Crystal structures of FolM alternative dihydrofolate reductase 1 from Brucella suis and Brucella canis reveal prototypical NADPH-dependent short-chain reductases with structural similarity to protozoan pteridine reductases that are potential drug targets.
Keywords: oxidoreductases, short-chain dehydrogenase/reductase family, dihydrofolate reductases, NADPH, Brucella suis, Brucella canis, Seattle Structural Genomics Center for Infectious Disease, SSGCID
Abstract
Members of the bacterial genus Brucella cause brucellosis, a zoonotic disease that affects both livestock and wildlife. Brucella are category B infectious agents that can be aerosolized for biological warfare. As part of the structural genomics studies at the Seattle Structural Genomics Center for Infectious Disease (SSGCID), FolM alternative dihydrofolate reductases 1 from Brucella suis and Brucella canis were produced and their structures are reported. The enzymes share ∼95% sequence identity but have less than 33% sequence identity to other homologues with known structure. The structures are prototypical NADPH-dependent short-chain reductases that share their highest tertiary-structural similarity with protozoan pteridine reductases, which are being investigated for rational therapeutic development.
1. Introduction
Brucellosis is the most common bacterial zoonotic disease and is caused by the bacterial genus Brucella, which infects humans who consume contaminated animal products, or through contact with infected animals and their secretions (Ducrotoy et al., 2016 ▸; Godfroid, Al Dahouk et al., 2013 ▸). Brucella are classified as category B infectious agents that can be aerosolized (de Figueiredo et al., 2015 ▸). Serological evidence suggests that human brucellosis is misdiagnosed as malaria or other febrile diseases in sub-Saharan Africa (Ducrotoy et al., 2017 ▸). Brucellosis is highly contagious and affects economically important livestock and wild animals globally (Ducrotoy et al., 2017 ▸; Godfroid, Garin-Bastuji et al., 2013 ▸; Godfroid et al., 2011 ▸; Megersa et al., 2011 ▸). While brucellosis has been eradicated in cattle and small ruminants in a few countries, it remains endemic globally within a wide range of animal hosts (Moreno, 2014 ▸).
Current control approaches for brucellosis include vaccination, education and basic hygiene; however, these strategies have not effectively reduced the disease burden due to cost and other issues (Ariza et al., 2007 ▸). Notably, current vaccines are species-specific and are devastating to pregnant livestock, and cultural practices among rural dwellers and nomadic groups that rear animals are often incompatible with disease control (Ducrotoy et al., 2017 ▸; Godfroid, Al Dahouk et al., 2013 ▸). There is a continued need to develop new cost-effective approaches to treat infected animals, including the rational design or repurposing of small molecules that target enzymes that are vital for bacterial survival. The Seattle Structural Genomics Center for Infectious Disease (SSGCID) has determined the crystal structures of many target enzymes, including FolM alternative dihydrofolate reductase 1 from two Brucella species, B. suis and B. canis. Dihydrofolate reductase reduces dihydrofolic acid to tetrahydrofolic acid using reduced nicotinamide adenine dinucleotide phosphate (NADPH) as the electron donor. While this reaction is catalyzed by the enzyme dihydrofolate reductase (DHFR) in mammals and other organisms, some bacteria have an alternative pathway for reduced folate biosynthesis using FolM alternative dihydrofolate reductase 1 (Levin et al., 2004 ▸). Here, we present the crystal structures of FolM alternative dihydrofolate reductase 1 from two Brucella species, B. suis (BsFolM) and B. canis (BcFolM).
BsFolM and BcFolM are 95% identical in sequence. BLAST alignment of the protein sequences against the Protein Data Bank (PDB) reveals the most similar proteins to be Tt0495 from Thermus thermophilus HB8 (Pampa et al., 2014 ▸) with ∼32% sequence identity and ∼85% coverage; Leishmania major pteridine reductase (Schüttelkopf et al., 2005 ▸) with ∼30% sequence identity and ∼90% coverage; Mycobacterium smegmatis short-chain reductase (Blaise et al., 2017 ▸) with ∼33% sequence identity and ∼85% coverage; and Trypanosoma cruzi pteridine reductase 2 (Schormann et al., 2005 ▸) with ∼30% sequence identity and ∼88% coverage. The reported crystal structures of BsFolM and BcFolM are the first steps towards identifying new therapeutics for brucellosis.
2. Materials and methods
2.1. Macromolecule production
Cloning, expression and purification were conducted as part of the Seattle Structural Genomics Center for Infectious Disease (SSGCID) following standard protocols described previously (Myler et al., 2009 ▸; Stacy et al., 2011 ▸; Bryan et al., 2011 ▸; Choi et al., 2011 ▸; Serbzhinskiy et al., 2015 ▸). The full-length FolM genes from B. suis (UniProt A0A0H3G2T6) and B. canis (UniProt A9MA73) were PCR-amplified from genomic DNA using the primers shown in Tables 1 ▸ and 2 ▸, respectively. The resultant amplicons were cloned into the ligation-independent cloning (LIC; Aslanidis & de Jong, 1990 ▸) expression vector pBG1861 encoding a noncleavable 6×His fusion tag (MAHHHHHHM-ORF). The plasmids containing A0A0H3G2T6 and A9MA73 were tested for expression and 2 l of culture was grown using auto-induction medium (Studier, 2005 ▸) in a LEX Bioreactor (Epiphyte Three). The expression clones for BrsuA.00010.a.B1.GE36748 and BrcaA.00010.a.B1.GE38297 are available at https://www.ssgcid.org/available-materials/expression-clones/.
Table 1. Production of FolM alternative dihydrofolate reductase 1 from B. suis .
| Source organism | Brucella suis 1330 |
| DNA source | Dr Jean-Jacques Letesson (University of Namur, Belgium) |
| Forward primer | 5′-CTCACCACCACCACCACCATATGGTGTTGAATGATCCCGAAGC-3′ |
| Reverse primer | 5′-ATCCTATCTTACTCACTTATTCGGTAATTCCTGCAATGTCGG-3′ |
| Expression vector | pBG1861 |
| Expression host | E. coli BL21(DE3)R3 Rosetta cells |
| Complete amino-acid sequence of the construct produced | MAHHHHHHMLNDPEARMVANCPVLVTGGARRIGKAIVEDLASHGFPVAIHCNRSLDEGEAIANRINDSGGNACVVQADLEGDVRGLVKQASDRIGPIRLLVNNASLFQEDKVGALDMALWDRHFAVHLKTPVILAEDMRKALPEDQDGLVVNIIDQRVWKLNPQFFSYTLSKSALWNATRTLAQALAPRIRVNAIAPGPTLPSERQRPEDFERQVSKLPLQRAPELPEFGRTVRYFWENRSITGQMIALDGGQHLAWETPDIAGITE |
Table 2. Production of FolM alternative dihydrofolate reductase 1 from B. canis .
| Source organism | Brucella canis RM-666 (NCTC 10854) |
| DNA source | ATCC 23365 |
| Forward primer | 5′-CTCACCACCACCACCACCATATGGTGTTGAATGATCCCGAAGC-3′ |
| Reverse primer | 5′-ATCCTATCTTACTCACTTATTCGGTAATTCCTGCAATGTCGG-3′ |
| Expression vector | pBG1861 |
| Expression host | E. coli BL21(DE3)R3 Rosetta cells |
| Complete amino-acid sequence of the construct produced | MAHHHHHHMVLNDPEARMVANCPVLVTGGARRIGKAIVEDLASHGFPVAIHCNRSLDEGEAIANRINDSGGNACVVQADLEGDVRGLVKQASDRIGPIRLLVNNASLFQEDKVGALDMALWDRHFAVHLKTPVILAEDMRKALPEDQDGLVVNIIDQRVWKLNPQFFSYTLSKTALWNATRTLAQALAPRIRVNAIAPGPTLPSERQRPEDFERQVSKLPLQRAPELPEFGRTVRYFWENRSITGQMIALDGGQHLAWETPDIAELPNK |
His-BsFolM and His-BcFolM were purified in a two-step protocol consisting of an Ni2+-affinity chromatography step and size-exclusion chromatography (SEC). All chromatography runs were performed on an ÄKTApurifier 10 (GE) using automated IMAC and SEC programs according to previously described procedures (Bryan et al., 2011 ▸). Thawed bacterial pellets were lysed by sonication in 200 ml lysis buffer [25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 0.5% CHAPS, 30 mM imidazole, 10 mM MgCl2, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 250 µg ml−1 4-benzenesulfonyl fluoride hydrochloride (AEBSF), 0.025% azide]. After sonication, the crude lysate was clarified with 20 µl (25 units µl−1) benzonase and incubated while mixing at room temperature for 45 min. The lysate was then clarified by centrifugation at 10 000 rev min−1 for 1 h using a Sorvall centrifuge (Thermo Scientific). In the IMAC step, the clarified supernatant was passed over an Ni–NTA HisTrap FF 5 ml column (GE Healthcare) pre-equilibrated with loading buffer (25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 30 mM imidazole, 1 mM TCEP, 0.025% sodium azide). The column was washed with 20 column volumes (CV) of loading buffer and eluted with a linear gradient over 7 CV of loading buffer plus 250 mM imidazole. Peak fractions, as determined by UV at 280 nm, were pooled and concentrated. A SEC column (Superdex 75, GE Healthcare) was equilibrated with running buffer (25 mM HEPES pH 7.0, 500 mM NaCl, 5% glycerol, 2 mM DTT, 0.025% azide). The peak fractions were collected and analyzed by SDS–PAGE. The SEC peak fractions eluted as a single large peak at a molecular mass of ∼77 kDa, suggesting an oligomer, most likely dimeric, trimeric or tetrameric enzyme. The peak fractions were pooled and concentrated to 28.5 mg ml−1 (His-BsFolM) or 32.3 mg ml−1 (His-BcFolM) as assessed by the OD280 using an Amicon concentration system (Millipore). Aliquots of 200 µl were flash-frozen in liquid nitrogen and stored at −80°C until use for crystallization.
2.2. Crystallization
Purified His-BsFolM and His-BcFolM were screened for crystallization in 96-well sitting-drop plates against the JCSG++ HTS (Jena Bioscience) and MCSG1 (Molecular Dimensions) crystallization screens. Equal volumes of protein solution (0.4 µl) and precipitant solution were set up at 290 K against 80 µl reservoir in sitting-drop vapor-diffusion format. Before crystallization, NADPH was added to the protein solution to a final concentration of 4 mM (BsFolM) or 6 mM (BcFolM). The precipitant solution was MCSG-1 condition A1 (Tables 3 ▸ and 4 ▸). The crystals were harvested and cryoprotected with crystallization solution supplemented with 20% ethylene glycol before flash-cooling in liquid nitrogen.
Table 3. Crystallization of FolM alternative dihydrofolate reductase 1 from B. suis (BsFolM).
| Method | Vapor diffusion, sitting drop |
| Plate type | Rigaku Reagents XJR |
| Temperature (K) | 290 |
| Crystallization | BsFolM (19 mg ml−1) incubated with 4 mM NADPH, mixed 1:1 with MCSG1 condition A1 [20%(w/v) PEG 8000, 100 mM HEPES pH 7.5] |
| Composition of reservoir solution | 20%(w/v) PEG 8000, 100 mM HEPES pH 7.5 |
| Volume and ratio of drop | 0.4 µl:0.4 µl |
| Volume of reservoir (µl) | 80 |
Table 4. Crystallization of FolM alternative dihydrofolate reductase 1 from B. canis (BcFolM).
| Method | Vapor diffusion, sitting drop |
| Plate type | Rigaku Reagents XJR |
| Temperature (K) | 290 |
| Crystallization | BcFolM (32.3 mg ml−1) incubated with 6 mM NADPH, mixed 1:1 with 20%(w/v) PEG 8000, 100 mM HEPES pH 7.5 |
| Composition of reservoir solution | 20%(w/v) PEG 8000, 100 mM HEPES pH 7.5 |
| Volume and ratio of drop | 0.4 µl:0.4 µl |
| Volume of reservoir (µl) | 80 |
2.3. Data collection and processing
Data were collected at 100 K at the Advanced Photon Source, Argonne National Laboratory (Table 5 ▸). The data were reduced with XSCALE (Kabsch, 2010 ▸). Raw X-ray diffraction images are available at the Integrated Resource for Reproducibility in Macromolecular Crystallography at https://www.proteindiffraction.org/.
Table 5. Data-collection and processing statistics for FolM alternative dihydrofolate reductase 1 from B. suis (PDB entry 5tgd, BsFolM) and B. canis (PDB entry 5bt9, BcFolM).
| PDB code | 5tgd | 5bt9 |
|---|---|---|
| Diffraction source | APS beamline 21-ID-F | APS beamline 21-ID-F |
| Wavelength (Å) | 0.97872 | 0.97872 |
| Temperature (K) | 100 | 100 |
| Detector | RayoniX MX-300 CCD | MAR Mosaic 225 mm CCD |
| Crystal-to-detector distance (mm) | 220 | 130 |
| Rotation range per image (°) | 1 | 1 |
| Total rotation range (°) | 200 | 220 |
| Space group | P21 | P21 |
| a, b, c (Å) | 76.35, 76.52, 98.26 | 76.57, 75.60, 99.18 |
| α, β, γ (°) | 90, 109.47, 90 | 90, 109.23, 90 |
| Mosaicity (°) | 0.180 | 0.168 |
| Resolution range (Å) | 50–1.70 (1.74–1.70) | 50.0–1.50 (1.54–1.50) |
| Total No. of reflections | 491527 (36146) | 783894 (57336) |
| No. of unique reflections | 116233 (8502) | 164992 (11908) |
| Completeness (%) | 99.0 (98.4) | 96.4 (94.6) |
| Multiplicity | 4.22 (4.25) | 4.8 (4.8) |
| 〈I/σ(I)〉 | 19.84 (2.87) | 18.26 (3.39) |
| R r.i.m. † | 0.050 (0.486) | 0.053 (0.553) |
| Overall B factor from Wilson plot (Å2) | 18.85 | 15.63 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
Both structures were solved by molecular replacement. BcFolM was solved with BALBES (Long et al., 2008 ▸) with PDB entry 2uvd, a 3-oxoacyl-(acyl carrier protein) reductase (Ba3989) from Bacillus anthracis (Zaccai et al., 2008 ▸), as the search model. BsFolM was solved with MoRDa (Vagin & Lebedev, 2015 ▸) using BcFolM (PDB entry 5bt9) as the search model. Both structures were refined using iterative cycles of refinement in Phenix (Liebschner et al., 2019 ▸) followed by manual structure-rebuilding cycles in Coot (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸). The quality of both structures was checked using MolProbity (Chen et al., 2010 ▸). All data-reduction and refinement statistics are shown in Table 6 ▸. The BsFolM structure was refined to a resolution of 1.70 Å, while that of BcFolM was refined to 1.50 Å resolution. Figures depicting the structure were analyzed and prepared using PyMOL (version 1.5; Schrödinger). Multiple sequence alignments were performed using Clustal Omega (Li, 2003 ▸; Sievers et al., 2011 ▸). Coordinates and structure factors have been deposited in the Protein Data Bank (https://www.rcsb.org/) as entries 5tgd and 5bt9 for BsFolM and BcFolM, respectively.
Table 6. Structure-solution and refinement of FolM alternative dihydrofolate reductase 1 from B. suis (PDB entry 5tgd) and B. canis (PDB entry 5bt9).
| PDB code | 5tgd | 5bt9 |
|---|---|---|
| Resolution range (Å) | 50–1.70 (1.74–1.70) | 36.15–1.50 (1.51–1.50) |
| Completeness (%) | 99.1 | 96.2 |
| σ Cutoff | F > 1.34σ(F) | F > 1.35σ(F) |
| No. of reflections, working set | 116170 (8678) | 156072 (4573) |
| No. of reflections, test set | 1785 (153) | 8084 (235) |
| Final R cryst | 0.163 (0.2816) | 0.169 (0.2486) |
| Final R free | 0.198 (0.2825) | 0.188 (0.2877) |
| No. of non-H atoms | ||
| Protein | 7503 | 7542 |
| Ligand | 214 | 192 |
| Solvent | 748 | 724 |
| Total | 8465 | 8460 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.006 | 0.006 |
| Angles (°) | 0.828 | 1.132 |
| Average B factors (Å2) | ||
| Protein | 31.9 | 27.7 |
| Ligand | 33.5 | 26.6 |
| Water | 40.0 | 35.1 |
| Ramachandran plot | ||
| Most favored (%) | 96 | 95 |
| Allowed (%) | 4 | 5 |
3. Results and discussion
The structures of FolM alternative dihydrofolate reductase 1 from B. suis (BsFolM) and B. canis (BcFolM) were determined in the monoclinic space group P21 with four monomers in the asymmetric unit (Fig. 1 ▸). PDBsum analysis (http://www.ebi.ac.uk/pdbsum/) indicates that each monomer interacts with three other monomers, with two large interactions and one smaller interaction. The buried surface areas of the interactions are ∼1400, ∼1300 and ∼770 Å2 per monomer. These surface areas involve 31, 25 and 14 interface amino acids per monomer, respectively. The interface interactions are mostly hydrogen bonds and other nonbonded contacts. The tetramers are similar and superpose with an r.m.s.d. of ∼0.5 Å (Fig. 1 ▸ c). The tetramer is the prototypical short-chain dehydrogenase/reductase (SDR) tetramer, suggesting that the single SEC peak may indeed correspond to a tetramer.
Figure 1.
(a) BsFolM and (b) BcFolM assemble as prototypical FolM alternative dihydrofolate reductase 1 tetramers. (c) The BsFolM and BcFolM tetramers are almost identical based on their structural alignment.
Each monomer has the extended double-Rossmann fold of NADPH-dependent SDRs with a central seven-stranded parallel β-sheet sandwiched between two pairs of three α-helices. Both the BsFolM and BcFolM structures were co-crystallized with a cofactor (NADPH). The monomers are virtually identical, with an r.m.s.d. of ∼0.17 Å on superposing all main-chain atoms of both structures (Fig. 1 ▸).
The most similar structures to BsFolM and BcFolM were identified by PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm) analysis using the default threshold cutoffs of 70% for the percentage of the secondary structure of the target chain identified in the query protein and of the secondary structure of the query chain (Krissinel & Henrick, 2004 ▸). The most similar structures are protozoan pteridine reductases (Khalaf et al., 2014 ▸; Tulloch et al., 2010 ▸; Schormann et al., 2005 ▸). These structures share ∼29% sequence identity with BsFolM and BcFolM, and their main-chain Cα atoms align with an r.m.s.d. of ∼1.5 Å. These protozoan pteridine reductases are more similar to BsFolM and BcFolM than to the structures from Bacillus anthracis (Zaccai et al., 2008 ▸), Streptomyces (Wang et al., 2014 ▸), Serratia marcescens (Liu et al., 2018 ▸), Thermus thermophilus (Asada et al., 2009 ▸) or other bacteria.
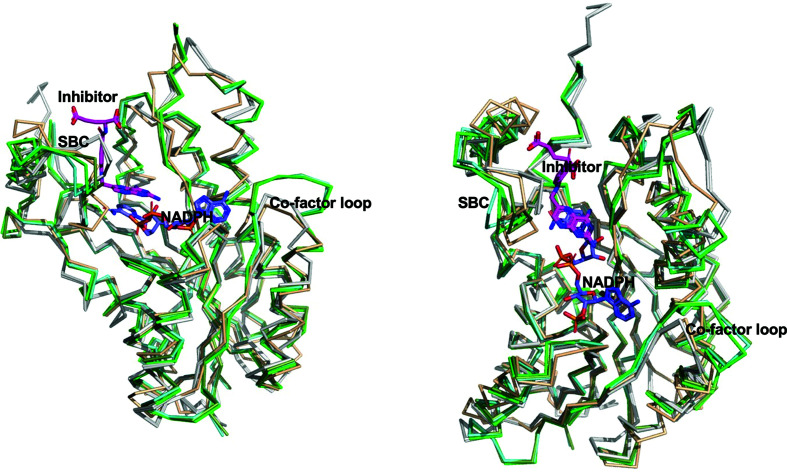
The BsFolM and BcFolM structures are in the closed conformation with ordered substrate-binding loops, as observed in protozoan pteridine reductases (Khalaf et al., 2014 ▸; Tulloch et al., 2010 ▸; Schormann et al., 2005 ▸; Schüttelkopf et al., 2005 ▸). Despite being identified as the closest structures by PDBeFold, the Trypanosoma proteins share a lower sequence identity to BsFolM and BcFolM than the MR search model from B. anthracis (Zaccai et al., 2008 ▸), which shares ∼30% sequence identity with both proteins (Fig. 2 ▸). Both structures have structural differences from the molecular-replacement search model, the 3-oxoacyl-(acyl carrier protein) reductase (Ba3989) from Bacillus anthracis, and have an r.m.s.d. of ∼2.12 Å on superposing all main-chain atoms (Fig. 3 ▸).
Figure 2.
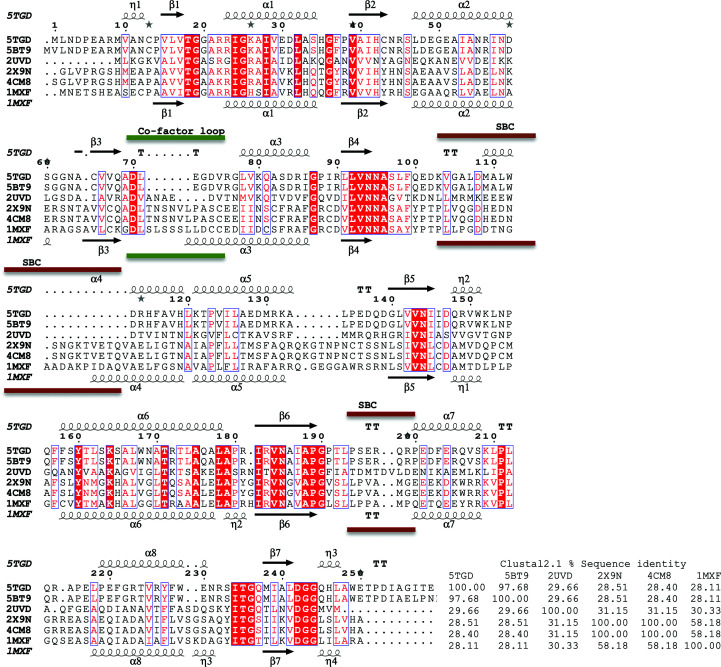
Structural and primary-sequence alignment of FolM alternative dihydrofolate reductase 1 from B. suis (PDB entry 5tgd) and B. canis (PDB entry 5bt9) with the molecular-replacement search model 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (PDB entry 2uvd) and protozoan structures (Trypanosoma brucei pteridine reductase with cyromazine, PDB entry 2x9n; T. brucei pteridine reductase ternary complex with cofactor and inhibitor, PDB entry 4cm8; T. cruzi pteridine reductase, PDB entry 1mxf). The secondary-structure elements are shown as follows: α-helices are shown as large coils, 310-helices are shown as small coils labeled η, β-strands are shown as arrows labeled β and β-turns are labeled TT. Identical residues are shown on a red background, with conserved residues in red and conserved regions in blue boxes. Regions of greatest variability within the core of the protein are identified with brown lines and labeled SBC due to their proximity to the substrate-binding cavity.
Figure 3.
Two views comparing BsFolM and BcFolM monomers with similar structures. The BsFolM and BcFolM monomers (gray) have the prototypical double-Rossmann fold of NADPH-dependent short-chain dehydrogenase/reductases observed in the molecular-replacement search model (tan) and protozoan pteridine reductase (green). The superposed protozoan structures are Trypanosoma brucei pteridine reductase with cyromazine (PDB entry 2x9n; cyan green), T. brucei pteridine reductase ternary complex with cofactor and inhibitor (PDB entry 4cm8; dark green) and T. cruzi pteridine reductase (PDB entry 1mxf; light green). The cofactor NADPH is shown in blue sticks, while the inhibitor from PDB entry 1mxf is shown as magenta sticks in the substrate-binding cavity. As in Fig. 2 ▸, SBC stands for substrate-binding cavity.
While the cofactor-binding cavities of BsFolM, BcFolM and the Trypanosoma proteins are well conserved, there is a loop insertion (labeled in green; Fig. 2 ▸). This loop (labeled the cofactor loop in Fig. 3 ▸) points away from the cofactor (NADPH) and aligns well in both BsFolM and BcFolM. Interestingly, this loop is conserved in the protozoan enzymes and forms a 6.5 Å larger cavity than that observed in the Brucella enzymes (both BsFolM and BcFolM; Fig. 3 ▸). Apart from this loop region, the cofactor-binding cavity is very similar in these enzymes. Furthermore, the residues involved in NADPH binding are well conserved (Fig. 4 ▸).
Figure 4.
LIGPLOT diagrams reveal well conserved NADPH-binding cavities in FolM alternative dihydrofolate reductase 1 from B. suis (PDB entry 5tgd) and B. canis (PDB entry 5bt9), Trypanosoma brucei pteridine reductase with cyromazine (PDB entry 2x9n) T. brucei pteridine reductase in a ternary complex with cofactor and inhibitor (PDB entry 4cm8) and T. cruzi pteridine reductase (PDB entry 1mxf). Identical amino-acid residues are circled.
As expected, the substrate-binding cavity of each protein shows the greatest structural difference (Figs. 2 ▸ and 3 ▸). This structural variability is believed to allow substrate specificity among SDRs. While the substrates of BsFolM and BcFolM are unknown, their substrate-binding cavities are large enough to accommodate the inhibitors identified by rational therapeutics discovery for human African trypanosomiasis and Chagas disease. There are >150 structures of complexes of protozoan pteridine reductases with unique inhibitors deposited in the Protein Data Bank (Khalaf et al., 2014 ▸; Tulloch et al., 2010 ▸; Schormann et al., 2005 ▸) that can serve as starting points for the discovery of therapeutics for brucellosis.
4. Conclusions
The high-resolution structures of FolM alternative dihydrofolate reductase 1 from B. suis and B. canis have prototypical NADPH-dependent short-chain reductase topology and structural similarity to the well characterized protozoan pteridine reductases. Despite their low sequence identity, their structural similarity to the protozoan pteridine reductases may accelerate drug-repurposing efforts.
Supplementary Material
PDB reference: BsFolM, 5tgd
PDB reference: BcFolM, 5bt9
Acknowledgments
The SSGCID consortium is directed by Dr Peter Myler (principal investigator) and comprises many different scientists working at multiple centers towards determining the three-dimensional structures of proteins from biodefense organisms and emerging infectious diseases. In particular, we would like to thank the SSGCID cloning, protein production and X-ray crystallography groups at the Center for Global Infectious Disease Research, the University of Washington and UCB.
Funding Statement
This work was funded by National Institute of Allergy and Infectious Diseases grants HHSN272201700059C, HHSN272201200025C, and HHSN272200700057C to Peter Myler and Peter Myler; National Institute of General Medical Sciences grant 1U01GM138433 to Oluwatoyin A. Asojo.
References
- Ariza, J., Bosilkovski, M., Cascio, A., Colmenero, J. D., Corbel, M. J., Falagas, M. E., Memish, Z. A., Roushan, M. R., Rubinstein, E., Sipsas, N. V., Solera, J., Young, E. J., Pappas, G., International Society of Chemotherapy & Institute of Continuing Medical Education of Ioannina (2007). PLoS Med. 4, e317. [DOI] [PMC free article] [PubMed]
- Asada, Y., Endo, S., Inoue, Y., Mamiya, H., Hara, A., Kunishima, N. & Matsunaga, T. (2009). Chem. Biol. Interact. 178, 117–126. [DOI] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Blaise, M., Van Wyk, N., Banères-Roquet, F., Guérardel, Y. & Kremer, L. (2017). Biochem. J. 474, 907–921. [DOI] [PubMed]
- Bryan, C. M., Bhandari, J., Napuli, A. J., Leibly, D. J., Choi, R., Kelley, A., Van Voorhis, W. C., Edwards, T. E. & Stewart, L. J. (2011). Acta Cryst. F67, 1010–1014. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Choi, R., Kelley, A., Leibly, D., Nakazawa Hewitt, S., Napuli, A. & Van Voorhis, W. (2011). Acta Cryst. F67, 998–1005. [DOI] [PMC free article] [PubMed]
- Ducrotoy, M., Bertu, W. J., Matope, G., Cadmus, S., Conde-Álvarez, R., Gusi, A. M., Welburn, S., Ocholi, R., Blasco, J. M. & Moriyón, I. (2017). Acta Trop. 165, 179–193. [DOI] [PubMed]
- Ducrotoy, M. J., Conde-Álvarez, R., Blasco, J. M. & Moriyón, I. (2016). Vet. Immunol. Immunopathol. 171, 81–102. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Figueiredo, P. de, Ficht, T. A., Rice-Ficht, A., Rossetti, C. A. & Adams, L. G. (2015). Am. J. Pathol. 185, 1505–1517. [DOI] [PMC free article] [PubMed]
- Godfroid, J., Al Dahouk, S., Pappas, G., Roth, F., Matope, G., Muma, J., Marcotty, T., Pfeiffer, D. & Skjerve, E. (2013). Comp. Immunol. Microbiol. Infect. Dis. 36, 241–248. [DOI] [PubMed]
- Godfroid, J., Garin-Bastuji, B., Saegerman, C. & Blasco, J. M. (2013). Rev. Sci. Tech. OIE, 32, 27–42. [DOI] [PubMed]
- Godfroid, J., Scholz, H. C., Barbier, T., Nicolas, C., Wattiau, P., Fretin, D., Whatmore, A. M., Cloeckaert, A., Blasco, J. M., Moriyon, I., Saegerman, C., Muma, J. B., Al Dahouk, S., Neubauer, H. & Letesson, J. J. (2011). Prev. Vet. Med. 102, 118–131. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Khalaf, A. I., Huggan, J. K., Suckling, C. J., Gibson, C. L., Stewart, K., Giordani, F., Barrett, M. P., Wong, P. E., Barrack, K. L. & Hunter, W. N. (2014). J. Med. Chem. 57, 6479–6494. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Levin, I., Giladi, M., Altman-Price, N., Ortenberg, R. & Mevarech, M. (2004). Mol. Microbiol. 54, 1307–1318. [DOI] [PubMed]
- Li, K. B. (2003). Bioinformatics, 19, 1585–1586. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Liu, J.-S., Kuan, Y.-C., Tsou, Y., Lin, T.-Y., Hsu, W.-H., Yang, M.-T., Lin, J.-Y. & Wang, W.-C. (2018). Sci. Rep. 8, 2316. [DOI] [PMC free article] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Megersa, B., Biffa, D., Abunna, F., Regassa, A., Godfroid, J. & Skjerve, E. (2011). Trop. Anim. Health Prod. 43, 651–656. [DOI] [PubMed]
- Moreno, E. (2014). Front. Microbiol. 5, 213. [DOI] [PMC free article] [PubMed]
- Myler, P. J., Stacy, R., Stewart, L., Staker, B. L., Van Voorhis, W. C., Varani, G. & Buchko, G. W. (2009). Infect. Disord. Drug Targets, 9, 493–506. [DOI] [PMC free article] [PubMed]
- Pampa, K. J., Lokanath, N. K., Kunishima, N. & Rai, R. V. (2014). Acta Cryst. D70, 994–1004. [DOI] [PubMed]
- Schormann, N., Pal, B., Senkovich, O., Carson, M., Howard, A., Smith, C., DeLucas, L. & Chattopadhyay, D. (2005). J. Struct. Biol. 152, 64–75. [DOI] [PubMed]
- Schüttelkopf, A. W., Hardy, L. W., Beverley, S. M. & Hunter, W. N. (2005). J. Mol. Biol. 352, 105–116. [DOI] [PubMed]
- Serbzhinskiy, D. A., Clifton, M. C., Sankaran, B., Staker, B. L., Edwards, T. E. & Myler, P. J. (2015). Acta Cryst. F71, 594–599. [DOI] [PMC free article] [PubMed]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Stacy, R., Begley, D. W., Phan, I., Staker, B. L., Van Voorhis, W. C., Varani, G., Buchko, G. W., Stewart, L. J. & Myler, P. J. (2011). Acta Cryst. F67, 979–984. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Tulloch, L. B., Martini, V. P., Iulek, J., Huggan, J. K., Lee, J. H., Gibson, C. L., Smith, T. K., Suckling, C. J. & Hunter, W. N. (2010). J. Med. Chem. 53, 221–229. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Lebedev, A. (2015). Acta Cryst. A71, s19.
- Wang, H., Zhang, H., Zou, Y., Mi, Y., Lin, S., Xie, Z., Yan, Y. & Zhang, H. (2014). PLoS One, 9, e97996. [DOI] [PMC free article] [PubMed]
- Zaccai, N. R., Carter, L. G., Berrow, N. S., Sainsbury, S., Nettleship, J. E., Walter, T. S., Harlos, K., Owens, R. J., Wilson, K. S., Stuart, D. I. & Esnouf, R. M. (2008). Proteins, 70, 562–567. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: BsFolM, 5tgd
PDB reference: BcFolM, 5bt9