Abstract
MS is the most effective method to directly identify peptides presented on human leukocyte antigen (HLA) molecules. However, current standard approaches often use 500 million or more cells as input to achieve high coverage of the immunopeptidome, and therefore, these methods are not compatible with the often limited amounts of tissue available from clinical tumor samples. Here, we evaluated microscaled basic reversed-phase fractionation to separate HLA peptide samples offline followed by ion mobility coupled to LC–MS/MS for analysis. The combination of these two separation methods enabled identification of 20% to 50% more peptides compared with samples analyzed without either prior fractionation or use of ion mobility alone. We demonstrate coverage of HLA immunopeptidomes with up to 8107 distinct peptides starting with as few as 100 million cells. The increased sensitivity obtained using our methods can provide data useful to improve HLA-binding prediction algorithms as well as to enable detection of clinically relevant epitopes such as neoantigens.
Keywords: HLA, immunopeptidomics, FAIMS, basic reversed-phase fractionation, ion mobility
Abbreviations: ACN, acetonitrile; APC, antigen-presenting cell; BCS, backbone cleavage score; bRP, basic reversed-phase; CE, collision energy; CV, compensation voltage; FA, formic acid; FAIMS, high field asymmetric waveform ion mobility spectrometry; GBM, glioblastoma; HCD, higher energy collisional dissociation; HLA, human leukocyte antigen; HLA-I, human leukocyte antigen class I; IL, interleukin; IP, immunoprecipitation; MEL, melanoma; MRM, multiple reaction monitoring; PBMC, peripheral blood mononuclear cell; PIP, precursor isolation purity; PSM, peptide spectrum match; RCC, renal cell carcinoma; SM, Spectrum Mill; TCR, T-cell receptor
Graphical Abstract

Highlights
-
•
Deep immunopeptidome coverage using liquid and gas phase separation.
-
•
Up to 50% more HLA-I peptides using microscaled basic reversed-phase fractionation.
-
•
Ion mobility separation (FAIMS) increases HLA-I peptide identifications by up to 58%.
-
•
Increased sensitivity provided by these methods enables detection of neoantigens.
In Brief
Here, we evaluated off-line microscaled basic reversed-phase fractionation as well as the use of ion mobility coupled to LC–MS/MS for analysis of peptides presented on HLA-I. The two separation methods enabled identification of 20% to 50% more peptides compared with samples analyzed without either prior fractionation or use of ion mobility alone starting with as few as 100 million cells. The increased sensitivity obtained using our methods can enable detection of low abundant but clinically relevant epitopes such as neoantigens.
Antigen presentation and subsequent recognition by cytotoxic T cells is a central component of the adaptive immune response. Therapies harnessing the power of the immune system are showing great potential in personalized cancer treatment (1, 2, 3, 4). Because these therapies rely on recognition of tumor-associated antigens, viral antigens, or neoantigens by cytotoxic T cells, discovery and identification of these antigens is crucial to impact patient treatment outcomes. Cell surface antigens presented on highly polymorphic human leukocyte antigen class I (HLA-I) molecules are short peptide sequences that are predominantly nine amino acids long. Which antigens are presented by an individual's HLA allele is often predicted computationally or evaluated using biochemical binding assays with a biased set of selected synthetic peptides. MS enables direct and untargeted identification of thousands of endogenously processed and presented peptide antigens and therefore provides a deeper insight into the immunopeptidome (5, 6, 7, 8). In particular, MS-based detection of potential tumor antigens can confirm the presentation of specific epitopes by tumor cells, identify new antigens with therapeutic potential, or monitor changes in antigen presentation in response to treatment (9, 10, 11, 12).
At present, the identification of HLA-I eluted peptides by MS (immunopeptidomics) is hampered by multiple factors. First, HLA-bound peptides span a diverse repertoire of sequences as each HLA allele has a unique peptide-binding motif. Second, despite their different sequences, these peptides are very similar to each other in length and amino acid composition. Thus, effective separation using standard LC–MS/MS methods optimized on tryptic proteome digests remains challenging. Third, thousands of peptides are estimated to be presented by each cell, each with relatively low abundance, and therefore, a large amount of input material is needed to achieve high sensitivity. Fourth, algorithmic interpretation of HLA-I eluted peptide spectra is much more challenging than for trypsin-derived peptide spectra typical of proteomic studies because (a) the narrow length distribution of the complex HLA-I eluted peptides mixture hampers chromatographic separation leading to more coisolated precursor ions producing mixed MS/MS spectra, (b) charge bearing basic residues (e.g., Lys, Arg) are not always at the C terminus and may be absent altogether, leading to substantially altered distributions of fragment ion series, and (c) the lack of restricted enzyme specificity inflates the sequence database search space by ~100× (13).
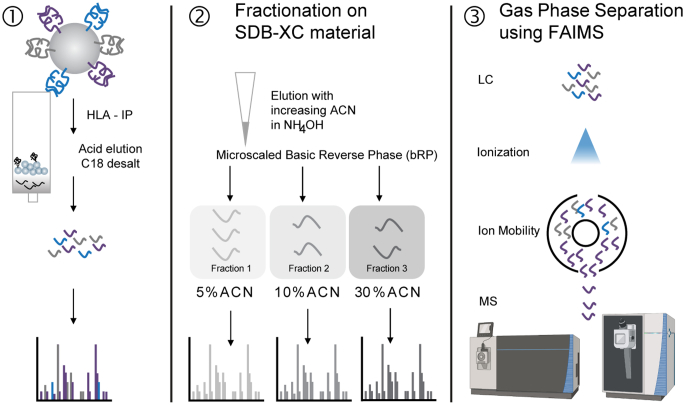
While recent improvements in MS instrumentation have made HLA-I eluted peptide identification more feasible, the detection of promising tumor rejection antigens such as neoantigens remains particularly challenging. The amount of HLA-eluted peptide material derived from small amounts of tumor tissue obtained in a clinical setting is often limited. Moreover, extensive sample handling following enrichment of HLA-I eluted peptides can lead to sample loss. Successful tissue-based approaches have typically utilized billions of cells and grams of material for analysis (9, 14). Previously, we developed and optimized methods for isolation of immunoprecipitated HLA–peptide complexes from as few as 50 million cells and 0.2 g of clinical specimens yielding 1000 to 2000 peptides per sample (Fig. 1, left panel) (5, 8, 15, 16). For subsequent analysis by LC–tandem MS (LC–MS/MS), 2 × 3 HLA:peptide immunoprecipitations (IPs, 2× 150 × 106 cells) were pooled, desalted, and analyzed in technical replicates. The method has yielded sufficient depth (1000–2000 peptides per sample) for profiling of monoallelic HLA-expressing cell lines and HLA-bound antigens from patient tumor specimens (8). While the depth of coverage obtained using such small amounts of sample was high by historical standards, it has generally been inadequate to detect low-frequency peptides (i.e., like neoantigens) consistently. Furthermore, we observed that with each repeat injection, we uncovered more of the peptidome; around 20% of peptides were only detected in one of four injections suggesting that there is far more of the peptidome to be discovered (supplemental Fig. S1, A and B).
Fig. 1.
Schematic overview of HLA sample preparation approaches. (1) Prior standard workflow in which HLA-I peptides are acid eluted and desalted before LC–MS/MS analysis with repeat injection. (2) Offline basic reversed-phase separation of eluted peptides, further separated into three fractions using SDB-XC material using increasing ACN concentrations in ammonium hydroxide (5%, 10%, and 30% ACN in 0.1% NH4OH, pH 10). (3) Separation of ions in the gas phase using FAIMS coupled to the mass spectrometer. ACN, acetonitrile; FAIMS, high field asymmetric waveform ion mobility spectrometry; HLA, human leukocyte antigen.
Recent improvements in proteome profiling protocols for small sample input amounts and the introduction of ion mobility for gas phase fractionation online with the MS system have the potential to improve the depth of HLA-I eluted peptide analysis. Stage-Tip–based basic reversed-phase (bRP) fractionation using alternative non-C18 solid phases has already shown benefits for proteomic analysis of low cell numbers after flow cytometry (17). In addition, use of high field asymmetric waveform ion mobility spectrometry (FAIMS) on hybrid quadrupole-orbitrap instruments has been demonstrated to increase peptide and protein identifications in single-shot proteomics experiments (18, 19, 20). The advantage of FAIMS for gas phase separation of low input and post-translationally modified peptide samples has also been reported (21).
Here, we investigated the impact of using offline Stage-Tip fractionation and online gas phase separation by FAIMS to increase the depth of immunopeptidome coverage for cell line and patient tissue samples with relatively low amounts of input HLA-I eluted peptides. We present an optimized HLA-I immunopeptidomics workflow for the analysis of HLA-I eluted peptides derived from multiple tumor types, revealing that a combination of fractionation and FAIMS significantly improves the profiling depth of the HLA-I immunopeptidome. We demonstrate that this new workflow enables detection of neoantigens and further extends large-scale data generation capabilities that can motivate the retraining of epitope prediction algorithms.
Experimental Procedures
Generation of Single-Allele Cell Lines and Primary Human Samples
HLA class I–deficient B721.221 cell lines expressing a single HLA-I allele each were generated as previously described (8). Cell lines with stable surface HLA-I expression were generated through selection using 800 μg/ml G418 (Thermo Scientific), followed by enrichment of HLA-I positive cells through up to two serial rounds of fluorescent-activated cell sorting and isolation using a pan-HLA-I antibody (W6/32; Santa Cruz) on a FACSAria II instrument (BD Biosciences). All human tissues were obtained through Dana-Farber Cancer Institute–approved or Partners Healthcare–approved Institutional Review Board protocols. Conditions for growth and in vitro propagation of melanoma (MEL) and glioblastoma (GBM) tumor cell lines and of monocyte-derived dendritic cells were described previously (8).
For primary tumors and patient-derived cell lines, HLA–peptide complexes were immunoprecipitated from 0.1 to 0.2 g tissue or up to 50 million cells. Solid tumor samples were dissociated using a tissue homogenizer (Fisher Scientific 150 Homogenizer package; Fisher Scientific), and HLA complexes were enriched as described later.
HLA-I Peptide Enrichment and Peptide Elution
Soluble lysates from up to 50 million cells and up to 0.2 g from tumor tissue or tumor-derived cell lines were immunoprecipitated with W6/32 antibody (sc-32235; Santa Cruz) as described previously (8) (supplemental Table S1). Iodoacetamide (10 mM) was added to the lysis buffer to alkylate cysteines. For each experiment, two IPs were pooled (equivalent to 100 × 106 cells), HLA-I bound peptides were acid eluted on 1 cc 50 mg tC18 SepPak Cartridges (WAT054960; Waters) on a vacuum manifold 1 to 2 days prior to analysis on the instrument. SepPaks are equilibrated with 200 μl MeOH 2×, 100 μl 50% acetonitrile (ACN)/1% formic acid (FA), and 500 μl 1% FA 4×, respectively. Beads with HLA-I bound peptides were resuspended in 3% ACN/5% FA and transferred to the equilibrated cartridge. Peptides were eluted from HLA-I proteins with 10% acetic acid for 5 min twice and then desalted with 4× 500 μl 1% FA. Finally, peptides were eluted into the same tube from the desalt matrix using 250 μl 15% ACN/1% FA followed by 250 μl 50% ACN/1% FA and dried down. This step was performed for every sample described here. Patient samples were typically immunoprecipitated from the same lysate pool but eluted on different days to prevent/reduce peptide loss because of peptide degradation and adsorption to plastic while waiting for analysis. An overview of cell input and processing details for each sample is provided in supplemental Table S1.
For tumor and cancer cell line samples in the conventional or FAIMS approach, a second desalt was performed using a Stage-Tip with two disks of C18 material (Empore 2215). The material was washed and equilibrated with 100% methanol, followed by 50% ACN/1% FA and 1% FA. Dried peptides were resuspended in 3% FA/5% ACN and loaded onto the equilibrated Stage-Tip. Peptides were desalted with up to five washes of 1% FA before elution. Peptides were eluted with 15% ACN/1% FA followed by 30% ACN/1% FA and lyophilized in a vacuum concentrator. Samples were stored at −80 °C until LC–MS/MS data acquisition.
bRP Fractionation for HLA-I Peptides
For bRP fractionation, a Stage-Tip was prepared using two disks of SDB-XC material (Empore 2240), washed and equilibrated with 100% MeOH, followed by 50% ACN/1% FA and 1% FA. Dried peptides were resuspended in 3% FA/5% ACN and loaded onto the Stage-Tip. Peptides were desalted with up to five washes of 1% FA before elution in three fractions with increasing concentrations of ACN (5%, 10%, and 30%) in 0.1% (w/v) NH4OH (28% NH3 [w/v], 338818; Sigma–Aldrich), pH 10. Fractions were dried down in a vacuum concentrator and stored at −80 °C until LC–MS/MS data acquisition.
LC–MS/MS Data Acquisition
Samples were reconstituted in 7 μl 3% ACN/5% FA prior to LC–MS/MS analysis on a Fusion Lumos or Orbitrap Exploris 480 (Thermo Fisher Scientific). A pool of two IPs (equivalent to peptides from 100 × 106 cells) was injected twice (3 μl each) as a technical replicate for the conventional approach. In the case of one A∗02:01 sample, we also injected it as three technical replicates. Alternatively, for fractionated samples, peptides were resuspended in 4 μl, and the total fraction was injected. Peptides were loaded onto an analytical column (25–30 cm, 1.9 μm C18 [Dr Maisch HPLC GmbH], packed in-house PicoFrit 75 μm inner diameter, 10 μm emitter [New Objective]). Peptides were eluted with a linear gradient (EasyNanoLC 1200; Thermo Fisher Scientific) ranging from 6 to 30% solvent B (0.1% FA in 90% ACN) over 84 min, 30 to 90% B over 9 min and held at 90% B for 5 min at 200 nl/min. MS/MS were acquired in data-dependent acquisition. For the Fusion Lumos, settings were described previously (8). On the Orbitrap Exploris 480, MS1 spectra were collected until either 100% normalized automatic gain control target or a maximum injection time of 50 ms was reached. Monoisotopic peak detection was set to “peptide,” and “relax restrictions” was enabled; precursor fit filter was set to 50% fit threshold and 1.2 m/z fit window. Dynamic exclusion was 10 s. For HLA-I peptides, up to five precursors of charge 1+ between 800 and 1700 m/z or 10 precursors of charge 2 to 4+ were subjected to MS/MS acquisition. Precursors were isolated with a 1.1 m/z window, collected with 50% normalized automatic gain control target and 120 ms maximum injection time, fragmented at 30% higher energy collisional dissociation (HCD), and acquired at 15,000 resolution.
When FAIMS was attached, spray voltage was increased to 1900 V and FAIMS was set to standard resolution. FAIMS compensation voltages (CVs) were set to -50 and -70 with a cycle time of 1.5 s per FAIMS experiment. Further evaluated settings included CV combinations -40|-60. If CV -20 was used, MS2 spectra were triggered for the five most abundant MS1 precursors. MS2 fill time was set to 100 ms, and all other parameters were the same as in the no FAIMS setting. Instrument performance was evaluated using a 10 ng Jurkat proteome digest, and data were acquired above a peptide yield threshold of >9000 peptides for Fusion Lumos and >13,000 peptides for Orbitrap Exploris.
HLA Peptide Identification Using Spectrum Mill
Mass spectra were interpreted using the Spectrum Mill (SM) software package, version 7.00 (Broad Institute; proteomics.broadinstitute.org). MS/MS spectra were excluded from searching if they did not have a precursor sequence MH+ in the range 600 to 4000, had a precursor charge >5, or had a minimum of <5 detected peaks. Merging of similar spectra with the same precursor m/z acquired in the same chromatographic peak was disabled. Before searches, all MS/MS spectra had to pass the spectral quality filter with a sequence tag length >1 (i.e., minimum of three masses separated by the in-chain masses of two amino acids). The maximum sequence tag lengths were calculated in the SM Data Extractor module using the ESI-QEXACTIVE-HCD-v2 peak detection parameters. MS/MS spectra were searched against a protein sequence database that contained 98,298 entries, including all University of California Santa Cruz Genome Browser genes with hg19 annotation of the genome and its protein-coding transcripts (63,691 entries), common human virus sequences (30,181 entries), and recurrently mutated proteins observed in tumors from 26 tissues (4167 entries), as well as 259 common laboratory contaminants including proteins present in cell culture media and IP reagents. Mutation files for 26 tumor tissue types were obtained from the Broad GDAC portal (gdac.broadinstitute.org). Recurrent mutations in the coding region within each of the 26 tumor types (frequency = 3 for stomach adenocarcinoma, uterine corpus endometrial carcinoma; frequency = 5 for adrenocortical carcinoma, pancreatic adenocarcinoma, MEL; and frequency = 2 for rest) were included. For tumor samples, patient-specific mutations were appended to the database. The following are the MS/MS search parameters: digest: no-enzyme specificity; instrument: ESI-QEXACTIVE-HCD-HLA-v3; fixed modification: cysteinylation of cysteine; variable modifications: carbamidomethylation of cysteine, oxidation of methionine and pyroglutamic acid at peptide N-terminal glutamine; precursor mass tolerance of ±10 ppm; product mass tolerance of ±10 ppm; and a minimum matched peak intensity of 30%.
Peptide spectrum matches (PSMs) for individual spectra were automatically designated as confidently assigned using the SM autovalidation module to apply target-decoy–based false discovery rate estimation at the PSM level of <1% false discovery rate. Score threshold determination also required that peptides had a minimum sequence length of 7, and PSMs had a minimum backbone cleavage score (BCS) of 5. BCS is a peptide sequence coverage metric, and the BCS threshold enforces a uniformly higher minimum sequence coverage for each PSM, at least four or five residues of unambiguous sequence. The BCS score is a sum after assigning a 1 or 0 between each pair of adjacent amino acids in the sequence (maximum score is peptide length −1). To receive a score, cleavage of the peptide backbone must be supported by the presence of a primary ion type for HCD: b, y, or internal ion C terminus (i.e., if the internal ion is for the sequence PWN, then BCS is credited only for the backbone bond after the N). The BCS metric serves to decrease false positives associated with spectra having fragmentation in a limited portion of the peptide that yields multiple ion types.
PSMs were consolidated to the peptide level to generate lists of confidently observed peptides for each allele using the SM protein/peptide summary module's peptide-distinct mode with filtering distinct peptides set to case sensitive. A distinct peptide was the single highest scoring PSM of a peptide detected for each sample. Different modification states observed for a peptide were each reported when containing amino acids configured to allow variable modification; a lowercase letter indicates the variable modification (C-cysteinylated, c-carbamidomethylated).
The precursor isolation purity (PIP), precursor signal-to-noise metrics, and precursor averagine Chi-squared metrics were calculated by the SM Data Extractor module. PIP is the intensity of the precursor ion and its isotopes divided by the total peak intensity in the precursor isolation window used to generate an MS/MS spectrum (combined from the two MS spectra immediately before and after the MS/MS spectrum). PIP <50% indicates expected contamination by cofragmented peptides and constitutes the default threshold for disregarding reporter ion quantitation. The precursor signal/noise ratio—(root mean square) is calculated in the MS spectrum used to trigger an MS/MS spectrum where signal constitutes the sum of the peaks in the precursor isotope cluster, and noise constitutes the smallest 90% of the centroided peaks in the spectrum that are at least 3% the height of the base peak in the spectrum. Precursor averagine Chi squared, a measure of the precursor ion isotope cluster shape (1—chi-squared), is calculated from the combined isotope cluster shape of the two MS1 scans immediately before and after the MS/MS scan and then compared with the theoretical isotope cluster shape of averagine via a standard Chi-square statistic calculated on the peak intensities. Identification rate was calculated by dividing the number of identified MS/MS spectra by the number of filtered MS/MS spectra that passed initial quality assessment in the SM Data Extractor module.
Filtering of MS-Identified Peptides and Subsequent Data Analysis
The list of LC–MS/MS-identified peptides was filtered to remove potential contaminants in the following ways: (1) peptides observed in negative control runs (blank beads and blank immunoprecipitates); (2) peptides originating from the following species: “STRSG,” “HEVBR,” “ANGIO432,” “ANGIO394,” “ANGIO785,” “ANGIO530”, “ACHLY,” “PIG,” “ANGIO523,” “RABIT,” “STAAU,” “CHICK,” “Pierce-iRT,” “SOYBN,” “ARMRU,” and “SHEEP” as common laboratory contaminants including proteins present in IP reagents (note that “BOVINE” peptides derived from cell culture media were not excluded as they appear to have undergone processing and presentation and exhibit anchor residue motifs consistent with the human peptides observed for each allele); (3) peptides for which both the preceding and C-terminal amino acids were tryptic residues (R or K) as tryptic peptide contamination can easily be introduced when detaching cells in cell culture or by carryover contaminants on the LC–MS/MS instrumentation (8). Subsequent data analysis was performed in R using in-house scripts and the ggplot and Upset (22, 23) packages.
HLA Peptide Prediction
HLA peptide prediction was performed using HLAthena (hlathena.tools) (8). Unless otherwise specified, peptides were assigned to an allele using a percentile rank cutoff of ≤0.5. In some instances, peptides were also predicted with NetMHCpan 4.0 EL (http://www.cbs.dtu.dk/services/NetMHCpan-4.0) (24).
Nonmetric Multidimensional Scaling
A pairwise relative peptide distance matrix was computed between every pair of 9-mer peptide sequences identified in monoallelic datasets evaluated in the different approaches. The distance between two peptides is defined as the sum of AA residue dissimilarities at each position along the sequences. Dissimilarity is weighted by the entropy at the corresponding position (5). Nonmetric multidimensional scaling was used to reduce the dimensionality of the distance matrix so peptides can be visualized as points in 2D.
Multiple Reaction Monitoring for Neoantigen Detection
Tier 2 multiple reaction monitoring (MRM) assay configuration using heavy labeled synthetic peptides was done on TSQ Quantiva triple quadrupole mass spectrometer (Thermo Fisher) coupled with Easy-nLC 1200 ultra-high pressure liquid chromatography system (Thermo Fisher). Skyline Targeted Mass Spec Environment ([64 bit] 20.2.0.343 version) was used throughout assay configuration and all data analyses. First, a spectral library for the peptide YIHGRGWAL was generated on a Fusion Lumos (Thermo Fisher). Spectral library was uploaded to Skyline, and six most intense fragment ions (transitions) were selected for MRM assay configuration. Next, collision energies (CEs) were optimized for all the transitions by LC–MRM/MS on TSQ Quantiva using Skyline's CE optimization module. For every transition starting with the instrument-specific calculated CE, we tested 10 additional CEs (five below and five above the calculated CE) in increments of 2. The list of transitions with varying CE values was exported from Skyline and used for building MRM method in Xcalibur software (Thermo Scientific). Equimolar mixture of peptides at 50 fm/μl was analyzed by LC–MRM/MS on Quantiva using this method. Resulting data were analyzed on Skyline, which then selected the CE that resulted in the highest peak area for each transition. In the final step of CE optimization, MRM data were acquired with optimized CE values for every transition. Using this dataset in Skyline, the best two transitions were selected manually. After CE optimization, we analyzed three replicates of six IPs pooled with either 20 fmol or 40 fmol heavy peptide spiked in.
LC was performed on 75 μm ID picofrit columns packed in house to a length of 28 to 30 cm with Reprosil C18-AQ 1.9 μm beads (Dr Maisch GmbH) with solvent A of 0.1% FA/3% ACN and solvent B of 0.1% FA/90% ACN at 200 nl/min flow rate and the same gradient as described previously. MS parameters include 1.5 s cycle time, Q1 and Q3 resolution of 0.4 and 0.7, respectively, and real-time scheduling window of 10 min.
T-Cell Receptor Reconstruction and Expression in T Cells for Reactivity Screening
T-cell receptors (TCRs) selected from the study by Oliveira et al. were cloned and transduced in donor T cells (25). Briefly, the full-length TCRA and TCRB chains, separated by Furin SGSG P2A linker, were synthesized in the TCRB/TCRA orientation (Integrated DNA Technologies) and cloned into a lentiviral vector under the control of the pEF1α promoter using Gibson assembly (New England Biolabs, Inc). For generation of TCRs, full-length TCRA V–J regions were fused to optimized mouse TRA constant chain and the TCRB V–D–J regions to optimized mouse TRB constant chain to allow preferential pairing of the introduced TCR chains, enhanced TCR surface expression, and functionality (26, 27).
Donor T cells enriched from peripheral blood mononuclear cells (PBMCs) using PanT cell selection kit (Miltenyi) were activated with antiCD3/CD28 dynabeads (Thermo Fisher Scientific) in the presence of 5 ng/ml of interleukin 7 (IL-7) and IL-15 (PeproTech). After 2 days, activated cells were transduced with a lentiviral vector encoding the TCRB–TCRA chains. Briefly, lentiviral particles were generated by transient transfection of the lentiviral packaging Lenti-X 293T cells (Takahara) with the TCR encoding plasmids and packaging plasmids (VSVg and PSPAX2) (28) using Transit LT-1 (Mirus). Lentiviral supernatant was harvested 2 days later, for two subsequent days, and used to transduce activated T cells. Lentiviral transductions were performed by inoculation of the virus at 2000 rpm, 37 °C for 2 h, and cells were cultured on viral supernatant for 3 days. Six days after activations, the beads were removed from culture and expanded in media enriched with IL-7 and IL-15. Transduction efficiency was determined by flow cytometric analysis using the anti-mTCRB antibody. Transduced T cells were used at 14 days post-transduction for TCR reactivity tests. Briefly, 2.5 × 105 TCR-transduced T cells were put in contact with the following targets: (i) patient-derived MEL cell lines (0.25 × 105 cells); (ii) patient PBMCs (2.5 × 105 cells); (iii) patient Epstein–Barr virus– lymphoblastoid cell lines (2.5 × 105 cells/well) alone or pulsed with peptides; (iv) medium, as negative control; and (v) phorbol myristate acetate (50 ng/ml; Sigma–Aldrich) and ionomycin (10 μg/ml; Sigma–Aldrich) as positive controls. Peptide pulsing of target cells was performed by incubating Epstein–Barr virus–lymphoblastoid cells in fetal bovine serum–free medium at a density of 5 × 106 cells/ml for 2 h in the presence of individual peptides (107 pg/m; Genscript). After an overnight incubation, TCR reactivity was measured through detection of CD137 surface expression (PE-antihuman CD137; Biolegend) on CD8+ TCR transduced (mTCRB+) T cells using a Fortessa flow cytometer (BD Biosciences).
Experimental Design and Statistical Rationale
For MS experiments, sample size was n = 1 for each method evaluation per monoallelic cell line, tumor-derived cell line, or primary tumor. The conventional sample preparation and data acquisition method was used as control for evaluation of increase in identifications using other approaches. For conventional analysis, a sample was injected as a technical replicate, but results were aggregated across technical repeat injections for analysis. No statistical testing was used.
For A∗02:01, we evaluated another biological replicate. Peptide intensity correlation was evaluated using Pearson R.
For correlation of observed and synthetic peptide fragment ion spectra, spectral contrast angle was calculated using crossproduct of vectors of fragment ion masses and intensities for each of the spectra sp1 and sp2 (29).
Results
Approaches for HLA Peptidome Isolation
In an effort to improve sensitivity for detection of HLA-I eluted peptides, we evaluated the impact of microscaled bRP fractionation (Fig. 1, middle panel) as well as gas phase separation using FAIMS (Fig. 1, right panel). The effect of microscaled fractionation and FAIMS on the coverage of the immunopeptidome was evaluated individually and in combination, as described later. For all evaluations, we enriched HLA-I peptide complexes from 100 × 106 cells (i.e., 2 IPs, each at 50 × 106 cells) that each expressed a single allele (5, 8) or from up to 0.2 g of primary tumor or tumor-derived cell line material using the pan-HLA class I antibody W6/32 (supplemental Table S1). HLA-bound peptides were isolated from the protein using acid elution. In what we refer to as the “conventional” approach, peptides eluted from monoallelic or multiallelic samples were desalted on C18 SepPaks using a stepped elution of 15% and 50% ACN in 0.1% FA. Multiallelic (e.g., tumor) samples, if not subjected to fractionation (see later), were further subjected to a second desalt with a 30% ACN as the highest elution concentration (8, 9). All eluted monoallelic or multiallelic peptides were analyzed as two technical replicate injections without FAIMS in the conventional approach. The fractionation approach used the same first desalt step, followed by fractionation on SDB-XC and injecting each fraction as a single run on the instrument. The FAIMS approach included both desalt steps and analysis of two technical replicate injections. The tested tumor-derived cell lines originated from MEL (3), GBM (2), or clear cell renal cell carcinoma (RCC) tumor specimens.
bRP Fractionation Increases Peptidome Coverage
Although the value of sample prefractionation to increase coverage of the immunopeptidome has been reported by several groups (30, 31, 32), most studies utilize at least 500 million cells as input for HLA peptide enrichment. We aimed to decrease the amount of input sample required to the more readily obtainable amount of 100 × 106 cells by testing microscaled fractionation on bRP on Stage-Tips with two punches of SDB-XC material (see Experimental Procedures section). We observed a 25 to 50% gain in peptide identifications for monoallelic cell lines as well as multiallelic samples by Stage-Tip fractionation using the SDB-XC bRP material compared with our conventional approach (Fig. 2A). For example, peptides derived from 100 × 106 B721.221 cells only expressing HLA-A∗02:01 injected in two technical replicates yielded 3483 unique peptides on the Orbitrap Exploris 480. Stage-Tip fractionation of the same amount of HLA-I eluted peptides nearly doubled the yield to 6084 unique peptides (74% increase), of which 2936 peptides were identified by both workflows. For a biological replicate of the same allele, we also injected the nonfractionated sample three times to demonstrate that this increase in peptide observations was not attributed to measurement time alone (supplemental Fig. S2A). While three replicate injections did identify 1723 peptides more than two replicate injections in this analysis, fractionation identified an additional 2509 peptides and a total of 6939 peptides similar to the number of peptides identified in the first replicate. Hence, while measurement time influenced peptide yield, fractionation still added increased depth. Fractionation also outperformed elution with 30% ACN on the SDB-XC bRP material (supplemental Fig. S2B). The overall peptide yields for MEL tumor-derived cell lines (MEL1 and MEL6) as well as tissue samples of renal cell cancer (RCC1 and RCC4M) increased by 30% relative to our prior approach (Fig. 2A).
Fig. 2.
Basic reversed-phase separation increases peptide yield.A, the number of unique peptides identified by LC–MS/MS obtained without off-line fractionation (empty squares, “conventional” on x-axis) and using fractionation (filled black squares) across monoallelic and multiallelic samples. Unfilled bars correspond to peptides identified in both approaches; blue-filled bars are peptides unique to unfractionated analyses, whereas purple-filled bars are peptides unique to fractionated samples. Numbers in bars indicate the percent of total peptides per approach. B, motif of 9-mer peptides identified in the unfractionated (top) and fractionated (bottom) acquisition of the monoallelic HLA-A∗02:01 sample. C, nonmetric multidimensional scaling (NMDS) plot (see Experimental procedures section) showing clusters of peptides identified in both (gray), unfractionated (blue), and fractionated (purple) samples of A∗02:01 cells. D, UpSet plot showing peptide spectrum matches (PSMs) identified in the three fractions of the A∗02:01 peptidome. Horizontal bars at bottom left indicate the number of total PSMs per fraction; dots and lines under the vertical bars identify peptides identified in only one (single dot) or multiple fractions (dots with lines). E, Same as D but for RCC1T. F, allele assignment of peptides identified in the individual fractions of RCC1T (top left, fraction of assigned peptides at HLAthena rank <0.5) as well as in conventional and fractionated RCC1T (bottom left, fraction of peptides at HLAthena rank <0.5). Top right box plot shows peptide pI of peptides per fraction, allele motifs, and median pI of all peptides assigned to each allele are depicted at the bottom. HLA, human leukocyte antigen.
To confirm that the peptides newly identified using bRP fractionation were bona fide HLA-I binding peptides, we evaluated how well their sequences matched known HLA-I binding motifs and submotifs. For the A∗02:01 eluted peptides, the motifs of technical replicate injection and fractionation revealed the same anchor residues (Fig. 2B), and the submotifs from these two approaches overlapped when clustered based on their amino acid sequences by nonmetric multidimensional scaling (Fig. 2C; see Experimental procedures section) suggesting that the additional peptide identifications obtained from fractionation were likely to bind to HLA-A∗02:01 without introducing novel peptide motifs.
To better understand why we obtained a distinct subset of HLA-I eluted peptides using the fractionation approach, we evaluated the peptides' physiochemical properties in our fractionated dataset. The number of unique peptide identifications per fraction indicated a reasonable separation of peptides based on hydrophobicity and retention on the Stage-Tip in basic conditions. For the HLA-A∗02:01 monoallelic line, 3077 peptides were identified in the first fraction, 3064 peptides in the second fraction, and 1956 peptides in the third fraction. Because peptides were still quite similar in overall properties, 648 and 680 peptides eluted in both fractions 1 and 2 or in fractions 2 and 3, respectively, and 328 peptides were identified in all three fractions (Fig. 2D). Intensities of peptides detected in biological replicate fractionation experiments of A∗02:01 correlated well (Pearson R = 0.7; supplemental Fig. S2C). Peptides identified in both experiments also typically elute in the same fraction, with some peptides overlapping between replicates and two fractions (supplemental Fig. S2D).
The gain in identifications upon bRP fractionation was even more pronounced for multiallelic samples, particularly when their peptidomes were derived from binding motifs with a diverse set of physiochemical characteristics (hydrophobicity and pI). For example, the majority of newly identified peptides for the bRP fractionated human renal cancer tissue sample RCC1 was found in fractions 1 and 2 (5% and 10% cutoff; Fig. 2E, supplemental Fig. S2E for MEL1). Using the HLA peptide presentation prediction algorithm HLAthena (8), each identified peptide was scored against each patient allele and assigned to a specific allele with a rank cutoff of <0.5. This analysis revealed that under bRP conditions at pH 10, peptides separated based on their interaction with the mobile and stationary phase dependent on allele-binding motif, hydrophobicity, and peptide pI (Fig. 2F and supplemental Fig. S2F for MEL1). The median pI of identified peptides in fraction 1 across all alleles was 4.5 and 89% (n = 1111) of B∗40:01 binders eluted in fraction 1 (median pI = 4.37). Since the binding motif of B∗40:01 has a preference for glutamic acid (E) in position 2, E-containing peptides were likely to be net-negatively charged in our basic buffer conditions and thus not well retained on the SDB-XC bRP stationary phase. Conversely, peptides associated with alleles bearing basic amino acid residues in the anchor positions, A∗66:01 and C∗06:02, will tend to be positively charged, retained better, and contributed more peptides to the later fractions. Similarly, peptides associated with alleles having hydrophobic residues in their anchor positions, such as B∗57:01, tended to preferentially contribute to later bRP fractions (Fig. 2F). Taken together, bRP fractionation separated peptides in an orthogonal manner to the acidic reversed-phase packing material used in the online gradient and yielded significantly more HLA-I eluted peptide identifications in only 6 h of measurement time compared with technical replicate analyses that take 4 h.
HLA Peptide Identification Benefits From Separation of Ions in the Gas Phase
Next, we investigated how the separation of ions by their ion mobility in the gas phase can benefit HLA-I eluted peptide analysis. For these studies, we used the recently introduced FAIMSpro interface from Thermo Scientific that fits orbitrap instruments (e.g., Fusion Lumos and Orbitrap Exploris 480). We tested a range of CVs and optimized the acquisition method for HLA-I eluted peptides using two CVs -50 and -70 (CV), triggering MS2 for 1.5 s at each CV. Each sample set was acquired on the same instrument (either Fusion Lumos or Exploris; supplemental Table S1) with and without FAIMS and two replicate injections of the same sample amount (100 × 106 cells). The use of FAIMS increased the number of HLA-I eluted peptides identified by an average of 58% across all samples evaluated (Fig. 3A). For the peptidome acquisition of the monoallelic B∗53:01-expressing cells and the multiallelic MEL1 cells, FAIMS acquisition doubled the number of peptides identified compared with no FAIMS. In the case of the RCC1 tumor samples, FAIMS did not increase the overall peptide identifications but rather revealed 655 new HLA-I eluted peptides not identified using the conventional approach.
Fig. 3.
Ion mobility separation increases peptide yield.A, the number of HLA-I eluted peptide identifications obtained using FAIMS (filled black squares) or without FAIMS (empty squares) across monoallelic and multiallelic samples. Unfilled bars correspond to peptides identified in both approaches; blue filled bars are peptides unique to no-FAIMS (conventional), whereas red-filled bars are peptides unique to samples analyzed with FAIMS. Numbers in bars indicate the percent of peptides per approach. B, motif of 9-mer peptides identified in without FAIMS (top) and with FAIMS (bottom) acquisition of the monoallelic HLA-B∗53:01 sample on the Orbitrap Exploris. C, NMDS plot of peptides identified in both (gray), conventional (blue), and with FAIMS (red) samples of B∗53:01. D, precursor isolation purity of peptides in FAIMS (red) and conventional (blue) acquisition. E, number of identifications in all samples across the chromatographic gradient colored by detection in normal acquisition (no FAIMS, blue), + FAIMS acquisition (red), and both acquisition modes (gray, bin size = 30). FAIMS, high field asymmetric waveform ion mobility spectrometry; HLA, human leukocyte antigen.
The main peptide motif of identified peptides using FAIMS matched the motif obtained using conventional approaches (Fig. 3B). They overlapped in their submotif clusters as illustrated by A∗02:01 and B∗53:01 (Fig. 3C) and had similar length distributions compared with the traditional injection (supplemental Fig. S3A). Altogether, these characterizations indicate that the new identifications were credible HLA-I binders.
The increased number of identified HLA-I eluted peptides appeared to be related to the overall improved data quality obtained using FAIMS, which facilitated spectral interpretation and identification. FAIMS increased the PIP (see Experimental procedures section) from a median between 74 to 93% and 89 to 97% (Fig. 3D). Hence, fewer coisolated peptide precursor ions competed for one PSM, yielding fewer chimeric spectra and more identifications overall. FAIMS separation facilitated the identification of more precursor ions throughout the whole run but especially increased the yield in the middle of the gradient, when most of the peptides eluted from the column (Fig. 3E). Total ion chromatograms of individual CV values showed that FAIMS helped to separate ions before they enter the MS (supplemental Fig. S3B), and therefore, purer MS/MS spectra can be acquired and identified. While FAIMS acquisition with two CVs triggered acquisition of fewer MS/MS spectra than without FAIMS, a higher percentage (identification rate) of the acquired spectra led to an identified peptide sequence in the FAIMS run for all samples (e.g., A∗02:01: 30.5% versus 11.5%; supplemental Fig. S3C). For samples acquired on the Orbitrap Exploris, we observed up to a 3-fold increase in the percent of identified spectra relative to no FAIMS (supplemental Fig. S3C). Further analysis on a A∗02:01 and MEL6 sample showed sample-dependent improved precursor signal-to-noise scores, PIP, and isotopic envelope metrics when using FAIMS (supplemental Fig. S3, D and E). This was particularly pronounced for low-intensity peptides in a multiallelic sample.
Hydrophobic HLA-I eluted peptides lacking basic residues can be observed as singly charged precursor species (0–40%) in the conventional acquisition (supplemental Table S2) but were lost using the two CV settings for FAIMS described previously despite allowing for z = 1 during precursor selection in the instrument method. To improve the transmission of singly charged peptides, we tested adding an additional CV of -20, which should be better suited to stabilize z = 1 precursors (18). Peptides derived from monoallelic cell lines expressing either A∗11:01 or B∗07:02 were used to evaluate the effect. While A∗11:01 features a tryptic-like motif with lysine in the C-terminal position, B∗07:02 is representative of a nonpolar peptide pool with a xPxxxxxxL motif. FAIMS experiments were run on peptides from both alleles using CV settings of -50|-70 and CVs of -20|-45|-65. Both methods yielded a similar number of identifications, but while less than 0.1% of PSMs were identified with a single charge in A∗11:01, CV-20 increased the percentage of singly charged PSMs for B∗07:02 to 4% (supplemental Fig. S3D). In another comparison, we evaluated CV combinations -20|-40|-60 and -20|-50|-70 and found that for B∗07:02, 6% of singly charged ions led to a peptide identification at CV -20. Since most HLA-I eluted peptides were identified at CV-50 or CV-60, one of these two CVs should be used for data acquisition in combination with a second CV that stabilizes multiply charged ions. If the sample composition includes alleles with hydrophobic anchor residues known to yield high proportions of singly charged precursors (supplemental Table S2), a third FAIMS experiment at CV -20 and precursor filter for z = 1 can be included (supplemental Fig. S3E). In multiallelic samples, different FAIMS CVs could stabilize slightly different peptide allele fractions likely because of charge state or peptide length. Applying multiple CVs per MS experiment reduced potential allele under representation when using FAIMS (supplemental Fig. S3F).
In summary, these studies demonstrate that use of the FAIMSpro interface leads to the identification of novel and valid peptides because of improved precursor separation/selection and generation of purer MS/MS spectra.
Each Individual Workflow Variation Can Add a Distinct Set of Peptides
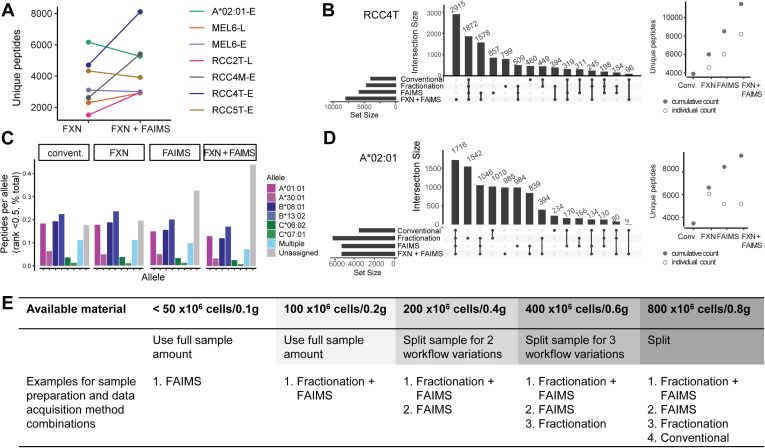
Given the increases in depth of HLA-I peptidome coverage obtained using optimized and microscaled bRP fractionation and ion mobility independently, we next evaluated the effect of combining these two approaches. We found that for four of seven samples tested, combining fractionation plus FAIMS yielded more peptides than fractionation alone (Fig. 4A). However, this was not universally the case, and even in cases where it was true, each method employed also often revealed distinct subsets of peptides.
Fig. 4.
Combination of fractionation and FAIMS for greater coverage of the immunopeptidome.A, number of unique 8–11-mer identifications in fractionated samples acquired with and without FAIMS. “L” and “E” indicate the instrument, Fusion Lumos or Orbitrap Exploris, used for acquisition. B, peptides identified across all modes of acquisition of RCC4T. Horizontal bars at bottom left indicate the number of total unique peptides per acquisition; dots and lines under the vertical bars identify peptides identified in only one (single dot) or multiple acquisitions (dots with lines). Each acquisition (empty circle) adds unique peptide identifications to the total cumulative peptidome (filled circle). C, allele assignment of peptides identified in RCC4T (HLAthena, rank <0.5) across four different acquisitions. D, same as B but for HLA-A∗02:01 sample. E, the table with recommended sample preparation and data acquisition strategies based on input available. FAIMS, high field asymmetric waveform ion mobility spectrometry.
In the case of the multiallelic patient sample RCC4T, off-line Stage-Tip fractionation plus FAIMS added the most unique peptides found in that experiment, followed by using FAIMS alone (Fig. 4B). In total, 11,392 8- to 11-mers were identified for this patient sample using only eight IPs originating from 0.1 g of tumor material each. Peptides were assigned to the allele to which they were most likely bound (A∗01:01, A∗30:01, B∗08:01, B∗13:02, C∗06:02, and C∗07:02) using HLAthena (8) and a rank cutoff of 0.5. The relative contribution per allele to the total peptidome was similar across different acquisition schemes with the majority of peptides presented on A∗01:01, B∗08:01, and B∗13:02 (Fig. 4C). NetMHC prediction revealed similar results (supplemental Fig. S4A).
A relatively high percentage of the HLA-I eluted peptides in the FAIMS (37%) and fractionated + FAIMS (52%) samples of RCC4T could not be assigned to an allele at rank <0.5. Half of those peptides were weak binders with rank <2 (supplemental Fig. S4A). Closer investigation of the motif of the nonbinders at rank <2 suggested potential additional binders for HLA A∗30:01 and HLA B∗13:02 (supplemental Fig. S4B). These alleles have a low positive predictive value in HLAthena (8), and hence, binding prediction models can still be improved once more peptides are identified.
For the RCC2 tumor sample, a total of 3936 HLA-I peptides were detected across all four experiments, whereas only 773 peptides were commonly identified in all acquisitions (supplemental Fig. S4C). The lower yield compared to RCC4T is likely because of plentiful necrotic tissue and diminished HLA peptide presentation. Fractionation in combination with FAIMS alone identified 75% of all peptides and had similar contribution per allele to the total peptidome as the other acquisition schemes.
Applying all variations of the four workflows to a common pool of the monoallelic HLA-A∗02:01 sample yielded a total of 9439 unique peptides (Fig. 4D) with varying peptide identifications in each acquisition mode. Of those, only 1716 were identified across all four conditions. Acquisitions without FAIMS (conventional and fractionation) added 2786 (1542 + 1010 + 234) peptides, whereas 2808 additional identifications were found only in acquisitions that used FAIMS (985 + 984 + 839). HLA-I eluted peptide yield was greatest for a fractionated sample acquired without FAIMS. In this particular case, the gap in performance between FAIMS and no FAIMS on a QC standard sample (10 ng Jurkat proteome tryptic digest) was less pronounced than usual. Hence, maintaining optimal instrument performance is an important consideration.
As each method identified another distinct set of peptides presented on HLA-I, we suggest that greater coverage of the immunopeptidome can be achieved by applying multiple peptide separation approaches rather than performing repeated injection, if sufficient input material is available. Based on our results, when aiming at achieving greatest coverage, we propose a decision tree to guide through sample preparation and acquisition methods (Fig. 4E). For most tumor samples evaluated in this study, fractionation in combination with FAIMS acquisition resulted in the highest yield of peptides from one sample and enabled neoantigen identifications (see the following paragraphs), and hence, it was chosen as the preferred sample preparation and data collection method. However, depending on the amount of sample available, FAIMS acquisition without fractionation added another larger set of distinct peptides and can therefore be chosen as an alternative to technical replicates. This was followed by fractionation without FAIMS. Although our conventional approach was outperformed by all other strategies, it still revealed additional peptides not observed in any of the other acquisitions.
Increased Coverage Can Enable the Detection of Clinically Relevant Neoantigens
Increased numbers of identified peptides from lower amounts of tumor sample input can also increase the likelihood of detecting clinically relevant epitopes such as neoantigens. Using either fractionation, FAIMS or both, we profiled different samples from primary tumor as well as from tumor-derived cell lines on the Fusion Lumos or the Orbitrap Exploris. The underlying tumor types had different mutational burdens, with MEL harboring more mutations compared with GBM or RCC.
In two MEL samples, MEL1 and MEL6, we were able to detect four and three neoantigens, respectively, starting with only 100 × 106 cells or 0.2 g of input per sample using fractionation (Exploris) or FAIMS acquisition (Fusion Lumos) (Fig. 5A). These neoantigens were missed in analyses of these samples using the conventional workflow as well as prior runs of the same tumor-derived cell lines (8). For GBM and RCC, no neoantigens were detected using these input amounts.
Fig. 5.
Optimized data acquisition strategies facilitate neoantigen discovery.A, neoantigens identified in tumor-derived cell lines MEL 1 and MEL 6 acquired with fractionation ± FAIMS, their protein of origin, detection method, transcript abundance level (TPM), and the most likely allele the peptide would bind to as predicted by HLAthena using MSi (peptide sequence–only model) and MSiCE (cleavability and expression integrative model). B, mirror plots for the MS spectra of two neoantigen sequences (LLHTELERFL and NSKKKWFLF) in the discovery experiment (top) and the corresponding synthetic peptide spectrum (bottom, red: y-ion, blue: b-ion, and green: internal ion). C, targeted MS experiment for mutated peptide YIHGRGWAL using a heavy peptide. After optimization, transitions of y6- and y7-ion were acquired of the heavy peptide alone (left, blank) and of the heavy peptide spiked into a peptidome sample of MEL6. Endogenous peptide detection is shown on the top, and detection of the heavy peptide is shown at the bottom. D, CD137 signal of neoantigen-specific TCRs in different target backgrounds (PMA/ionomycin = positive control; MEL= tumor cell line, PBMCs from the corresponding patient, APCs pulsed with DMSO, mutant, or WT peptide). Percent upregulation is shown for each panel. APC, antigen-presenting cell; DMSO, dimethyl sulfoxide; FAIMS, high field asymmetric waveform ion mobility spectrometry; MEL, melanoma; PBMC, peripheral blood mononuclear cell; PMA, phorbol myristate acetate; TCR, T-cell receptor.
Four of the detected neoantigens were predicted as strong binders to one of the patient's respective alleles (HLAthena MSi and MSiCE, rank <0.5), and one was predicted as a weak binder (rank <2). For the two peptides identified in MEL 1 with predicted binding above the rank threshold (LLHTELERFL, MSiCE rank 4.07 and NSKKKWFLF, MSiCE rank 3.73), we confirmed that our identifications were correct using synthetic peptides (spectral contrast angle LLHTELERFL: 0.805, NSKKKWFLF: 0.964; Fig. 5B). The neoantigen YIHGRGWAL from NCEH1 detected in MEL 6 was also evaluated using targeted MS with heavy-labeled peptide. We observed the selected MS2 transitions for this peptide in an HLA peptide pool injection (six IPs, ~300 × 106 cells) but not in a blank injection (Fig. 5C). Triplicate analysis across HLA IP samples from different cell culture batches confirmed the presentation of this neoantigen.
We then tested the reactivity of peptide-specific TCR clones against three of these MS-detected neoantigens using the CD137 signal as a marker for TCR transduction and activation. When cultivated with patient-derived MEL cells that presented the antigens KRPTPTFHF, YIHGRGWAL, and QLPDFVNQL at physiological levels as well as with antigen-presenting cells (APCs) pulsed with mutant peptides, TCRs were activated (Fig. 5D, MEL track (red) and APC-Mut pep track). In contrast, incubation of PBMCs or APCs pulsed with dimethyl sulfoxide or WT peptides did not lead to TCR activation. In summary, our improved workflow enabled the detection of clinically relevant antigens that can initiate a tumor-specific T-cell response.
Discussion
To gain depth in the assessment of the HLA peptidome, most studies have used at least 500 million cells as input, numbers that typically cannot be obtained from clinical specimens, whether they are tumor specimens procured through standard biopsy or surgery, or even from cell line experiments where the number of obtainable cells is limited. Therefore, more sensitive approaches are needed to enable routine HLA peptidome analysis for clinical samples.
Here, we demonstrated the ability to achieve deep coverage of the HLA peptidome from cells and tissues using input amounts of as little as 100 × 106 cells or 0.15 g wet weight of tumor. Further increases in peptide yield relative to repeat injection of the same sample were obtained by fractionating the peptide specimen offline into three fractions using microscaled bRP on Stage-Tips. The use of SDB-XC material separated the peptidome in an allele- (and hence amino acid composition) dependent manner. By evaluating this method across a range of diverse HLA-I alleles in both monoallelic and multiallelic settings, we observed that, although individual fractions reflected peptide characteristics obtained when binding and eluting under basic conditions, the total peptidome per sample was not biased to any particular allele. Offline bRP fractionation using other materials such as C18 would also be feasible but may lead to the loss of the orthogonality to the on-line acidic RP separation afforded by the SDB-XC material. We noted that only three fractions were required per experiment, and thus, measurement time remained comparable to our standard approach using two replicate injections.
Gas phase separation of ions using FAIMS increased the HLA-I eluted peptide yield compared with acquisition without FAIMS by reducing coisolation of peptide precursors and mitigated instances of (i) one precursor would not be triggered, (ii) the ions would cofragment and lead to mixed spectra without FAIMS, and (iii) removal of singly and highly charged contaminants introduced by the IP in one or the other CV. The usage of a combination of CVs (-50 and -70 on the Thermo FAIMS Pro device) yielded the widest range of peptides. While singly charged ions are likely not acquired with these settings, the fraction of HLA-I eluted peptides lost was generally negligible compared with the overall gain in identifications. When foreseeing a sample composition lacking in basic residues and dominated by potentially nonpolar side chains likely to lead to a substantial proportion of singly charged peptide precursors (e.g., allele B∗07:02), we propose adding a third experiment with a CV of -20 (supplemental Fig. S3, F and G) (18). We expect similar benefits for use of ion mobility for HLA-I eluted peptide analysis with other types of instruments such as the TimsTOFpro from Bruker.
We also observed that variation in sample preparation, as well as data acquisition methods for the same sample, could affect the overall peptide yield and reveal unique peptides not found with more than one of the methods used. In addition, for some samples run on the Orbitrap Exploris, fractionation alone yielded more peptides compared with FAIMS approaches. In general, we observed that instrument performance greatly influences peptide yield. Especially when adding FAIMS, tip shape, and positioning, spray stability and the proper connection of the FAIMS unit to the instrument are crucial, and best performance is not always easily achieved. Such variability in the identified peptide population because of sample preparation and data acquisition have been observed by others (19, 30, 33, 34) but is not yet fully understood.
The increase in sensitivity for detection of HLA-I eluted peptides obtained by either using bRP fractionation, FAIMS acquisition, or a combination of both enabled the identification of neoantigens from tumor-derived cell line samples that were not found in conventional experiments. We confirmed that MS-detected neoantigens can induce immune responses and are instrumental for the identification of cognate antigens of TCRs as well as selection of potential vaccine targets (25).
Based on these findings, we developed a decision tree for different scenarios, dependent on the amount of available sample (Fig. 4E). Specifically, if only material for a single acquisition is available, our current preferred methods are FAIMS and bRP fractionation together with FAIMS-based acquisition, given our success in using these approaches to detect neoantigens. If limiting instrument time is a concern, we still recommend using FAIMS while giving consideration to reducing the number of fractions or simply injecting the same sample twice using different CV sets for each acquisition (21). In our experience, repeat technical injections provided the lowest yield of HLA-I eluted peptides; thus, we recommend varying acquisitions by using bRP fractionation and FAIMS alone and in combination in cases where sufficient sample is available.
Future improvements to these sample preparation and data acquisition methods may include but are not limited to (i) the use of automation for HLA peptide enrichment (35), (ii) online LC columns of smaller inner diameter (36), or (iii) shorter gradients in combination with FAIMS acquisition methods (37). Chemical labeling has also been shown to increase the number of peptide identifications but may potentially change the overall allele representation in the sample (19). Furthermore, a recent study suggests that higher concentrations of ACN during peptide elution and desalting could enable detection of more hydrophobic and immunogenic peptides missed in current sample preparation approaches (38).
In conclusion, offline separation and gas phase fractionation increased the yield of HLA-I–presented peptides analyzed by LC–MS/MS and allowed for detection of low abundant neoantigens. We foresee that similar observations will hold true for HLA class II peptides and that these acquisition approaches will benefit antigen discovery not only in cancer but also in other fields, including autoimmune and infectious diseases.
Data availability
The original mass spectra, PSMs, and the protein sequence databases used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000087743/. Raw files and skyline documents for the MRM experiment have been deposited in the ProteomeXchange Consortium (PXD027165) via Panorama Public https://panoramaweb.org/0HgG5Q.url.
Supplemental data
This article contains supplemental data (8).
Conflict of interest
D. A. B. reported nonfinancial support from Bristol-Myers Squibb, honoraria from LM Education/Exchange Services, and personal fees from Octane Global, Defined Health, Dedham Group, Adept Field Solutions, Slingshot Insights, Blueprint Partnerships, Charles River Associates, Trinity Group, and Insight Strategy, outside the submitted work. P. A. O. has received research funding from and has advised Neon Therapeutics, Bristol-Meyers Squibb, Merck, CytomX, Pfizer, Novartis, Celldex, Amgen, Array, AstraZeneca/MedImmune, Armo BioSciences, and Roche/Genentech, outside the submitted work. D. B. K. has previously advised Neon Therapeutics and has received consulting fees from Neon Therapeutics. D. B. K. owns equity in Agenus, Armata Pharmaceuticals, Breakbio, BioMarin Pharmaceutical, Bristol Myers Squibb, Celldex Therapeutics, Chinook Therapeutics, Editas Medicine, Exelixis, Gilead Sciences, IMV, Lexicon Pharmaceuticals, Moderna, and Regeneron Pharmaceuticals. BeiGene, a Chinese biotech company, supports unrelated research at TIGL. C. J. W. holds equity in BioNTech, Inc and receives research funding from Pharmacyclics, Inc. S. A. C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, and Seer and an ad hoc scientific advisor to Pfizer and Biogen. All the other authors declare no competing interests.
Acknowledgments
The authors thank Sebastian Vaca and Pierre M. Jean-Beltran for computational assistance. The authors thank Scott Goulding for experimental discussions. This work was made possible by a grant from BroadIgnite at the Broad Institute of MIT and Harvard (to S. K.). This work was further supported by The G. Harold and Leila Y. Mathers Foundation.
Funding and additional information
S. K. was supported by the Cancer Research Institute as a Hearst Foundation fellow. D. B. K. was supported by the Emerson Collective. This work was also supported in part by grants from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (to S. A. C). This work was supported in part from grants from the National Institutes of Health (NCI-1R01CA155010, NCI-U24CA224331 [to C. J. W.]; NIH/NCI R21 CA216772-01A1, NCI-R01 CA229261 [to P. A. O.]; and NCI-SPORE-2P50CA101942-11A1 [to D. B. K.], NCI-SPORE P50CA101942-15 [to D. A. B.]), Department of Defense's Congressionally Directed Medical Research Programs (KC190128) (to D. A. B), and a Team Science Award from the Melanoma Research Alliance (to C. J. W. and P. A. O.). G. O. was supported by the American Italian Cancer Foundation fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
S. K. and S. A. C. conceptualization; S. K. methodology; S. K., A. A., K. R. C., S. S., and H. K. formal analysis; S. K., A. A., S. R., G. O., P. M. L., A. T., V. C., H. K, and D. B. K. investigation; S. K., K. R. C., S. S., J. G. A., D. A. B., P. A. O., N. H., C. J. W., and S. A. C. data curation; S. K., K. R. C., and S. A. C writing–original draft; S. K., K. R. C., C. J. W., and S. A. C. writing–review and editing.
Contributor Information
Susan Klaeger, Email: sklaeger@broadinstitute.org.
Steven A. Carr, Email: scarr@broad.mit.edu.
Supplemental data
References
- 1.Bilusic M., Madan R.A. Therapeutic cancer vaccines: the latest advancement in targeted therapy. Am. J. Ther. 2012;19:e172–e181. doi: 10.1097/MJT.0b013e3182068cdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E., Shukla S.A., Hu Z., Li L., Le P.M., Allesøe R.L., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., Chen C., Olive O., Carter T.A., Li S., Lieb D.J., et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., Omokoko T., Vormehr M., Albrecht C., Paruzynski A., Kuhn A.N., et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 5.Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M., Clauser K.R., Hacohen N., Rooney M.S., Carr S.A., Wu C.J. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Admon A., Bassani-Sternberg M. The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.O111.011833. O111.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt D., Henderson R., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A., Appella E., Engelhard V. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 8.Sarkizova S., Klaeger S., Le P.M., Li L.W., Oliveira G., Keshishian H., Hartigan C.R., Zhang W., Braun D.A., Ligon K.L., Bachireddy P., Zervantonakis I.K., Rosenbluth J.M., Ouspenskaia T., Law T., et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020;38:199–209. doi: 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M.E., Werner A., Hein R., H Busch D., Peschel C., et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 2016;7:13404. doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong C., Marino F., Pak H., Racle J., Daniel R.T., Müller M., Gfeller D., Coukos G., Bassani-Sternberg M. High-throughput and sensitive immunopeptidomics platform reveals profound interferonγ-mediated remodeling of the human leukocyte antigen (HLA) ligandome. Mol. Cell Proteomics. 2018;17:533–548. doi: 10.1074/mcp.TIR117.000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löffler M.W., Kowalewski D.J., Backert L., Bernhardt J., Adam P., Schuster H., Dengler F., Backes D., Kopp H.G., Beckert S., Wagner S., Königsrainer I., Kohlbacher O., Kanz L., Königsrainer A., et al. Mapping the HLA ligandome of colorectal cancer reveals an imprint of malignant cell transformation. Cancer Res. 2018;78:4627–4641. doi: 10.1158/0008-5472.CAN-17-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shraibman B., Barnea E., Kadosh D.M., Haimovich Y., Slobodin G., Rosner I., López-Larrea C., Hilf N., Kuttruff S., Song C., Britten C., Castle J., Kreiter S., Frenzel K., Tatagiba M., et al. Identification of tumor antigens among the HLA peptidomes of glioblastoma tumors and plasma. Mol. Cell Proteomics. 2018;17:2132–2145. doi: 10.1074/mcp.RA118.000792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Escobar H., Reyes-Vargas E., Jensen P.E., Delgado J.C., Crockett D.K. Utility of characteristic QTOF MS/MS fragmentation for MHC class I peptides. J. Proteome Res. 2011;10:2494–2507. doi: 10.1021/pr101272k. [DOI] [PubMed] [Google Scholar]
- 14.Marcu A., Bichmann L., Kuchenbecker L., Kowalewski D., Freudenmann L., Backert L., Mühlenbruch L., Szolek A., Lübke M., Wagner P., Engler T., Matovina S., Wang J., Hauri-Hohl M., Martin R., et al. HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T-cell-based cancer immunotherapy. J. Immunother. Cancer. 2021;9:e002071. doi: 10.1136/jitc-2020-002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keskin D.B., Reinhold B.B., Zhang G.L., Ivanov A.R., Karger B.L., Reinherz E.L. Physical detection of influenza A epitopes identifies a stealth subset on human lung epithelium evading natural CD8 immunity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2151–2156. doi: 10.1073/pnas.1423482112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keskin D.B., Reinhold B., Lee S.Y., Zhang G., Lank S., O'Connor D.H., Berkowitz R.S., Brusic V., Kim S.J., Reinherz E.L. Direct identification of an HPV-16 tumor antigen from cervical cancer biopsy specimens. Front. Immunol. 2011;2:75. doi: 10.3389/fimmu.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers S.A., Rhoads A., Cocco A.R., Peckner R., Haber A.L., Schweitzer L.D., Krug K., Mani D.R., Clauser K.R., Rozenblatt-Rosen O., Hacohen N., Regev A., Carr S.A. Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated murine immune cells. Mol. Cell Proteomics. 2019;18:995–1009. doi: 10.1074/mcp.RA118.001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebert A.S., Prasad S., Belford M.W., Bailey D.J., McAlister G.C., Abbatiello S.E., Huguet R., Wouters E.R., Dunyach J.J., Brademan D.R., Westphall M.S., Coon J.J. Comprehensive Single-Shot Proteomics with FAIMS on a Hybrid Orbitrap Mass Spectrometer. Anal. Chem. 2018;90:9529–9537. doi: 10.1021/acs.analchem.8b02233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfammatter S., Bonneil E., Lanoix J., Vincent K., Hardy M.P., Courcelles M., Perreault C., Thibault P. Extending the comprehensiveness of immunopeptidome analyses using isobaric peptide labeling. Anal. Chem. 2020;92:9194–9204. doi: 10.1021/acs.analchem.0c01545. [DOI] [PubMed] [Google Scholar]
- 20.Pfammatter S., Bonneil E., McManus F.P., Prasad S., Bailey D.J., Belford M., Dunyach J.J., Thibault P. A novel differential ion mobility device expands the depth of proteome coverage and the sensitivity of multiplex proteomic measurements. Mol. Cell Proteomics. 2018;17:2051–2067. doi: 10.1074/mcp.TIR118.000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Udeshi N.D., Mani D.C., Satpathy S., Fereshetian S., Gasser J.A., Svinkina T., Olive M.E., Ebert B.L., Mertins P., Carr S.A. Rapid and deep-scale ubiquitylation profiling for biology and translational research. Nat. Commun. 2020;11:359. doi: 10.1038/s41467-019-14175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickham H. Springer Science & Business Media; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 24.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giacomo O., Stromhaug K., Klaeger S., Kula T., Frederick D., Le P., Forman J., Huang T., Li S., Zhang W., Xu Q., Cieri N., Clauser K.R., Shukla S.A., Neuberg D., et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature. 2021;596:119–125. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen C.J., Li Y.F., El-Gamil M., Robbins P.F., Rosenberg S.A., Morgan R.A. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 2007;67:3898–3903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haga-Friedman A., Horovitz-Fried M., Cohen C.J. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T cell functional avidity. J. Immunol. 2012;188:5538–5546. doi: 10.4049/jimmunol.1103020. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z., Anandappa A.J., Sun J., Kim J., Leet D.E., Bozym D.J., Chen C., Williams L., Shukla S.A., Zhang W., Tabbaa D., Steelman S., Olive O., Livak K.J., Kishi H., et al. A cloning and expression system to probe T-cell receptor specificity and assess functional avidity to neoantigens. Blood. 2018;132:1911–1921. doi: 10.1182/blood-2018-04-843763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabb D.L., MacCoss M.J., Wu C.C., Anderson S.D., Yates J.R., 3rd Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal. Chem. 2003;75:2470–2477. doi: 10.1021/ac026424o. [DOI] [PubMed] [Google Scholar]
- 30.Demmers L.C., Heck A.J.R., Wu W. Pre-fractionation extends but also creates a bias in the detectable HLA class Ι ligandome. J. Proteome Res. 2019;18:1634–1643. doi: 10.1021/acs.jproteome.8b00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mommen G.P., Frese C.K., Meiring H.D., van Gaans-van den Brink J., de Jong A.P., van Els C.A., Heck A.J. Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD) Proc. Natl. Acad. Sci. U. S. A. 2014;111:4507–4512. doi: 10.1073/pnas.1321458111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell A.W., Ramarathinam S.H., Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 2019;14:1687–1707. doi: 10.1038/s41596-019-0133-y. [DOI] [PubMed] [Google Scholar]
- 33.Nicastri A., Liao H., Muller J., Purcell A.W., Ternette N. The choice of HLA-associated peptide enrichment and purification strategy affects peptide yields and creates a bias in detected sequence repertoire. Proteomics. 2020;20 doi: 10.1002/pmic.201900401. [DOI] [PubMed] [Google Scholar]
- 34.Sturm T., Sautter B., Wörner T.P., Stevanović S., Rammensee H.G., Planz O., Heck A.J.R., Aebersold R. Mild acid elution and MHC immunoaffinity chromatography reveal similar albeit not identical profiles of the HLA class I immunopeptidome. J. Proteome Res. 2021;20:289–304. doi: 10.1021/acs.jproteome.0c00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., McAlpine P.L., Heberling M.L., Elias J.E. Automated ligand purification platform accelerates immunopeptidome analysis by mass spectrometry. J. Proteome Res. 2021;20:393–408. doi: 10.1021/acs.jproteome.0c00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong Y., Liang Y., Motamedchaboki K., Huguet R., Truong T., Zhao R., Shen Y., Lopez-Ferrer D., Zhu Y., Kelly R.T., Kelly Improved single-cell proteome coverage using narrow-bore packed NanoLC columns and ultrasensitive mass spectrometry. Anal. Chem. 2020;92:2665–2671. doi: 10.1021/acs.analchem.9b04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bekker-Jensen D.B., Martínez-Val A., Steigerwald S., Rüther P., Fort K.L., Arrey T.N., Harder A., Makarov A., Olsen J.V. A compact quadrupole-orbitrap mass spectrometer with FAIMS interface improves proteome coverage in short LC gradients. Mol. Cell Proteomics. 2020;19:716–729. doi: 10.1074/mcp.TIR119.001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klatt M., Mack K., Bai Y., Aretz Z., Nathan L., Mun S.S., Dao T., Scheinberg D. Solving an MHC allele-specific bias in the reported immunopeptidome. JCI Insight. 2020;5:e141264. doi: 10.1172/jci.insight.141264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra, PSMs, and the protein sequence databases used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000087743/. Raw files and skyline documents for the MRM experiment have been deposited in the ProteomeXchange Consortium (PXD027165) via Panorama Public https://panoramaweb.org/0HgG5Q.url.