Abstract
Granulosa cells (GCs) are decisive players in follicular development. In this study, the follicle tissues and GCs were isolated from the goose during the peak-laying period to perform hematoxylin-eosin staining and RNA-seq, respectively. Moreover, the dynamic mRNA and lncRNA expression profiles and mRNA-lncRNA network analysis were integrated to identify the important genes and lncRNAs. The morphological analysis showed that the size of the GCs did not significantly change, but the thickness of the granulosa layer cells differed significantly across the developmental stages. Subsequently, 14,286 mRNAs, 3,956 lncRNAs, and 1,329 TUCPs (transcripts with unknown coding potential) were detected in the GCs. We identified 37 common DEGs in the pre-hierarchical and hierarchical follicle stages, respectively, which might be critical for follicle development. Moreover, 3,089 significant time-course DEGs (Differentially expressed genes) and 13 core genes in 4 clusters were screened during goose GCs development. Finally, the network lncRNA G8399 with CADH5 and KLF2, and lncRNA G8399 with LARP6 and EOMES were found to be important for follicular development in GCs. Thus, the results would provide a rich resource for elucidating the reproductive biology of geese and accelerate the improvement of the egg-laying performance of geese.
Keywords: goose, ovary development, follicle selection, differentiation, proliferation
Introduction
Granulosa cells (GCs) not only regulate follicular development in poultry through proliferation, differentiation, apoptosis, and steroidogenesis but also provide an essential microenvironment for follicular growth by synthesizing and secreting various hormones (Johnson, 2015; Hu et al., 2017). In poultry, a highly efficient follicular development is essential for excellent egg-laying (Niu et al., 2021). Although goose is one of the economically important waterfowls, its imperfect reproductive performance seriously dampens the potential development of the goose industry (Gao G. et al., 2015). To improve reproductive performance, the molecular mechanism of GCs in regulating follicular development has been extensively studied (Liu et al., 2015). For example, some genes play important roles in goose follicular development, such as AMH (Anti-Mullerian Hormone) (Johnson, 2012), FSHR (Follicle Stimulating Hormone Receptor) (Kang et al., 2009), CYP17 (Cytochrome P450 Family 17) (Huang et al., 2018), Smad9 (SMAD Family Member 9) (Yu et al., 2017), and BMP4 (Bone Morphogenetic Protein 4) (Yuan et al., 2019).
Unlike mammals, the follicles in poultry possess unique characteristics. The follicles are classified according to the size, as pre-hierarchical or hierarchical follicles. The pre-hierarchical follicles differ from the hierarchical follicles in undergoing a process of raising and selection; under normal circumstances, with no follicular selection, the number of hierarchical follicles remains unchanged (Meng et al., 2019; Shen et al., 2019). Moreover, the hierarchical follicles mature to ovulate, and eventually, when the single follicle is formed, the follicle selection is repressed (Johnson, 2015). Clear differences in the histological character and steroidogenesis between the pre-hierarchical and hierarchical follicles have been identified, conferring the physical and functional readiness for follicle development (Deng et al., 2018). Besides, significant changes in histomorphology and steroid synthesis and secretion occur during follicular development (Ocn-Grove et al., 2012). After being selected to develop into hierarchical follicles, the thickness of the granular layer is gradually decreased to a single layer, while that of the membrane layer is rapidly increased (Kim et al., 2013).
This study investigated the phenotypic differences in the size of the GCs, and the thickness of the granulosa layer cells at these developmental stages by HE staining. Then, the temporal expression profiles of the differentially expressed genes (DEGs) and differentially expressed long noncoding RNAs (DElncRNAs) were determined across the GCs, and the genes or lncRNA (Long non-coding RNA) in GCs crucial during the follicular development were screened. Furthermore, the cis-and trans-target genes were predicted and the mRNA-lncRNA network was constructed to analyze the important genes and lncRNAs. Our findings thus aim to broaden our understanding of follicle development, providing a new robust resource for gene discovery and validation in goose.
Materials and Methods
Experimental Animals and Sample Collection
All the animal experiments were carried out strictly according to the guidelines of the Animal Care and Welfare Committee of Chongqing Academy of Animal Science (CAAS), China. The Sichuan White goose population was reared individually in the cages (600 × 800 × 900 mm) and fed rice grains ad libitum (Wang et al., 2017) under natural environmental conditions in the waterfowl experimental base of the waterfowl-breeding base in Rongchang County, Chongqing City, China (105.48°N, 29.34°E). The egg-laying performance of the individuals was recorded and statistically analyzed twice a day. Ten individuals that consequently laid eggs during the peak egg-laying period (45 weeks) were selected as experimental animals. These geese were slaughtered 2 h before egg-laying to obtain the follicle tissue samples (SWF, LWF, SYF, LYF, F5 to F1) that were representatives of the end of each of the developmental stages. Of these ten individuals, five were used to survey the changes in the histomorphology of the follicle tissues, and the remaining five were used to obtain a dynamic gene expression atlas of the goose GCs. The follicles were classified according to the diameter and color into the small white follicle (SWF, 2–4 mm), large white follicle (LWF, 4–6 mm), the small yellow follicle (SYF, 6–8 mm), the large yellow follicle (8–10 mm, LYF), the fifth-largest follicle (F5), the fourth-largest follicle (F4), the third-largest follicle (F3), the second-largest follicle (F2), and the largest follicle (F1).
Morphometry of the Follicular Tissues
The follicular tissues were fixed at 4°C with paraformaldehyde (4%). After 24 h of fixation, the tissues were buried in paraffin and sliced by a fully automatic dehydrator along the axes for histological staining using hematoxylin-eosin (HE). The follicle samples were analyzed using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with dry lenses and a microscope digital camera, Olympus DP70 (Olympus, Tokyo, Japan). After staining each slice with hematoxylin and eosin (H&E staining), the structure of the follicular tissues at 100× and 400× magnifications, including the thickness of the granular layer and the size of GCs in each stage were analyzed using the Image-Pro Plus software (version 6.0). More than five follicles at each stage were randomly selected, with more than five different regions being randomly selected from each follicle for slicing, while more than five different regions were also randomly selected from the follicles at each of F5–F1 for slicing.
Total RNA Extraction, Library Preparation, and Sequencing
The GCs were collected from the follicular tissues in the phosphate-buffered saline (PBS) and immediately frozen in liquid nitrogen, followed by storage at −80°C for RNA-seq and real-time PCR. The total RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, United States) and further purified using an RNeasy column (Qiagen, Valencia, CA, United States), according to the manufacturer’s instructions. The NanoDrop ND-1000 (Nanodrop Technologies, Wilmington, DE, United States) and 1% agarose gel electrophoresis were employed to analyze the concentration and integrity of RNA, respectively. Finally, a total of 45 total RNA samples (4 μg each) were isolated from the nine stages of the five individuals. The mRNA isolated from the total RNA by binding oligo (dT) magnetic beads was cleaved into fragments (approximately 155 bp). The 45 cDNA libraries were constructed using these fragments, as described previously (Gao G. L. et al., 2015), and then sequenced using Illumina Hiseq X Ten (Novogene Bioinformatics Technology Co., Ltd., Beijing, China; http://www.novogene.com).
Data Analysis
After removing the adaptors, the sequences with uncertain bases, low-quality sequences, and sequences of less than 50 bp, as well as the high-quality data from the 45 libraries were mapped to the goose genome, which was assembled by our lab (Li et al., 2020) by alignment tool STAR (version 2.6.0c) (Dobin and Gingeras, 2015; Dobin and Gingeras, 2016) with the parameters of ENCODE standard RNA-seq pipeline. Then, the transcripts were reconstructed with parameters of -g -u -b --library-type fr-first strand using the Cufflinks (2.2.1) (Pollier et al., 2013; Ghosh and Chan, 2016). Then, transcripts were filtered by length (<250 bp), an expression level (FPKM <0.1), and clipped-exons (first or last exons <15 bp were clipped) for each library, and the Assemblyline utility (https://code.google.com/archive/p/assemblyline/) was used to filter the transfrags produced by the background noise (Iyer et al., 2015). Next, all the filtered libraries were merged and compared with the reference annotation by TACO(Niknafs et al., 2017) to remove the transcripts annotated as mRNAs. The remaining transcripts were considered to be non-coding RNAs and were subjected to lncRNA and TUCP (transcripts with unknown coding potential) identification. For the followed identification, the CPC2 (http://cpc2.cbi.pku.edu.cn/) (Kang et al., 2017) was run to analyze the coding potential, and Pfamscan (v.1.6) was applied to check the domain hits of the nucleic acid sequences against the Pfam (release31) database after the open reading frames were obtained using EMBOSS (version 6.5.7). A domain hit by mRNAs and putative lncRNAs were checked by Fisher’s exact test, and those domains with an odds ratio of less than 10.0 or p-value greater than 0.05 were considered as likely artifacts. After removing the transcripts in all likely domains and with coding potential (CPC score >0), the remaining transcripts were identified to be long non-coding RNAs. We classified the lncRNAs into five types (intergenic lncRNAs; antisense lncRNAs; sense-overlapping lncRNAs; divergent lncRNAs and convergent lncRNAs) as stated previously (Jin et al., 2018). To identify the expression profiles of the mRNA, lncRNA, and TUCP gene, the hierarchical clustering was performed using hclust function in R (stats package v3.5.3) (Team, 2013). Also, the dimensional reduction analysis was performed and the result was visualized using the Rt-SNE package (Van Der Maaten and Hinton, 2008).
The Analysis of Differentially Expressed Genes (DEGs) and Cluster Analysis Time-Series
We employed the EdgeR software to analyze the DEGs between the adjacent pairs of developmental stages using the TPM normalization method. The fold changes were also estimated within this package, and the false discovery rate (FDR) was obtained by adjusting the p-value. A 0.05 FDR and 1.5-fold change were set as the threshold to define DEGs. Moreover, the next-maSigPro method of the maSigPro program R package was used to identify the genes with differential temporal expression profiles from SWF to F1 GCs (Nueda et al., 2014). The differentially expressed targets were classified into four clusters using the hclust function of maSigPro, and the median profiles of the resulting clusters were plotted using the maSigPro to visualize their expression patterns.
The Prediction of the Cis-target and Trans-target Genes
The cis-target genes of lncRNA were defined as the mRNA located within 100 kb, whereas the trans-target genes were those having a Pearson correlation coefficient more than 0.95 or less than −0.95 with p-value < 0.05. The cis-target genes and trans-target genes of lncRNA, and the four gene expression patterns were integrated. Then, the co-expression mRNA-lncRNA network was generated by analyzing using the weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008). The genes with the number of edges equal or more than ten were defined as the hub genes, and then the networks were visualized by the Cystoscope software (version 3.6.0) (Kohl et al., 2011).
Quantitative Real-Time PCR
To confirm the results of the RNA-seq, six of the DEGs were randomly selected for qRT-PCR. The total RNAs were isolated from the same samples used for the RNA-seq of the 45 individuals. The goose glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was selected as an internal control gene for normalization. The primers of the six genes were designed for qRT-PCR analyses (Supplementary Table S1) and synthesized by Invitrogen (Shanghai, China). Three independent qPCR runs were performed using the Applied Biosystems 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA, United States) in 20 μl reaction mixtures using the SYBR Premix Ex Taq system (Takara, Co., Dalian, China). The following qPCR program was applied: a single cycle of 95°C for 1 min; 40 cycles of 95°C for 5 s and 60°C for 34 s; and one cycle for analyzing the melting curve analysis. The relative expression levels of the candidate genes relative to GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase) were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Results
Morphometry of the Follicular Tissues
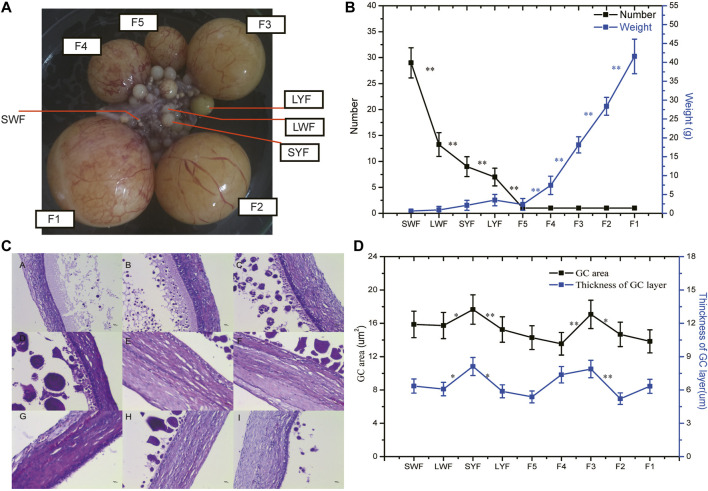
In this study, five sustainable egg-laying Sichuan White goose (three consecutive laid eggs in a week) during the peak-laying period (45 weeks) were selected as experimental animals (Figure 1A). Our results revealed that the number of pre-hierarchical follicles gradually decreased over time, showing extremely significant differences between the SWF vs. LWF, LWF vs. SYF, SYF vs. LWF, and LWF vs. F5, respectively (p < 0.01) (Figure 1B). There was a single follicle for the hierarchical follicles (F5–F1) during every developmental stage, but the weight gradually increased from SWF to F1 (Figure 1B). The HE staining results showed that the granular cell area of the nine follicles (Figure 1C) showed no differences between any developmental stages. However, the thickness of the granulosa cell layer increased and then decreased to its nadir in the LYF follicles (Figure 1D).
FIGURE1.
The weights and phenotypic differences in the various sizes of the GCs. Note (A), The phenotypic of goose follicles; (B), The numbers and weight of the goose follicles; (C), Histological staining for follicular tissues using hematoxylin-eosin (HE); (D), The granular cell area and thickness of the granulosa layer cells.
Data Statistics
A total of 315.98 Gb raw reads were obtained from these 45 libraries ranging from 4.74 to 9.74 Gb. After quality control, we acquired 311.84 Gb clean data (∼6.93 Gb per each) of all samples ranging between 4.68 and 9.63 Gb, and the qualities and alignment ratios were shown in Supplementary Table S2. In this study, the 8,199 lncRNA transcripts (from 5,531 gene locus) were identified, of which 7,045 were classified into intergenic lncRNAs; antisense lncRNAs; sense-overlapping lncRNAs; divergent lncRNAs, and convergent lncRNAs (Supplementary Figure S1). Besides, we identified 2,664 TUCPs (from 1,444 gene locus).
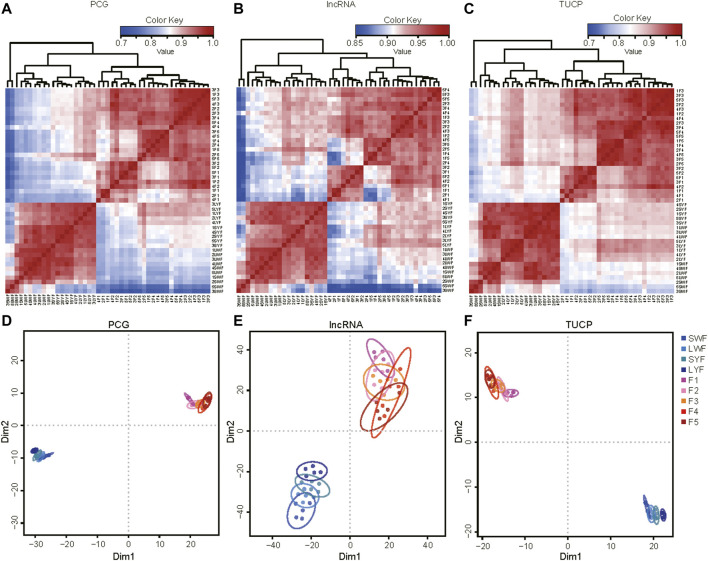
Then, the expression level of PCG, lncRNA, and TUCP were normalized to TPM (Transcripts Per Kilobase of exon model per Million mapped reads) with the software Kallisto (Bray et al., 2016), and PCG with TPM >0.5 in at least three replicates of one group, the lncRNA or TUCP with TPM >0.1 in at least three replicates of one group were considered to be expressed and were subjected to the continuing analysis. In total, 14,286 mRNAs, 3,956 lncRNAs, and 1,299 TUCPs were substantially expressed in our research. Furthermore, the results of Pearson matrix correlations of mRNA (Figure 2A), lncRNA (Figure 2B) and TUCP (Figure 2C), and t-SNE of mRNA (Figure 2D), lncRNA (Figure 2E) and TUCP (Figure 2F) showed that the 45 samples could be differentiated at both PCG, lncRNA, and TUCP of GCs in the expression levels of the pre-hierarchal follicles and hierarchal follicles.
FIGURE 2.
The common DEGs in the pre-hierarchical follicles and hierarchical follicle GCs.
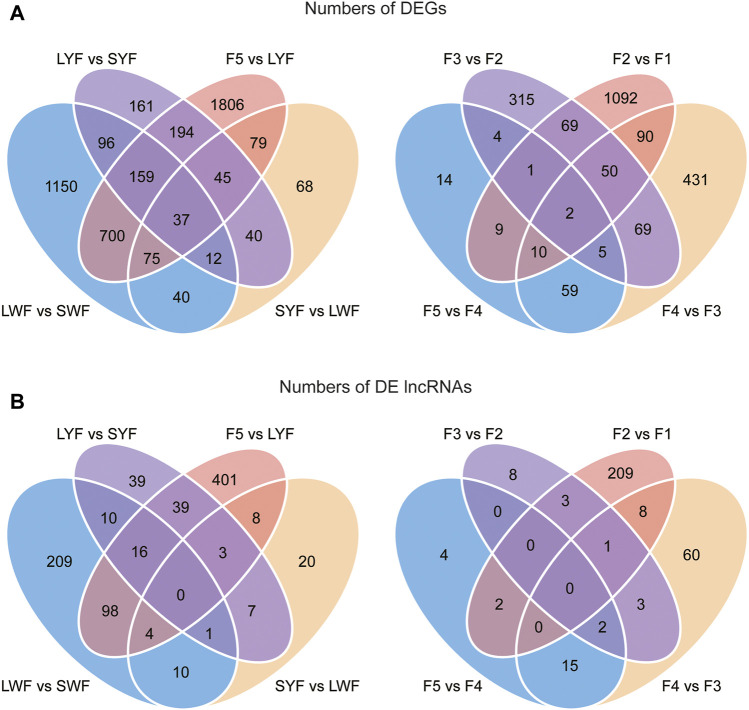
In this study, 2,269, 396, 744, 3,095, 104, 716, 515, and 1,323 DEGs (Supplementary Table S3) were detected by comparing the adjacent pairs of developmental stages (i.e., SWF vs. LWF, LWF vs. SYF, SYF vs. LYF, LYF vs. F5, F5 vs. F4, F4 vs. F3, F3 vs. F2, and F2 vs. F1 groups, respectively). To investigate the important DEGs from the SWF to F1 developmental stages, we screened the common DEGs with vital roles in regulating the follicular developmental stage. The results revealed 37 and 2 genes commonly and differentially expressed in the pre-hierarchical and hierarchical follicles (Figure 3A), suggesting that the 37 and 2 genes be important for the pre-hierarchical GCs (SWF, LWF, SYF, and LYF) and hierarchical GCs (F5, F4, F3, F2, and F1), respectively.
FIGURE 3.
The common DEGs and DElncRNAs detected by comparing the adjacent pairs of developmental stages. Note (A): The common DEGs in the pre-hierarchical follicles and hierarchical follicle GCs; (B): The common DElncRNAs in the pre-hierarchical follicles and hierarchical follicle GCs.
In this study, 348, 53, 115, 569, 22, 88, 16, and 223 DElncRNAs were also identified between the adjacent stages (SWF vs. LWF, LWF vs. SYF, SYF vs. LYF, LYF vs. F5, F5 vs. F4, F4 vs. F3, F3 vs. F2, and F2 vs. F1 groups, respectively) (Figure 3A; Supplementary Table S4). We found no common DElncRNAs in the pre-hierarchical follicles or hierarchical follicles (Figure 3B). To evaluate the accuracy of the results from RNAseq, the quantitative real-time PCR method was employed to detect the genes expression patterns of WASL (Wiskott-Aldrich syndrome protein), GPR63 (G-protein coupled receptor 63), FST (Follistatin precursor), AtTTP (At1g68200), NNT [NAD(P) transhydrogenase, mitochondrial], and P2RY6 (Pyrimidinergic Receptor P2Y6) genes (Supplementary Figure S2). The results were consistent with the RNA-seq data, demonstrating that the results were robust and credible.
Cluster Analysis Time-Series Analysis
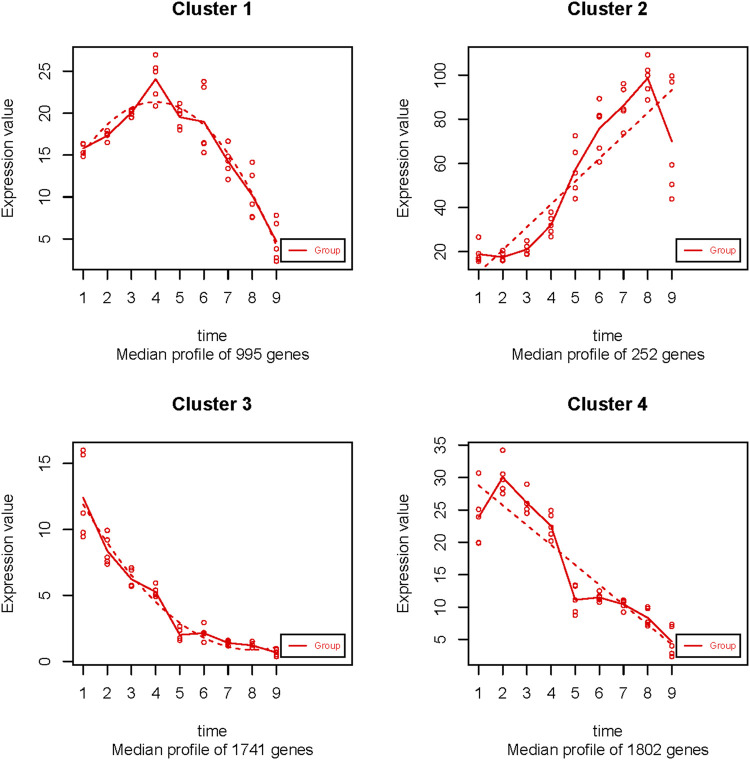
To investigate the genome-wide mRNA and lncRNA expression pattern profiles during the development of the follicular GCs, the nine-stage (SWF, LWF, SYF and LYF, F5 to F1) of goose GCs were considered with temporal changes to determine the progression patterns, similar to that of the GCs development. The maSigPro method was employed to identify 3,089 significant time-course DEGs (Supplementary Table S5) and were divided into 4 clusters at the nine-time point of GCs (Figure 4; Supplementary Figure S3).
FIGURE 4.
The 4 temporal gene expression profiles.
Overall, the 995 gene expression in cluster 1 reached the top point in LYF, and then consistently decreased to the lowest point at the SWF stage (Figure 4). The expression of 252 genes in cluster 2 was relatively lower in the pre-hierarchical period than that in the hierarchical period, which consistently increased and reached the peak in the F2 GC stage (Figure 4). On the contrary, the expression of 1741 genes in cluster 3 was similar to that of cluster 4, containing 1802 genes, which were expressed at a comparatively higher level during the pre-hierarchical period, and drooped to the bottom in the F1 GC stage, but the difference between the two clusters was that the top of genes expression at SWF in the cluster 3, rather than that in the cluster 4 in LWF (Figure 4).
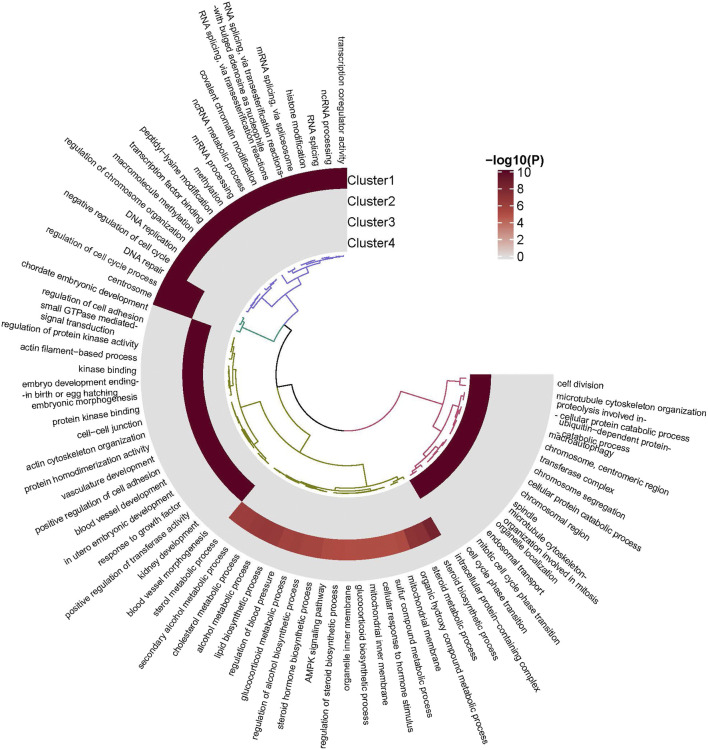
The gene functional enrichment analysis was performed by the Metascape website to investigate the biological characteristics of the genes in the 4 clusters during the GC development. 1) The gene function analysis showed that the genes in cluster 1 were significantly enriched in mRNA processing (GO:0006397), ncRNA metabolic process (GO:0034660), covalent chromatin modification (GO:0016569). 2) The result of functional enrichment showed that the genes in cluster 2 significantly enriched in the steroid biosynthetic process (GO:0006694), steroid metabolic process (GO:0008202), organic hydroxy compound metabolic process (GO:1901615). 3) The functional analysis showed that the genes in cluster 3 were significantly enriched in the regulation of protein kinase activity (GO:0045859), regulation of cell adhesion (GO:0030155), regulation of cell adhesion (GO:0043009). 4) The results of the functional enrichment showed that the genes in cluster 4 were significantly enriched in cell division (GO:0051301), microtubule cytoskeleton organization (GO:0000226), mitotic cell cycle phase transition (GO:0044772). The DNA repair (GO:0006281), regulation of cell cycle process (GO:0010564), the centrosome (GO:0005813), cell projection assembly (GO:0030031), plasma membrane-bounded cell projection assembly (GO:0120031) were common in clusters 1 and 2 (Figure 5).
FIGURE 5.
The gene enrichment for the 4 clusters.
Core Genes Screening
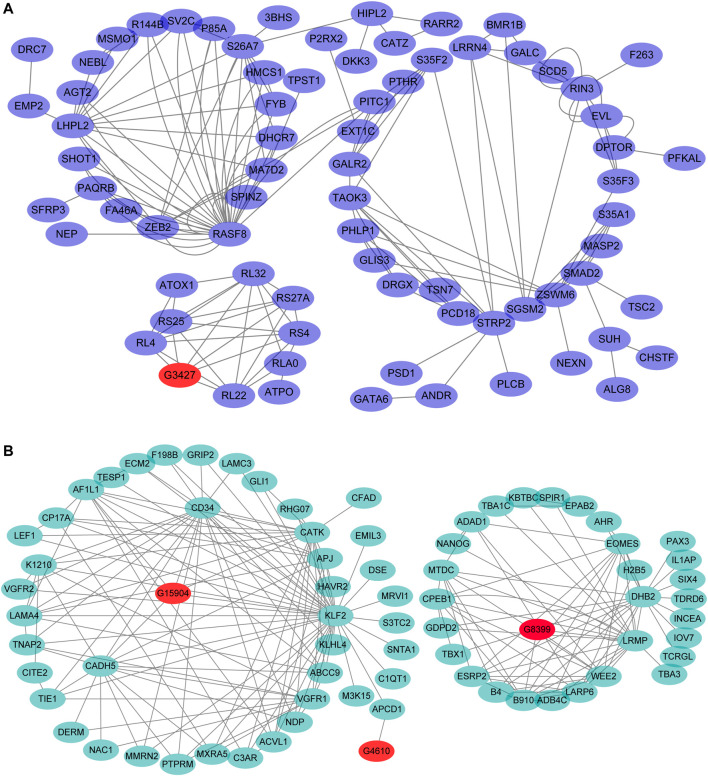
Thirteen hub genes were screened from the four clusters (Figure 6; Supplementary Figure S4). There were three hub genes in the cluster1: GON4L (GON-4-like protein), NUMA1 (Nuclear mitotic apparatus protein 1), SAFB1 (scaffold attachment factor B1) (Supplementary Figure S4); In cluster2, the core gene was RASF8 (Ras association domain-containing protein 8) (Figure 6). There were six core genes in the cluster3: APJ (Apelin receptor), CATK (Cathepsin K), CD34 (Hematopoietic progenitor cell antigen CD34), EOMES (Eomesodermin), KLF2 (Krueppel-like factor 2), VGFR1 (Vascular endothelial growth factor receptor 1) (Figure 6). There were three genes in the cluster4: PLPL7(Patatin-like phospholipase domain-containing protein 7), RFA2 (Replication Protein A2), and TMOD1 (Tropomodulin-1) (Supplementary Figure S4).
FIGURE 6.
The cis-and trans-target genes were predicted and the mRNA-lncRNA network in cluster 2 and 3. Note: (A), The mRNA-lncRNA network in cluster 2; (B), The mRNA-lncRNA network in cluster 3.
Exploration of the Genes That Continuously Regulated the Development of GCs in Goose
It is difficult to predict the function of the three types of non-coding RNAs owing to the lacing of the annotation for lncRNAs in goose. Here, we explored the functional relatedness between the mRNAs and cis-lncRNAs or trans-lncRNAs using the co-expression analysis or the WGCNA method in the 4 clusters. In cluster 2, the G3427 co-expressed with RL4 (Ribosomal Protein L4), RS25 (Ribosomal Protein S25), RL32 (Ribosomal Protein L32), RS27A (Ribosomal Protein S27a), RS4 (Ribosomal protein S4), and RL22 (Ribosomal Protein L22) (Figure 6). In cluster 3, the G15904 co-expressed with CADH5 (Cadherin 5), KLF2 (Kruppel Like Factor 2), CD34 (CD34 Molecule), and the G4610 were associated with APCD1 (APC Down-Regulated 1). The G8399 co-expressed with ESRP2 (Epithelial Splicing Regulatory Protein 2), ADAD1 (Adenosine Deaminase Domain Containing 1), EOMES (Eomesodermin), DHB2 (Estradiol 17-beta-dehydrogenase 2), LRMP (Lymphoid-restricted membrane protein), and LARP6 (La Ribonucleoprotein 6, Translational Regulator) and B910 (Maternal B9.10 protein) genes (Figure 6).
Discussion
Morphometry of the Follicular Tissues
The pre-hierarchal and preovulatory follicles clusters in goose are subjected to stages like initiation, development, and selection. The number of follicles in the goose was found to decrease from SWF to LYF, while the diameters and the weights increased simultaneously (Figure 1A), which is consistent with that of the other poultry, (Johnson, 2015; Gao et al., 2019). These results suggested that follicle selection occurs in the pre-hierarchical follicles rather than in the hierarchical ones, and the number of hierarchical follicles instead remains unchanged suppressing the follicle selection consistent with that in other poultry, along with the increase in the follicular diameters and weight (Ghanem and Johnson, 2018). Moreover, the thickness of the granulosa cell layer was found to increase and then decrease in the LYF follicles (Figures 1C,D), which were consistent with the findings of a previous study on the histological study of the pre-hierarchical follicles in goose (Dong et al., 2014).
The DEGs and DElncRNAs Between the Adjacent Pairs of Developmental Stages
The PCA analysis showed that the nine GCs groups were clustered into two groups: pre-hierarchical and hierarchical follicles (Figure 2). These results suggested a high correlation between the gene expression levels of the biological replicates and showed the excellent reliability of our experiment. Interesting, there were most DEGs in the LYF vs. F5 (3,095) and most DElncRNAs 569) (Supplementary Tables S3, S4). However, no differences were identified in the granular cell area or the thickness of the granulosa cell layer between LYF and F5, or F5 and F4, suggesting that the F5 developmental stage were likely to show independent and unstable transition (Onagbesan et al., 2009; Kim et al., 2016; Kim et al., 2018). Some of these genes (Figure 3), such as the FST gene, have multiple roles in follicular development, steroid hormone synthesis, and granulose cell proliferation (Liu et al., 2015); these genes also included the WASL gene, which modulates the Wnt signaling and is necessary for the hair follicle cycling in mice (Lyubimova et al., 2010).
Cluster Analysis Time-Series Analysis
The growth and proliferation of the GCs from the pre-hierarchical follicles are the key to initiation (Eppig, 2001), and the apoptosis of GCs leads to follicular atresia (Rolaki et al., 2005; Manabe et al., 2008). The common feature in cluster 1, cluster 3, and cluster 4 are that the genes were consistently increased in the pre-hierarchical stags (including SWF, LWF, SYF, and LYF) (Figure 4; Supplementary Table S6), and the genes expression showed a continuous decline in the hierarchical stages (F5, F4, F3, F2, and F1). Previous studies demonstrated that the GCs have crucial roles in follicular development. Interestingly, the genes belonging to cluster 1 were significantly enriched in the cell differentiation pathways clusters, regulation of cell cycle process (GO:0010564), DNA replication (GO:0006260), cell division (GO:0051301), regulation of telomere maintenance (GO:0032204) (Supplementary Table S6). 2) Functional analysis showed that the genes in cluster 3 were significantly enriched in the embryonic development clusters (Supplementary Table S6), such as the chordate embryonic development (GO:0043009), urogenital system development (GO:0001655), cell morphogenesis involved in differentiation (GO:0000904), cell division (GO:0051301), gland development (GO:0048732). 3) The results of the functional enrichment showed that the genes in cluster 4 were significantly enriched in the cell division clusters (GO:0051301), autophagy clusters (GO:0006914), apoptotic signaling pathway clusters (GO:0097190) (Supplementary Table S6). These results have shown that the GCs regulated the follicular development and selection process through the genes from the pre-hierarchical follicular GCs enriched in the processes of proliferation and apoptosis, embryonic development. However, the detailed molecular mechanisms still needed further investigations.
The Core Genes in GC Development
The hub genes identified in cluster 1 were Gon4l, NUMA1, and SAFB1 (Supplementary Figure S4A). Gon4l was conserved within the animal species (Tsai et al., 2020), with crucial roles in cell proliferation, cell differentiation, cell cycle, cell viability, embryonic patterning, cardiomyocyte proliferation, and ventricular fate maintenance in worms, flies, mice, and fish (Friedman et al., 2000; Liu et al., 2007; Bulchand et al., 2010; Lu et al., 2010; Colgan et al., 2021). Moreover, Gon4l has been reported to participate in cell proliferation, cell survival, and mesoderm-derived tissue specification including those of the blood and somites in the zebrafish, (Budine et al., 2020). Deficiency of Gon4l in zebrafish results in cellular apoptosis and failure of the cell to differentiate, blocking the mitotic cell division in the developing embryos (Friedman et al., 2000; Liu et al., 2007). One of the hub genes in cluster1, NUMA1, is involved in the maintenance and formation of the spindle poles and mitotic spindle organization during meiotic cell division.(Chu et al., 2016; Torii et al., 2020). The absence of NUMA1 resists cell death in the PTC-318 cell (Gisler et al., 2020), and the alternative splicing of the gene is associated with the cellular proliferation and centrosome amplification in the epithelial cells (Sebestyén et al., 2016). SAFB1 regulates RNA processing and neuronal function rendering the chromatin permissive for DNA damage signaling and myogenic differentiation (Hernandez-Hernandez et al., 2013; Rivers et al., 2015). Previous studies have shown the SAFB1 absence in mice to reduce the mice growth by affecting the IGF-1 level, affecting the reproductive ability of the female mice by reducing the levels of estradiol and progesterone. On the contrary, SAFB1 overexpression tends to shorten the S phase of the cell cycle, thereby accelerating cell apoptosis (Townson et al., 2004; Ivanova et al., 2005). In conclusion, these three genes have vital roles in embryonic development and are involved in multiple roles in goose GCs development.
In cluster 2, RASSF8 is provital for embryonic development in Drosophila, where it is involved in pupal wing cell hexagonal packing promoting wing elongation and epithelial ordering (Chan et al., 2021). RASSF8 plays a role in cell growth, regulation of the Wnt and NFκB pathways, and cell-cell adhesion (Lock et al., 2010). Moreover, the RASSF8 overexpression has been found to affect the process of cell growth, cell apoptosis (Karthik et al., 2018).
VEGFR1, in cluster 3, is vital for angiogenesis, blood vessel patterning on the retinal astrocytes, neuronal precursors during fetal and retinal development (Chappell et al., 2019; Marini et al., 2019). One of the main functions of CatK is to regulate bone resorption in mammals (Dai et al., 2020), Previous studies have shown CatK to be associated with diseases, including skeletal diseases, renal disease, cardiovascular system, central nervous system, respiratory system, autoimmune diseases. (Bühling et al., 2004; Yang et al., 2008; Hua et al., 2015; Zhao et al., 2019; Dai et al., 2020; Dauth et al., 2020). Moreover, CatK is also associated with thyroid development, brain development, lipid homeostasis, and metabolism (Dauth et al., 2020).
In cluster 4, TMOD1 critically mediates the actin dynamics, participating in the cell migration, motility, proliferation, cycle, morphology, neurite outgrowth, and spine formation (Fischer et al., 2003; Moyer et al., 2010; Fath et al., 2011; Lu et al., 2020). TMOD1 deficiency in mice is known to lead to embryonic lethality with cardiac defects (Fritz-Six et al., 2003). In conclusion, we speculate that TMOD1 still play an important role during goose GCs development.
The Core Genes and lncRNA in GC Development
In cluster 3, the lncRNA G15904 co-expressed with CADH5, CD34 (Figure 6B). Interestingly, CADH5 has a multi-functional biological function in the endothelial cells, such as regulating cell proliferation, apoptosis, cell survival, cell adhesion, cell shape, cell motility (Dejana and Vestweber, 2013; Sauteur et al., 2014; Grimsley-Myers et al., 2020). The CDH5-null mice have severe cardiovascular defects resulting in premature embryonic death (Carmeliet et al., 1999). Moreover, another gene associated with the lncRNA, KLF2, is essential for the development and differentiation of the lungs (Anderson et al., 1995; Maity et al., 2020), inhibiting the endothelial cell apoptosis and promoting metabolic quiescence, thus, resulting in embryonic hemorrhage and death in the KLF2 null mice (Doddaballapur et al., 2015). Taken together, the lncRNA G15904 may regulate the expression of the genes CADH5 and KLF2 by participating in the granulosa cells in embryonic development, cell proliferation, and apoptosis. However, further investigations are required for an in-depth understanding of the molecular mechanisms for the lncRNA G15904in the future.
LARP6, one of the co-expressed genes associated with the lncRNA G8399 encodes a protein that prevents cell death until it is triggered by dephosphorylation and degraded (Sheel et al., 2020). We also found that EOMES, one of the target genes for the lncRNA G8399, is involved in the trophoblast differentiation as well as gastrulation, regulating both the mesodermal delamination, endodermal specification, which are essential for developing the tissues extraembryonic to the uterus during the mammalian embryogenesis (Kidder and Palmer, 2010; Probst and Arnold, 2017). Thus, the lncRNA G8399 may participate in the cell death, development, and reproduction process, but the gene function of this lncRNA demands further investigations.
In conclusion, this study measured the expression of the genes in the nine stages of development of goose GCs. Furthermore, the genome-wide mRNA and lncRNA expression pattern profiles were investigated during the development of the follicular GCs and the function of the genes of four clusters was explored during the nine developmental stages. Furthermore, two lncRNA-gene regulatory networks were also identified in goose GCs that could be explored in the future.
Data Availability Statement
These high-quality datasets have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSM3927138 to GSM3927181 (the detailed information is listed in Supplementary Table S2).
Ethics Statement
The animal study was reviewed and approved by Animal Care and Welfare Committee of Chongqing Academy of Animal Science, China.
Author Contributions
GG, QW, and XZ conceived the finding; GG designed and supervised the project. GG, SH, and HW performed the bioinformatic analysis. GG, CZ, and RW wrote the article. GG, KZ, CZ, HZ, and YX collected the samples. GG, KZ, HZ, and YX checked, reviewed, and edited the article. GG, KZ, HZ, CZ, and XZ participated in the experiments. All of the authors have read and approved the final article.
Funding
This work was supported by grants from the Chongqing Scientific Research Institution Performance incentive project (cstc2019jxjl80009), the Science Foundation of Chongqing Project (cstc2019jcyj-msxmX0072), the Earmarked Fund for China Agriculture Research System (CARS-43-51, CARS-43-15), the Chongqing Key Project of Technology Innovation and Application Development (grant number cstc2019jscx-gksbX0097) and the Chongqing Scientific Research Institution Performance incentive project (cstc2018jxjl80022).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.786287/full#supplementary-material
References
- Anderson K. P., Kern C. B., Crable S. C., Lingrel J. B. (1995). Isolation of a Gene Encoding a Functional Zinc finger Protein Homologous to Erythroid Krüppel-like Factor: Identification of a New Multigene Family. Mol. Cel Biol 15, 5957–5965. 10.1128/mcb.15.11.5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 34, 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Budine T. E., de Sena-Tomás C., Williams M. L. K., Sepich D. S., Targoff K. L., Solnica-Krezel L. (2020). Gon4l/Udu Regulates Cardiomyocyte Proliferation and Maintenance of Ventricular Chamber Identity during Zebrafish Development. Developmental Biol. 462, 223–234. 10.1016/j.ydbio.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühling F., Röcken C., Brasch F., Hartig R., Yasuda Y., Saftig P., et al. (2004). Pivotal Role of Cathepsin K in Lung Fibrosis. Am. J. Pathol. 164, 2203–2216. 10.1016/s0002-9440(10)63777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S., Menon S. D., George S. E., Chia W. (2010). Muscle Wasted: a Novel Component of theDrosophilahistone Locus Body Required for Muscle Integrity. 123 , 2697–2707. 10.1242/jcs.063172 [DOI] [PubMed] [Google Scholar]
- Carmeliet P., Lampugnani M.-G., Moons L., Breviario F., Compernolle V., Bono F., et al. (1999). Targeted Deficiency or Cytosolic Truncation of the VE-Cadherin Gene in Mice Impairs VEGF-Mediated Endothelial Survival and Angiogenesis. Cell 98, 147–157. 10.1016/s0092-8674(00)81010-7 [DOI] [PubMed] [Google Scholar]
- Chan E. H., Zhou Y., Aerne B. L., Holder M. V., Weston A., Barry D. J., et al. (2021). RASSF8-mediated Transport of Echinoid via the Exocyst Promotes Drosophila wing Elongation and Epithelial Ordering. Development. 148 , dev199731. 10.1242/dev.199731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. C., Darden J., Payne L. B., Fink K., Bautch V. L. (2019). Blood Vessel Patterning on Retinal Astrocytes Requires Endothelial Flt-1 (VEGFR-1). Jdb 7, 18. 10.3390/jdb7030018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X., Chen X., Wan Q., Zheng Z., Du Q. (2016). Nuclear Mitotic Apparatus (NuMA) Interacts with and Regulates Astrin at the Mitotic Spindle. J. Biol. Chem. 291, 20055–20067. 10.1074/jbc.m116.724831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D. F., Goodfellow R. X., Colgan J. D. (2021). The Transcriptional Regulator GON4L Is Required for Viability and Hematopoiesis in Mice. Exp. Hematol. 98, 25–35. 10.1016/j.exphem.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R., Wu Z., Chu H. Y., Lu J., Lyu A., Liu J., et al. (2020). Cathepsin K: The Action in and beyond Bone. Front. Cel Dev. Biol. 8, 433. 10.3389/fcell.2020.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauth S., Rakov H., Sîrbulescu R. F., Ilieş I., Weber J., Batbajar Dugershaw B., et al. (2020). Function of Cathepsin K in the central Nervous System of Male Mice Is Independent of its Role in the Thyroid Gland. Cell Mol Neurobiol 40, 695–710. 10.1007/s10571-019-00765-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana E., Vestweber D. (2013). The Role of VE-Cadherin in Vascular Morphogenesis and Permeability Control. Prog. Mol. Biol. translational Sci. 116, 119–144. 10.1016/b978-0-12-394311-8.00006-6 [DOI] [PubMed] [Google Scholar]
- Deng Y., Gan X., Chen D., Huang H., Yuan J., Qiu J., et al. (2018). Comparison of Growth Characteristics of In Vitro Cultured Granulosa Cells from Geese Follicles at Different Developmental Stages. Biosci. Rep. 38. 10.1042/BSR20171361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Gingeras T. R. (2015). Mapping RNA-Seq Reads with STAR. Curr. Protoc. Bioinformatics 51, 1111–1911. 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Gingeras T. R. (2016). “Optimizing RNA-Seq Mapping with STAR,” in Data Mining Techniques for the Life Sciences (Springer; ), 245–262. 10.1007/978-1-4939-3572-7_13 [DOI] [PubMed] [Google Scholar]
- Doddaballapur A., Michalik K. M., Manavski Y., Lucas T., Houtkooper R. H., You X., et al. (2015). Laminar Shear Stress Inhibits Endothelial Cell Metabolism via KLF2-Mediated Repression of PFKFB3. Arterioscler Thromb. Vasc. Biol. 35, 137–145. 10.1161/atvbaha.114.304277 [DOI] [PubMed] [Google Scholar]
- Dong X., Liu H. H., Wang J. W., Xiao Q. H., Yuan X., Li L., et al. (2014). Histological and Developmental Study of Prehierarchical Follicles in Geese. Folia Biol. (Krakow) 62, 171–177. 10.3409/fb62_3.171 [DOI] [PubMed] [Google Scholar]
- Eppig J. (2001). Oocyte Control of Ovarian Follicular Development and Function in Mammals. Reproduction 122, 829–838. 10.1530/rep.0.1220829 [DOI] [PubMed] [Google Scholar]
- Fath T., Fischer R. S., Dehmelt L., Halpain S., Fowler V. M. (2011). Tropomodulins Are Negative Regulators of Neurite Outgrowth. Eur. J. Cel Biol. 90, 291–300. 10.1016/j.ejcb.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R. S., Fritz-Six K. L., Fowler V. M. (2003). Pointed-end Capping by Tropomodulin3 Negatively Regulates Endothelial Cell Motility. 161 , 371–380. 10.1083/jcb.200209057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L., Anna-Arriola S. S., Hodgkin J., Kimble J. (2000). gon-4, a Cell Lineage Regulator Required for Gonadogenesis in Caenorhabditis elegans . Developmental Biol. 228, 350–362. 10.1006/dbio.2000.9944 [DOI] [PubMed] [Google Scholar]
- Fritz-Six K. L., Cox P. R., Fischer R. S., Xu B., Gregorio C. C., Zoghbi H. Y., et al. (2003). Aberrant Myofibril Assembly in Tropomodulin1 Null Mice Leads to Aborted Heart Development and Embryonic Lethality. 163 , 1033–1044. 10.1083/jcb.200308164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Li Q., Zhao X., Ding N., Han Q., Su J., et al. (2015a). Transcriptome Profiling of the Hypothalamus during Prelaying and Laying Periods in Sichuan white Geese (Anser cygnoides). Anim. Sci. J. 86, 800–805. 10.1111/asj.12356 [DOI] [PubMed] [Google Scholar]
- Gao G. L., Wang H. W., Zhao X. Z., Li Q., Li J., Li Q. R., et al. (2015b). Feeding Conditions and Breed Affect the Level of DNA Methylation of the Mitochondrial Uncoupling Protein 3 Gene in Chicken Breast Muscle1. J. Anim. Sci. 93, 1522–1534. 10.2527/jas.2014-8431 [DOI] [PubMed] [Google Scholar]
- Gao S., Gan X., He H., Hu S., Deng Y., Chen X., et al. (2019). Dynamic Characteristics of Lipid Metabolism in Cultured Granulosa Cells from Geese Follicles at Different Developmental Stages. Biosci. Rep. 39, BSR20192188. 10.1042/BSR20192188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem K., Johnson A. L. (2018). Follicle Dynamics and Granulosa Cell Differentiation in the Turkey Hen Ovary. Poult. Sci. 97, 3755–3761. 10.3382/ps/pey224 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Chan C.-K. K. (2016). “Analysis of RNA-Seq Data Using TopHat and Cufflinks,” in Plant Bioinformatics (Springer; ), 339–361. 10.1007/978-1-4939-3167-5_18 [DOI] [PubMed] [Google Scholar]
- Gisler S., Maia A. R. R., Chandrasekaran G., Kopparam J., Van Lohuizen M. (2020). A Genome-wide Enrichment Screen Identifies NUMA1-Loss as a Resistance Mechanism against Mitotic Cell-Death Induced by BMI1 Inhibition. PLoS ONE 15, e0227592. 10.1371/journal.pone.0227592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley-Myers C. M., Isaacson R. H., Cadwell C. M., Campos J., Hernandes M. S., Myers K. R., et al. (2020). VE-cadherin Endocytosis Controls Vascular Integrity and Patterning during Development. J. Cel Biol 219. 10.1083/jcb.201909081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez J. M., Mallappa C., Nasipak B. T., Oesterreich S., Imbalzano A. N. (2013). The Scaffold Attachment Factor B1 (Safb1) Regulates Myogenic Differentiation by Facilitating the Transition of Myogenic Gene Chromatin from a Repressed to an Activated State. Nucleic Acids Res. 41, 5704–5716. 10.1093/nar/gkt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Duggavathi R., Zadworny D. (2017). Expression and Regulation of Prolactin-like Protein Messenger RNA in Undifferentiated Chicken Granulosa Cells. Gen. Comp. Endocrinol. 240, 191–197. 10.1016/j.ygcen.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Hua Y., Robinson T. J., Cao Y., Shi G. P., Ren J., Nair S. (2015). Cathepsin K Knockout Alleviates Aging‐induced Cardiac Dysfunction. Aging Cell 14, 345–351. 10.1111/acel.12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Chen D., Hu S., Wang J., Liu H., Hu J., et al. (2018). Molecular Characterization, Expression and Cellular Localization of CYP17 Gene during Geese ( Anser cygnoides ) Follicular Development. Gene 658, 184–190. 10.1016/j.gene.2018.03.029 [DOI] [PubMed] [Google Scholar]
- Ivanova M., Dobrzycka K. M., Jiang S., Michaelis K., Meyer R., Kang K., et al. (2005). Scaffold Attachment Factor B1 Functions in Development, Growth, and Reproduction. Mol. Cel Biol 25, 2995–3006. 10.1128/mcb.25.8.2995-3006.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., et al. (2015). The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 47, 199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Hu S., Tu T., Huang Z., Tang Q., Ma J., et al. (2018). Global Long Noncoding RNA and mRNA Expression Changes between Prenatal and Neonatal Lung Tissue in Pigs. Genes 9, 443. 10.3390/genes9090443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. L. (2015). Ovarian Follicle Selection and Granulosa Cell Differentiation. Poult. Sci. 94, 781–785. 10.3382/ps/peu008 [DOI] [PubMed] [Google Scholar]
- Johnson P. (2012). Follicle Selection in the Avian Ovary. Reprod. Domest. Anim. 47, 283–287. 10.1111/j.1439-0531.2012.02087.x [DOI] [PubMed] [Google Scholar]
- Kang B., Jiang D. M., Zhou R. J., Yang H. M. (2009). Expression of Follicle-Stimulating Hormone Receptor (FSHR) mRNA in the Ovary of Zi Geese during Developmental and Egg Laying Stages. Folia Biol. (Krakow) 58, 61–66. 10.3409/fb58_1-2.61-66 [DOI] [PubMed] [Google Scholar]
- Kang Y.-J., Yang D.-C., Kong L., Hou M., Meng Y.-Q., Wei L., et al. (2017). CPC2: a Fast and Accurate Coding Potential Calculator Based on Sequence Intrinsic Features. Nucleic Acids Res. 45, W12–W16. 10.1093/nar/gkx428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik I. P., Desai P., Sukumar S., Dimitrijevic A., Rajalingam K., Mahalingam S. (2018). E4BP4/NFIL3 Modulates the Epigenetically Repressed RAS Effector RASSF8 Function through Histone Methyltransferases. J. Biol. Chem. 293, 5624–5635. 10.1074/jbc.ra117.000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder B. L., Palmer S. (2010). Examination of Transcriptional Networks Reveals an Important Role for TCFAP2C, SMARCA4, and EOMES in Trophoblast Stem Cell Maintenance. Genome Res. 20, 458–472. 10.1101/gr.101469.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Ocón-Grove O., Johnson A. L. (2013). Bone Morphogenetic Protein 4 Supports the Initial Differentiation of Hen (Gallus gallus) Granulosa Cells. Biol. Reprod. 88 (161), 161–167. 10.1095/biolreprod.113.109694 [DOI] [PubMed] [Google Scholar]
- Kim D., Johnson A. L. (2018). Differentiation of the Granulosa Layer from Hen Prehierarchal Follicles Associated with Follicle‐stimulating Hormone Receptor Signaling. Mol. Reprod. Dev. 85, 729–737. 10.1002/mrd.23042 [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J., Johnson A. L. (2016). Vascular Endothelial Growth Factor and Angiopoietins during Hen Ovarian Follicle Development. Gen. Comp. Endocrinol. 232, 25–31. 10.1016/j.ygcen.2015.11.017 [DOI] [PubMed] [Google Scholar]
- Kohl M., Wiese S., Warscheid B. (2011). “Cytoscape: Software for Visualization and Analysis of Biological Networks,” in Data Mining in Proteomics (Springer; ), 291–303. 10.1007/978-1-60761-987-1_18 [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. J. B. B. (2008). WGCNA: an R Package for Weighted Correlation Network Analysis. BMC Bioinformatics 9, 1–13. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gao G., Lin Y., Hu S., Luo Y., Wang G., et al. (2020). Pacific Biosciences Assembly with Hi-C Mapping Generates an Improved, Chromosome-Level Goose Genome. GigaScience 9, giaa114. 10.1093/gigascience/giaa114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang W., Li Q., Liu J., Zhang T., Zhou T., et al. (2015). The Comprehensive Mechanisms Underlying Nonhierarchical Follicular Development in Geese (Anser cygnoides). Anim. Reprod. Sci. 159, 131–140. 10.1016/j.anireprosci.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Liu Y., Du L., Osato M., Teo E. H., Qian F., Jin H., et al. (2007, The Zebrafish Udu Gene Encodes a Novel Nuclear Factor and Is Essential for Primitive Erythroid Cell Development. 110 , 99–106. 10.1182/blood-2006-11-059204 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lock F. E., Underhill-Day N., Dunwell T., Matallanas D., Cooper W., Hesson L., et al. (2010). The RASSF8 Candidate Tumor Suppressor Inhibits Cell Growth and Regulates the Wnt and NF-Κb Signaling Pathways. Oncogene 29, 4307–4316. 10.1038/onc.2010.192 [DOI] [PubMed] [Google Scholar]
- Lu F., Cui D., Mu B., Zhao L., Mu P. (2020). Downregulation of TMOD1 Promotes Cell Motility and Cell Proliferation in Cervical Cancer Cells. Oncol. Lett. 19, 3339–3348. 10.3892/ol.2020.11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Hankel I. L., Knisz J., Marquardt A., Chiang M.-Y., Grosse J., et al. (2010). The Justy Mutation Identifies Gon4-like as a Gene that Is Essential for B Lymphopoiesis. Blood 207, 1359–1367. 10.1084/jem.20100147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimova A., Garber J. J., Upadhyay G., Sharov A., Anastasoaie F., Yajnik V., et al. (2010). Neural Wiskott-Aldrich Syndrome Protein Modulates Wnt Signaling and Is Required for Hair Follicle Cycling in Mice. J. Clin. Invest. 120, 446–456. 10.1172/jci36478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity J., Deb M., Greene C., Das H. (2020). KLF2 Regulates Dental Pulp-Derived Stem Cell Differentiation through the Induction of Mitophagy and Altering Mitochondrial Metabolism. Redox Biol. 36, 101622. 10.1016/j.redox.2020.101622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe N., Matsuda-Minehata F., Goto Y., Maeda A., Cheng Y., Nakagawa S., et al. (2008). Role of Cell Death Ligand and Receptor System on Regulation of Follicular Atresia in Pig Ovaries. Reprod. Domest. Anim. 43, 268–272. 10.1111/j.1439-0531.2008.01172.x [DOI] [PubMed] [Google Scholar]
- Marini M., Manetti M., Sgambati E. (2019). Immunolocalization of VEGF/VEGFR System in Human Fetal Vomeronasal Organ during Early Development. Acta Histochem. 121, 94–100. 10.1016/j.acthis.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Meng B., Cao Z., Gai Y., Liu M., Gao M., Chen M., et al. (2019). Effects of Recombinant Goose Adiponectin on Steroid Hormone Secretion in Huoyan Geese Ovarian Granulosa Cells. Anim. Reprod. Sci. 205, 34–43. 10.1016/j.anireprosci.2019.03.019 [DOI] [PubMed] [Google Scholar]
- Moyer J. D., Nowak R. B., Kim N. E., Larkin S. K., Peters L. L., Hartwig J., et al. (2010). Tropomodulin 1-null Mice Have a Mild Spherocytic Elliptocytosis with Appearance of Tropomodulin 3 in Red Blood Cells and Disruption of the Membrane Skeleton. Blood 116, 2590–2599. 10.1182/blood-2010-02-268458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niknafs Y. S., Pandian B., Iyer H. K., Chinnaiyan A. M., Iyer M. K. (2017). TACO Produces Robust Multisample Transcriptome Assemblies from RNA-Seq. Nat. Methods 14, 68–70. 10.1038/nmeth.4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C., Zhang S., Mo G., Jiang Y., Li L., Xu H., et al. (2021). Effects of ODC on Polyamine Metabolism, Hormone Levels, Cell Proliferation and Apoptosis in Goose Ovarian Granulosa Cells. Poult. Sci. 100, 101226. 10.1016/j.psj.2021.101226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda M. J., Tarazona S., Conesa A. (2014). Next maSigPro: Updating maSigPro Bioconductor Package for RNA-Seq Time Series. Bioinformatics 30, 2598–2602. 10.1093/bioinformatics/btu333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocn-Grove O., Poole D., Johnson A. (2012). Bone Morphogenetic Protein 6 Promotes FSH Receptor and Anti-mllerian Hormone mRNA Expression in Granulosa Cells from Hen Prehierarchal Follicles. Reproduction 143, 825. [DOI] [PubMed] [Google Scholar]
- Onagbesan O., Bruggeman V., Decuypere E. (2009). Intra-ovarian Growth Factors Regulating Ovarian Function in Avian Species: a Review. Anim. Reprod. Sci. 111, 121–140. 10.1016/j.anireprosci.2008.09.017 [DOI] [PubMed] [Google Scholar]
- Pollier J., Rombauts S., Goossens A. (2013). “Analysis of RNA-Seq Data with TopHat and Cufflinks for Genome-wide Expression Analysis of Jasmonate-Treated Plants and Plant Cultures,” in Jasmonate Signaling (Springer; ), 305–315. 10.1007/978-1-62703-414-2_24 [DOI] [PubMed] [Google Scholar]
- Probst S., Arnold S. J. (2017). Eomesodermin-At Dawn of Cell Fate Decisions during Early Embryogenesis. Curr. Top. Dev. Biol. 122, 93–115. 10.1016/bs.ctdb.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Rivers C., Idris J., Scott H., Rogers M., Lee Y. B., Gaunt J., et al. (2015). iCLIP Identifies Novel Roles for SAFB1 in Regulating RNA Processing and Neuronal Function. BMC Biol. 13, 111–115. 10.1186/s12915-015-0220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolaki A., Drakakis P., Millingos S., Loutradis D., Makrigiannakis A. (2005). Novel Trends in Follicular Development, Atresia and Corpus Luteum Regression: a Role for Apoptosis. Reprod. Biomed. Online 11, 93–103. 10.1016/s1472-6483(10)61304-1 [DOI] [PubMed] [Google Scholar]
- Sauteur L., Krudewig A., Herwig L., Ehrenfeuchter N., Lenard A., Affolter M., et al. (2014). Cdh5/VE-cadherin Promotes Endothelial Cell Interface Elongation via Cortical Actin Polymerization during Angiogenic Sprouting. Cel Rep. 9, 504–513. 10.1016/j.celrep.2014.09.024 [DOI] [PubMed] [Google Scholar]
- Sebestyén E., Singh B., Miñana B., Pagès A., Mateo F., Pujana M. A., et al. (2016). Large-scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks. Genome Res. 26, 732–744. 10.1101/gr.199935.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheel A., Shao R., Brown C., Johnson J., Hamilton A., Sun D., et al. (2020). Acheron/LARP6 Is a Survival Protein that Protects Skeletal Muscle from Programmed Cell Death during Development. Front. Cel Dev. Biol. 8, 622. 10.3389/fcell.2020.00622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Li T., Zhang G., Wu P., Chen F., Lou Q., et al. (2019). Dynamic Expression and Functional Analysis of circRNA in Granulosa Cells during Follicular Development in Chicken. BMC genomics 20, 96–15. 10.1186/s12864-019-5462-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R. C. (2013). R: A Language and Environment for Statistical Computing. [Google Scholar]
- Torii T., Ogawa Y., Liu C.-H., Ho T. S.-Y., Hamdan H., Wang C.-C., et al. (2020). NuMA1 Promotes Axon Initial Segment Assembly through Inhibition of Endocytosis. J. Cel Biol 219, e201907048. 10.1083/jcb.201907048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson S. M., Kang K., Lee A. V., Oesterreich S. (2004). Structure-Function Analysis of the Estrogen Receptor α Corepressor Scaffold Attachment Factor-B1. J. Biol. Chem. 279, 26074–26081. 10.1074/jbc.m313726200 [DOI] [PubMed] [Google Scholar]
- Tsai S. M., Chu K. C., Jiang Y. J. (2020). Newly Identified Gon4l/Udu-Interacting Proteins Implicate Novel Functions. Sci. Rep. 10, 14213–14214. 10.1038/s41598-020-70855-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Maaten L., Hinton G. J. J. O. M. L. R. (2008). Visualizing Data Using T-SNE. JMLR 9, 2579–2605. [Google Scholar]
- Wang C., Gao G. L., Huang J. X., Zhang K. S., Zhong H., Wang H. W., et al. (2017). Nutritive Value of Dry Citrus Pulp and its Effect on Performance in Geese from 35 to 70 Days of Age. J. Appl. Poult. Res. 26, 253–259. 10.3382/japr/pfw069 [DOI] [Google Scholar]
- Yang M., Sun J., Zhang T., Liu J., Zhang J., Shi M. A., et al. (2008). Deficiency and Inhibition of Cathepsin K Reduce Body Weight Gain and Increase Glucose Metabolism in Mice. Atvb 28, 2202–2208. 10.1161/atvbaha.108.172320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Chen F., Zhang L., Wang H., Chen J., Zhang Z., et al. (2017). Smad9 Is a Key Player of Follicular Selection in Goose via Keeping the Balance of LHR Transcription. bioRxiv , 213546. [Google Scholar]
- Yuan J., Deng Y., Zhang Y., Gan X., Gao S., Hu H., et al. (2019). Bmp4 Inhibits Goose Granulosa Cell Apoptosis via PI3K/AKT/Caspase-9 Signaling Pathway. Anim. Reprod. Sci. 200, 86–95. 10.1016/j.anireprosci.2018.11.014 [DOI] [PubMed] [Google Scholar]
- Zhao R., He X.-W., Shi Y.-H., Liu Y.-S., Liu F.-D., Hu Y., et al. (2019). Cathepsin K Knockout Exacerbates Haemorrhagic Transformation Induced by Recombinant Tissue Plasminogen Activator after Focal Cerebral Ischaemia in Mice. Cel Mol Neurobiol 39, 823–831. 10.1007/s10571-019-00682-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These high-quality datasets have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSM3927138 to GSM3927181 (the detailed information is listed in Supplementary Table S2).