Abstract
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase widely expressed in many cancers such as non-small cell lung cancer (NSCLC), pancreatic cancer, breast cancer, and head and neck cancer. Mutations such as L858R in exon 21, exon 19 truncation (Del19), exon 20 insertions, and others are responsible for aberrant activation of EGFR in NSCLC. First-generation EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib have clinical benefits for EGFR-sensitive (L858R and Del19) NSCLC patients. However, after 10-12 months of treatment with these inhibitors, a secondary T790M mutation at the gatekeeper position in the kinase domain of EGFR was identified, which limited the clinical benefits. Second-generation EGFR irreversible inhibitors (afatinib and dacomitinib) were developed to overcome this T790M mutation. However, their lack of selectivity toward wild-type EGFR compromised their clinical benefits due to serious adverse events. Recently developed third-generation irreversible EGFR TKIs (osimertinib and lazertinib) are selective toward driving mutations and the T790M mutation, while sparing wild-type EGFR activity. The latest studies have concluded that their efficacy was also compromised by additional acquired mutations, including C797S, the key residue cysteine that forms covalent bonds with irreversible inhibitors. Because second- and third-generation EGFR TKIs are irreversible inhibitors, they are not effective against C797S containing EGFR triple mutations (Del19/T790M/C797S and L858R/T790M/C797S). Therefore, there is an urgent unmet medical need to develop next-generation EGFR TKIs that selectively inhibit EGFR triple mutations via a non-irreversible mechanism.
Keywords: EGFR, NSCLC, Acquired resistance, T790M, C797S, TKIs
INTRODUCTION
One-quarter of global cancer deaths are caused by lung cancer (Sharma et al., 2007). Approximately 235,000 new cases of lung cancer patients were reported in the USA in 2020. The two major sub-categories of lung cancer are small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC accounts for almost 85% of all lung cancers (Finlay et al., 2014). The subtypes of NSCLC are adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. In advanced NSCLC, the 5-year survival rate is less than 20% (Midha et al., 2015). Epidermal growth factor receptor (EGFR) is one of the most investigated drug targets to treat NSCLC.
ERBB family
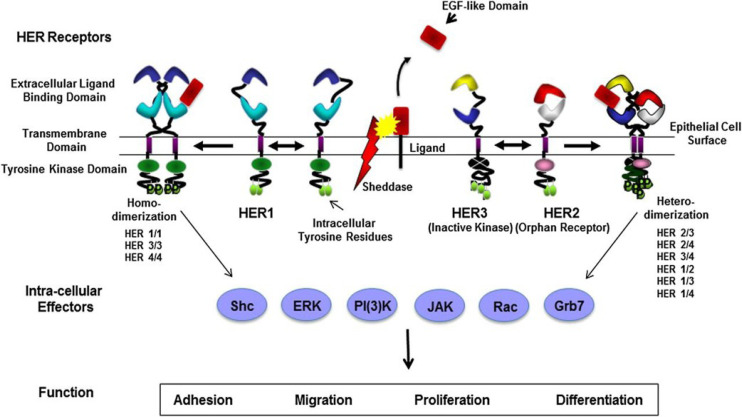
EGFR belongs to the ERBB gene family (Wee and Wang, 2017). ERBB family kinases consist of four types i) EGFR/ERBB1/HER1, ii) NEU/ERBB2/HER2, iii) ERBB3/HER3, and iv) ERBB4/HER4, and are activated by ligand binding to the receptor to form homo-dimer or hetero-dimers (Table 1) (Finigan et al., 2012). ERBB2 has no evidence of direct ligand binding; however, it is a heterodimeric binding partner to its family members (Tzahar et al., 1996). All family members of ERBB have intrinsic tyrosine kinase activity, except for ERBB3. ERBB family kinases are inactive when they exist in the monomeric form. On the ligand binding, such as with EGF and transforming growth factor-α (TGF-α), the receptors undergo conformational changes that enable the formation of homo-dimerization or hetero-dimerization, which activates the kinase domain (Singh et al., 2016).
Table 1.
ERBB family receptors and ligands
| Receptor | Ligand |
|---|---|
| ERBB1/HER1/EGFR | EGF |
| TGF-α | |
| Amphiregulin | |
| Betacellulin | |
| Epiregulin | |
| ERBB2/HER2 | No ligand |
| ERBB3/HER3 | Neuregulin 1, 2 |
| ERBB4/HER 4 | Betacellulin |
| Heparin-binding EGF | |
| Epiregulin | |
| Neuregulin 1-4 |
Structure of EGFR
EGFR is a glycoprotein with 170 kDa and is a member of the receptor tyrosine kinase. EGFR contains three domains: i) an extracellular ligand-binding domain, ii) a transmembrane domain, and iii) an intracellular domain that contains an ATP-binding active site with a tyrosine moiety (Roskoski, 2014). The extracellular domain comprises 621 amino acids and is subdivided into four regions (exons 1-16). EGFR activation begins with ligand binding, followed by homo- or hetero-dimerization, resulting in tyrosine phosphorylation which is located in the intracellular tyrosine kinase domain (Carpenter et al., 1991). The transmembrane domain consists of 23 amino acids (Ile622 to Met644). The intracellular domain consists of three major regions: flexible juxtamembrane (exons 16 and 17), tyrosine kinase domain (exons 18-24), and C-terminal tail (exons 25-28) (Morrow and Grant, 2000). The tyrosine kinase domain is further divided into two parts: the N-lobe and C-lobe. The ATP-binding active site is located between these two lobes. Kinase downstream signaling is activated by trans autophosphorylation interactions of the N-lobe of the receptor to the C-lobe of the other receptor (Tanner and Kyte, 1999). The kinase region also contains lysine rich residue, which is known as the main site of receptor ubiquitination (Zhang et al., 2006). In addition, the C-terminal tail contains several tyrosine residues that undergo autophosphorylation after activation of the receptor and serve as docking sites for effector proteins that transmit the signal further downstream. These proteins also regulate signal transduction (Walton et al., 1990).
EGFR downstream signaling pathways
EGF binds to EGFR and forms homo- or hetero-dimerization with other ERBB members, leading to EGFR receptor phosphorylation and the activation of downstream pathways (Ferguson et al., 2003) (Fig. 1). Ultimately, it drives the phosphorylation of downstream effectors such as RAS–RAF–MEK–ERK–MAPK and PI3K–AKT–mTOR (Batzer et al., 1994). The activation of these kinases stimulates the complete signaling network with many effects, such as cell proliferation, differentiation, migration, growth, and apoptosis inhibition (Olsen et al., 2006).
Fig. 1.
EGFR cellular signaling pathway (Finigan et al., 2012) (Permission granted by the American Thoracic Society).
Role of EGFR in NSCLC
EGFR plays a significant role in stimulating lung cancer generation and propagation (Ferlay et al., 2007). Lung cancer development proceeds from deregulated cellular proliferation that stimulates normal cells into malignant transformation. EGFR plays a significant role in initiating and triggering signaling events in NSCLC and SCLC. Within NSCLC, there are three types of sub-categories: i) adenocarcinomas, ii) squamous cell carcinomas, and iii) large cell carcinomas (Kenfield et al., 2008). EGFR overexpression has been identified among these subtypes of NSCLC, with the highest rates seen in squamous tumors (90%) and the lowest rates in adenocarcinomas (40%) and very little in large cell adenocarcinoma. Research has shown that EGFR can be a drug target in NSCLC therapy. The inhibition of EGFR function can be targeted by two major strategies: i) intracellular kinase signaling inactivation by TKIs and ii) neutralizing EGFR and ligands with antibodies (Ciardiello and Tortora, 2008).
Mutations in EGFR
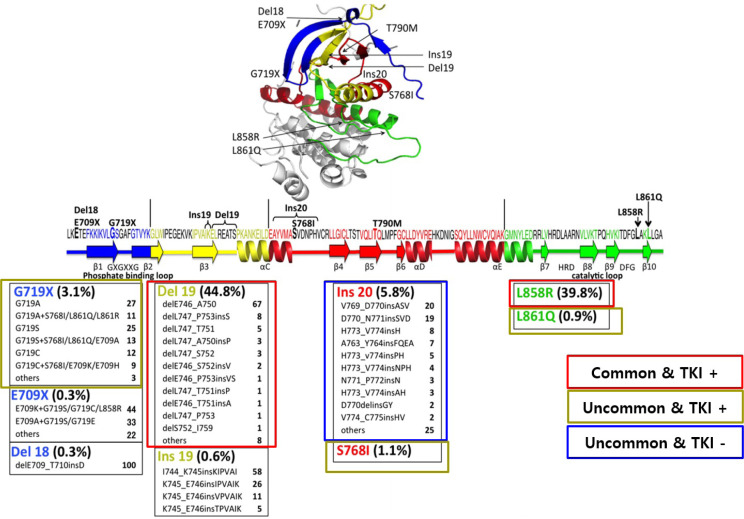
EGFR L858R and Del19 mutations are the two major driving mutations, and account for somatic mutations in the kinase domain of the EGFR gene related to lung adenocarcinoma. A total of 35% of Asians and 15% of Caucasians suffering from lung adenocarcinoma have EGFR-related mutations (Kobayashi and Mitsudomi, 2016). According to the COSMIC database, nearly 590 types of epithermal growth factor receptor mutations have been reported to date. The majority of these genes are present in the first four exons (18-21) of the tyrosine kinase domain (Fig. 2) (Kosaka et al., 2004). Early studies simplified the complexity of tumor genotypes by dividing them into mutant or wild-type. The most common mutations (sensitizing mutations) are Del19 (exon 19 deletions) and L858R (mutation in exon 21) (Tokudome et al., 2020). These sensitizing mutations or driving mutations account for 85% of EGFR mutations in NSCLC (Li et al., 2008). In addition, EGFR exon 20 insertion mutations are heterogeneous and are the third most common EGFR mutation, but no TKI is available yet (Goldberg et al., 2018).
Fig. 2.
Frequency of EGFR mutations in lung cancer and availability of targeted tyrosine kinase inhibitors (modified from the original figure, Kobayashi and Mitsudomi, 2016) [Permission granted from the publisher (John Wiley and Sons)].
Mechanism of EGFR activation by mutations
After EGF ligand binding to the EGFR, the C-helix conformation changes from outward to inward. This conformational change activates the enzyme from an inactive form to an active conformation (Purba et al., 2017). This active conformation forms important interactions with the p-loop of the active site, which is located between the cleft of the N-lobe and C-lobe (Yun et al., 2007). Earlier research confirmed that the driving mutations (Del19 and L858R) induced a conformational change, which disrupted the inactive form of EGFR, causing an equilibrium swing toward an active state over an inactive state. This change allowed EGFR to become ligand-independent and form homo- and hetero-dimerization, resulting in the proliferation and activation of downstream signaling pathways (Vyse and Huang, 2019). This variance in conformational change between the wild-type and mutant receptors resulted in the dependency of tumor cells on EGFR signaling. By targeting these classical activating EGFR mutations, there is room to treat NSCLC patients by making EGFR TKIs (Harrison et al., 2020).
CHRONICLES OF EGFR TKIS
During the early 1970s, lung cancer was considered as one unit arising from the lung. Later, chemotherapy combination studies of all patients with lung cancer, irrespective of subtypes such as SCLC or NSCLC, resulted in a minimum benefit to the patients, with the overall survival rate less than seven months (Pao and Chmielecki, 2010). During the early 2000s, a groundbreaking chemotherapy trial with platinum doublets studied in all patients with NSCLC revealed no particular benefit in the overall survival rate of the patients (Sandler et al., 2006), irrespective of histological subtype. Nonetheless, subdividing lung cancers into more clinically related subsets has continued. From this research analysis, scientists confirmed the main histological differences among lung cancer types, such as SCLC and NSCLC. NSCLC consists of multiple subtypes, such as adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (Travis et al., 2011). Today, additional subcategorization is driven by the understanding that tumors can be defined by different molecular criteria. The discovery that separate subsets of cancers harbor precise driver mutations in genes encoding signaling enzymes that are essential for cellular proliferation and survival led to one of the most promising treatment approaches. In 2004, the first EGFR mutant tumors were discovered, which presented the best-studied model of oncogene dependence in lung cancer (Gazdar, 2009).
First-generation EGFR TKIs
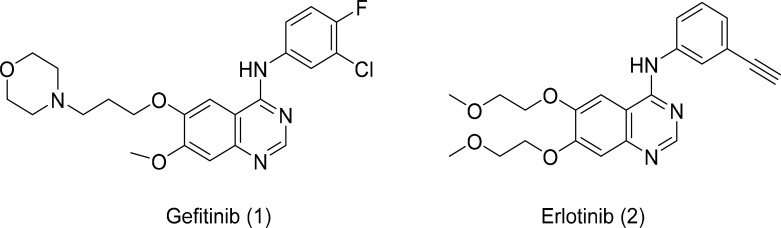
Research has confirmed that almost 85% of EGFR mutations in NSCLC consist of Del19 (deletions in exon 19) and L858R point mutations (within exon 21). Several small molecule inhibitors have been designed and evaluated for their inhibitory activity against EGFR mutant kinases. These kinase inhibitors belong to competitive inhibitors as they bind to the intracellular active region of the tyrosine kinase domain, where ATP binds to the kinase activation (Du and Lovly, 2018). Anilinoquinazoline-based derivatives, such as gefitinib and erlotinib (Fig. 3), are first-generation EGFR inhibitors and are reversible inhibitors. Gefitinib and erlotinib exhibit significant potency against driving mutations (Del19 and L858R) (Eck and Yun, 2010). The treatment of NSCLC patients with first-generation EGFR inhibitors led to a positive response rate of approximately 60%-80%. Although these small molecule inhibitors showed promising results, most of the patients who responded to this treatment faced drug resistance after nearly 10-12 months of treatment (Karachaliou et al., 2018). The major resistance mechanism is the replacement of threonine 790 with methionine residue (T790M) in nearly 60% of patients. This mutation increases the affinity for ATP and decreases the affinity to the first-generation EGFR inhibitors (Ko et al., 2017).
Fig. 3.
First-generation EGFR inhibitors.
Second-generation EGFR TKIs
To overcome the T790M mutation arising from the treatment of first-generation inhibitors, several irreversible 4-anilinoquinazoline analogs have been developed.
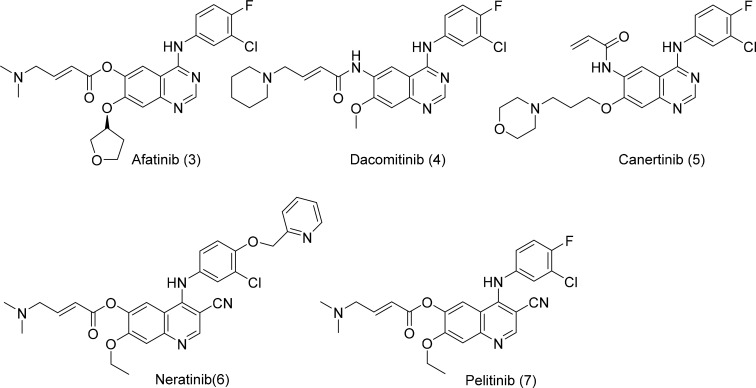
The second-generation EGFR TKIs such as afatinib, dacomitinib, neratinib, canertinib, and pelitinib (Fig. 4) have electrophilic acrylamide to form covalent interactions with Cys797 at the ATP-binding site of EGFR (Yu and Riely, 2013). The anilinoquinazoline moiety forms a hinge binding within the ATP-binding domain, providing a potential interaction with the gatekeeper Met790 residue and shows nanomolar potency toward EGFR Del19/T790M and EGFR L858R/T790M. However, the clinical benefits were compromised due to poor selectivity toward wild-type EGFR. Second-generation EGFR TKIs suffer intolerable low maximum tolerable dose due to dose-limiting toxicity, resulting from targeting wild-type EGFR and the repulsion created by the gatekeeper mutation T790M (Lin et al., 2014).
Fig. 4.
Second-generation EGFR inhibitors.
Third-generation EGFR TKIs
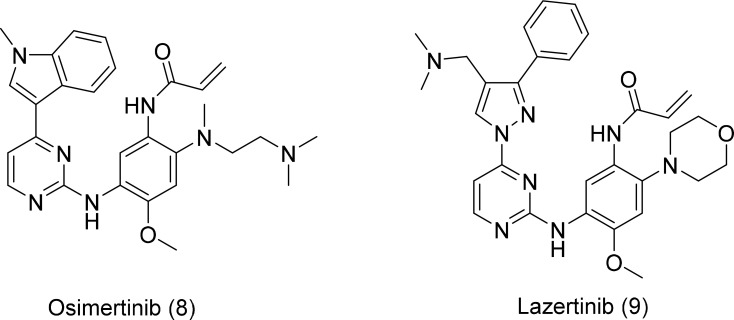
Third-generation irreversible EGFR inhibitors, namely osimertinib and lazertinib, are potent against mutated EGFR and display encouraging efficacy in NSCLC patients who are resistant to first- and second-generation inhibitors. They selectively target Del19 and L858R mutations, as well as the gatekeeper T790M-resistance mutation, by forming a covalent bond with the C797 residue, located in the ATP-binding site of mutant EGFR (Grabe et al., 2018). Both osimertinib and lazertinib (Fig. 5) exhibit superior activity against T790M mutants, with minimal off-target effects and fewer adverse events related to less inhibition of wild-type EGFR. Due to the excellent efficacy and better safety profile, osimertinib was approved to treat T790M-positive patients who have advanced EGFR mutations after first- or second-generation EGFR TKIs (Gao et al., 2016). The high clinical efficiency of osimertinib was compromised by modifying nucleophilic cysteine to serine in the active site, the C797S mutation. The point mutation of C797S led to acquired resistance, limiting the use of third-generation EGFR inhibitors (Lategahn et al., 2019).
Fig. 5.
Third-generation EGFR inhibitors.
Lazertinib showed promising antitumor activity in patients with tumors that had activated EGFR mutations and T790M resistance mutations. Lazertinib displayed less efficacy against wild-type EGFR than osimertinib, indicating that lazertinib might have fewer off-target side effects than osimertinib. Moreover, in a murine brain metastasis model, lazertinib inhibited intracranial tumor growth more efficiently than osimertinib. Clinical data have shown that lazertinib might be more effective in treating lung cancer with brain metastasis (Lategahn et al., 2019).
Resistance mechanism caused by the C797S mutation
The C797S mutation in EGFR is one of the mechanisms of drug resistance to third-generation EGFR TKIs such as osimertinib and lazertinib. Recent study findings have shown that C797S/T790M is in cis conformation in 82% of patients, C797S/T790M is in trans conformations in 10% of patients, C797S alone without T790M is in 6% of patients, and two co-existing C797S clones (one in cis with T790M and one in trans) are in 2% of patients. In addition, 84% of patients showed additional resistance mechanisms with C797S, including EGFR amplification (48%), MET amplification (16%), BRAF V600E (5%), and PIK3CA mutation (15%) (Piotrowska et al., 2015). The context in which C797S develops regarding the other EGFR alleles influences the efficacy of subsequent treatments. Niederst et al. (2015) emphasized that the allelic context of C797S acquired resistance after treatment with third-generation EGFR TKIs affects subsequent treatment strategies. This study concluded that if C797S and T790M mutations were in trans, resistant cells will be resistant to the third-generation EGFR TKIs but respond to a combination of the first- and third-generation TKIs. If the mutations are in cis, they are unresponsive to any EGFR TKI alone or in combination. In addition, if C797S develops along with the driving mutations of EGFR (when third-generation TKIs are administered in the first-line setting), the cells become resistant to third-generation EGFR TKIs but show a response to first-generation EGFR TKIs (Niederst et al., 2015).
Small molecule against EGFR-TM NSCLC
The C797S point mutation is one of the major problems for NSCLC patients who are treated with currently available EGFR TKIs. According to updated clinical studies, approximately 40% of patients have developed C797S mutations after treatment with third-generation EGFR TKIs. Globally, many pharmaceutical companies are focused on the development of small molecules to treat EGFR triple mutant NSCLC (Table 2).
Table 2.
Fourth-generation EGFR inhibitors
| Compound | Structure | Clinical trails | Reference |
|---|---|---|---|
| BLU-945 | Undisclosed | Phase 1 | Schalm et al., 2020 |
| BBT-176 | Undisclosed | Phase 1 |
Next-generation allosteric EGFR TKIs
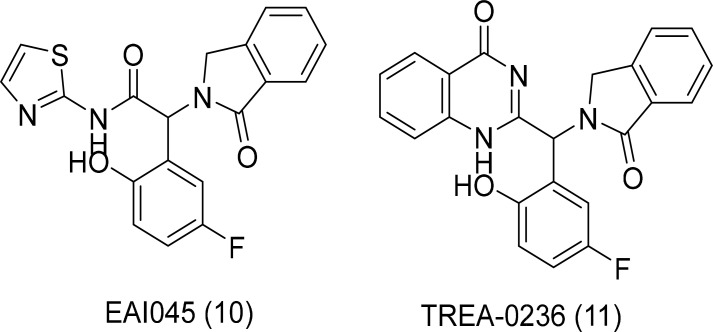
The appearance of the C797S point mutation in EGFR prevented clinical treatment with third-generation inhibitors that are effective against EGFR T790M mutation. The C797S mutation prohibited the formation of covalent bonds with the Michael acceptor of the third-generation inhibitors. Because of this drawback, there is an immediate requirement for the development of next-generation EGFR inhibitors targeting the triple mutant L858R/T790M/C797S and Del19/T790M/C797S (Lu et al., 2018). As third-generation EGFR TKIs are irreversible inhibitors, novel treatment modalities are required in a non-irreversible fashion against EGFR T790M/C797S mutations (Akazawa et al., 2020). Recent studies have indicated that EAI045 and TREA-0236 (Fig. 6) inhibit EGFR mutants in an allosteric manner (Lee et al., 2018).
Fig. 6.
Allosteric EGFR inhibitors.
These two small molecules showed activity in biochemical assays against L858R/T790M and L858R/T790M/C797S mutations (Jia et al., 2016). Both these compounds bind to the inactive conformation of mutated EGFR in the allosteric pocket. These are not active against wild-type EGFR because of the steric clash with L858 and L861. Combination therapy and the EGFR antibody cetuximab led to a significant increase in cellular potency. The development of allosteric inhibitors can be a significant step to treat EGFR mutant NSCLC. EAI045 showed high inhibition levels to L858R/T790M and L858R/T790M/C797S mutations with a novel mechanism.
Mutant-selective allosteric EGFR degraders
Allosteric EGFR inhibitors successfully inhibited the L858R/T790M/C797S mutation in an in vitro biochemical assay. However, EAI045 is not effective in blocking EGFR-driven proliferation in cells as a single agent because of its differential potency on the two subunits of the dimeric receptor, which interact asymmetrically in the active state (Tinivella and Rastelli, 2018). Thus, it requires cetuximab for improved potency in cells and in vivo. Allosteric inhibitors were only effective in combination with antibodies, such as cetuximab. Toxicity-related concerns arose from the off-target issues of cetuximab, which binds to all EGFR forms, including the wild-type. This limited the potential of allosteric inhibitors for therapeutic use (Bhullar et al., 2018).
As a promising alternative to inhibition, targeted protein degradation (TPD) using small molecules has emerged as a novel therapeutic approach. The TPD mechanism depends on degrader molecules that bind the protein target of interest and recruit it to an E3 ubiquitin ligase. This contiguity with E3 ligase affects target polyubiquitination and subsequent proteasomal degradation (Lai and Crews, 2017).
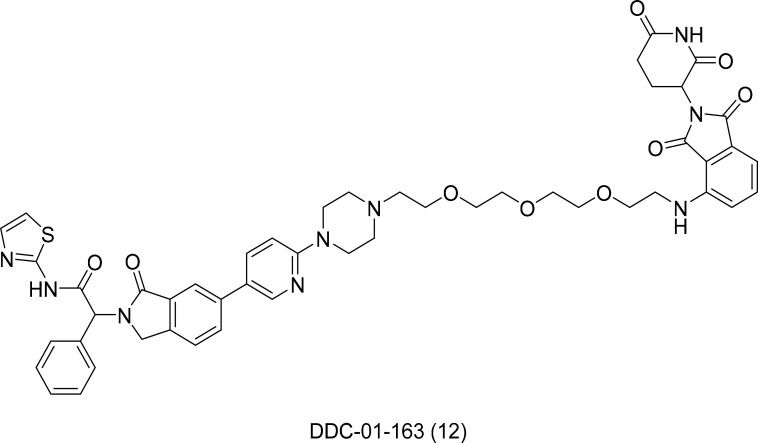
DDC-01-163 (Fig. 7) was developed as an allosteric degrader selective for EGFR triple mutants. It successfully inhibited the proliferation of EGFR mutant Ba/F3 cells (L858R/T790M) with no activity against the wild-type. Furthermore, the mechanism of allosteric degrader activity is independent of cetuximab. This result represents a noteworthy advance over existing EGFR allosteric inhibitors (De Clercq et al., 2019).
Fig. 7.
Structure of DDC-01-163 an allosteric EGFR degrader.
DDC-01-163 induced the selective degradation and inhibition of mutant EGFR cells in a dose-dependent manner and was especially inactive toward wild-type EGFR. The human cancer cell line, H1975, also displayed consistent results. These encouraging results suggest that the development of allosteric PROTACs for EGFR could assist a wide range of patients with EGFR L858R/T790M/C797S mutations (Jang et al., 2020).
Summary and future prospects
Gefitinib and erlotinib are first-generation reversible EGFR TKIs. They are quinazoline-based derivatives that act as ATP competitive inhibitors binding reversibly to the tyrosine kinase pocket of EGFR. Both drugs exhibited high inhibitory activity against wild-type and mutant EGFR. However, they ultimately develop resistance due to secondary mutations (Wang et al., 2016).
Afatinib and dacomitinib are irreversible EGFR TKIs. Although their structures are similar to gefitinib or erlotinib with a quinazoline backbone, their unique acrylate side chain feature binds covalently to the C797 of EGFR, creating irreversible inhibition of the EGFR tyrosine kinase. Afatinib and dacomitinib demonstrated in vitro activity against activating EGFR mutations and wild-type EGFR, which causes dose-limiting toxicity (Abdallah and Hirsh, 2018).
Osimertinib is an irreversible third-generation EGFR TKI. The key core structure changed from quinazolin to pyrimidine. Its acrylate side chain was introduced toward the solvent region to form covalent bonds with C797. Osimertinib potently inhibits T790M-positive tumors and spares wild-type EGFR. Osimertinib effectively inhibited EGFR-sensitizing and T790M-resistance mutations. It has also been approved as a first-line treatment for patients with EGFR mutation-positive NSCLC. Treatment was limited by generating third mutation C797S, which causes resistance to osimertinib (Kishikawa et al., 2020) (Table 3).
Table 3.
Summary of EGFR inhibitors
| EGFR wt. |
EGFR mutations | Inhibition mode | |||
|---|---|---|---|---|---|
| del_19 L858R |
del_19/T790M L858R/T790M |
del_19/T790M/C797S L858R/T790M/C797S |
|||
| Gefitinib (First Gen.) | Sensitive | Sensitive | Resistant | Resistant | Reversible |
| Afatinib (Second Gen.) | Sensitive | Sensitive | Sensitive | Resistant | Irreversible |
| Osimertinib (Third Gen.) | Resistant | Sensitive | Sensitive | Resistant | Irreversible |
| (Fourth Gen.) | Resistant | Sensitive | Sensitive | Sensitive | Not irreversible |
TKIs are standard treatment options for NSCLC with EGFR mutations (Zarogoulidis et al., 2016). Increasing clinical investigations have explored the value of fourth-generation EGFR TKIs plus antiangiogenic drugs as the first-line treatment for EGFR-mutated NSCLC. Fourth-generation EGFR TKIs should be reversible or allosteric mechanisms (Maity et al., 2020). Recent studies have proposed TPD, an alternative inhibition modality using small molecules.
CONCLUSION
NSCLC patients treated with first-generation EGFR TKIs have shown a dramatic response. Retrospective analyses of first-generation EGFR TKI trials have reported response rates of 77% in patients with sensitizing EGFR mutations (L858R and Del19). However, resistance was acquired by developing the secondary mutation T790M. The development of second-generation TKIs has shown encouraging results for secondary mutations. Conversely, inhibition of wild-type EGFR caused adverse events such as rash, acne, and diarrhea.
Irreversible third-generation EGFR TKIs have better efficacy and safety profiles by sparing wild-type EGFR than other inhibitors. However, the treatment was limited by the third mutation C797S. The mechanism of resistance to third-generation EGFR TKIs is more heterogeneous than that of other inhibitors. In many cases, the putative mechanism of resistance has not yet been identified.
In conclusion, treatment options for third-generation EGFR TKIs resistant are not available. Patients with EGFR triple mutations may receive subsequent therapies, such as immunotherapy and cytotoxic chemotherapy. Nonetheless, several areas of unmet medical needs remain to be addressed to provide treatment options for EGFR-related NSCLC patients.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the generous financial support provided by the Korea Research Institute of Chemical Technology (KK2131-30).
REFERENCES
- Abdallah S. M., Hirsh V. Irreversible tyrosine kinase inhibition of epidermal growth factor receptor with afatinib in EGFR activating mutation-positive advanced non-small-cell lung cancer. Curr. Oncol. 2018;25:S9–S17. doi: 10.3747/co.25.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa Y., Saito Y., Yoshikawa T., Saito K., Nosaka K., Shimomura M., Mizuno S., Nakamoto Y., Nakatsura T. Efficacy of immunotherapy targeting the neoantigen derived from epidermal growth factor receptor T790M/C797S mutation in non-small cell lung cancer. Cancer Sci. 2020;111:2736–2746. doi: 10.1111/cas.14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer A. G., Rotin D., Ureña J. M., Skolnik E. Y., Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol. Cell. Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192-5201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhullar K. S., Lagarón N. O., McGowan E. M., Parmar I., Jha A., Hubbard B. P., Rupasinghe H. P. V. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol. Cancer. 2018;17:48. doi: 10.1186/s12943-018-0804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C. D., Ingraham H. A., Cochet C., Walton G. M., Lazar C. S., Sowadski J. M., Rosenfeld M. G., Gill G. N. Structural analysis of the transmembrane domain of the epidermal growth factor receptor. J. Biol. Chem. 1991;266:5750–5755. doi: 10.1016/S0021-9258(19)67659-3. [DOI] [PubMed] [Google Scholar]
- Ciardiello F., Tortora G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- De Clercq D. J. H., Heppner D. E., To C., Jang J., Park E., Yun C. H., Mushajiang M., Shin B. H., Gero T. W., Scott D. A., Jänne P. A., Eck M. J., Gray N. S. Discovery and optimization of dibenzodiazepinones as allosteric mutant-selective EGFR inhibitors. ACS Med. Chem. Lett. 2019;10:1549–1553. doi: 10.1021/acsmedchemlett.9b00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Lovly C. M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer. 2018;17:58. doi: 10.1186/s12943-018-0782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck M. J., Yun C. H. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim. Biophys. Acta. 2010;1804:559–566. doi: 10.1016/j.bbapap.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson K. M., Berger M. B., Mendrola J. M., Cho H. S., Leahy D. J., Lemmon M. A. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/S1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Ferlay J., Autier P., Boniol M., Heanue M., Colombet M., Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann. Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- Finigan J. H., Downey G. P., Kern J. A. Human epidermal growth factor receptor signaling in acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;47:395–404. doi: 10.1165/rcmb.2012-0100TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay M. R., Anderton M., Ashton S., Ballard P., Bethel P. A., Box M. R., Bradbury R. H., Brown S. J., Butterworth S., Campbell A., Chorley C., Colclough N., Cross D. A., Currie G. S., Grist M., Hassall L., Hill G. B., James D., James M., Kemmitt P., Klinowska T., Lamont G., Lamont S. G., Martin N., McFarland H. L., Mellor M. J., Orme J. P., Perkins D., Perkins P., Richmond G., Smith P., Ward R. A., Waring M. J., Whittaker D., Wells S., Wrigley G. L. Discovery of a potent and selective EGFR inhibitor (AZD9291) of both sensitizing and T790M resistance mutations that spares the wild type form of the receptor. J. Med. Chem. 2014;57:8249–8267. doi: 10.1021/jm500973a. [DOI] [PubMed] [Google Scholar]
- Gao X., Le X., Costa D. B. The safety and efficacy of osimertinib for the treatment of EGFR T790M mutation positive non-small-cell lung cancer. Expert Rev. Anticancer Ther. 2016;16:383–390. doi: 10.1586/14737140.2016.1162103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar A. F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. E., Montesion M., Young L., Suh J., Greenbowe J., Kennedy M., Giaccone G., Akerley W. L., Dowlati A., Creelan B. C., Hicks J. K., Hesketh P. J., Kelly K. L., Riess J. W., Miller V. A., Stephens P. J., Frampton G. M., Ali S., Gregg J. P., Albacker L. A. Multiple configurations of EGFR exon 20 resistance mutations after first- and third-generation EGFR TKI treatment affect treatment options in NSCLC. PLoS ONE. 2018;13:e0208097. doi: 10.1371/journal.pone.0208097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe T., Lategahn J., Rauh D. C797S resistance: the undruggable EGFR mutation in non-small cell lung cancer? ACS Med. Chem. Lett. 2018;9:779–782. doi: 10.1021/acsmedchemlett.8b00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. T., Vyse S., Huang P. H. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin. Cancer Biol. 2020;61:167–179. doi: 10.1016/j.semcancer.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J., To C., De Clercq D. J. H., Park E., Ponthier C. M., Shin B. H., Mushajiang M., Nowak R. P., Fischer E. S., Eck M. J., Jänne P. A., Gray N. S. Mutant-selective allosteric EGFR degraders are effective against a Broad Range of drug-resistant mutations. Angew. Chem. 2020;59:14481–14489. doi: 10.1002/anie.202003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Yun C. H., Park E., Ercan D., Manuia M., Juarez J., Xu C., Rhee K., Chen T., Zhang H., Palakurthi S., Jang J., Lelais G., DiDonato M., Bursulaya B., Michellys P. Y., Epple R., Marsilje T. H., McNeill M., Lu W., Harris J., Bender S., Wong K. K., Jänne P. A., Eck M. J. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachaliou N., Fernandez-Bruno M., Bracht J. W., Rosell R. EGFR first- and second-generation TKIs-there is still place for them in EGFR-mutant NSCLC patients. Transl. Cancer Res. 2018;8:S23–S47. doi: 10.21037/tcr.2018.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenfield S. A., Wei E. K., Stampfer M. J., Rosner B. A., Colditz G. A. Comparison of aspects of smoking among the four histological types of lung cancer. Tob. Control. 2008;17:198–204. doi: 10.1136/tc.2007.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa T., Kasai T., Okada M., Nakachi I., Soda S., Arai R., Takigami A., Sata M. Osimertinib, a third-generation EGFR tyrosine kinase inhibitor: a retrospective multicenter study of its real-world efficacy and safety in advanced/recurrent non-small cell lung carcinoma. Thorac. Cancer. 2020;11:935–942. doi: 10.1111/1759-7714.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko B., Paucar D., Halmos B. EGFR T790M: revealing the secrets of a gatekeeper. Lung Cancer. 2017;8:147–159. doi: 10.2147/LCTT.S117944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: perspectives for individualized treatment strategy. Cancer Sci. 2016;107:1179–1186. doi: 10.1111/cas.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T., Yatabe Y., Endoh H., Kuwano H., Takahashi T., Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- Lai A. C., Crews C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lategahn J., Keul M., Klövekorn P., Tumbrink H. L., Niggenaber J., Müller M. P., Hodson L., Flaßhoff M., Hardick J., Grabe T., Engel J., Schultz-Fademrecht C., Baumann M., Ketzer J., Mühlenberg T., Hiller W., Günther G., Unger A., Müller H., Heimsoeth A., Golz C., Blank-Landeshammer B., Kollipara L., Zahedi R. P., Strohmann C., Hengstler J. G., van Otterlo W. A. L., Bauer, Rauh D. Inhibition of osimertinib-resistant epidermal growth factor receptor EGFR-T790M/C797S. Chem. Sci. 2019;10:10789–10801. doi: 10.1039/C9SC03445E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim J., Duggirala K. B., Go A., Shin I., Cho B. C., Choi G., Chae C. H., Lee K. Allosteric inhibitor TREA-0236 containing non-hydrolysable quinazoline-4-one for EGFR T790M/C797S mutants inhibition. Bull. Korean Chem. Soc. 2018;39:895–898. doi: 10.1002/bkcs.11491. [DOI] [Google Scholar]
- Li A. R., Chitale D., Riely G. J., Pao W., Miller V. A., Zakowski M. F., Rusch V., Kris M. G., Ladanyi M. EGFR mutations in lung adenocarcinomas: clinical testing experience and relationship to EGFR gene copy number and immunohistochemical expression. J. Mol. Diagn. 2008;10:242–248. doi: 10.2353/jmoldx.2008.070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wang X., Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am. J. Cancer Res. 2014;4:411–435. [PMC free article] [PubMed] [Google Scholar]
- Lu X., Yu L., Zhang Z., Ren X., Smaill J. B., Ding K. Targeting EGFR(L858R/T790M) and EGFR(L858R/T790M/C797S) resistance mutations in NSCLC: current developments in medicinal chemistry. Med. Res. Rev. 2018;38:1550–1581. doi: 10.1002/med.21488. [DOI] [PubMed] [Google Scholar]
- Maity S., Pai K. S. R., Nayak Y. Advances in targeting EGFR allosteric site as anti-NSCLC therapy to overcome the drug resistance. Pharmacol. Rep. 2020;72:799–813. doi: 10.1007/s43440-020-00131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha A., Dearden S., McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Am. J. Cancer Res. 2015;5:2892–2911. [PMC free article] [PubMed] [Google Scholar]
- Morrow M. R., Grant C. W. The EGF receptor transmembrane domain: peptide-peptide interactions in fluid bilayer membranes. Biophys. J. 2000;79:2024–2032. doi: 10.1016/S0006-3495(00)76450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederst M. J., Hu H., Mulvey H. E., Lockerman E. L., Garcia A. R., Piotrowska Z., Sequist L. V., Engelman J. A. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin. Cancer Res. 2015;21:3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Pao W., Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska Z., Niederst M. J., Karlovich C. A., Wakelee H. A., Neal J. W., Mino-Kenudson M., Fulton L., Hata A. N., Lockerman E. L., Kalsy A., Digumarthy S., Muzikansky A., Raponi M., Garcia A. R., Mulvey H. E., Parks M. K., DiCecca R. H., Dias-Santagata D., Iafrate A. J., Shaw A. T., Allen A. R., Engelman J. A., Sequist L. V. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purba E. R., Saita E. I., Maruyama I. N. Activation of the EGF receptor by ligand binding and oncogenic mutations: the "rotation model". Cells. 2017;6:13. doi: 10.3390/cells6020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Sandler A., Gray R., Perry M. C., Brahmer J., Schiller J. H., Dowlati A., Lilenbaum R., Johnson D. H. Paclitaxel-carboplatin alone or with bevacizumab for Non-small-cell lung cancer. N. Engl. J. Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- Schalm S. S., Dineen T., Lim S. M., Park C. W., Hsieh J., Woessner R., Zhang Z., Wilson K., Eno M., Wilson D., Williams B., Campbell J., De Savi C., Stevison F., Utt C., Guzi T., Dorsch M., Hoeflich K., Chul Cho B. C. 1296P BLU-945, a highly potent and selective 4th generation EGFR TKI for the treatment of EGFR T790M/C797S resistant NSCLC. Ann. Oncol. 2020;31:S839. doi: 10.1016/j.annonc.2020.08.1610. [DOI] [Google Scholar]
- Sharma S. V., Bell D. W., Settleman J., Haber D. A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Singh B., Carpenter G., Coffey R. J. EGF receptor ligands: recent advances. F1000Res. 2016;5:2270. doi: 10.12688/f1000research.9025.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K. G., Kyte J. Dimerization of the extracellular domain of the receptor for epidermal growth factor containing the membrane-spanning segment in response to treatment with epidermal growth factor. J. Biol. Chem. 1999;274:35985–35990. doi: 10.1074/jbc.274.50.35985. [DOI] [PubMed] [Google Scholar]
- Tinivella A., Rastelli G. Investigating the selectivity of allosteric inhibitors for mutant T790M EGFR over wild type using molecular dynamics and binding free energy calculations. ACS Omega. 2018;3:16556–16562. doi: 10.1021/acsomega.8b03256. [DOI] [Google Scholar]
- Tokudome N., Koh Y., Akamatsu H., Fujimoto D., Okamoto I., Nakagawa K., Hida T., Imamura F., Morita S., Yamamoto N. Differential significance of molecular subtypes which were classified into EGFR exon 19 deletion on the first line afatinib monotherapy. BMC Cancer. 2020;20:103. doi: 10.1186/s12885-020-6593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W. D., Brambilla E., Noguchi M., Nicholson A. G., Geisinger K. R., Yatabe Y., Beer D. G., Powell C. A., Riely G. J., Van Schil P. E., Garg K., Austin J. H., Asamura H., Rusch V. W., Hirsch F. R., Scagliotti G., Mitsudomi T., Huber R. M., Ishikawa Y., Jett J., Sanchez-Cespedes M., Sculier J. P., Takahashi T., Tsuboi M., Vansteenkiste J., Wistuba I., Yang P. C., Aberle D., Brambilla C., Flieder D., Franklin W., Gazdar A., Gould M., Hasleton P., Henderson D., Johnson B., Johnson D., Kerr K., Kuriyama K., Lee J. S., Miller V. A., Petersen I., Roggli V., Rosell R., Saijo N., Thunnissen E., Tsao M., Yankelewitz D. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahar E., Waterman H., Chen X., Levkowitz G., Karunagaran D., Lavi S., Ratzkin B. J., Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 1996;16:5276–5287. doi: 10.1128/MCB.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyse S., Huang P. H. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct. Target. Ther. 2019;4:5. doi: 10.1038/s41392-019-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton G. M., Chen W. S., Rosenfeld M. G., Gill G. N. Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. J. Biol. Chem. 1990;265:1750–1754. doi: 10.1016/S0021-9258(19)40080-X. [DOI] [PubMed] [Google Scholar]
- Wang Y., Guo Z., Li Y., Zhou Q. Development of epidermal growth factor receptor tyrosine kinase inhibitors against EGFR T790M. Mutation in non small-cell lung carcinoma. Open Med. (Wars.) 2016;11:68–77. doi: 10.1515/med-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee P., Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:52. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. A., Riely G. J. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in lung cancers. J. Natl. Compr. Canc. Netw. 2013;11:161–169. doi: 10.6004/jnccn.2013.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C. H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., Eck M. J. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarogoulidis P., Lampaki S., Chinelis P., Lazaridis G., Baka S., Rapti A. Tyrosine kinase inhibitors for non-small cell lung cancer and eye metastasis: disease relapse or a new entity? Med. Hypothesis Discov. Innov. Ophthalmol. 2016;5:132–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]