 44). Because the Tat portion of the fusion protein also contains its own RNA-binding domain, it was possible to indirectly assess the expression levels of this and all other fusions described in this study by independently measuring activation levels on a HIV-1 TAR reporter. By this criterion, all fusion proteins were expressed at similar levels, showing less than a twofold variation in activity (Fig. 2C). All activities shown were normalized accordingly.
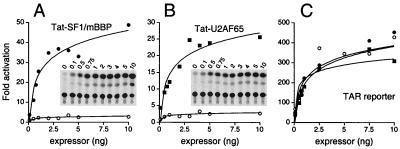
FIG. 2.
Activation of the BPS reporter by Tat-fused SF1/mBBP and Tat-fused U2AF65. (A) Titration of the Tat-fused SF1/mBBP expressor plasmid (filled circles) and unfused Tat (open circles) on the BPS reporter. Tat-expressing and BPS reporter plasmids were cotransfected into HeLa cells, and CAT activities were measured after 44 h. (Insets) CAT assays, with expressor plasmid amounts (nanograms) indicated. Fold activation was determined relative to the activity of the reporter alone. (B) Titration of the Tat-fused U2AF65 expressor plasmid (filled squares) and unfused Tat (open circles) on the BPS reporter. (C) Titration of the Tat-fused SF1/mBBP (filled circles), Tat-fused U2AF65 (filled squares), and unfused Tat (open circles) expressor plasmids on the HIV-1 TAR reporter. All fusions contain the RNA-binding domain of Tat and thus are able to activate transcription via the TAR element. Relative activities of the fusion proteins on the TAR reporter were used to normalize for fusion protein expression levels.
We constructed a BPS reporter in which the optimal mammalian BPS (identical to the conserved yeast BPS, UACUAAC) (63) was cloned in place of TAR followed by a PPT derived from the adenovirus major-late pre-mRNA and an AG dinucleotide (BPS reporter; Fig. 1C). In principle, the PPT and AG dinucleotide could recruit the endogenous U2AF heterodimer (the 65- and 35-kDa subunits, respectively) cooperatively to the RNA along with the Tat-fused SF1/mBBP protein. Cotransfection of the Tat-fused SF1/mBBP expressor and BPS reporter plasmids into HeLa cells resulted in strong (∼45-fold), dose-dependent activation of CAT activity, whereas the unfused Tat protein showed no activation (Fig. 2A). The observed interaction is dependent on the BPS, as mutating the sequence to poorly match the mammalian YNCURAY consensus reduced activity more than fivefold [see the BPS(m) (ACAGUCA) reporter; Fig. 1C and 3A]. The effects of other BPS mutations are described below.
If endogenous U2AF65 and U2AF35 were being recruited to the BPS reporter as hypothesized, we reasoned that activation also should be observed if U2AF65 was fused to Tat, with recruitment of endogenous SF1/mBBP and U2AF35. The U2AF65 subunit of the U2AF heterodimer recognizes the PPT and interacts cooperatively with SF1/mBBP; the interaction is mediated by an interaction between an N-terminal region of SF1/mBBP and the third RNP domain of U2AF65 (8, 9, 44). Indeed, cotransfection of a Tat-fused U2AF65 expressor plasmid (Fig. 1B) with the wild-type BPS reporter plasmid resulted in strong (∼25-fold), dose-dependent activation of CAT activity (Fig. 2B), consistent with the inference that SF1/mBBP and U2AF65 both bind to the BPS reporter.
Cooperative binding of the SF1/mBBP-U2AF65-U2AF35 complex.
To further test whether the endogenous U2AF subunits are recruited to the BPS reporter, we next measured activation of a mutant reporter, deficient for binding to U2AF65, in which several pyrimidines in the PPT were replaced by purines (Fig. 1C) (60). Activation of the PPT(m) reporter by both the Tat-fused SF1/mBBP and Tat-fused U2AF65 proteins was reduced 5- to 10-fold (Fig. 3), suggesting that the interaction of Tat-fused U2AF65 with the 3′ splice site requires an intact PPT and that the U2AF65-PPT interaction stabilizes the SF1/mBBP-BPS interaction, as observed in vitro (8). The cooperative nature of the protein-protein and protein-RNA interactions is further supported by the observation that activation by the Tat-fused U2AF65 is reduced more than fivefold for the BPS(m) reporter (Fig. 3B).
We next deleted amino acids 1 to 61 of SF1/mBBP (SF1/mBBPΔN in Fig. 1B), which eliminates U2AF65 binding but not RNA binding (44). Activation of the wild-type BPS-PPT-AG reporter was reduced 5- to 10-fold (Fig. 3A), further suggesting that the interaction between SF1/mBBP and U2AF65 is required for efficient RNA binding in this system. As expected, the SF1/mBBPΔN mutant also was inactive on the PPT(m) reporter.
It recently has been reported that U2AF35 binds to the AG dinucleotide at the intron-exon boundary, stabilizing the U2AF65-PPT interaction, particularly if the PPT is not optimal (24, 38, 60, 64). To test whether the U2AF35 subunit also binds to our BPS reporter, we measured the activities of Tat-fused SF1/mBBP and Tat-fused U2AF65 on a reporter in which the AG dinucleotide was changed to CA (Fig. 1C) and observed a three- to fourfold decrease in activity in both cases (Fig. 3). We next constructed a Tat-fused U2AF65 deletion mutant (U2AF65Δ95–138; Fig. 1B) that does not interact with U2AF35 (22, 62) and observed a similar decrease in activity on the wild-type BPS-PPT-AG reporter (Fig. 3B). Thus, U2AF35 appears to be recruited to the complex, stabilizing the U2AF65-PPT interaction and consequently the SF1/mBBP-BPS interaction. The activity of the U2AF65Δ95–138 mutant on the BPS(m) reporter is reduced even further (Fig. 3B), suggesting that both the SF1/mBBP-BPS and U2AF65-U2AF35 interactions contribute to stabilizing the U2AF65-PPT interaction. It is interesting that the splicing of the adenovirus major-late pre-mRNA, from which our strong PPT is derived, is not dependent on the AG dinucleotide (24, 60) whereas our BPS reporter appears to be at least partially dependent on the U2AF35-AG interaction. It seems plausible that the increased dependence on the AG dinucleotide in our system may reflect the lack of additional protein-protein or protein-RNA interactions in the spliceosome that help stabilize the U2AF65 interaction in the absence of the U2AF35-RNA interaction. Alternatively, our results may reflect differences in the intrinsic RNA-binding affinity that are not rate limiting for splicing.
BPS binding specificity.
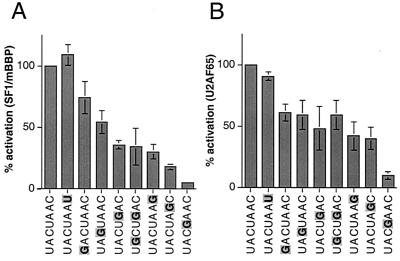
As described above, mammalian BPSs typically are defined by the broad consensus YNCURAY sequence (14) and show rates of splicing that differ by more than 1 order of magnitude (63). Because the SF1/mBBP-BPS interaction is likely replaced later in spliceosome assembly by the base pairing of U2 snRNA, effects of BPS variation on splicing efficiency might reflect either one or both binding events. Our reporter system appears to accurately reflect assembly of SF1/mBBP-BPS complexes and therefore provides a reasonable tool to monitor the effect of BPS variation on the initial protein binding events, although we cannot exclude the influence of other factors (see Discussion). We constructed a series of BPS variant reporters with single nucleotide changes to the UACUAAC sequence (Fig. 1C) and measured activation by Tat-fused SF1/mBBP. A 20-fold range of activities was observed, with the yeast UACUAAC sequence and a conservative C-to-U change at the last position producing the highest activities and changes of the conserved branch point adenosine at the sixth position (UACUAGC) and of the conserved uridine at the fourth position (UACGAAC) producing the lowest activities (Fig. 4A). Mutations at the fourth and sixth positions also strongly decrease SF1/mBBP RNA-binding affinity in vitro (9). Changing the last position of the YNCURAY consensus sequence to a purine decreases activity about threefold, as does changing the fifth partially degenerate position (R) from A to G (Fig. 4A). Changing the third conserved C or the first partially degenerate position (Y) produces modest decreases of less than twofold. Changing the second nucleotide at the degenerate position (N) along with changing the fifth position, which creates the normal adenovirus major-late pre-mRNA 3′ splice site region (encompassing the BPS, PPT, and AG), produces the same activity as changing the fifth position alone. It is interesting that in vitro binding assays were relatively insensitive to changes at the degenerate positions (9), suggesting that our reporter system, in which other proteins are recruited, may be more sensitive to small differences in RNA-binding affinity. Given the tight correspondence between SF1/mBBP complex formation and the known splicing efficiencies of BPS variants (see Discussion), our results support an important role for the SF1/mBBP-BPS interaction in determining 3′ splice site utilization.
FIG. 4.
Activation of BPS variant reporters by Tat-fused SF1/mBBP (A) and Tat-fused U2AF65 (B). Percentages of activation and standard deviations (bars) were calculated as for Fig. 3, with activities normalized to the Tat-fused SF1/mBBP-BPS reporter and Tat-fused U2AF65-BPS reporter interactions, respectively. BPS substitutions are highlighted.
We also measured the activities of Tat-fused U2AF65 on the same series of BPS variant reporters and generally observed a good correlation to the activities of Tat-fused SF1/mBBP although, as one might expect, altering the BPS had a greater effect on SF1/mBBP than on U2AF65 (Fig. 4B). Thus, U2AF65 binding to the PPT (and even to the “strong” PPT used here) also reflects the “strength” of the BPS, further demonstrating the interdependence of SF1/mBBP and U2AF in forming the spliceosome commitment complex.
Mutagenesis and modeling of the KH domain.
Relatively little is known about how the maxi-KH domains of STAR proteins recognize RNA, and we wished to use our reporter system to help define the RNA-binding surface of SF1/mBBP by mutagenesis. To identify amino acids potentially involved in BPS recognition, we chose the structures of several KH domains previously solved by nuclear magnetic resonance (NMR) or crystallography (34, 39; Musco et al., letter) and aligned their sequences with that of the KH domain of SF1/mBBP (Fig. 5A). Focusing largely on residues previously implicated in RNA binding (34), but prior to determination of the Nova-2 cocrystal structure, and focusing also on charged and conserved residues, we selected 23 positions in SF1/mBBP to generate alanine mutants (Fig. 5A and B). These positions were scattered throughout the different putative secondary structures and included no residues in the hydrophobic core. The expression levels of the mutants were similar, as assessed indirectly by measuring the activity of the Tat fusion on a HIV-1 TAR reporter as described above, and activities on the BPS reporter were normalized accordingly. Eleven mutations in Tat-fused SF1/mBBP reduced activity by at least twofold on the BPS reporter (Fig. 5C). All residues shown to be important, with the exception of Phe152, are highly conserved between SF1/mBBP and yBBP (Fig. 6A), and, conversely, almost all residues that do not appear to be important for binding are not conserved. Most of the important residues were grouped in adjacent structural elements, namely, the putative first and second α helices, the conserved GXXG loop between the two helices, and the second β strand.
FIG. 6.
(A) Sequence alignment of SF1/mBBP, BBP from Saccharomyces cerevisiae (yBBP), and KH domain 3 from Nova-2. Black boxes, identical residues; shaded boxes, conserved residues. The corresponding units of secondary structure are indicated. An extra C-terminal region is shown, compared to the sequence shown in Fig. 5A; this region corresponds to part of the Nova-2 structure. Asterisks and numbers, positions in SF1/mBBP important for binding based on the mutagenesis data (Fig. 5); minuses, positions where mutations have little or no effect. The numbering of Nova-2 residues discussed in the text (and not shown) corresponds to that described for the cocrystal structure (35). The region corresponding to the beginning of QUA2 is indicated. (B) Structure of the Nova-2 KH domain, taken from the cocrystal structure of the protein-RNA complex (35). Amino acids involved in RNA binding are shown in yellow. (C) Structure of the Nova-2 KH domain complexed to RNA (35), with the RNA tetranucleotide specifically recognized by the protein shown in yellow. The 5′ and 3′ ends of the RNA are indicated. (D) Homology model of SF1/mBBP maxi-KH domain, based on the structure of the RNA-bound Nova-2 domain and the alignment shown in panel A. The approximate locations of residues important for RNA binding, as defined by mutagenesis, are shown in yellow. The large β1/α1 and β2/β3 loops found in the SF1/mBBP maxi-KH domain were positioned arbitrarily by the homology modeling. (E) Proposed RNA-binding orientation of the SF1/mBBP-U2AF complex, based on similarities to the Nova-2 protein-RNA complex (see Discussion). The schematic drawing is not intended to indicate the relative orientations of the U2AF65 and U2AF35 subunits or of the RS domain of U2AF65, which also may contact the RNA (56).
Based on the recent Nova-2 KH domain–RNA cocrystal structure and the sequence alignment shown in Fig. 6A, we generated a homology model of the KH domain of SF1/mBBP (Fig. 6D). In the Nova-2 KH domain–RNA complex (Fig. 6C), nucleotides from an RNA hairpin loop are observed to bind in a hydrophobic pocket of the KH domain formed by the first two α helices and an edge of the second β strand and flanked by the conserved GXXG loop and a variable loop. Our mutagenesis data imply a similar RNA-binding surface for the maxi-KH domain of SF1/mBBP; amino acids presumed to be important for BPS recognition are located on our structural model of SF1/mBBP (Fig. 6D) at several positions analogous to those observed to contact the RNA in the Nova complex (Fig. 6B). With the exception of one lysine in α3 that cannot be aligned readily with the Nova-2 sequence, these residues define a relatively contiguous binding surface. NMR chemical shift mapping experiments using a KH domain from hnRNP K and a DNA oligonucleotide (4) also imply a similar binding region. Taken together, these results suggest that many or all KH domains, whether from a multi-KH domain protein or a STAR protein, may use similar faces for interacting with RNA. The specific interactions used to recognize RNA undoubtedly will differ among the various RNA-protein complexes.
DISCUSSION
Assembly of multiprotein complexes using the Tat hybrid system.
We have described a reporter assay based on the Tat hybrid system that monitors the cooperative binding of SF1/mBBP, U2AF, and perhaps other splicing factors to 3′ intronic sequences in vivo. An important advantage of this system is the ability to recruit additional endogenous nuclear proteins, in addition to the Tat fusion protein, to RNA sites engineered into the reporter. A related system utilizing the equine anemia virus Tat protein has been used to study a poliovirus protein-RNA interaction that also requires a host protein for RNA binding (11). With the BPS reporter, we have shown that Tat can activate transcription when fused to different components of the same multiprotein complex, SF1/mBBP or U2AF65, suggesting that Tat can act from multiple locations of a large RNA-protein complex, potentially even if tethered to proteins associated with RNA only indirectly via protein-protein interactions. This degree of flexibility is consistent with previous findings that a variety of RNA-binding domains can be functionally fused to Tat and many types of RNA sites can be accommodated in place of TAR (33). We do not know yet the limitations of the Tat hybrid system including, for example, possible steric restrictions imposed by the transcriptional machinery required for Tat activation. Nevertheless, the Tat hybrid system seems to be a useful tool for dissecting large and complex ribonucleoprotein complexes, such as the spliceosome, and for identifying novel interacting proteins from cDNA libraries fused to Tat (33, 54). Although we only have tested recruitment of SF1/mBBP and U2AF to the BPS, it is possible that other proteins such as UAP56, which interacts with U2AF (22), or even U2 snRNA or other snRNP components are stably or transiently bound. It also is possible that addition of a 5′ splice site may allow assembly of higher-order complexes that bridge the 5′ and 3′ splice sites.
Assembly of SF1/mBBP-RNA complexes.
Spliceosome assembly is highly dynamic and involves an ordered set of binding and rearrangement events utilizing different protein and RNA components. Recognition of the BPS by SF1/mBBP is an early step in the assembly pathway, and SF1/mBBP forms part of a complex that includes U1 snRNP bound near the 5′ splice site and U2AF bound to the PPT near the 3′ splice site. Later, SF1/mBBP is thought to be displaced through an unidentified ATP-dependent mechanism resulting in U2 snRNA base pairing to the BPS (40, 52). BPS recognition is likely to be regulated, and indeed it is known that splicing efficiency is strongly influenced by variations in the BPS (45, 63). Our results support a direct role for the SF1/mBBP-BPS interaction in determining the efficiency of 3′ splice site usage. We observed a 20-fold range of activities on a set of BPS variant reporters, with the UACUAAC conserved yeast sequence and UACUAAU (changed residues are in boldface) having the highest activities, GACUAAC, UAGUAAC, UACUGAC, and UACUAAG having intermediate activities, and UACUAGC and UACGAAC having the lowest activities. These preferences correlate well with in vitro SF1/mBBP binding experiments (8) and splicing assays that show a >10-fold range in splicing efficiency (UACUAAC > UACUGAC ≫ UACGGAC) (63). Additional in vitro splicing assays have shown that a BPS with a U-to-A change at the fourth position strongly decreases splicing efficiency and also alters 3′ splice site selection in vivo (45). Our corresponding U-to-G mutation shows the weakest activity (Fig. 4). In contrast, changes to the last position have relatively little effect on splicing efficiency (45) and either had no effect (C to U) or produced a modest decrease (C to G) in our assays. Compensatory mutations between the BPS and U2 snRNA have shown that the differences in splicing efficiency can only be partially explained by effects on base pairing (42). Thus it seems likely that the strength of the SF1/mBBP-BPS interaction contributes directly to 3′ splice site selection.
The situation at yeast introns is somewhat different in that the BPS is highly conserved whereas the PPT is less well conserved. Mud2p is the apparent U2AF65 yeast homolog and interacts with yBBP (1, 44), but, unlike U2AF65, Mud2p is not essential for splicing. In vitro, yBBP binds the UACUAAC BPS with higher affinity and specificity than does SF1/mBBP, leading to the suggestion that mammalian BPS complexes are more highly dependent on cooperative interactions between SF1/mBBP and U2AF65 than are yeast complexes (8, 9). Our results, however, suggest that SF1/mBBP recognizes the BPS with substantially higher specificity in vivo than in vitro, presumably due to its assembly into a larger, and probably more stable, RNA-protein complex. It is interesting that a Tat-fused yBBP was completely inactive on our BPS reporter (data not shown) despite the high specificity of the binary yBBP-BPS interaction in vitro. yBBP and U2AF65 do not interact in a two-hybrid assay (44), and we presume that the lack of Tat-fused yBBP activity reflects the requirement for U2AF65 binding. Our results emphasize how the specificity of an RNA-protein interaction can be influenced by its neighbors (2, 43, 50, 57) and underscore the value of studying the interactions in an in vivo context.
As mentioned above, not all relevant features of an intron are present in our BPS reporter, e.g., the 5′ splice site required to recruit U1 snRNP or enhancer sites required to recruit important SR proteins (59), both of which are needed for the formation of the commitment complex. Thus, we do not know how interactions with other components of the splicing machinery missing in our system may affect the SF1/mBBP interaction or whether factors that may later help displace SF1/mBBP from the complex to allow U2 snRNP binding are recruited to our BPS reporters. Later steps in the spliceosome assembly pathway apparently weaken the binding of U2AF65 (16), and it is possible that components of this process also influence the interactions observed in our reporter system. There clearly exists a large set of interdependent, cooperative interactions that ultimately determines the efficiency of 3′ splice site usage. In this regard it should be noted that depletion of yBBP from nuclear extracts has only a mild effect on splicing, and significant effects on splicing efficiency are seen only by combining mutations in the BPS or 5′ splice site with mutations in the yBBP protein (25, 47, 48). Thus, the essential role of SF1/mBBP may only be unmasked in the context of a suboptimal arrangement of components or in the pre-mRNAs of some essential genes. Furthermore, our results indicate that not only is the SF1/mBBP-BPS interaction highly dependent on U2AF65 binding, as previously observed (44), but also that the U2AF65-PPT interaction is dependent on SF1/mBBP binding (Fig. 3B). The binding of U2AF65 to a “weak” PPT is stabilized by U2AF35 binding to the AG dinucleotide at 3′ splice sites (24, 38, 60, 64), and it is possible that the SF1/mBBP-BPS interaction will play an even more important role in the context of a weakened PPT.
Given that recognition of the highly degenerate BPS in the early stages of spliceosome assembly provides an attractive regulatory target, it is particularly interesting that SF1/mBBP, like other STAR family members, contains sequence motifs in its C-terminal domain that often are used to interact with signaling proteins. It has been shown that proline-rich regions of one alternatively spliced form of SF1/mBBP interact with the WW motif of FBP11 and with the SH3 domain of Abl, whereas another splice variant interacts with the WW motif of FBP21 (6, 7). FBP11 is related to yeast U1 snRNP protein Prp40p, previously shown to interact with yBBP (1). It is interesting to speculate that interactions with these or other proteins, perhaps some involved in signaling, may regulate the BPS recognition properties of SF1/mBBP and that different protein-protein interactions involving alternatively spliced forms of SF1/mBBP may regulate alternative splicing of some pre-mRNAs (31).
RNA recognition by the maxi-KH domain.
Based on our mutagenesis of the maxi-KH domain of SF1/mBBP, a presumptive RNA-binding surface emerged (Fig. 6D), which is consistent with structural studies of non-STAR protein KH domains (4, 35). The protein-RNA interface seen in the Nova-2 KH domain–RNA complex (35) shows extensive van der Waals contacts from an aliphatic platform of the KH domain and hydrogen bonds between hydrophilic side chains and the Watson-Crick faces of single-stranded bases. We identified a number of charged and hydrophilic side chains within α1, α2, β2, and the GXXG loop of SF1 that appear to contribute to RNA binding (Fig. 5), and these correspond to regions of Nova-2 that contact the RNA (compare Fig. 6B and D). We did not examine residues in the aliphatic platform because many also help form the hydrophobic core of the domain. The β2/β3 loop of Nova-2 also contacts the RNA, and it is interesting that the large β1/α1 and β2/β3 loops found in maxi-KH domains seem positioned to form additional interactions (Fig. 6D). These large loops also have been implicated in protein-protein interactions in maxi-KH domain proteins (17, 39). Residues from the flanking QUA1 and QUA2 regions seem positioned to further extend the binding surface and might enable recognition of the longer seven-nucleotide BPS (versus the four-nucleotide recognition site for the Nova-2 KH domain). Deletion of QUA1 of the Qk1 protein was found to abolish RNA binding, and deletion of QUA2 had a modest effect (18). In yBBP, a mutation in QUA2 contributes to a mutant phenotype deficient in forming commitment complexes (48). Although not conserved in sequence, the regions flanking simple KH domains also may be important for RNA recognition. In Nova-2, an arginine located in an extended C-terminal helix and corresponding to the beginning of QUA2 (Fig. 6A) is required for high-affinity RNA binding (35). Additional mutagenesis of Tat-fused SF1/mBBP may help test whether these other regions directly contact the RNA, particularly if mutants with altered RNA-binding specificities can be found, or whether they mediate protein-protein interactions.
In addition to the apparent similarities of the SF1/mBBP and Nova-2 RNA-binding interfaces, similarities between the BPS and the UCAY tetranucleotide site recognized by the Nova domain permit us to propose a binding orientation for the entire SF1/mBBP-U2AF complex (Fig. 6E). We noticed that the last four nucleotides of the consensus BPS (URAY) are similar to those of the Nova-2 domain binding site (UCAY), differing only in the second position. Given that several KH domains appear to recognize tetranucleotide sequences (28, 36, 41), we asked whether analogous amino acids in SF1/mBBP and Nova-2 might be used to contact the RNA. Key residues of Nova-2 used to recognize uracil at the first tetranucleotide position and adenosine at the third position are well conserved in the SF1/mBBP maxi-KH domain. Gly18 and Ala19 of Nova-2 form van der Waals interactions with the uracil (35), and the equivalent positions in SF1/mBBP are Gly154 and Leu155 (Fig. 6A), both of which are important for BPS binding, as shown by our mutagenesis data (Fig. 5). Backbone atoms from Ile41, Leu21, and Leu28 of Nova-2 contact the adenosine at the third tetranucleotide position (35), and the equivalent positions in SF1/mBBP are conserved hydrophobic residues Ile177, Ile157, and Leu164. Given these similarities, we tentatively assign a binding orientation for the RNA as in the Nova-2 complex, with the 3′ end of the BPS positioned near the N terminus of SF1/mBBP (Fig. 6E). This model is consistent with the location of the PPT 3′ to the BPS and the U2AF65 binding domain at the N terminus of SF1/mBBP (Fig. 6E). More structural data clearly are needed to identify the specific contacts to the RNA and to establish the relative juxtaposition of the subunits, but it will be particularly interesting if cooperative interactions between SF1/mBBP and U2AF require a discrete spatial arrangement on the RNA, as seen with other multiprotein or multidomain complexes (2, 20, 26, 43, 57), and if BPS recognition can be influenced by altering the arrangement of the complex.
ACKNOWLEDGMENTS
We thank Michael Green for communicating results prior to publication, Stephen Burley for providing coordinates of Nova-2 KH domain structures, Alan Cheng for help with the molecular modeling, Tom Blumenthal, Mark Bedford, Amy Kistler, Christine Guthrie, Don Rio, and membes of the Frankel laboratory for helpful discussions, and Valerie Calabro, Donna Campisi, Chandreyee Das, Rob Nakamura, and Ralph Peteranderl for comments on the manuscript.
This work was supported by a Human Frontiers postdoctoral fellowship and an NIH postdoctoral training grant (to H.P-.Z.) and by grants from the National Institutes of Health. J.A.B. is a Burroughs Wellcome Fund Fellow of the Life Sciences Research Foundation.
REFERENCES
- 1.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Agalarov S C, Prasad G S, Funke P M, Stout C D, Williamson J R. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–112. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 3.Arning S, Grüter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 4.Baber J L, Levens D, Libutti D, Tjandra N. Chemical shift mapped DNA-binding sites and 15N relaxation analysis of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K. Biochemistry. 2000;39:6022–6032. doi: 10.1021/bi000105e. [DOI] [PubMed] [Google Scholar]
- 5.Baber J L, Libutti D, Levens D, Tjandra N. High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J Mol Biol. 1999;289:949–962. doi: 10.1006/jmbi.1999.2818. [DOI] [PubMed] [Google Scholar]
- 6.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bedford M T, Reed R, Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund J A, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 10.Berglund J A, Fleming M L, Rosbash M. The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA. 1998;4:998–1006. doi: 10.1017/s1355838298980499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair W S, Parsley T B, Bogerd H P, Towner J S, Semler B L, Cullen B R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 12.Buckanovich R J, Darnell R B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 14.Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 15.Carlo T, Sierra R, Berget S M. A 5′ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol Cell Biol. 2000;20:3988–3995. doi: 10.1128/mcb.20.11.3988-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champion-Arnaud P, Gozani O, Palandjian L, Reed R. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol Cell Biol. 1995;15:5750–5756. doi: 10.1128/mcb.15.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Damaj B B, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen T, Richard S. Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deo R C, Bonanno J B, Sonenberg N, Burley S K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 21.Draper D E. Themes in RNA-protein recognition. J Mol Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 22.Fleckner J, Zhang M, Valcárcel J, Green M R. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 23.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 24.Guth S, Martinez C, Gaur R K, Valcarcel J. Evidence for substrate-specific requirement of the splicing factor U2AF35 and for its function after polypyrimidine tract recognition by U2AF65. Mol Cell Biol. 1999;19:8263–8271. doi: 10.1128/mcb.19.12.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guth S, Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J Biol Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 26.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 27.Jan E, Motzny C K, Graves L E, Goodwin E B. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen K B, Musunuru K, Lewis H A, Burley S K, Darnell R B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanamori H, Dodson R E, Shapiro D J. In vitro genetic analysis of the RNA binding site of vigilin, a multi-KH-domain protein. Mol Cell Biol. 1998;18:3991–4003. doi: 10.1128/mcb.18.7.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krämer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krämer A, Quentin M, Mulhauser F. Diverse modes of alternative splicing of human splicing factor SF1 deduced from the exon-intron structure of the gene. Gene. 1998;211:29–37. doi: 10.1016/s0378-1119(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 32.Krämer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landt S G, Tan R, Frankel A D. Screening RNA-binding libraries using a Tat-fusion system in mammalian cells. Methods Enzymol. 2000;318:350–363. doi: 10.1016/s0076-6879(00)18062-0. [DOI] [PubMed] [Google Scholar]
- 34.Lewis H A, Chen H, Edo C, Buckanovich R J, Yang Y Y, Musunuru K, Zhong R, Darnell R B, Burley S K. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 35.Lewis H A, Musunuru K, Jensen K B, Edo C, Chen H, Darnell R B, Burley S K. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 36.Lin Q, Taylor S J, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 37.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 38.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 39.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 40.Nilsen T W. RNA-RNA interactions in nuclear pre-mRNA splicing. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 41.Ostareck-Lederer A, Ostareck D H, Hentze M W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biol Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 42.Parker R, Siliciano P G, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 43.Price S R, Evans P R, Nagai K. Crystal structure of the spliceosomal U2B"-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 44.Rain J-C, Rafi Z, Rhani Z, Legrain P, Krämer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed R, Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988;2:1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- 46.Rubin G M, et al. Comparative genomics of eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith C A, Calabro V, Frankel A D. An RNA-binding chameleon. Mol Cell. 2000;6:1067–1076. doi: 10.1016/s1097-2765(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 51.Smith C A, Crotty S, Harada Y, Frankel A D. Altering the context of an RNA bulge switches the binding specificities of two viral Tat proteins. Biochemistry. 1998;37:10808–10814. doi: 10.1021/bi980382+. [DOI] [PubMed] [Google Scholar]
- 52.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 53.Steitz T A. RNA recognition by proteins. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 427–450. [Google Scholar]
- 54.Tan R, Frankel A D. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc Natl Acad Sci USA. 1998;95:4247–4252. doi: 10.1073/pnas.95.8.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao J, Frankel A D. Electrostatic interactions modulate the RNA-binding and transactivation specificities of the human immunodeficiency virus and simian immunodeficiency virus Tat proteins. Proc Natl Acad Sci USA. 1993;90:1571–1575. doi: 10.1073/pnas.90.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valcarcel J, Gaur R K, Singh R, Green M R. Interaction of U2AF65 RS region with pre-mRNA of branch point and promotion base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 57.Varani L, Gunderson S I, Mattaj I W, Kay L E, Neuhaus D, Varani G. The NMR structure of the 38 kDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol. 2000;7:329–335. doi: 10.1038/74101. [DOI] [PubMed] [Google Scholar]
- 58.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 59.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 60.Wu S, Romfo C M, Nilsen T W, Green M R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 61.Zamore P D, Green M R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Zamore P D, Carmo-Fonseca M, Lamond A I, Green M R. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhuang Y, Goldstein A M, Weiner A M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci USA. 1989;86:2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zorio D A R, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]