Introduction
The Pharmacogene Variation Consortium (PharmVar) was founded in 2017 to provide the clinical and research communities a repository and standardized nomenclature of genes contributing to variability in drug metabolism and response. Over the past four years, PharmVar has provided the research and clinical pharmacogenetics/genomics communities with essential information for flagship pharmacogenes with well-established drug-gene relationships and published clinical guidelines such as CYP2C9, CYP2C19 and CYP2D6. In this perspective we highlight recent milestones and standardization efforts.
PharmVar: A Global Resource
The number of PharmVar users has steadily increased demonstrating its uniqueness and importance for the scientific community. As shown in Figure 1, PharmVar has grown into a global resource that is regularly visited by members of the scientific community from countries around the world. Over the past year, PharmVar recorded over 28,500 users (33.3% from the US) and over 60,000 and 78,000 sessions and page views, respectively. Furthermore, a survey conducted in August 2020 (n=103 respondents; over 74% of users self-identified as ‘academic, 37% conducting research and 39% performing pharmacogenetic testing) indicated that users access PharmVar daily (8.7%), weekly (48%) or monthly (40%) and an overwhelming majority (94%) indicted that PharmVar is a critical resource for their work. Not surprisingly, CYPs 2D6, 2C19, 2C9, 3A4, 3A5 and 2B6 were the most accessed (in the listed order). Users indicated that PharmVar is the “Best resource for looking up P450 variants”, “PharmVar is vital to my work as a clinician, educator and researcher” and “It is an invaluable resource for identifying important haplotypes in drug metabolism”.
Figure 1. PharmVar User Base.
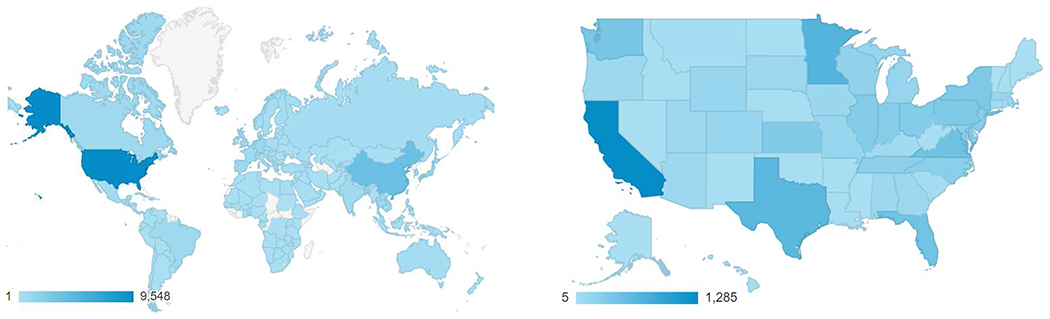
PharmVar is a resource that is globally (A) and nationally (B) utilized (data reflect usage between April 1, 2020 through March 29, 2021). The graded bar indicates number of users; user density is indicated by color intensity. User demographics excludes authors’ institutions to avoid counting PharmVar and PharmGKB site activities towards site use data.
PharmVar Genes
Clinically important pharmacogenes for which Clinical Pharmacogenetics Implementation Consortium (CPIC©) guidelines are available such as CYP2B6, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 have extensively been curated by PharmVar gene experts (Suppl Table 1). These efforts are described in detail in the PharmVar GeneFocus series of review articles which have been published for CYP2B6 (1), CYP2C19 (2) and CYP2D6 (3). These reviews provide a plethora of other information which not only complements CPIC guidelines, but serve as a guide to the gene’s nomenclature, information displayed on the PharmVar gene page, and information regarding the characterization of novel alleles, and more, which is critical information whether one is a clinician or basic scientist.
Several genes including CYP3A4 and CYP2A6 are currently undergoing extensive curation and revisions necessary to make their nomenclature compliant with PharmVar standards (4). Others, including some CYP2C genes and CYP4F2, have been transitioned with minimal curation efforts to allow users to take advantage of PharmVar database features. However, the alleles currently displayed for these genes are likely only a fraction of those existing among populations, which poses a knowledge gap. Several genes including CYP1A1, CYP1A2, CYP1B1 and CYP2E1 are still awaiting curation and require major updates and revisions before their transition into the PharmVar database.
PharmVar has added two important pharmacogenes, NUDT15 (5) and DPYD. For the latter, PharmVar designed a rsID-based gene page format to accommodate the listing of variants rather than haplotype-based star alleles as this was the expert-preferred format moving forward. In addition, PharmVar is introducing SLCO1B1, an important drug transporter gene for which star allele-based nomenclature has been utilized for many years on a self-assignment basis but has never been systemically captured. SLCO1B1 experts have collected and reviewed published information and updated star allele definitions to conform to PharmVar rules and standards. Furthermore, sequence data available to the PharmVar team are being utilized to confirm numerous allele definitions and/or complement incomplete information, as well as discover novel haplotypes. This effort is essential as the PharmVar-developed nomenclature is being used for the CPIC update on SLCO1B1/statin dose recommendations.
Standardization
PharmVar Rules:
One major tenet of PharmVar is to standardize allele definitions across genes. To that end, PharmVar has developed standards (4, 6) and allele definition criteria (6) including a rule-based system that allows all suballeles to be collapsed into a single ‘core’ allele definition, i.e. a core allele represents all suballeles categorized under a star number. Applying these rules may require revising allele definitions. A prime example is CYP2C19*1. As described in more detail in the CYP2C19 GeneFocus (2), one suballele, CYP2C19*1.001 (legacy name CYP2C19*1A), differed from all other *1 suballeles by not having c.991A>G (p.I331V) and thus, did not conform to the PharmVar allele definition criteria. This allele was therefore renamed as CYP2C19*38. This change also triggered the CYP2C19*1 core allele definition to gain c.991A>G (p.I331V) as now all *1 subvariants have this variant. Since p.I331V is believed to not impact function, this variant is typically not genotyped and therefore, in the absence of testing CYP2C19*38, may be defaulted as CYP2C19*1 allele and reported as such. This example highlights that a gene’s genomic reference sequence does not necessarily represent the most commonly observed haplotype, i.e. CYP2C19*1 is considerably more common compared to CYP2C19*38 which matches the genomic NG_008384.3 reference sequence (RefSeq) issued by the NCBI (7).
Revisions of star allele definitions may also be triggered by RefSeq updates. Current genomic and transcript reference sequences often differ from previous version initially used for allele definitions in their 5’ and 3’-portions and intron sequences and occasionally exhibit differences in their coding regions. Such changes may affect variant positions especially if the genomic reference sequence is used for variant reporting as opposed to transcript positions. This is exemplified by CYP2D6 for which variant positions shifted due to intronic insertions/deletions between the legacy sequence and the genomic RefSeq. While PharmVar still provides the M33388 legacy sequence, the field has moved toward using positions according to the current RefSeq. Changes in a gene’s genomic RefSeq may cause ‘variant-switching’ which is exemplified by CYP3A4. Specifically, c.−392 was defined as A>G in the past (i.e., the now discontinued reference sequence AF280107.01 had c.−392A) but is now defined as c.−392G>A since the current RefSeq (NG_008421.1) has c.−392G. Thus, all alleles which had the c.−392A>G SNV in the past now match the RefSeq and consequently no longer show the variant, while all other alleles gained c.−392G>A.
To facilitate keeping track of changes, PharmVar is cross-referencing legacy star allele names on its gene pages to allow backtracking. ‘Variant-switching’ as exemplified above for CYP3A4 and other notable revisions such as alleles that have been merged or retired, are documented in the gene’s ‘Change Log’ file (Tables 1 and 2 therein). Revisions are also itemized and discussed in detail in the PharmVar GeneFocus review series.
Updates such as those described above are often viewed as ‘painful’ or ‘interruptive’ and met with resistance. They are however inevitable and necessary, to not only provide consistent nomenclature across pharmacogenes, but also reflect current knowledge and align our efforts with other accepted standards in the field of genetics.
Locus Reference Genomic (LRG) sequences:
PharmVar has utilized LRG sequences (8) for CYP2B6, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 as these were deemed to be stable references and therefore superior for clinical reporting purposes. However, in the wake of the MANE (Matched Annotation from the NCBI and EMBL-EBI (9)) project, “MANE Select” sequences (based on one well-supported transcript of the per protein-coding locus) are now viewed as the reference standard. A MANE Select sequence perfectly aligns to GRCh38 and represents 100% identity (5’UTR, coding sequence, 3’UTR). Since current genomic RefSeqs are based on their respective MANE Select transcripts, the new reference standard, LRGs are likely not to be issued in the future and are being replaced by MANE Select-derived genomic RefSeqs. Consequently, PharmVar is replacing the LRGs for the above-mentioned genes with their genomic RefSeqs to align itself with the global MANE standardization efforts. Since all LRGs are 100% concordant with their RefSeqs, this update did not affect any star allele annotations.
Human Genome Variation Society nomenclature:
Display of sequence variants on the PharmVar gene pages is not fully compliant with the nomenclature that is authorized by the Human Genome Variation Society (HGVS), the Human Variome Project (HVP) and the Human Genome Organization (HUGO). This nomenclature was developed to describe sequence variations in a uniform and unequivocal manner (10) and has become the global standard, often referred to as “HGVS”. To address this shortcoming, PharmVar now provides HGVS annotations in its Variant Window. As shown and detailed in Figure 2, variant annotations are provided according to HGVS, but also in the more traditional ‘PharmVar’ style many users are accustomed to and wish to maintain. While HGVS may not specify the nucleotide that is deleted or inserted, the PharmVar annotations do provide this information. In addition, a position provided by HGVS may also deviate from its PharmVar counterpart which is typically due to different sequence alignment modes (e.g., if the reference is GATTGAT and the variation is GATTTGAT, the first or last ‘T’ may be annotated as ‘duplicated’ or ‘inserted’). HGVS annotations are easily retrieved through PharmVar’s Application Programming Interface (API).
Figure 2. Variation Window Redesigned to include HGVS annotations.
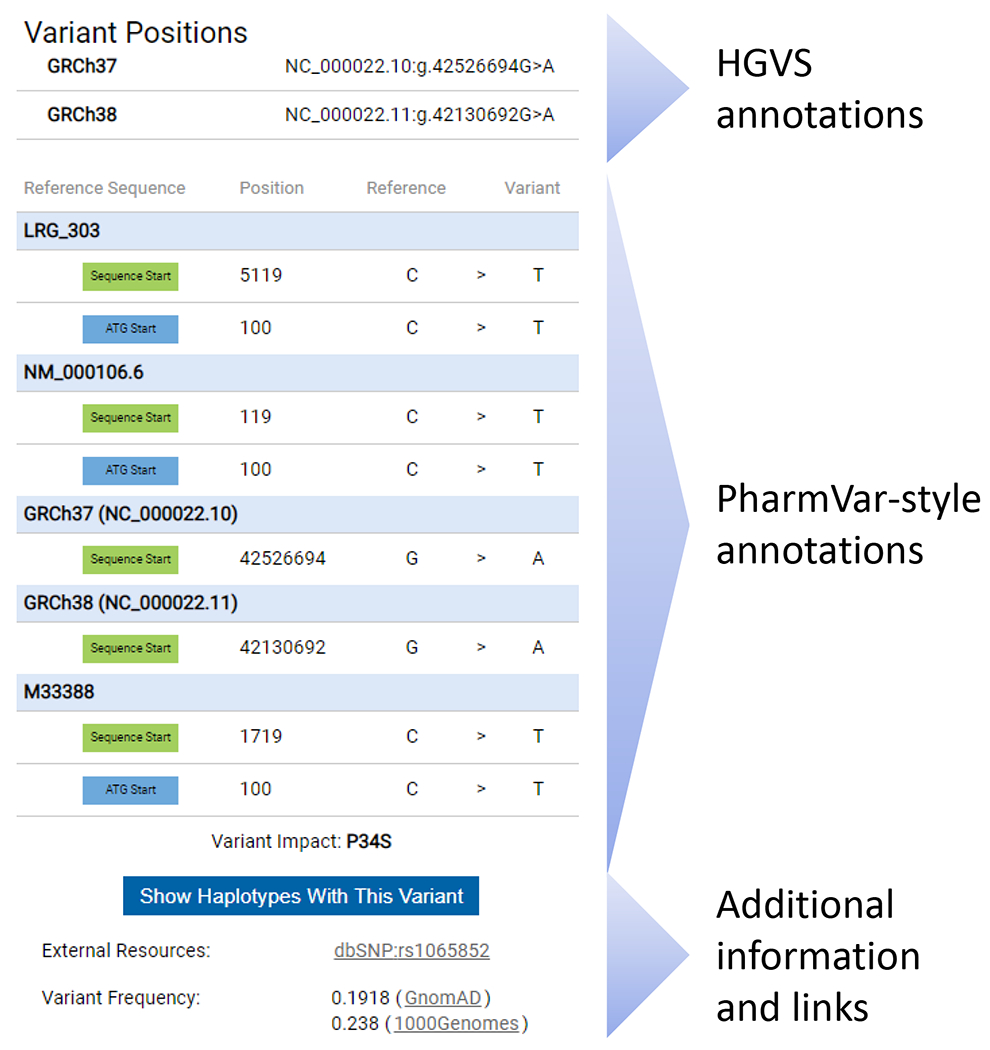
The new Variation Window is exemplified for a variant that is present on numerous CYP2D6 haplotypes, or star alleles. The Variation Window is activated by clicking on a SNV on a gene page. The top portion of the Variation Window displays SNV coordinates according to HGVS nomenclature on the gene, transcript and genome (GRCh37 and GRCh38) levels. Coordinates are displayed as obtained through the NCBI Variation Services and are only manually curated if no results are returned. The middle portion displays SNV positions ‘PharmVar-style’ on the gene, transcript and genome (GRCh37 and GRCh38) levels listing positions for both count modes (sequence start and the ‘A’ of the ATG’ start codon being +1). Note that PharmVar consistently uses the 3’ Rule to determine the positions of inserted or deleted nucleotide(s) and thus, HGVS and ‘PharmVar-style’ positions may differ for insertion/deletion variants in some instances which is most likely explained by differences in sequence alignments. PharmVar also displays single nucleotide insertions as ‘ins’ while HGVS displays them as duplications or ‘dup’. Additional details and examples are provided in the PharmVar ‘Standards document’ (4). The bottom portion of the variation window provides a filter option to display all variants with the selected SNV, the link to dbSNP (if a rsID exists), as well as SNV frequency information. There is also a bar providing the option to display all haplotypes with the selected variant.
Looking Forward
PharmVar is critical to the foundations of pharmacogenomics/genetics resources such as PharmGKB and CPIC, but also to other genetic resources such as ClinGen, ClinVar and the Genetic Testing Registry. For the integration of genetics, encompassing not only disease-causing genes but also pharmacogenes that play crucial roles in drug metabolism and response, to everyday medical treatment, it is critical that standards and nomenclature of genes (alleles, haplotypes, etc.) are defined. It is not useful to clinicians for example, to get a report from one clinical laboratory that refers to a gene and its variants one way, and a report from another clinical laboratory reporting the variants in a different way. Inconsistent nomenclature is not only a challenge for reporting, but potentially also affects how function is defined and genotype is translated to phenotype. As a basic resource for the medical genomics community, it is essential that funding of these foundational resources continues. Otherwise, the scientific community will continue to find itself speaking different dialects without knowing whether they are referring to the same or a different genetic component.
Supplementary Material
Acknowledgements
We like to thank all gene experts for serving PharmVar; we could not have done any of it without their continuous support.
Funding
PharmVar the NIH National Institute of General Medical Sciences (R24 GM123930; PI AG) from August 2017 - July 2021. We also acknowledge the indirect support of funding for PharmGKB (U24 HG010615; PI TEK).
References
- (1).Desta Z et al. PharmVar GeneFocus: CYP2B6. Clin Pharmacol Ther, Epub Jan 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Botton MR et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther 109, 352–66 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Nofziger C et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther 107, 154–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).PharmVar Standards https://www.pharmvar.org/genes.
- (5).Yang JJ et al. Pharmacogene Variation Consortium Gene Introduction: NUDT15. Clin Pharmacol Ther 105, 1091–4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).PharmVar Criteria https://www.pharmvar.org/criteria.
- (7).NCBI Reference Sequence Database https://www.ncbi.nlm.nih.gov/refseq/.
- (8).Locus Reference Genomic (LRG) Records https://www.lrg-sequence.org/.
- (9).NCBI Matched Annotation from NCBI and EMBL-EBI (MANE) https://www.ncbi.nlm.nih.gov/refseq/MANE/.
- (10).Sequence Variant Nomenclature per the Human Genome Variation Society https://varnomen.hgvs.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


