Summary
Early erythroid progenitors are transit-amplifying cells with high proliferative capacity committed to undergoing red cell differentiation. CD71/CD24low progenitors are less mature and have greater proliferative capacity than CD71/CD24high. We present protocols for isolation of CD71/CD24low progenitors from mouse fetal liver using both fluorescence-activated cell sorting (FACS) and immunomagnetic enrichment. CD71/CD24low progenitors isolated with both approaches show similar transcriptomes at single-cell resolution and exhibit characteristic proliferative responses to glucocorticoids.
For complete details on the use and execution of this protocol, please refer to Li et al. (2019).
Subject areas: Cell Biology, Cell isolation, Single Cell, Flow Cytometry/Mass Cytometry, Developmental biology, Cell Differentiation
Graphical abstract
Highlights
-
•
FACS isolation of early erythroid progenitor cells from mouse fetal liver
-
•
Immunomagnetic isolation of early erythroid progenitor cells from mouse fetal liver
-
•
Both approaches show similar transcriptomics at single-cell resolution
-
•
Both approaches show similar proliferative responses to glucocorticoids
Early erythroid progenitors are transit-amplifying cells with high proliferative capacity committed to undergoing red cell differentiation. CD71/CD24low progenitors are less mature and have greater proliferative capacity than CD71/CD24high. We present protocols for isolation of CD71/CD24low progenitors from mouse fetal liver using both fluorescence-activated cell sorting (FACS) and immunomagnetic enrichment. CD71/CD24low progenitors isolated with both approaches show similar transcriptomes at single-cell resolution and exhibit characteristic proliferative responses to glucocorticoids.
Before you begin
CD71/CD24low mouse erythroid progenitors have been used extensively to model erythroid transit amplification and differentiation (Li et al., 2014, 2019; Flygare et al., 2011; Pop et al., 2010; Harandi et al., 2010; Sjogren et al., 2015; Trivedi et al., 2019). Here we detail isolation protocols for obtaining CD71/CD24low primary erythroid progenitors from mouse fetal liver. These protocols are contingent upon the investigator receiving appropriate institutional approval for working with mice and performing mouse breeding.
Institutional approval to generate and necropsy E14.5 pregnant C57BL/6J mice
Timing: Institution-dependent
-
1.
Obtain Institutional Animal Care and Use Committee (IACUC) approval and oversight to purchase, breed, and necropsy embryonic day 14.5 (E14.5) pregnant C57BL/6J mice in order to obtain fetal livers necessary for this protocol.
Generating E14.5 pregnant C57BL/6J mice
Timing: 14.5 days before the isolation
-
2.
Mate C57BL/6 mice to generate E14.5 pregnant mice. Place males and females together in the afternoon 15 days prior to isolation, and separate them the following morning.
CRITICAL: Each pregnant mouse should yield an average of 8 fetuses with approximately 20,000 CD71/CD24low progenitor cells per fetus. As such, breed an appropriate number of mice for the number of cells you intend to isolate.
Note: This protocol works best when 3–4 pregnant mice (about 24–32 fetuses) are used.
Making PNEG (PBS, 2% neonatal calf serum, 20 μM EDTA, 10 mM glucose) buffer
Timing: 20 min
-
3.
Combine all PNEG buffer components in a sterile tissue culture hood as specified in the materials and equipment section.
-
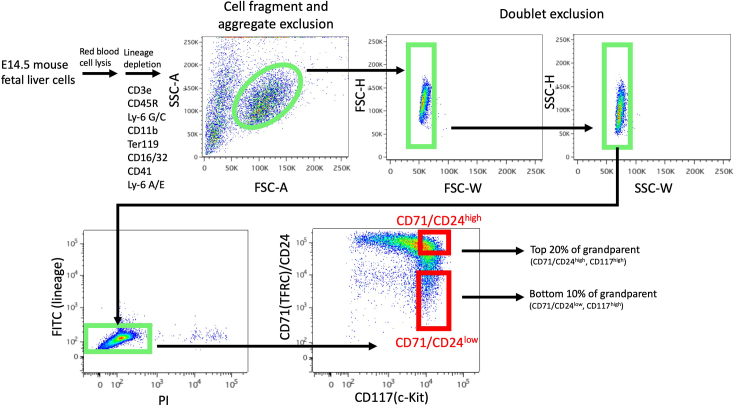
4.
Sterile filter and aliquot into 50 mL conical tubes. Store at 4°C until use. PNEG buffer can be stored at 4°C for a maximum of 2 weeks.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 1× Phosphate buffered saline (PBS) | Sigma-Aldrich | SKU: D8537-100ML |
| 0.5 M EDTA (pH 8.0) | Invitrogen | Ref#AM9260G |
| Newborn Calf Serum (NCS) | Thermo Fisher Scientific | Cat#16010167 |
| 40% w/v glucose solution | Fisher Scientific | Cat#50-841-706 |
| Ammonium chloride solution | STEMCELL Technologies | Cat#07850 |
| Streptavidin magnetic beads | BD-Pharmingen | Cat#557812 |
| Propidium Iodide (PI), 1 mg/mL | Sigma-Aldrich | Cat#P4864; CAS#25535-16-4 |
| Recombinant murine stem cell factor (SCF) | PeproTech | Cat#250-03C |
| Recombinant murine insulin-like growth factor 1 (IGF-1) | PeproTech | Cat#250-19 |
| Erythropoietin (Epo) | Amgen | CAS#113427-24-0 |
| Penicillin/Streptomycin (100×) | Gibco | Cat#15140122 |
| Dexamethasone (Dex), USP | Sigma-Aldrich | D9184; CAS#:50-02-2 |
| Trypan Blue | Gibco | Cat#15250061 |
| Stemspan Serum-free expansion media (SFEM) II | Stem Cell Technologies | Cat#:09655 |
| Methanol | Fisher Scientific | Model:A412-500; CAS#67-56-1 |
| Antibodies | ||
| Biotin mouse lineage panel ([working]: 0.55 test/mL per antibody) | BD-Pharmingen | Cat#559971 |
| Biotinylated CD16/32 (clone 93) antibody ([working]: 0.55 μg/mL) | eBioscience | Cat#13-0161-86; RRID: AB_466379 |
| Biotinylated CD41 (clone eBioMWReg30) antibody ([working]: 0.55 μg/mL) | eBioscience | Cat#13-0411-85; RRID: AB_763489 |
| Biotinylated Ter119 antibody (clone Ter119) ([working]: 0.3 μg/mL) | eBioscience | Cat#13-5921-81; RRID: AB_469756 |
| Biotinylated Ly-6A/E (i.e., Sca-1) (clone D7) antibody ([working]: 0.55 μg/mL) | eBioscience | Cat#13-5981-85; RRID: AB_466835 |
| Streptavidin-FITC ([working]: 1 μg/mL) | eBioscience | Cat#11-4317-87 |
| APC anti-mouse CD117 (c-Kit) (clone 2B8) ([working]: 5 μg/mL) | eBioscience | Cat#17-1172-83; RRID: AB_469429 |
| PE anti-mouse CD24 (clone M1/69) ([working]: 0.6 μg/mL ) | eBioscience | Cat#12-0242-82; RRID: AB_465598 |
| PE anti-mouse CD71 (clone R17217) ([working]: 0.6 μg/mL ) | eBioscience | Cat#12-0711-81; RRID: AB_465739 |
| Critical commercial assays | ||
| EasySep™ PE Positive Selection Kit II | STEMCELL Technologies | Cat#17684 |
| EasySep™ Magnet | STEMCELL Technologies | Serial #71053 |
| BD IMag™ magnetic stand | BD Biosciences | Cat#552311 |
| Experimental models:Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock#000664; RRID:IMSR_JAX:000664 |
| Deposited data | ||
| Single cell RNA-sequencing and CITE-seq data | National Center for Biotechnology Information Gene Expression Omnibus | NCBI GEO accession number GSE188287 |
| Other | ||
| Cell culture plate, 24-well | Eppendorf | Cat#0030722019 |
| Round-bottom polypropylene tubes, 5 mL | STEMCELL Technologies | Cat#38056 |
| 50 mL conical tubes | CELLTREAT | Part#229422 |
| 15 mL conical tubes | Fisher Scientific | Cat#339650 |
| 5 mL serological pipette | Fisher Scientific | Cat#1367522 |
| Dumont #4 Forceps Inox Tip Size 0.06 × 0.02 mm | Roboz Surgical Store | Part#RS-4904 |
| 4” Long Serrated Slight Curve 0.8 mm Tip Forceps | Roboz Surgical Store | Part#RS-5135 |
| 70 μm Nylon Cell Strainer | Corning | Ref#352350 |
| 0.1 μm aPES membrane filter (500 mL) | Thermo Fisher Scientific | Cat#566-0010 |
| BD FACSAria 2 | BD Biosciences | Cat#650110 |
Materials and equipment
| PNEG buffer | Final concentration | Amount |
|---|---|---|
| 1× phosphate buffered saline (PBS) | n/a | 500 mL |
| 0.5 M EDTA | 20 μM | 200 μL |
| Newborn calf serum | 2% | 10 mL |
| 40% w/v glucose stock solution in water | 10 mM | 2 mL |
| Total | n/a | 500 mL |
CRITICAL: Sterile-filter PNEG buffer using a 0.1 μm membrane filter.
Note: PNEG buffer can be stored at 4°C for a maximum of 1 month.
| Lineage depletion antibody mix | Amount of manufacturer supplied stock solution (per 8 fetuses) |
|---|---|
| BD-Pharmingen CD3e antibody (manufacturer reported stock concentration of 500 tests/mL) | 11 μL |
| BD-Pharmingen CD45R antibody (manufacturer reported stock concentration of 500 tests/mL) | 11 μL |
| BD-Pharmingen Ly-6G & Ly-6C antibody (manufacturer reported stock concentration of 500 tests/mL) | 11 μL |
| BD-Pharmingen CD11b antibody (manufacturer reported stock concentration of 500 tests/mL) | 11 μL |
| BD-Pharmingen Ter119 antibody (manufacturer reported stock concentration of 500 tests/mL) | 11 μL |
| E-Biosciences biotinylated CD16/32 (clone 93) antibody (manufacturer reported stock concentration of 0.5 mg/mL) | 11 μL |
| E-Biosciences biotinylated CD41 (clone eBioMWReg30) antibody (manufacturer reported stock concentration of 0.5 mg/mL) | 11 μL |
| E-Biosciences biotinylated Ly-6A/E (Clone D7) antibody (manufacturer reported stock concentration of 0.5 mg/mL) | 11 μL |
| E-Biosciences biotinylated Ter119 antibody (manufacturer reported stock concentration of 0.5 mg/mL) | 6 μL |
| Total | 94 μL |
Note: Freshly prepare lineage depletion antibody mix by combining antibodies in a 1.5 mL tube. Keep on ice until use. The stock solutions of all antibodies are at the original concentrations supplied by the manufacturers.
Note: All BD-Pharmingen antibodies from the Biotin mouse lineage panel (CAT#:559971) are only specified in tests per μ7 by the manufacturer.
| Serum-free erythroid liquid expansion media (± 100 nM Dex) | Final concentration | Amount |
|---|---|---|
| Stemspan SFEM II | n/a | 50 mL |
| Recombinant murine SCF (stock conc. 100 mcg/mL) | 100 ng/mL | 50 μL |
| Recombinant murine IGF-1 (stock conc. 80 mcg/mL) | 40 ng/mL | 25 μL |
| Recombinant human Epo (stock conc. 20,000 U/mL) | 2 U/mL | 5 μL |
| Penicillin/Streptomycin (100×) | 1% | 500 μL |
| Dexamethasone (Dex) (1 mM in methanol) | 100 nM | 5 μL |
| Total | n/a | 50 mL |
CRITICAL: Sterile-filter serum-free erythroid liquid expansion media using a 0.1 μM membrane filter prior to adding cytokines, Dex or methanol. Stemspan SFEM II is the only media our lab has tested for erythroid proliferation assays, although additional serum-free media from other sources may potentially also be acceptable.
Note: Serum-free erythroid liquid expansion media can be stored at 4°C for a maximum of 2 weeks.
Step-by-step method details
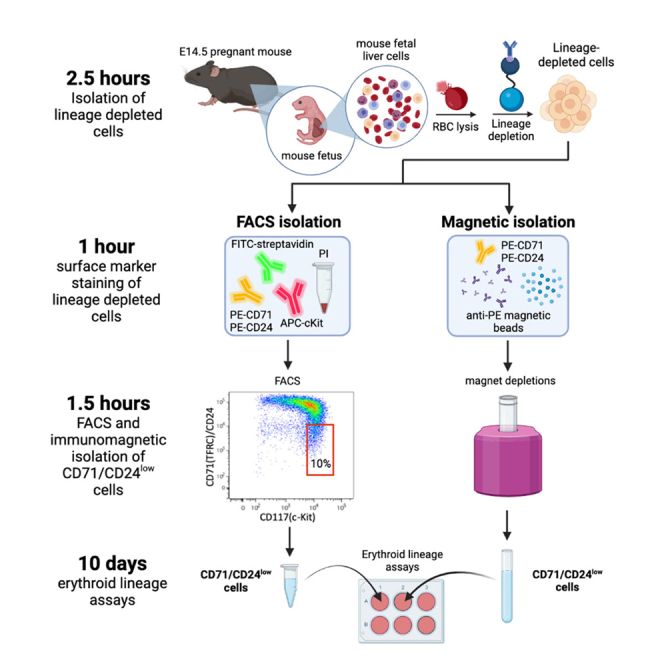
The following protocol outlines steps for isolating both CD71/CD24low and CD71/CD24high early erythroid progenitor cells from mouse fetal livers extracted from E14.5 pregnant C57BL/6J mice. Two methods for isolation, FACS and immunomagnetic isolation using the EasySep PE Positive Selection Kit II, are described. For both methods, fetal liver cells are isolated from fetuses from E14.5 pregnant mice, and subsequently lineage depleted. Lineage depleted cells are either stained for FACS isolation or immunomagnetic isolation using an EasySep PE Positive Selection Kit II. Isolated CD71/CD24low cells from each method can then be cultured and utilized for downstream applications including growth curve analysis, genomic DNA isolation, or retroviral transduction.
Before beginning, prepare ten 50 mL aliquots (500 mL total) of PNEG buffer.
Isolation of lineage depleted cells
Timing: 2.5 h
Described here are detailed steps to isolate lineage depleted cells from fetal livers from E14.5 pregnant mice.
-
1.
Euthanize E14.5 pregnant mice using CO2 asphyxiation followed by an approved secondary method such as cervical dislocation
Note: To minimize any cell death after euthanasia is performed, conduct CO2 asphyxiation in a vicinity close to a tissue culture hood, if possible.
-
2.
Necropsy mice and place fetuses in a sterile, 10 cm petri dish on ice containing 5–10 mL PNEG. Count the total number of fetuses (Methods video S1).
-
3.
In a sterile tissue culture hood, dissect out all fetal livers and place in 50 mL conical tube containing 5 mL PNEG
Note: Two small pairs of forceps are most useful for extracting fetal livers. Gently clamp one pair above the liver and under the arms of the fetus, and gently but with increasing pressure clamp the other pair below the liver at the hips of the fetus. To further facilitate excision of the liver, tilt the two clamped forceps towards one another, angling towards the posterior side of the fetus (Methods video S2).
-
4.
Vigorously create a single cell suspension from the fetal livers using a well-charged 5 mL serological pipette
-
5.Using a P1000 pipette, apply the cell suspension to a 70 μm filter on top of 50 mL conical
-
a.Scrape the filter with the pipette tip to disrupt any clotted tissue
-
a.
-
6.Take any remaining connective tissue aggregates back into the first conical, add 5 mL PNEG to this conical tube, resuspend this mixture, and reapply to the filter, this time using a 5 mL serological pipette
-
a.Repeat this step until a total of 50 mL of filtered cell suspension is collected in the second conical
-
a.
-
7.
Centrifuge cells at 500 × g for 5 min at 4°C and aspirate off the supernatant
-
8.
Add 4 mL Ammonium Chloride solution to lyse red blood cells
Note: While Ficoll-Pacque separation media can also be used to obtain mononuclear cells, the process for doing so takes significantly longer in this context than conducting RBC lysis, and often requires RBC lysis after Ficoll-Paque separation of cells. Therefore, RBC lysis is our preferred method for mononuclear cell isolation in this protocol.
-
9.
Incubate on ice for 6 min, then add PNEG to a total volume of 50 mL
-
10.Centrifuge at 500 × g for 5 min at 4°C and aspirate off the media
-
a.The pellet should be much whiter this time; if not, repeat the red blood cell lysis
-
a.
-
11.
Vigorously resuspend the pellet in 5 mL PNEG using a 5 mL serological pipette
-
12.
Apply this cell suspension to a 70 μM filter on top of a 50 mL conical
-
13.
Take any remaining red cell membrane aggregates (will appear similar in nature to connective tissue from step 6 above) from the filter back into the original 50 mL conical, resuspend vigorously with an additional 5 mL of PNEG, and apply to the same 70 μM filter
-
14.
Based on the number of fetuses, add the appropriate amount of Lineage depletion antibody mix to the 10 mL cell suspension. Refer to the materials and equipment for the appropriate amount of each lineage depletion antibody to add for the number of fetuses isolated.
-
15.
Incubate on ice for 30 min, then add PNEG to a total volume of 50 mL
-
16.
Centrifuge at 500 × g for 5 min at 4°C and aspirate off the media
-
17.
Resuspend the pellet in 2 mL PNEG
-
18.
Vortex streptavidin magnetic beads for 20 s
-
19.
Add streptavidin magnetic beads to the cells and mix well. For FACS isolation, add 125 μL of streptavidin magnetic beads per 8 fetuses. For immunomagnetic isolation, add 187.5 μL streptavidin magnetic beads per 8 fetuses.
Note: Isolating CD71/CD24low cells via FACS includes a FITC-lineage gating step when sorting, whereas no such additional lineage removal step exists for the immunomagnetic isolation strategy; therefore, we recommend using 1.5× the amount of streptavidin beads when doing immunomagnetic isolation to ensure purity of lineage depleted cells.
Note: Vortexing briefly works well for this
-
20.Incubate in a 4°C refrigerator for 20 min. No tapping of the tube is required during incubation.
-
a.During this time, set up two 5 mL polypropylene tubes in BD Imag magnetic stand
-
a.
-
21.
Briefly mix the cell and bead mixture before adding the mix into the 1st tube on magnetic stand
-
22.Incubate on the magnet in a 4°C refrigerator for 5 min
-
a.Check that the beads have moved to the magnet side of the tube when the incubation is finished
-
a.
-
23.Carefully pipette the supernatant into a second polypropylene tube that is on the same magnetic stand
-
a.Be sure not to move the position of the first tube, or the magnetic beads will fall off and contaminate the bead-free supernatant.
-
a.
-
24.
Incubate the magnet in a 4°C refrigerator for 5 min
-
25.
Carefully pipette the supernatant into a 15 mL conical and fill to a total volume of 15 mL with PNEG
-
26.
Centrifuge at 500 × g for 5 min at 4°C
-
27.
Resuspend the pellet in 1mL PNEG and keep on ice
-
28.
Optional verification step: Count the cells on a hemocytometer
Note: On average, expect a total of at least 2–3 million lineage depleted cells per 8 fetuses used.
CRITICAL: To minimize loss of any cells from this rare population, keep cells on ice at all times when not actively manipulating cells.
Surface marker staining of lineage depleted cells for both FACS and magnetic isolation
Timing: 1 h
The following steps describe how to stain lineage depleted cells for FACS and immunomagnetic isolation of CD71/CD24low and CD71/CD24high erythroid progenitor cells.
-
29.Add 200 μL PNEG to each of 6 polypropylene FACS tubes and to each add the following antibody stains:
-
a.Main Sample tube: 10 μL APC-CD117 (clone 2B8; [stock]: 0.5 mg/mL; [working]: 5 μg/mL), 3 μL PE-CD71 (clone R127217; [stock]: 0.2 mg/mL; [working]: 0.6 μg/mL), 3 μL PE-CD24 (clone M1/69; [stock]: 0.2 mg/mL; [working]: 0.6 μg/mL), and 2 μL FITC-streptavidin ([stock]: 0.5 mg/mL; [working]: 1 μg/mL)
-
b.Tubes ultimately used for compensation controls:
-
i.Propidium Iodide (PI) single stain tube: no stain added yet at this step
-
ii.FITC single stain tube: 0.5 μL FITC-streptavidin ([working]: 1.25 μg/mL)
-
iii.APC single stain tube: 2 μL APC-CD117 (clone 2B8; [working]: 5 μg/mL)
-
iv.PE single stain tube: 0.7 μL PE-CD71 (clone R127217; [working]: 0.7 μg/mL) and 0.7 μL PE-CD24 (clone M1/69; [working]: 0.7 μg/mL)
-
v.Unstained control tube: no stain needs to be added to unstained control
-
i.
-
a.
-
30.
Add 10 μL lineage depleted cells to each compensation control tube.
Note: Lineage depleted cells are used to set up voltages during compensation so that voltages and gating are most accurate for this particular cell type. We have observed that some of these markers (in particular CD117, CD71, and CD24) have much higher surface expression density than any other surface markers that can be commonly used for staining compensation controls (e.g. anti-B220 staining of splenocytes). Thus, in order to correctly compensate for the high fluorescence intensities in our system, we must use single stains of the actual lineage depleted fetal liver cells.
Note: Add 20 μL lineage depleted cells to each single-stain tube if only 1–2 pregnant mice are used.
-
31.
Add the rest of the cells to Main Sample tube
-
32.Vortex all tubes and incubate on ice and in the dark for 20 min
-
a.During this incubation, prepare 1× PI solution by adding 2 μL PI (1 mg/mL) to 2 mL PNEG and leave on ice until use
-
b.During this incubation, also aliquot 2 mL PNEG into 15 mL conical tubes labeled with either CD71/CD24low or CD71/CD24high for FACS collection
-
a.
-
33.
After the incubation, fill FACS tubes to near top with PNEG to wash
-
34.
Centrifuge at 500 × g for 5 min at 4°C and pour off the supernatant
Note: Gently pour off the supernatant rather than aspirate so as to not risk losing any cells. When pouring off the supernatant, pour in one swift and gentle motion and do not shake the tube so as to not dislodge the pellet.
-
35.
Resuspend all samples, except for the PI single stain and Main Sample, in 200 μL PNEG
-
36.
Resuspend the Main Sample in 1000 μL 1× PI solution and PI single stain sample in 200 μL 1× PI solution
Note: Isolate cells by sorting or immunomagnetic purification purify as soon as possible after surface marker staining is complete. Cells can be left for a maximum 3–4 h after surface marker staining is completed, but the longer the elapsed time, the higher the percentage of dead cells is observed, and thus a lower cell yield.
FACS isolation of CD71/CD24low and CD71/CD24high erythroid progenitor cells
Timing: 1.5 h
Note: FACS for this protocol has always been conducted on a BD FACSAria using an 85 μm nozzle.
-
37.
Set the voltages for each stain (APC, FITC, PE, and PI) using unstained and single-stain controls.
-
38.
Set gates to exclude cell fragments and multicellular aggregates and include only forward-scatter (FSC) singlets, side-scatter (SSC) singlets, live (PI negative) cells, and lineage-negative (FITC negative) cells.
-
39.
When running the Main Sample, plot PE (CD24&CD71) versus APC (c-Kit) and gate on CD117-high and CD24&CD71-low cells that make up 10% of the population to isolate CD71/CD24low erythroid progenitors (Figure 1).
-
40.
To isolate CD71/CD24high progenitors, gate on CD117-high and CD24&CD71-high cells that make up 20% of the population.
Note: For optimal sorting results, an 85 μm nozzle and a flow rate of 1.5 should be used.
CRITICAL: Given the sensitivity of liver cells, once removed from mice, cells should be sorted into PNEG buffer and kept on ice or at 4°C whenever possible to ensure that the appropriate conditions and nutrients for short-term cell survival are maintained.
Figure 1.
CD71/CD24low and CD71/CD24high FACS sorting schematic
Immunomagnetic isolation of CD71/CD24low
Timing: 1.5 h
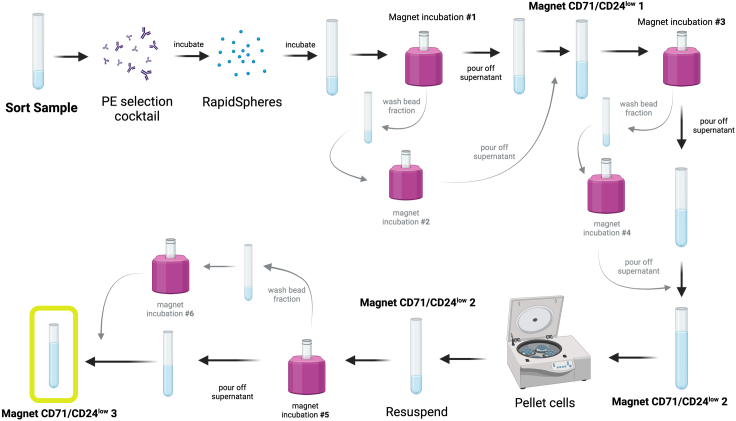
If a FACS facility is not easily accessible, CD71/CD24low progenitor cells can be isolated using the EasySep PE Positive Selection Kit II. Following surface marker staining, lineage depleted cells are stained with anti-PE antibodies. CD71/CD24low cells exhibit low expression of CD24 and CD71, and therefore can be isolated by depleting cells stained with PE.
Note: If isolating cells using both FACS and immunomagnetic isolation methods on the same day, a good tip is to conduct the magnetic isolation protocol during the sort, as both take about 1.5 h. This is only possible, however, if two different people are available to carry out each protocol.
Note: Reference Figure 2 as a helpful guide for this protocol.
-
41.
Centrifuge the Main Sample cells at 500 × g for 5 min at 4°C
-
42.
Count cells on a hemocytometer
-
43.If less than 10,000,000 cells total, resuspend cells in 100 μL PNEG
-
a.If more than 10,000,000 cells, resuspend in a volume of PNEG to make a final concentration of 100,000,000 cells/mL
-
a.
-
44.
Add 100 μL of EasySep PE Selection Cocktail to sample per mL of resuspended cells
Note: If <10,000,000 cells total, add 10 μL EasySep PE Selection Cocktail
-
45.
Incubate mixture at 4°C for 15 min. No tapping of the tube is required during the incubation.
-
46.
Vortex EasySep RapidSpheres for 30 s, then add 50 μL /mL of sample
Note: If <10,000,000 cells total, add 5 μL EasySep RapidSpheres
-
47.
Incubate at 4°C for 10 min
-
48.
Add 1.25 mL PNEG to the sample and mix gently by pipetting up and down 2–3 times
CRITICAL: Before every magnetic incubation, thoroughly wash the sides of the tube with the PNEG that was added to the sample, and briefly mix the solution by pipetting to get all cells in solution and to break up any cell-bead clumps.
-
49.
Incubate solution in the EasySep magnet at 4°C in a refrigerator for 5 min
Note: This is the first out of six magnet incubation steps. It is a good idea to mark down each step so that one does not lose track.
-
50.
Pour off the supernatant into a new polystyrene tube and label “magnet CD71/CD24low 1”
Note: Keep all cell samples, such as the “magnet CD71/CD24low 1” sample, on ice whenever not in direct use.
-
51.
Thoroughly wash the bead-bound fraction in the first tube with 1.25 mL PNEG
-
52.
Incubate the bead-bound fraction in the EasySep magnet at 4°C in a refrigerator for 5 min
Note: This is the second out of six CD71/CD24low magnet incubation steps.
-
53.
Pour off the supernatant into the “magnet CD71/CD24low 1” tube
-
54.
Incubate the “magnet CD71/CD24low 1” tube in EasySep magnet for 5 min at 4°C in a refrigerator
Note: This is the third out of six CD71/CD24low magnet incubation steps.
-
55.
Pour off supernatant and label a new polystyrene tube “magnet CD71/CD24low 2”
-
56.
Wash the bead-bound fraction with 1.25 mL PNEG
-
57.
Incubate this fraction in the EasySep magnet at 4°C for 5 min in a refrigerator
Note: This is the fourth out of six CD71/CD24low magnet incubation steps.
Note: There should be a total volume of 3.75 mL in the polystyrene tube at this point. A maximum of 3 mL can be used for each magnetic incubation, which is why the cells must be pelleted in the next step before another magnetic incubation.
-
58.
Centrifuge “magnet CD71/CD24low 2” at 500 × g for 5 min at 4°C
-
59.
Carefully pour off the supernatant and resuspend the cell pellet in 1.25 mL PNEG
Note: When pouring off the supernatant, pour in one gentle motion so as to not dislodge the cell pellet.
-
60.
Incubate the “magnet CD71/CD24low 2” tube in EasySep magnet at 4°C for 5 min in a refrigerator
Note: This is the fifth out of six CD71/CD24low magnet incubation steps.
-
61.
Pour off the supernatant into a new tube labeled “magnet CD71/CD24low 3”
-
62.
Wash the bead-bound fraction with 1.25 mL PNEG
-
63.
Incubate the bead-bound fraction in the EasySep magnet at 4°C for 5 min in a refrigerator
Note: This is the sixth and final CD71/CD24low magnet incubation step.
-
64.
Pour off the supernatant into the “magnet CD71/CD24low 3” tube
Note: This is the final sample of magnetically-isolated CD71/CD24low cells. The total volume of the sample should be 2.5 mL.
-
65.
Optional verification step: Save a small amount of the Main Sample and of the “magnet CD71/CD24low 3” sample (minimum of 10,000 cells each) and conduct flow cytometry using the same gating strategy as specified in the FACS isolation of CD71/CD24low and CD71/CD24high cells section. Ideally, the cells should fall within the CD71/CD24low gate.
Figure 2.
Flow chart depicting the protocol for magnetic isolation of CD71/CD24low erythroid progenitor cells
All illustrations created with BioRender.com.
Culturing CD71/CD24low and CD71/CD24high for growth curves
Timing: 10 days
-
66.
Pellet FACS- or immunomagnetically-isolated CD71/CD24low and CD71/CD24high cells at 500 × g for 5 min at 4°C and aspirate off the supernatant
-
67.
Resuspend cells in 100 μL PNEG and keep on ice
-
68.
Count cells on a hemocytometer
-
69.
Add 1 mL Serum-free erythroid liquid expansion media per well of a 24-well plate. Seed the appropriate number of cells in each well.
Note: We perform 3 technical replicates (3 wells) per treatment condition and seed 10,000 cells per well. Two treatments, using Dex-supplemented and Dex-free Serum free erythroid liquid expansion media, were used for our growth curves.
Note: Refer to materials and equipment for Serum-free erythroid liquid expansion media recipe.
-
70.
Incubate cells at 37°C and 5% CO2
-
71.
If conducting a growth curve, count cells on days 1, 2, 3, 6, 8, and 10 after seeding. Refer to Tables 1 and 2 for examples of calculating expansion per single cell from raw cell counts.
CRITICAL: Re-seed cells at 50,000 cells per well on days 3 and 6.
CRITICAL: For each biological replicate, use the same stock solution of Serum-free erythroid liquid expansion media (with the appropriate 100 nM Dex or methanol supplements) for all replicates.
Table 1.
Sample of raw data collected from three technical replicates from Dex-supplemented and no-Dex media on days 2 and 6 after isolation and seeding
| Day after initial seeding | Well | Media supplement | Cells per mL | Well volume (mL) | Total number of cells |
|---|---|---|---|---|---|
| 2 | A1 | no Dex | 465,000 | 0.92 | 427,800 |
| 2 | B1 | Dex | 810,000 | 0.92 | 685,400 |
| 2 | A2 | no Dex | 675,000 | 0.92 | 621,000 |
| 2 | B2 | Dex | 535,000 | 0.92 | 492,200 |
| 2 | A3 | no Dex | 600,000 | 0.89 | 534,000 |
| 2 | B3 | Dex | 685000 | 0.91 | 623,350 |
| 6 | A1 | no Dex | 330,000 | 0.95 | 313,500 |
| 6 | B1 | Dex | 450,000 | 0.93 | 418,500 |
| 6 | A2 | no Dex | 165,000 | 0.92 | 151,800 |
| 6 | B2 | Dex | 595,000 | 0.93 | 553,350 |
| 6 | A3 | no Dex | 190,000 | 0.91 | 172,900 |
| 6 | B3 | Dex | 495,000 | 0.88 | 435,600 |
Table 2.
Average number of cells counted from three technical replicates, scaled averages, and average expansion per single cell on days 2 and 6 after initial seeding
| Day after initial seeding | Media supplement | Average number of cells | Scaled average number of cells | Average expansion per single cell |
|---|---|---|---|---|
| 2 | No Dex | 527,600 | 527,600 | 53 |
| 2 | Dex | 600,317 | 600,317 | 60 |
| 6 | No Dex | 212,733 | 7,764,767 | 776 |
| 6 | Dex | 469,150 | 30,003,081 | 3000 |
Note: From the raw cell count data (Table 1), the average number of cells for each biological replicate was generated (Table 2). To generate scaled averages, which accounts for re-seeding cells at 50,000 cells per well on days 3 and 6 after initial seeding, cell count averages on days 6, 8, and 10 were divided by the number of cells re-seeded (50,000 cells) and multiplied by the most recent scaled average value before re-seeding (Table 2). Average expansion per single cell was calculated by dividing each scaled average by the number of cells seeded on day 0 (10,000 cells).
Expected outcomes
FACS isolation of CD71/CD24low cells typically yields 3%–5% of the number of lineage depleted cells used as input, and immunomagnetic isolation of CD71/CD24low cells typically yields 2%–6% of lineage depleted cells applied used as input. FACS isolation of CD71/CD24high cells is quite variable, yielding 2%–10% of the number of lineage depleted cells used as input. As an example, a successful isolation when using four E14.5 pregnant mice yielded 35 fetuses and 22,100,000 lineage depleted cells. 7,300,000 of these cells were used for FACS sorting and 14,600,000 cells were used for immunomagnetic isolation (the other 200,000 cells were used as FACS control samples). Immunomagnetic isolation yielded 850,000 CD71/CD24low cells (5.8% yield). FACS isolation yielded 350,000 CD71/CD24low cells (4.8% yield) and 595,000 CD71/CD24high cells.
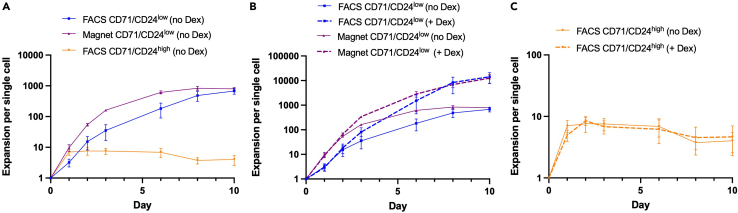
CD71/CD24low and CD71/CD24high cells demonstrate distinct growth properties in ex vivo proliferation assays in serum free erythroid liquid expansion media consisting of 100 ng/mL recombinant murine stem cell factor (SCF), 40 ng/mL recombinant murine insulin-like growth factor 1 (IGF-1), 2 units/mL recombinant human erythropoietin (EPO), and 1% penicillin/streptomycin in Stemspan serum free expansion medium II (Li et al., 2019; Flygare et al., 2011; Zhang et al., 2013; Lee et al., 2015; Gao et al., 2016). FACS CD71/CD24low cells should have approximately 30× the maximum proliferation of CD71/CD24high cells (Figure 3A). Additionally, in the presence of 100 nM Dexamethasone (a glucocorticoid that increases proliferative capacity of early erythroid progenitor cells), Magnet and FACS CD71/CD24low cells should have an approximately 15× and 20× increase in maximum proliferation, respectively (Figure 3B), whereas CD71/CD24high cells should have a minimal increase in maximum proliferation in the presence of 100 nM Dexamethasone (Figure 3C).
Figure 3.
Proliferative capacity of isolated early erythroid progenitor populations
(A) Ex vivo growth curves of FACS-isolated CD71/CD24low (n = 4) and CD71/CD24high (n = 3) cells, and magnetically isolated CD71/CD24low cells (n = 4) grown in Dex-free erythroid expansion media. Comparing points of maximum proliferation, FACS CD71/CD24low vs Magnet CD71/CD24low p = 0.860; FACS CD71/CD24low vs FACS CD71/CD24high p < 0.001; Magnet CD71/CD24low vs FACS CD71/CD24high p < 0.001.
(B) Ex vivo growth curve of FACS-isolated and magnetically isolated CD71/CD24low cells (both n = 4) grown in media supplemented with and without 100 nM Dex. Comparing points of maximum proliferation, FACS CD71/CD24low (no Dex) vs FACS CD71/CD24low (+ Dex) p < 0.001; Magnet CD71/CD24low (no Dex) vs Magnet CD71/CD24low (+ Dex) p < 0.001.
(C) Ex vivo growth curve of FACS-isolated CD71/CD24high cells (both n = 3) grown in media supplemented with and without 100 nM Dex. Comparing points of maximum proliferation, FACS CD71/CD24high (no Dex) vs FACS CD71/CD24high (+ Dex) p = 0.813. All error bars represent standard error of the mean.
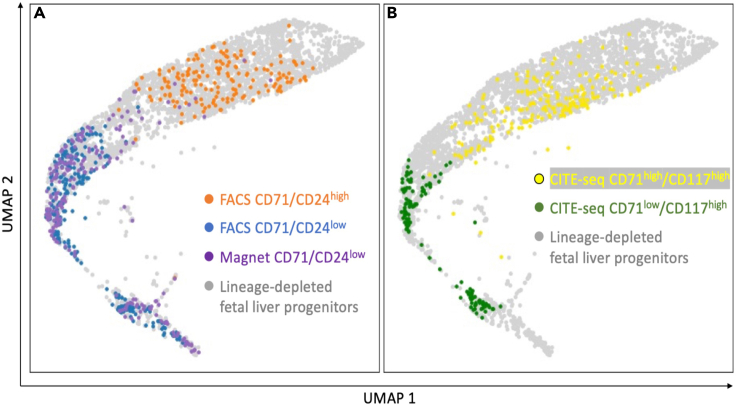
The transcriptome state of individual CD71/CD24low and CD71/CD24high cells can be assessed using single cell RNA sequencing (scRNAseq). We performed scRNAseq of FACS isolated CD71/CD24low and CD71/CD24high cells, immunomagnetic isolated CD71/CD24low cells, and lineage depleted cells. Dimensionality reduction analysis demonstrated transcriptomes of individual CD71/CD24low cells from both FACS and immunomagnetic isolation occupied a distinct distribution in dimensionality reduction space compared to individual CD71/CD24high cells from FACS isolation (Figure 4).
Figure 4.
Single-cell RNA-seq analysis of erythroid progenitor populations
(A) Single cell RNA sequencing was performed using the 10× Genomics 3′ Digital Gene Expression platform on lineage depleted fetal liver progenitor cells and (A) FACS-isolated CD71/CD24low and CD71/CD24high cells and magnetically isolated CD71/CD24low cells.
(B) For comparative reference, proteogenomic oligo-tagged CITE-seq (Stoeckius et al., 2017) analysis of CD71 and CD117 expression in lineage depleted fetal liver progenitors identifies transcriptome state of individual CD71low/CD117high and CD71high/CD117high erythroid progenitor cells. TotalSeqA0012 anti-mouse CD117 and TotalSeqA0441 anti-mouse CD71 antibodies (Biolegend) were used for CITE-seq staining. scRNAseq output was analyzed and visualized by the dimensionality reduction approach of Universal Manifold Approximation and Projection (UMAP) (Becht et al., 2018).
Quantification and statistical analysis
For proliferation assays, we perform three technical replicates, each starting with 10,000 cells in 1 mL of media in an individual well of a 24-well plate, for each individual biological replicate. We count cells on days 1, 2, 3, 6, 8, and 10 after initial seeding, with cells in each well diluted to 50,000 cells in 1 mL of fresh media on days 3 and 6 after initial seeding. When counting cells, we measure the total volume of media in each well to account for evaporation, and then assess cell concentration using a hemocytometer. Cell counts for the three technical replicates on each day are averaged to result in the cell count for the biological replicate. We then generate scaled cell counts by dividing by 10,000 to determine the proliferation of an individual cell. Student’s T-test comparing the maximum average expansion per single cell for each condition can be used to determine the significance of differences observed in maximum proliferation. After day 3 of culture, all cell counts should also factor in any of the dilutions performed on previous days.
Limitations
While FACS and immunomagnetic isolation of CD71/CD24low erythroid progenitor cells both yield cells with similar transcriptomes and phenotypic behavior in proliferation assays with and without Dex treatment, as mentioned above, the total yield of CD71/CD24low cells isolated by the immunomagnetic method is much more variable than the FACS method. Thus, when designing experiments using the immunomagnetic method, experimental plans should only anticipate 40,000 CD71/CD24low cells per 8 fetuses to ensure an adequate number of cells are available for downstream applications.
Absolute yield of CD71/CD24high erythroid progenitor cells from FACS isolation is also highly variable, and thus experimental plans should only anticipate 40,000 CD71/CD24high cells per 8 fetuses to ensure an adequate number of cells are available for downstream applications. An optimized method for immunomagnetic isolation of pure CD71/CD24high cells has not yet been developed.
While both FACS and immunomagnetic isolation methods yield between 3% and 10% progenitor cells from total Lineage depleted cells, these isolation methods may be better suited for different downstream applications. For instance, cells isolated using the immunomagnetic method grow more quickly initially compared to FACS-isolated CD71/CD24low cells (Figures 3A and 3B). This slight difference in initial growth is likely due to the increased stress that sorted cells experience from fluidic pressure during sorting, whereas immunomagnetically-isolated cells experience no such stress. Therefore, if an increase in cell growth during the first 5–7 days after isolation is a priority in an experiment, the immunomagnetic isolation method should be utilized. FACS isolation, on the other hand, should be utilized if certain quantifiable parameters, such as the percentage of dead cells or lineage-positive cells in the sample, are of interest. FACS isolation is also better suited for experiments in which optimal purity of the cells is a priority.
This protocol has not yet been tested for the isolation of erythroid progenitor cells from mouse bone marrow; however, utilization of this protocol to isolate CD71/CD24low cells from bone marrow and other tissues containing early erythroid progenitors is a potential future application of this work. While doing so will likely require modifications and optimizations to this protocol, expanding the utility of this protocol to other tissues in mice, or other model organisms, would be useful for the field and should be pursued.
Troubleshooting
Problem 1
Low yield of lineage depleted cells
There is a significantly lower yield of lineage depleted cells isolated when counted on a hemocytometer as specified in the optional verification step (step 28).
Potential solution
Ensure that the entire fetal liver is dissected out, and increase the amount of time spent homogenizing the fetal livers before applying to the 70 μM filter, making sure that there are no large clumps in the process. Ensure that washes of clotted tissue remaining on 70 μM filters are performed in a vigorous manner.
Problem 2
Low yield of FACS or magnetically-isolated CD71/CD24low cells
There is a significantly lower yield of CD71/CD24low cells from FACS (steps 37–40) or immunomagnetic isolation (steps 41–64) than specified in the expected outcomes.
Potential solution
If not done already, count the number of lineage depleted cells isolated as specified in the optional verification step (step 28). If fewer than 2–3 million lineage depleted cells per 8 fetuses are counted, pursue the potential solutions for problem 1.
If >50% of cells are dead when counting cells after FACS or immunomagnetic isolation, refer to problem 4 for troubleshooting.
If the low yield of CD71/CD24low cells is not explained by high cell death or a low lineage depleted cell yield, and cells are being isolated using FACS, pre-coat all FACS collection tubes with FBS by adding 20% FBS in PNEG buffer to tubes and incubating for at least 2 h at 4°C. After this incubation, replace the 20% FBS in PNEG buffer with regular PNEG buffer for cell collection. Pre-coating collection tubes in this way can prevent cell attraction to tube walls when sorted into collection tubes, which can result in cell rupture.
If a low cell yield is not explained by high cell death or low lineage depleted cell yield, and cells are being isolated using the immunomagnetic isolation method, increase the time spent washing the tube after each magnetic isolation to ensure all cells are washed back into solution before the next incubation.
Problem 3
Impure CD71/CD24low population from magnetic isolation
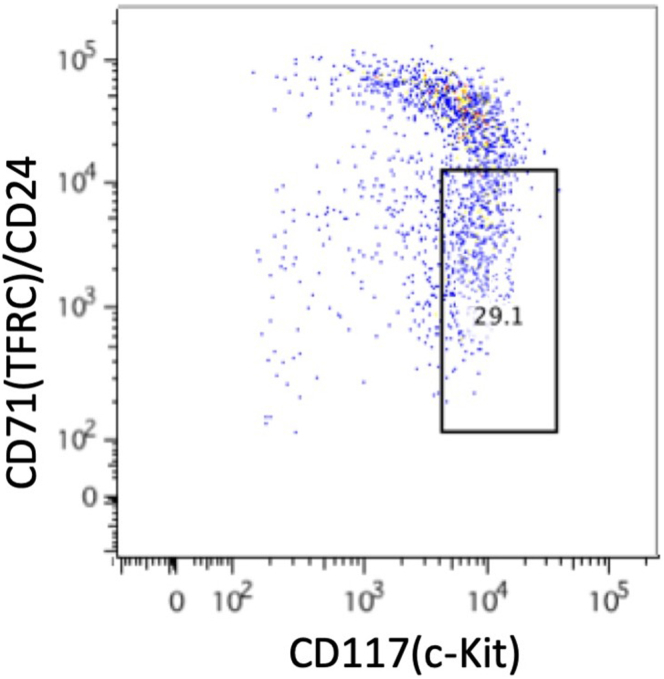
A majority of PE depleted cells do not fall within the CD71/CD24low gate when conducting flow cytometry as an optional verification step (step 65) for the magnetic isolation method.
Figure 5 shows a sample from an earlier iteration of this protocol in which only 3 magnet incubations were carried out and in which the bead and cell mixture was not thoroughly homogenized and washed off of the sides of the tube in between each incubation. Only 29.1% of cells fall within the CD71/CD24low gate, which was set using a small aliquot from the Sort Sample as outlined in the FACS isolation of CD71/CD24low cells and CD71/CD24high cells section (step 39).
Figure 5.
FACS plot of APC-CD117 versus PE-CD71/CD24 for an insufficiently PE depleted sample following use of the magnetic isolation protocol for CD71/CD24low cells
The displayed gate was obtained from a pre-enriched (not shown) sample gating on the bottom 10% of grandparent.
Potential solution
Increase the number of incubations and washes with the EasySep magnet. Additionally, when washing, make sure to wash all bead solution off of the sides of the tube and re-homogenize the solution thoroughly to break up any clumps and ensure all cells are in solution for the subsequent incubation.
Problem 4
High percentage of dead cells
When counting lineage depleted cells on a hemocytometer (step 28) or when conducting flow cytometry as an optional verification step (step 65), a high percentage (for instance, greater than 50% of cells) are stained with Trypan Blue or are PI positive.
Potential solution
Decrease the overall time of the isolation. This can be done by improving dissection efficiency, preparing reagents and tube set-up steps ahead of time or during incubation steps, and euthanizing mice in a nearby vicinity to where the isolation will be conducted. Additionally, during all 4°C incubations, ensure cells are always on fresh ice, and incubate samples in the fridge if possible. Lastly, several commercially available dead cell depletion kits can be purchased and applied to the final cell population as well.
Problem 5
Slow growth of CD71/CD24low cells in erythroid proliferation assays
When conducting a growth curve (step 70) or expanding CD71/CD24low cells for applications such as scRNAseq or genomic DNA isolation, cells grow significantly more slowly than expected based on the growth curves in Figure 3.
Potential solution
Ensure that all components used to make Serum-free erythroid expansion media have been freshly thawed within 2 weeks of use and have been stored at 4°C for the entire time from thaw to usage. Thaw new aliquots of cytokines, SFEM II media, Penicillin/Streptomycin, or Dexamethasone, make fresh media, and re-seed cells in this media if the 2-week time limit has been exceeded. If the problem persists with a new batch of freshly isolated CD71/CD24low cells and fresh media, conduct flow cytometry as an optional verification step (step 65) to determine whether the isolated cells fall within the appropriate CD71/CD24low cell gate. If cells do not fall within this gate, refer to problem 2.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the lead contact, Hojun Li (hojunli@mit.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was funded by NIH grants P01 HL032262 and P30 CA14051. H.L. is funded by NIH grant K08 DK123414, an American Society of Hematology (ASH) Scholar Award, a Charles W. (1955) and Jennifer C. Johnson Clinical Investigator Award, and Boston Children’s Hospital Office of Faculty Development Award. E.K. is funded by NIH grant K08 DK128571 and an ASH Scholar Award. The authors would like to acknowledge the Whitehead Institute Flow Cytometry Core Facility, the Massachusetts Institute of Technology BioMicro Center, and the Integrated Genomics and Bioinformatics Core of the Koch Institute for Integrative Cancer Research. Use of mice described in this study was approved by the Massachusetts Institute of Technology Committee for Animal Care.
Author contributions
T.W.B., E.K., and H.L. designed and optimized protocols and performed experiments. T.W.B., M.K.K., and H.L. analyzed data. All authors contributed to writing and editing the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.101070.
Contributor Information
Tatum W. Braun, Email: tbraun@mit.edu.
Hojun Li, Email: hojunli@mit.edu.
Data and code availability
All data and code are available by contacting the lead contact, Hojun Li (hojunli@mit.edu). Single cell RNA-sequencing and CITE-seq data can be downloaded from the National Center for Biotechnology Information Gene Expression Omnibus. The accession number for the sequencing data reported in this paper is NCBI GEO: GSE188287.
References
- Becht E., Mcinnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- Flygare J., Rayon Estrada V., Shin C., Gupta S., Lodish H.F. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Lee H.Y., da Rocha E.L., Zhang C., Lu Y.F., Li D., Feng Y., Ezike J., Elmes R.R., Barrasa M.I., et al. TGF-beta inhibitors stimulate red blood cell production by enhancing self-renewal of BFU-E erythroid progenitors. Blood. 2016;128:2637–2641. doi: 10.1182/blood-2016-05-718320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harandi O.F., Hedge S., Wu D.C., Mckeone D., Paulson R.F. Murine erythroid short-term radioprotection requires a BMP4-dependent, self-renewing population of stress erythroid progenitors. J. Clin. Invest. 2010;120:4507–4519. doi: 10.1172/JCI41291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Gao X., Barrasa M.I., Li H., Elmes R.R., Peters L.L., Lodish H.F. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015;522:474–477. doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Natarajan A., Ezike J., Barrasa M.I., Le Y., Feder Z.A., Yang H., Ma C., Markoulaki S., Lodish H.F. Rate of progression through a continuum of transit-amplifying progenitor cell states regulates blood cell production. Dev. Cell. 2019;49:118–129. doi: 10.1016/j.devcel.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hale J., Bhagia P., Xue F., Chen L., Jaffray J., Yan H., Lane J., Gallagher P.G., Mohandas N., et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood. 2014;124:3636–3645. doi: 10.1182/blood-2014-07-588806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop R., Shearstone J.R., Shen Q., Liu Y., Hallstrom K., Koulnis M., Gribnau J., Socolovsky M. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010;8:e1000484. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren S.E., Siva K., Soneji S., George A.J., Winkler M., Jaako P., Wlodarski M., Karlsson S., Hannan R.D., Flygare J. Glucocorticoids improve erythroid progenitor maintenance and dampen Trp53 response in a mouse model of Diamond-Blackfan anaemia. Br. J. Haematol. 2015;171:517–529. doi: 10.1111/bjh.13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi G., Inoue D., Chen C., Bitner L., Chung Y.R., Taylor J., Gonen M., Wess J., Abdel-Wahab O., Zhang L. Muscarinic acetylcholine receptor regulates self-renewal of early erythroid progenitors. Sci. Transl. Med. 2019;11:eaaw3781. doi: 10.1126/scitranslmed.aaw3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Prak L., Rayon-Estrada V., Thiru P., Flygare J., Lim B., Lodish H.F. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code are available by contacting the lead contact, Hojun Li (hojunli@mit.edu). Single cell RNA-sequencing and CITE-seq data can be downloaded from the National Center for Biotechnology Information Gene Expression Omnibus. The accession number for the sequencing data reported in this paper is NCBI GEO: GSE188287.