Abstract
Purpose:
The overall biological roles and clinical significance of most long noncoding RNAs(lncRNAs) in chemosensitivity are not fully understood. We investigated the biological function, mechanism and clinical significance of lncRNA NR_034085, which we termed miRNA processing related lncRNA(MPRL), in tongue squamous cell carcinoma(TSCC).
Experimental Design:
LncRNA expression in TSCC cell lines with cisplatin treatment was measured by lncRNA microarray and confirmed in TSCC tissues. The functional roles of MPRL were demonstrated by a series of in-vitro and in-vivo experiments. The miRNA profiles, RNA pull-down, RNA immunoprecipitation, serial deletion analysis and luciferase analyses were used to investigate the potential mechanisms of MPRL.
Results:
We found that MPRL expression was significantly upregulated in TSCC cell lines treated with cisplatin and transactivated by E2F1. MPRL controlled mitochondrial fission and cisplatin sensitivity through miR-483–5p. In exploring the underlying interaction between MPRL and miR-483–5p, we identified that cytoplasmic MPRL directly binds to pre-miR-483 within the loop region and blocks pre-miR-483 recognition and cleavage by TRBP-DICER-complex, thereby inhibiting miR-483–5p generation and upregulating miR-483–5p downstream target-FIS1 expression. Furthermore, overexpression or knockdown MPRL altered tumor apoptosis and growth in mouse xenografts. Importantly, we found that high expression of MPRL and pre-miR-483, and low expression of miR-483–5p were significantly associated with neoadjuvant chemosensitivity and better TSCC patients’ prognosis.
Conclusions:
We propose a model in which lncRNAs impair microprocessor recognition and efficient of pre-miRNA-cropping. In addition, our study reveals a novel regulatory network for mitochondrial fission and chemosensitivity and new biomarkers for predication of neoadjuvant chemosensitivity in TSCC.
Keywords: Long noncoding RNA, MPRL, mitochondrial fission, chemosensitivity, pre-miRNA processing, squamous cell carcinoma
Introduction
Cisplatin has been largely employed as a cornerstone treatment for a wide spectrum of solid neoplasms over the past 30 years. However, chemoresistance frequently develops and leads to therapeutic failure. The initial patient response to platinum-based therapies in oral squamous cell carcinoma (OSCC) is 80.6% (1); however, more than 70% of patients eventually relapse due to acquired resistance (2, 3). Numerous studies have focused on resistance mechanisms, but no substantive progress has been made in overcoming cisplatin resistance. Recent studies revealed that abnormal mitochondrial dynamics participate in the regulation of apoptosis (4, 5) and are linked to a variety of diseases(6). Recently, we demonstrated that miRNAs play an active role in mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma (TSCC)(3, 7).
miRNAs comprise a class of 19–25 nt small noncoding RNAs (ncRNAs) that fine-tune gene expression in eukaryotic organisms through inhibitory interactions based on partial complementarity with target mRNAs(8, 9). miRNA biogenesis is a multilayered process involving specific pathways to generate mature and functional molecules (10, 11). The vast majority of miRNAs are transcribed by RNA Polymerase II, which produces a long, capped, and polyadenylated primary molecule (pri-miRNA). The pri-miRNA folds into a hairpin structure and is a substrate for the RNase III family enzyme DROSHA, which releases an ~70 nt precursor miRNA (pre-miRNA) that is exported to the cytoplasm by XPO5. The pre-miRNA is further cropped by the TRBP-DICER complex generating an ~20 bp duplex, from which usually only one strand, the mature miRNA, is incorporated into the RNA-induced silencing complex (RISC)(12–15). Interestingly, a recent study reported a new regulatory mechanism for pri-miRNA processing at the posttranscriptional level(13); however, it is not yet clear whether miRNA biogenesis affects mitochondrial fission and cisplatin sensitivity.
Long noncoding RNAs (lncRNAs) are nonprotein-coding transcripts longer than 200 nt. A growing number of highly diverse ncRNAs have been revealed by global transcriptomic analysis, while only 2% of the genome is transcribed into protein-coding RNAs (16). LncRNAs with tissue- and development-specific expression patterns are associated with a variety of regulatory roles in different cell types, tissues and developmental conditions, such as chromatin modification(17), RNA processing(18), structural scaffolds(19) and modulation of apoptosis and invasion(20). Further, studies have demonstrated that lncRNAs can act as a competing endogenous RNA (ceRNA) to regulate miRNA expression (21, 22). A recent study reported that lncRNA could regulate pri-miRNA processing by DROSHA in the nucleus(13); however, the role of lncRNA in pre-miRNA cropping remains elusive.
Our present work revealed that the lncRNA MPRL participates in the regulation of mitochondrial fission and cisplatin sensitivity through the miR-483–5p-FIS1 axis. Moreover, our study found that MPRL can directly bind to pre-miR-483 and inhibit TRBP-DICER-mediated processing into mature miR-483. Remarkably, MPRL was further validated as a biomarker for predicting chemosensitivity and TSCC patient outcome. Our data reveal a novel role for lncRNA in promoting miRNA biogenesis in the cytoplasm, thus expanding the functions of lncRNA and miRNA in the mitochondrial network and chemosensitivity.
Materials and Methods
Cell culture
Three human TSCC cell lines, CAL-27, SCC-9 and SCC-25, were obtained from the American Type Culture Collection. Cisplatin was administered at its IC50(3, 7) at the indicated time. The stable cisplatin-resistant lines CAL-27-re and SCC-25-re were established by exposing CAL-27 or SCC-25 cells to cisplatin (Sigma, St Louis, MO, USA) in concentrations ranging from 10−7 M to 10−5 M(23). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. All cell lines were routinely tested for mycoplasma, and the genetic identity of the cell lines was confirmed by short tandem repeat (STR) profiling (ATCC). The cell lines were used for experiments within 10 passages after thawing.
LncRNA expression profiles
CAL-27 and SCC-9 cells were treated with cisplatin (Sigma, USA) at its IC50 (3, 7) for 24 h for lncRNA microarray assays. Sample labeling and array hybridization were performed by Arraystar Human lncRNA Microarray V3.0 according to the Arraystar microarray-based gene expression analysis protocol (Arraystar, Inc., Rockville, MD, USA).
miRNA expression profiles
CAL-27 and SCC-9 cells stably transduced with shRNA targeting MPRL were treated with cisplatin at the IC50 (Sigma, USA) for 24 h for miRNA microarray assays. Microarray experiments, target preparation and array hybridization were performed with an Agilent human miRNA microarray (8*60 K).
Rapid amplification of cDNA ends (RACE)
The 5′ and 3′-RACE experiments were performed using the Smart RACE CDNA Amplification Kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions. The primers used for mapping each end are listed in Table S1. The MPRL sequence from the RACE analyses is listed in Table S2.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as previously described(3, 7). Briefly, CAL-27 or SCC-9 cells (5×106) were washed with PBS and incubated for 10 min with 1% formaldehyde at room temperature. Crosslinking was halted by treatment with 0.1 M glycine for 5 min. The cells were washed twice with PBS, lysed for 1 h at 4°C in lysis buffer and then sonicated into chromatin fragments with an average length of 500–800 bp, as assessed via agarose gel electrophoresis. The samples were precleared with Protein-A agarose (Roche) for 1 h at 4°C on a rocking platform. Then, 5 μg of specific antibodies was added, and the samples were shaken overnight at 4°C. Immunoprecipitated DNA was purified using a QIAquick PCR purification kit (Qiagen) according to the manufacturer’s protocol. The final ChIP DNA was then used as a template in qPCR with the primers listed in Table S1. ChIP-grade anti-E2F1 antibody (Abcam, ab4070) and anti-IgG (Sigma, I5381) were used in this study.
Pre-miR-483 pull-down assay
The pre-miR-483 pull-down assay was performed as previously described with some modifications(22). CAL-27 cells were transfected with 50 nM wild-type (wt) MPRL or mutant (mut) MPRL vector and then harvested 24 h after transfection. The cells were washed with PBS, briefly vortexed, incubated in lysis buffer on ice for 10 min, and centrifuged for preclearing. Biotinylated DNA probes complementary to MPRL and a random probe (Table S3) were dissolved in 500 μl of wash/binding buffer. The probes were incubated with streptavidin-coated magnetic beads (Sigma) at 25°C for 2 h to generate probe-coated magnetic beads. Then, CAL-27 cell lysates were incubated with probe-coated beads, and after washing with the wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for qRT-PCR.
MPRL pull-down assay
The biotinylated DNA probe complementary to pre-miR-483 was synthesized and dissolved in 500 μl of wash/binding buffer (0.5 M NaCl, 20 mM Tris-HCl, pH 7.5, and 1 mM EDTA). The probes were incubated with streptavidin-coated magnetic beads (Sigma) at 25°C for 2 h to generate probe-coated magnetic beads. CAL-27 cell lysates were aliquoted for input, and the remaining lysates were incubated with probe-coated beads. After the beads were washed with wash/binding buffer, the RNA complexes bound to the beads were eluted and extracted for northern blot analysis. Biotin-labeled probes specific for pre-miR-483 and random pull-down probes (Table S3) were designed by and purchased from RiboBio (Guangzhou, China).
Electrophoretic mobility shift assay (EMSA)
The RNA substrate for the pre-miR-483 stem-loop sequence (76 nt) was obtained and labeled by random biotinylation during in vitro transcription from linearized DNA templates (pcDNA3.1(+)_pre-miR-483) using 0.25 mM biotin-16-UTP (Roche). MPRL RNA substrates (wt and mut, 298 nt) were transcribed in vitro from linearized DNA templates (MPRL-298 and mut-MPRL-298). All transcription reactions were performed using a T7 High Yield RNA synthesis kit (NEB #E2040). Binding reactions were carried out in 1x binding buffer (20 mM Tris-HCl, pH 8.0, 1 mM DTT, 1 mM MgCl2, 20 mM KCl, and 10 mM Na2HPO4-NaH2PO4, pH 8.0) with biotin-labeled pre-miR-483 alone or in the presence of increasing amounts (2.0–6.0 pmol) of unlabeled MPRL substrate. Each RNA mixture was preheated at 70ºC for 5 min, cooled and then maintained at 30ºC for 20 min. All reactions were then immediately separated in a native 5% polyacrylamide gel, transferred to a nylon membrane and developed using the BrightStar® BioDetect™ Kit System (Ambion).
RNA immunoprecipitation (RIP)
RIP was performed using a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (17–700, Millipore, Billerica, MA) according to the manufacturer’s instructions (24). Briefly, whole-cell extracts prepared in lysis buffer containing a protease inhibitor cocktail and RNase inhibitor were incubated on ice for 5 min and then centrifuged at 10,000 g and 4°C for 10 min. Magnetic beads were preincubated with 5 μg of IP-grade anti-DICER antibody (Abcam, ab14601) or anti-TRBP antibody (Abcam, ab180947) for 30 min at room temperature with rotation. The supernatant was added to bead-antibody complexes in immunoprecipitation buffer and incubated at 4°C overnight. Finally, RNA was purified and quantified by qRT-PCR. Input controls and normal IgG controls were assayed simultaneously to ensure that the signals were detected from RNA specifically bound to protein.
Serial deletion analysis and RNA pull-down in vitro
Serial deletion fragments of MPRL were inserted into pcDNA3.1 for in vitro transcription in the RNA pull-down assay. The RNA pull-down assay was performed as previously described with minor modifications (13). Fragmentation of in vitro transcribed biotin-labeled pre-miR-483 was generated by Pierce™ RNA 3’ End Biotinylation Kit (20160). Each reaction contained 80 pmol of pre-miR-483 and 320 pmol of sequentially deleted MPRL in the presence of cytoplasmic extracts from CAL-27 cells, and the relative amount of TRBP and DICER in each case was analyzed by western blot.
Luciferase assay
A luciferase assay was carried out as previously described with modifications(3, 7). We cloned the potential MPRL promoter region located −2000 to 200 from the MPRL transcriptional start site into the pGL3-Basic plasmid upstream of the luciferase reporter gene (pGL3-MPRL-wt). The predicted E2F1 binding site in pGL3-MPRL-wt was mutated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent, Catalog #200523) to obtain pGL3-MPRL-mut. In addition, the miR-483–5p response element in the 3’ UTR of FIS1 was cloned into the pGL3-control plasmid downstream of the luciferase reporter gene. Luciferase activity assays were performed as described previously (7, 25). Briefly, 1×105 cells were seeded into 24-well plates for 24 h. Then, CAL-27 cells with stable expression of wt-MPRL, mut-MPRL, MPRL shRNA or the corresponding negative control were transfected with pcDNA3.1_pre-miR-483, pGL3_FIS1–3’UTR or pGL3-Control empty vector using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Total amount of transfected DNA was maintained constant across transfections. After 24 h of transfection, cells were collected, and the assay was performed following the manufacturer’s instructions. Each luciferase assay was analyzed based on the ratio Renilla/Firefly. Luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Mitochondrial staining and analysis of mitochondrial fission
Mitochondrial staining was performed as described previously by us and other researchers with some modifications (3, 5, 7). Briefly, cells were plated onto coverslips and treated as described. Then, the cells were stained for 30 min with 0.1 μM MitoTracker Red CMXRos (Molecular Probes). Mitochondria were imaged using a laser scanning confocal TCS SP5 microscope (Leica, Solms, Germany). Mitochondrial morphology was assessed and quantified as described previously(26).
Apoptosis assay
Apoptosis was detected using TUNEL, flow cytometry and caspase-3/7 activity assays (7). TUNEL assays were performed using a kit from Roche (Cat. No. 11684795910) according to the user’s instructions. Sections were examined with an ImagerZ1 microscope (Zeiss, Jena, Germany). An investigator blinded to the treatment quantified 20 random fields of samples. Flow cytometry was performed using Annexin V and propidium iodide double staining (Sigma-Aldrich). Caspase-3/7 activity was determined using an Apo-ONE® Homogeneous Caspase-3/7 assay kit from Promega according to the manufacturer’s protocol.
Tumor xenografts
Male BALB/c nude mice aged 4 to 6 weeks were prepared for tumor implantation. CAL-27 cells (5×106/mouse) stably expressing MPRL or shRNA targeting MPRL were resuspended in 150 μL of PBS and injected subcutaneously into the flanks of the nude mice (n=6 in each group). One week after implantation, when the tumor became palpable at ~2 mm in diameter, cisplatin or saline was intraperitoneally injected at 5 mg/kg body weight every three days from days 8 to 32. At day 35, the primary tumors were carefully removed, imaged, and analyzed via western blotting (WB) and qRT-PCR.
Patient and tissue samples
Fresh tumor tissues from 23 TSCC patients for identification lncRNA profiles were obtained before and after neoadjuvant chemosensitivity, while specimens from 143 locally advanced TSCC patients were obtained before neoadjuvant chemosensitivity between Jan 1, 2004, and Dec 31, 2010. Patients with locally advanced resectable TSCC (stage III or IVA) underwent one or two cycles of neoadjuvant chemotherapy(1) (75 mg/m2 of cisplatin on day 1, 75 mg/m2 of docetaxel on day 1, and 750 mg/m2 of fluorouracil on days 1 to 5), and the tumor response to neoadjuvant chemotherapy was assessed by CT/MRI studies prior to radical resection.
Study approval
The patient study was conducted in accordance with the principles of the Declaration of Helsinki. Ethics approval was provided by the Sun Yat-sen University Committee for Ethical Review of Research Involving Human Subjects. Patients with TSCC were identified and provided written informed consent under 201460 of Sun Yat-sen Memorial Hospital. The animal experiments were performed in accordance with the Institutional Authorities’ guidelines and were formally approved by the Animal Ethics Committee of Sun Yat-sen University.
Statistical analysis
All data are presented as the mean ± SEM. Comparisons between 2 groups were performed by 2-tailed Student’s t tests using the SPSS 18.0 package (SPSS, Chicago, IL, USA). For multiple comparisons between groups, 1-way ANOVA followed by Dunnett’s multiple comparisons tests was performed. P values below 0.05 were considered significant.
Data availability
Data have been deposited in the Gene Expression Omnibus (GEO) DataSets (https://www.ncbi.nlm.nih.gov/gds) under the following accession numbers: GSE114929 and GSE115117.
Additional Materials and Methods are detailed in Supplementary Data.
Results
Differential expression of lncRNAs induced by cisplatin in TSCC cells and tumor tissues
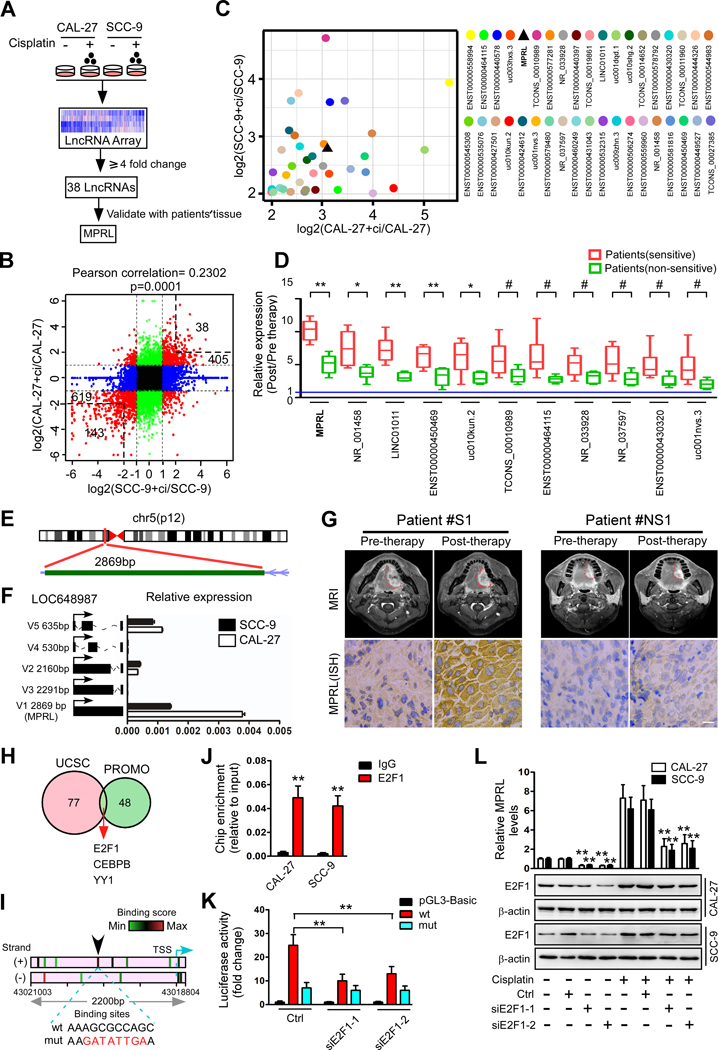
We have demonstrated that mitochondrial fission determines cisplatin sensitivity in TSCC (3, 7), and recent studies have reported that lncRNAs play active roles in regulating cancer biological properties (27). To understand whether lncRNAs are involved in regulating mitochondrial fission and cisplatin sensitivity in TSCC, we profiled lncRNA expression in two TSCC cell lines (CAL-27 and SCC-9) treated with or without cisplatin by LncRNA Array (Arraystar V3.0) (Figure 1A). There were 405 upregulated lncRNAs and 619 downregulated lncRNAs with significant differential expression (R2.0-fold) (Figure 1B and Figure S1A) in cisplatin-treated CAL-27 cells and SCC-9 cells compared to the corresponding untreated cells. To further narrow lncRNA candidates, we selected lncRNAs that showed at least a four-fold change in both groups of treated cells, yielding 38 upregulated lncRNAs and 143 downregulated lncRNAs (Figure 1B), and the 38 upregulated lncRNAs were further confirmed by qRT-PCR in both cell lines (Figure 1C). In an attempt to identify these 38 lncRNAs associated with chemosensitivity, we extracted RNA from TSCC tumors in chemosensitive and chemoresistant patients (Table S4) for further screening. From the 38 lncRNAs with significantly differential expression of in TSCC cell lines, 11 lncRNAs were found with significant upregulation in TSCC tissues from chemosensitive patients, comparing to the nonchemosensitive patients, while one lncRNA (RefSeq accession number NR_034085), here termed MPRL, was most upregulated in chemosensitive tumors (Figure 1D). Then, we confirmed that MPRL is located on chromosome 5 in humans and composed of one exon with a full length of 2869 nt in length by rapid amplification of cDNA ends (RACE) in the CAL-27 cell line (Figure 1E, Figure S1B, and Table S2). The noncoding nature of MPRL was confirmed by coding-potential analysis (Figure S1C and D). A previous study demonstrated that MPRL is one of the five splice variants of LOC648987 (28, 29). We found MPRL to be the most abundant transcript among all splice variants in both CAL-27 and SCC-9 cells (Figure 1F), implying that MPRL has potential regulatory activity in these cell lines. Importantly, the expression of MPRL determined by locked nucleic acid (LNA)-based in situ hybridization (ISH) was markedly increased in tumors from chemosensitive patients compared postchemotherapy with prechemotherapy (Figure 1G and Figure S2A). Analyses of GEO databases further showed that MPRL was significantly upregulated in chemosensitive breast cancer tumors (Figure S2B).
Figure 1. Differential expression of lncRNAs induced by cisplatin in TSCC cells and chemosensitive or nonsensitive tumors.
(A) Schematic flowchart depicting the strategy for microarray analysis and validation of lncRNAs. (B) Scatter plot of lncRNA expression in TSCC cells with or without cisplatin treatment. The red dots indicate differentially expressed lncRNAs (fold change ≥2); black dots indicate a fold change less than two in both cell lines, while green dots and blue dots indicate fold changes less than two in the SCC-9 and CAL-27 cells, respectively. X and y-axes, normalized sample signal values (log2 scale). (C) Thirty-eight lncRNAs upregulated by cisplatin treatment were identified by qRT-PCR in both CAL-27 and SCC-9 cells. (D) Eleven lncRNAs were significantly upregulated in TSCC tumors from chemosensitive and nonchemosensitive patients. (E) Schematic annotation of the MPRL genomic locus on chr5:43,016,152–43,019,003 in humans. Green rectangles represent exons. (F) The expression of five splice variants of LOC648987 in CAL-27 and SCC-9 cells. (G) Representative images (MRI scanning) of tumor response (upper panels) and MPRL expression (lower panels) in tissue specimens from patients with chemosensitive (S) and nonchemosensitive (NS) tumors. Scale bar, 20 μm. (H) Prediction of TFs of MPRL by overlaying ChIP-seq data from UCSC and the virtual laboratory of PROMO. (I) Binding motif analysis showing enriched E2F1 motifs in the MPRL promoter; the arrow indicates the binding sites with the highest score. The green to red color gradation is based on the ranking of each binding site from the minimum (green) to maximum (red) scores, which were analyzed by JASPAR. (J) ChIP-qPCR analysis of the E2F1 genomic occupancy in the MPRL promoter in CAL-27 and SCC-9 cells, as indicated. (K) Luciferase assay showing that E2F1 knockdown inhibits MPRL promoter activity in CAL-27 cells. (L) Cisplatin induced upregulation of E2F1 and MPRL expression, while silencing E2F1 decreased MPRL levels in both TSCC cell lines. #P<0.05, *P<0.01 and **P<0.001 versus control, 2-tailed Student’s t tests (D and J); **P<0.001, 1-way ANOVA followed by Dunnett’s tests (K and L).
To understand how MPRL levels are upregulated by cisplatin treatment in TSCC cells, we predicted putative transcription factors (TFs) by overlaying the ChIP-seq data from the UCSC Genome Browser and the virtual laboratory of PROMO (Figure 1H, Table S5). The intersection of data from the two databases showed three TFs, including E2F1, CEBPB and YY1. Among these TFs, E2F1 protein has been reported to accumulated in response to cisplatin treatment(30, 31), while E2F1 is frequently enriched at the MPRL promoter (Figure 1I). As expected, ChIP and a luciferase reporter assay also demonstrated the transcriptional functionality of E2F1 (Figure 1J and K, Figure S2C). Additionally, we found that E2F1 knockdown reduced the MPRL levels in CAL-27 and SCC-9 cells (Figure 1L), whereas overexpression of E2F1 upregulated the MPRL levels in both cell lines (Figure S2D).
MPRL enhances mitochondrial fission and cisplatin sensitivity and reverses cisplatin resistance in TSCC cells
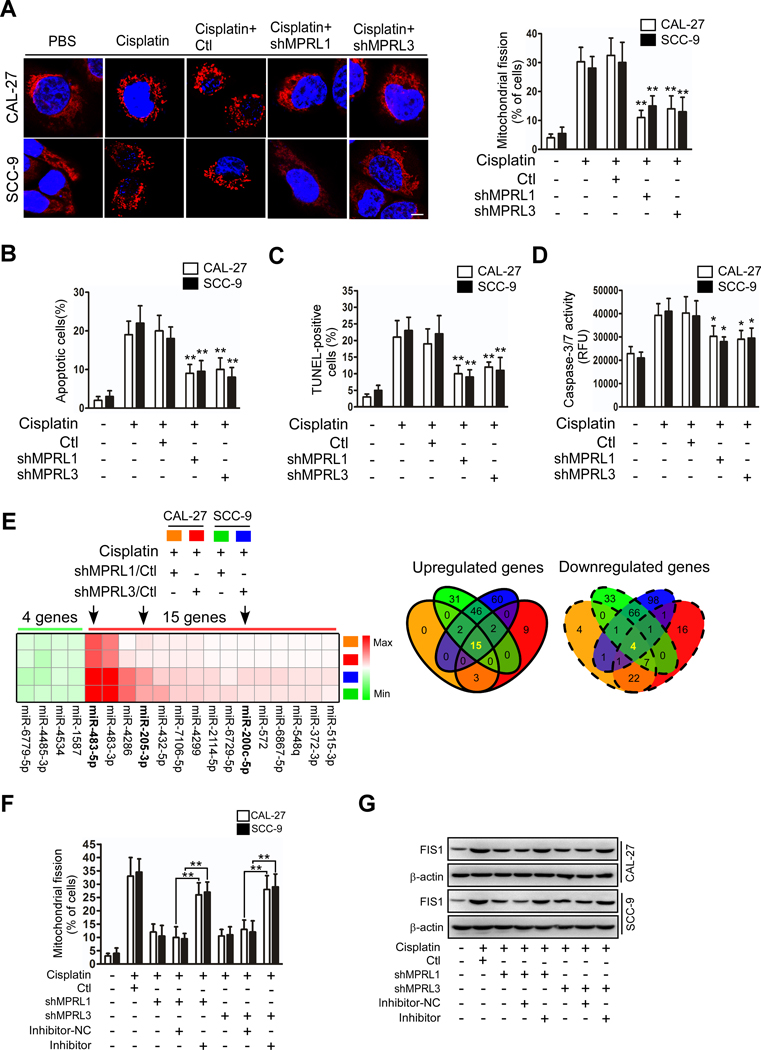
Our previous studies demonstrated that cisplatin induced apoptosis of TSCC cells through mitochondrial fission-mediated cytochrome c release and caspase-3/7 activation(3, 7). We wondered whether MPRL regulates mitochondrial fission and cisplatin sensitivity in TSCC cells. First, MPRL expression was silenced by four different shRNAs, while shRNA1 and shRNA3 showed better knockdown efficacy (Figure S3A) and were selected for further studies. As expected, we found that MPRL knockdown by shRNAs attenuated mitochondrial fission and apoptosis upon cisplatin treatment of TSCC cells (Figure 2A–D). In contrast, mitochondrial fission and apoptosis were augmented by enforced MPRL expression (Figure S3B–F) in TSCC cells under cisplatin treatment. Moreover, we wondered whether MPRL affects acquired cisplatin resistance. We found that MPRL was downregulated in two cisplatin-resistant TSCC cell lines (Figure S4A), which was consistent with the analysis of ovarian cancer cells from the GEO database (Figure S4B). Furthermore, cisplatin did not induce MPRL expression in cisplatin-resistant TSCC cells (Figure S4C), while overexpression of MPRL attenuated cisplatin resistance (Figure S4D). Collectively, these studies suggest that MPRL induces mitochondrial fission and cisplatin sensitivity in TSCC cells.
Figure 2. MPRL promotes mitochondrial fission and cisplatin sensitivity in TSCC through the miR-483–5p-FIS1 axis.
(A-D) Knockdown of MPRL attenuated mitochondrial fission and apoptosis in CAL-27 and SCC-9 cells. Mitochondrial fission was detected by staining with MitoTracker Red (left panel) and quantified (right) (A); Scale bar, 3 μm; cell apoptosis was detected using flow cytometry (B), TUNEL (C), and caspase-3/7 activity assays (D). (E) Target miRNAs of MPRL were screened by microarray in cells treated with cisplatin. Heat map (left panel) and Venn diagrams (right) depicted differentially expressed miRNAs in cisplatin-treated CAL-27 and SCC-9 cells stably expressing shMPRLs (fold change ≥1.5). (F) The inhibitory effect of MPRL knockdown on mitochondrial fission was attenuated after inhibiting miR-483–5p levels. Mitochondrial fission was detected by staining with MitoTracker Red. (G) Western blot analysis showed that the inhibitory effect of MPRL knockdown on FIS1 expression was attenuated after inhibiting miR-483–5p levels. *P<0.01 and **P<0.001, 2-tailed Student’s t tests.
MPRL regulates mitochondrial fission and apoptosis through miR-483–5p
Recent studies have suggested that lncRNAs may act as endogenous sponge RNAs by interacting with miRNAs, thereby influencing miRNA expression(21, 22, 32, 33). We previously found that miRNAs can regulate mitochondrial fission and cisplatin sensitivity in TSCC cells (3, 7). To explore the potential targets of lncRNAs regulating cisplatin sensitivity, we performed microarrays to detect miRNAs in both control and MPRL-silenced CAL-27 and SCC-9 cells under cisplatin treatment. Silencing of MPRL with both shRNA1 and shRNA3 upregulated 15 miRNAs and downregulated 4 miRNAs in CAL-27 and SCC-9 cells (Figure 2E). We applied qRT-PCR to identify all of these commonly changed miRNAs in both CAL-27 and SCC-9 cells (Figure S5A).
Interestingly, among the 15 upregulated miRNAs, miR-483–5p showed the greatest change and was demonstrated to inhibit mitochondrial fission and cisplatin sensitivity in TSCC in our previous study (7). We found that enforced expression of MPRL reduced mature miR-483–5p levels (Figure S5B). Therefore, we wondered whether MPRL regulates mitochondrial fission and cisplatin sensitivity in TSCC through miR-483–5p. To confirm the relationship between MPRL and miR-483–5p in the mitochondrial fission program and cisplatin sensitivity, we inhibited miR-483–5p(7) levels and observed that the inhibitory effect of MPRL knockdown on mitochondrial fission and apoptosis was attenuated in the presence of miR-483–5p inhibitors (Figure 2F, Figure S5C and D). Our previous report demonstrated that FIS1 is a downstream target of miR-483–5p. The decrease in FIS1 protein expression upon MPRL knockdown was also attenuated in the presence of miR-483–5p inhibitors (Figure 2G). In contrast, forced expression of miR-483–5p attenuated the mitochondrial fission and apoptosis induced by overexpression of MPRL in both TSCC cell lines (Figure S5E and F). Additionally, the analyses of miRNA-seq, RNA-seq and clinical data from the TCGA database also showed that higher miR-483–5p expression was significantly correlated with poor prognosis in multiple types of cancers (Figure S6A–E), further supporting the clinical relevance of the miR-483–5p signaling pathway. Taken together, these data suggest that miR-483–5p is a mediator of MPRL and that MPRL targets miR-483–5p in mitochondrial fission and apoptosis cascades in TSCC cells.
MPRL directly binds to the loop of pre-miR-483 and inhibits miR-483–5p generation
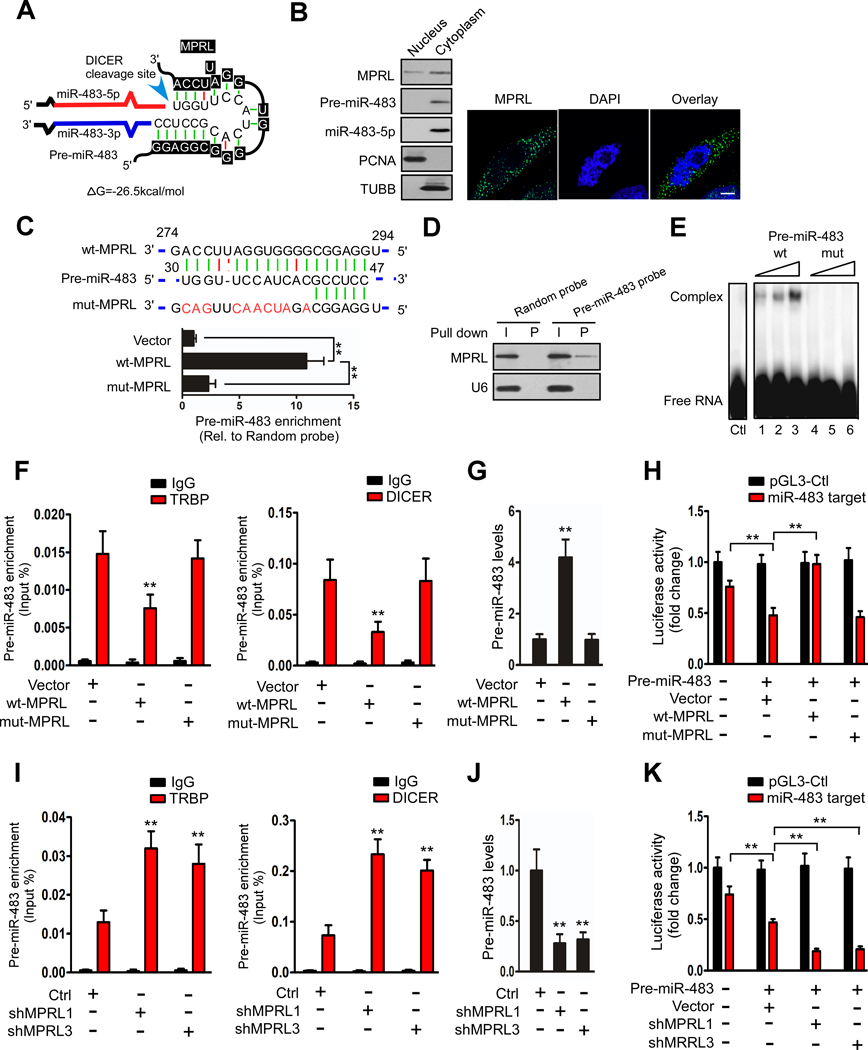
LncRNAs have been reported to act as endogenous sponge RNAs by interacting with miRNAs. To understand the mechanisms by which MPRL regulates the level of miR-483–5p, we tested whether MPRL interacts with miR-483–5p. The sequences of MPRL and miR-483–5p were compared using the bioinformatics programs miRPara and RNAhybrid, but no ideal target sites were identified. Notably, a recent study showed that lncRNAs can regulate pri-miRNA processing(13). Therefore, we compared the sequence of MPRL with that of pri-miR-483 and pre-miR-483 and found that MPRL contains a target site in miR-483 precursors with a free energy of −26.5 kcal/mol (Figure 3A). More importantly, the complementarity involves 19 nucleotides situated in the predicted DICER binding and cropping site within pre-miR-483, indicating the potential importance of the complementary sequence for the interplay of these two ncRNAs.
Figure 3. MPRL can directly bind to the loop of pre-miR-483.
(A) Predicted binding sites between MPRL and pre-miR-483; △G, free energy. (B) Northern blotting (left panel) and FISH (right panel) revealed that MPRL was located in both the nuclear and cytoplasm but was predominantly located in the cytoplasm, while pre-miR-483 and miR-483–5p were mainly located in the cytoplasm; scale bar, 3 μm. (C) MPRL can directly bind to pre-miR-483 in vivo. CAL-27 cells were transfected with wild-type or mutant MPRL (wt-MPRL or mut-MPRL) or empty vector. Twenty-four hours post-transfection, cells were harvested for biotin-based pull-down assays. The seed region of wt and mut-MPRL is shown (upper panel). Pre-miR-483 was analyzed by qRT-PCR (lower panel). (D) Pre-miR-483 can bind to MPRL in vivo. Cytoplasmic lysates of CAL-27 cells were incubated with magnetic beads coated with biotin-labeled probes specific to pre-miR-483 or random probes. After the beads were washed and the bead/RNA complexes were enriched, RNA was eluted from the streptavidin beads and analyzed by Northern blot. I, input (10% of each sample); P, pellet (100% of each sample). (E) EMSA showed that pre-miR-483 selectively binds in vitro to wt-MPRL (lanes 1–3) but not mut-MPRL (lanes 4–6). (F and G) Overexpression of wt-MPRL but not mut-MPRL reduced the coimmunoprecipitated pre-miR-483 with TRBP or DICER and increased pre-miR-483 levels in CAL-27 cells. (H) Luciferase reporter assays to test miR-483–5p functionality upon MPRL overexpression. CAL-27 cells were cotransfected with the indicated reporter vector, pre-miR-483, and with either wt-MPRL or mut-MPRL. Relative firefly luciferase/renilla activity was determined and enhanced by MPRL overexpression compared with the control vector (pGL3-Control). (I and J) Knockdown of MPRL increased the coimmunoprecipitated pre-miR-483 with TRBP or DICER and reduced pre-miR-483 levels in CAL-27 cells. (K) Luciferase activity was downregulated by silencing MPRL. **P<0.001, 1-way ANOVA followed by Dunnett’s tests for multiple comparisons.
To investigate the interaction of MPRL and pre-miR-483, we confirmed their subcellular location. Northern blotting and RNA fluorescence in situ hybridization (FISH) showed substantial amounts of MPRL in the nuclear compartment but also more abundantly in the cytosolic compartment (Figure 3B). Meanwhile, pre-miR-483 and miR-483–5p were mainly located in the cytoplasm (Figure 3B). To experimentally test the interaction between the two ncRNAs, we applied an RNA pull-down assay to test whether MPRL could pull down pre-miR-483. CAL-27 cells were transfected with wt-MPRL, mut-MPRL or empty vector (Figure S7A) and then harvested for biotin-based pull-down assays. Pre-miR-483 was pulled down by wt-MPRL, as analyzed by qRT-PCR, which was inhibited by the introduction of mutations that disrupt base pairing between MPRL and pre-miR-483 (Figure 3C), indicating sequence-specific recognition. We also employed an inverse pull-down assay to test whether pre-miR-483 could pull down endogenous MPRL using a biotin-labeled pre-miR-483-specific probe (Figure S7B). The results showed that pre-miR-483 and MPRL could be coprecipitated, although not all of the input MPRL was coprecipitated (Figure 3D). This finding suggests that a portion of MPRL directly interacts with pre-miR-483 in the cytoplasm. Furthermore, a 298-nt fragment of MPRL (MPRL-298) spanning the predicted pre-miR-483 binding site, including the wt and mut sequences, was transcribed in vitro after introducing substitutions in the predicted 10 complementary nucleotides and incubated in the presence of biotin-labeled pre-miR-483 (Figure S7C). Electrophoretic mobility shift assays (EMSAs) indicated that wt-MPRL formed a distinct complex with pre-miR-483 in vitro (Figure 3E, lanes 1–3), whereas this interaction was completely abolished by incubation with mut-MPRL (Figure 3E, lanes 4–6). Taken together, these data indicate that MPRL can directly bind to the pre-miR-483 loop sequence.
Moreover, enforced MPRL expression decreased the level of pre-miR-483 coimmunoprecipitated by TRBP and DICER (Figure 3F) along with the accumulation of pri-miR-483 (Figure S7D) and pre-miR-483 (Figure 3G) but reduction in miR-483–5p (Figure S7E) in CAL-27 cells. The MPRL that was mutated with 10 nucleotides had no effect on miR-483–5p generation or mitochondrial fission or cell apoptosis (Figure S7F and G). Furthermore, to test the implications at the level of mRNA target regulation, we used a luciferase-based reporter to test the miR-483–5p activity, with target sites in the 3’ UTR of FIS1 being cloned downstream of the luciferase reporter gene(7). Cotransfection with a construct for pre-miR-483 induced a reduction in luciferase activity, but overexpression of MPRL alleviated the repression of luciferase activity nearly to the levels of the empty vector, whereas mutant MPRL did not alter basal repression (Figure 3H). In contrast, silencing MPRL increased pre-miR-483 coimmunoprecipitation with TRBP and DICER (Figure 3I) and subsequently reduced pri-miR-483 (Figure S7H) and pre-miR-483 (Figure 3J) but unregulated miR-483–5p levels (Figure 2E and Figure S5A). A luciferase reporter assay further confirmed that knockdown of MPRL intensified the repression of luciferase activity (Figure 3K). Together, these results suggest that MPRL inhibits miR-483–5p expression and counteracts miR-483–5p repression of a functional target in vivo and that this effect requires the presence of complementary sequences.
MPRL masks pre-miR-483 recognition and cleavage by the TRBP-DICER complex
Next, we asked how MPRL inhibits miR-483–5p generation. It is well established that RNase III domains of DICER cleaves the pre-miRNA 3’ arm and 5’ arm while positioning loop residing within the RNase IIIA domain is essential for DICER activity(34). MPRL has been found to directly bind to the loop of pre-miR-483 that possibly causes inhibition of pre-miR-483 cleavage. Moreover, TRBP has been identified making an entry port for dsRNA in TRBP-DICER by interacting with the stem region of the dsRNA and also stimulates pre-miRNA processing by increasing substrate affinity to DICER and altering product length(35, 36). Given the disparity in length of two ncRNAs(pre-miR-483, 76 nt; MPRL, 2869 nt), we wonder whether MPRL blocked pre-miR-483 recognition by TRBP, thereby inhibiting the cleavage by TRBP-DICER complex(Figure 4A).
Figure 4. MPRL inhibits pre-miR-483 recognition and cleavage by the TRBP-DICER complex in CAL-27 cells.
(A) Schematic representation of inhibition of pre-miRNA recognition and cleavage by DICER-TRBP complex in the presence of MPRL. (B) Serial deletions of MPRL were used in RNA pull-down assays to identify valid length of MPRL that is required for physically masking the recognition of pre-miR-483 by the TRBP-DICER complex. (C) Site-directed mutagenesis of MPRL to 1100 nt resulted in the inability of MPRL to mask the recognition of pre-miR-483 by TRBP and DICER. (D and E) Overexpression of truncated isoforms of MPRL increased pre-miR-483 levels (D) and subsequently reduced miR-483–5p levels (E), while mutant MPRL (1–1100) had no effect. (F) Luciferase reporter assays showed that miR-483–5p function was inhibited by overexpression of truncated MPRL. (G) Forced expression of the truncated MPRL (1–1300) but not the MPRL (1–1100) abolished the increase in pre-miR-483 immunoprecipitated with TRBP and DICER by depletion of endogenous MPRL. (H and I) Overexpression of MPRL (1–1300) and MPRL (1–1100) increased pre-miR-483 levels (H) and reduced miR-483–5p levels (I). (J) Luciferase reporter assays demonstrated that MPRL(1–1300) and MPRL(1–1100) overexpression abolished the increase in miR-483–5p functionality by MPRL depletion. (K-L) qRT-PCR demonstrated that overexpressing MPRL or the truncated isoforms (1–1300) and (1–1100) prevented the reduction in pre-miR-484 (K) and attenuated the increase in miR-483–5p (L) induced by DICER, while silencing MPRL had the opposite effects. *P<0.01 and **P<0.001, 1-way ANOVA followed by Dunnett’s tests for multiple comparisons.
To ascertain how MPRL inhibits pre-miR-483 recognition, we used serial MPRL deletion analysis, in which pre-miR-483 incubated with truncated MPRL in the presence of cytoplasmic extract from CAL-27 cells. As expected, MPRL deletion to 1300 nt from its 5’-end preserved its ability to inhibit pre-miR-483 coimmunoprecipitated with TRBP and DICER, but deletion to 1100 nt abrogated such effect (Figure 4B), suggesting that MPRL size are critical to hide the close contact between the stem region of pre-miR-483 and TRBP that reduce pre-miR-483 recognition.
Moreover, RIP assays showed that the coimmunoprecipitation of pre-miR-483 was not downregulated in response to overexpression of MPRL(1–1100) (Figure 4C), but pre-miR-483 accumulated along with the reduction of mature miR-483, while transfection with mut-MPRL(1–1100) failed to take any effect (Figure 4D and E). Furthermore, enforced MPRL(1–1100) expression also greatly enhanced the luciferase expression in cells cotransfected with miR-483–5p target sites (Figure 4F) along with the increased mitochondrial fission and cisplatin sensitivity in both TSCC cell lines (Figure S8A and B). Thus, it suggested that 1100 nt deletion in 3’ end MPRL failed to block the pre-miR-483 recognition by TRBP but the truncated length reserved the suppressive function on pre-miR-483 cleavage by DICER, since MPRL(1–1100) included the binding sites with loop of pre-miR-483. Additionally, overexpression of truncated MPRL(1–1300) in MPRL-silenced cancer cells restored the function of MPRL in coimmunoprecipitation of pre-miR-483 with TRBP-DICER (Figure 4G), pre-miR-483 and mature miR-483–5p expression (Figure 4H–J), mitochondrial fission and cisplatin sensitivity (Figure S8C and D). Therefore, it suggested that MPRL mask pre-miR-483 recognition by TRBP required an appropriate length, which may hide the stem region of pre-miR-483 and caused the suppressive function as the full size. Finally, we verified the effect of MPRL on DICER-mediated pre-miR-483 cropping. Our data showed that enforced expression of MPRL or truncated isoforms (1–1300 and 1–1100) prevented the DICER-induced reduction in pre-miR-483 and increase in miR-483–5p, and knockdown of MPRL reversed this effect (Figure 4K and L, Figure S8E). Taken together, these data suggest that MPRL inhibit miR-483–5p generation through masking pre-miR-483 recognition and cleavage by TRBP-DICER complex which require a size-dependent manner.
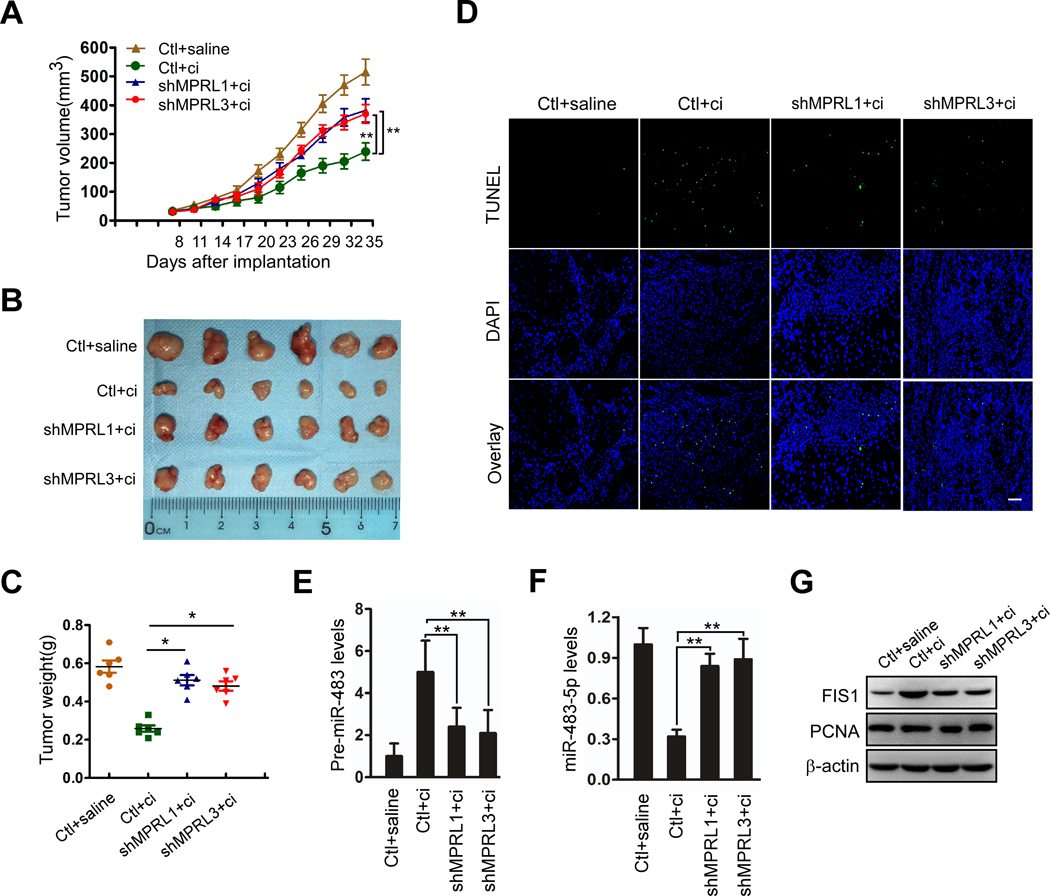
MPRL regulates cisplatin sensitivity in TSCC xenografts through the miR-483–5p signaling pathway
To further confirm the relationship between MPRL and pre-miR-483 in the regulation of cisplatin sensitivity, we established TSCC xenografts in vivo. CAL-27 cells with stable expression of MPRL shRNA (Figure S9A) showed enhanced tumor growth in the presence of cisplatin (Figure 5A, B and C). Moreover, apoptosis (Figure 5D and Figure S9B) and the expression of pre-miR-483 (Figure 5E) and pri-miR-483 (Figure S9C) were attenuated, and miR-483–5p levels were increased (Figure 5F) in cisplatin-treated xenografts that stably expressed MPRL shRNA. Meanwhile, FIS1, the direct target of miR-483–5p(7), was downregulated by silencing MPRL, but PCNA expression was not significantly changed in any group (Figure 5G), indicating that the influence of MPRL was not secondary to impaired proliferation.
Figure 5. MPRL knockdown inhibits apoptosis and cisplatin sensitivity in CAL-27 cell xenografts in BALB/c nude mice.
(A) Tumor growth curves for CAL-27 tumors. BALB/c nude mice bearing xenografts of CAL-27 cells with stable knockdown of MPRL or negative control (Ctl) were treated with saline or cisplatin (n=6 per group). The results are expressed as the mean ± SEM. (B) Photomicrographs of tumors from each group at day 35. (C) Tumor weight for each group. (D) TUNEL assays showed that apoptosis in response to cisplatin was attenuated by MPRL knockdown; Scale bar, 20 μm. (E and F) MPRL knockdown decreased pre-miR-483 expression (E) but upregulated miR-483–5p expression (F) in CAL-27 cell xenografts upon treatment with cisplatin. (G) Western blot showing the inhibitory effect of MPRL knockdown on FIS1 expression but not PCNA expression in cisplatin-treated tumors. (A) **P<0.001, 2-way ANOVA followed by Bonferroni’s post test; (C, E, F) *P<0.01 and **P<0.001, 1-way ANOVA followed by Dunnett’s tests for multiple comparisons.
In contrast, overexpression of MPRL inhibited tumor growth and enhanced cisplatin sensitivity (Figure S10A–D). Meanwhile, MPRL, pri-miR-483, pre-miR-483, miR-483–5p and miR-483–5p downstream gene FIS1 expression (Figure S10E–I) were also detected, and the results consistently supported that enforced MPRL expression inhibited pre-miR-483 processing in vivo. These results suggest that MPRL regulates cisplatin sensitivity and apoptosis of tumors by directly targeting pre-miR-483 processing.
High MPRL or pre-miR-483 expression and low miR-483–5p expression are associated with neoadjuvant chemosensitivity and good patient prognosis
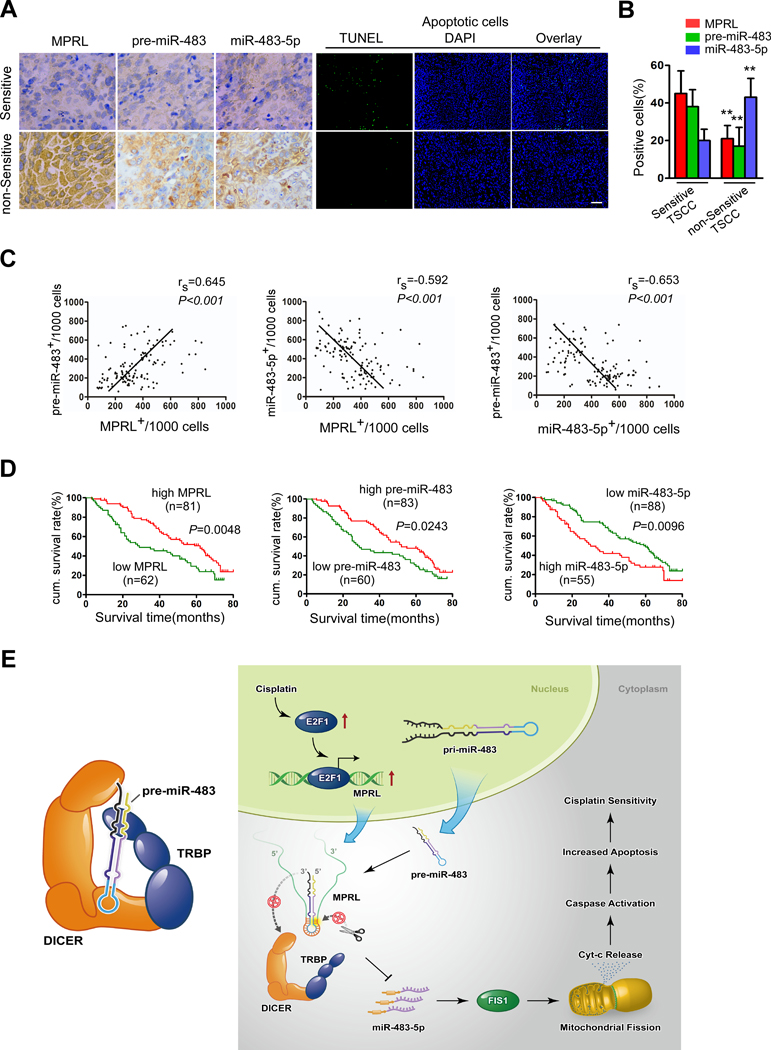
We evaluated the clinical significance of MPRL, pre-miR-483 and miR-483–5p with respect to chemosensitivity and prognosis of patients with TSCC. We performed a retrospective analysis of TSCC samples from 143 patients. In situ hybridization demonstrated that MPRL and pre-miR-483 expression were higher, while miR-483–5p expression was lower in neoadjuvant chemosensitive TSCC samples than in nonsensitive samples (Figure 6A). Consequently, neoadjuvant chemosensitive TSCC samples presented a higher percentage of apoptotic cells (Figure 6A), with a significant difference in the expression profiles, as determined by the percentage of positive cells (Figure 6B). Additionally, a Spearman rank order correlation analysis showed that MPRL expression correlated with pre-miR-483 levels (rs=0.645, P<0.001; Figure 6C); however, MPRL (rs=−0.592, P<0.001) and pre-miR-483 (rs=−0.653, P<0.001) expression in TSCC samples was inversely correlated with miR-483–5p levels.
Figure 6. MPRL, pre-miR-483 and miR-483–5p expression correlates with chemosensitivity and prognosis in TSCC patients.
(A) MPRL, pre-miR-483 and miR-483–5p expression and apoptosis were demonstrated in chemosensitive versus nonsensitive TSCC samples. MPRL, pre-miR-483 and miR-483–5p expression was analyzed using in situ hybridization (×200); apoptosis was detected using TUNEL assays; Scale bar, 20 μm. (B) Quantification of MPRL, pre-miR-483 and miR-483–5p expression in chemosensitive versus nonsensitive TSCC tumors. (C) Associations among MPRL, pre-miR-483 and miR-483–5p expression in TSCC were analyzed via Spearman rank order correlation. (D) Kaplan-Meier survival curves for TSCC patients were plotted for MPRL, pre-miR-483 and miR-483–5p expression, and survival differences were analyzed using a log-rank test. **P<0.001, 2-tailed Student’s t tests. (E) Schematic diagram depicting the proposed model in which MPRL regulates pre-miR-483 processing and determines mitochondrial fission and cisplatin sensitivity. (Left panel) TRBP recruits pre-miRNA-483 to the TRBP-DICER complex and ensures efficient pre-miRNA-483 processing. (Right panel) E2F1 transactivates MPRL expression under cisplatin treatment conditions. MPRL directly binds to the loop of pre-miR-483 and inhibits pre-miRNA recognition by physically masking the TRBP binding sites and blocking its stable association and cleavage by the TRBP-DICER complex, which compromises miRNA-483 generation and enhances FIS1 expression along with upregulated mitochondrial fission and cisplatin sensitivity.
Next, we analyzed the association of MPRL, pre-miR-483, and miR-483–5p expression with the clinicopathological status of TSCC patients (Table 1). No significant correlation between MPRL, pre-miR-483 or miR-483–5p expression with sex, age, lymph node status or clinical stage was observed; however, their expression was significantly associated with neoadjuvant chemosensitivity. Chemosensitive tumors expressed higher levels of MPRL and pre-miR-483 and lower levels of miR-483–5p. We also evaluated the correlation between MPRL, pre-miR-483 and miR-483–5p expression and patient overall survival (OS). A univariate Cox regression analysis indicated that patients with high MPRL and pre-miR-483 expression levels or low miR-483–5p levels had a longer OS (Table 1 and Figure 6D). The cumulative survival rate at 60 months was 46.91%, 44.58% and 46.59% among patients with high MPRL, high pre-miR-483 and low miR-483–5p expression, respectively; this rate was only 25.81%, 28.33% and 23.64% among those with low MPRL, low pre-miR-483 and high miR-483–5p expression, respectively (Table 1). Furthermore, a multivariate Cox regression analysis revealed that high MPRL expression and low miR-483–5p expression is an independent prognostic factor for good OS in patients with TSCC (Table 2). Together, these data suggest that MPRL and its direct targets pre-miR-483 and miR-483–5p correlate with neoadjuvant chemosensitivity and the OS of patients with TSCC.
Table 1.
Correlation among clinicopathological status and the expression of MPRL, pre-miR-483 or miR-483-5p in TSCC patients.
| Characteristics | MPRL(%) | P | pre-miR-483(%) | P | miR-483-5p(%) | P | |||
|---|---|---|---|---|---|---|---|---|---|
| No. of low Expression | No. of high Expression | No. of low Expression | No. of high Expression | No. of low Expression | No. of high Expression | ||||
| Sex | 0.584 | 0.916 | 0.131 | ||||||
| Male | 35(45.5) | 42(54.5) | 32(41.6) | 45(58.4) | 43(55.8) | 34(44.2) | |||
| Female | 27(40.9) | 39(59.1) | 28(42.4) | 38(57.6) | 45(68.2) | 21(31.8) | |||
| Age | 0.291 | 0.664 | 0.352 | ||||||
| <50 | 26(49.1) | 27(50.9) | 21(39.6) | 32(60.4) | 30(56.6) | 23(43.4) | |||
| ≥50 | 36(40.0) | 54(60.0) | 39(43.3) | 51(56.7) | 58(64.4) | 32(35.6) | |||
| Node metastasis | 0.951 | 0.073 | 0.084 | ||||||
| N0 | 28(43.1) | 37(56.9) | 22(33.8) | 43(66.2) | 45(69.2) | 20(30.8) | |||
| N+ | 34(43.6) | 44(56.4) | 38(48.7) | 40(51.3) | 43(55.1) | 35(44.9) | |||
| Clinical stage | 0.658 | 0.119 | 0.607 | ||||||
| Ⅲ | 39(44.8) | 48(55.2) | 41(47.1) | 46(52.9) | 55(63.2) | 32(36.8) | |||
| Ⅳ | 23(41.1) | 33(58.9) | 19(33.9) | 37(66.1) | 33(58.9) | 23(41.1) | |||
| Cisplatin | <0.001 | 0.028 | <0.001 | ||||||
| Sensitive | 21(28.0) | 54(72.0) | 25(33.3) | 50(66.7) | 62(81.6) | 14(18.4) | |||
| Non-sensitive | 41(60.3) | 27(39.7) | 35(51.5) | 33(48.5) | 26(38.8) | 41(61.2) | |||
| Status(60 months) | 0.010 | 0.048 | 0.006 | ||||||
| Survival | 16(29.6) | 38(70.4) | 17(31.5) | 37(68.5) | 41(75.9) | 13(24.1) | |||
| Death | 46(51.7) | 43(48.3) | 43(48.3) | 46(51.7) | 47(52.8) | 42(47.2) | |||
Table 2.
Univariate and multivariate analysis of factors associated with overall survival of patients with TSCC.
| Vavirable | Cases number | HR(95%CI) | P |
|---|---|---|---|
| Univariate analysis | |||
| Sex | |||
| Male vs Female | 77/66 | 0.949(0.483–1.864) | 0.746 |
| Age(years) | |||
| <50 vs ≥50 | 53/90 | 1.203(0.826–1.752) | 0.405 |
| Node metastasis | |||
| N0 vs N+ | 65/78 | 1.494(1.106–2.017) | 0.039 |
| Clinical stage | |||
| Ⅲ vs Ⅳ | 87/56 | 2.018(1.387–2.936) | <0.001 |
| Cisplatin | |||
| Sensitive vs Non-sensitive | 75/68 | 0.616(0.426–0.892) | 0.045 |
| MPRL | |||
| Low vs High | 62/81 | 1.721(1.215–2.438) | 0.005 |
| pre-miR-483 | |||
| Low vs High | 60/83 | 1.522(1.108–2.093) | 0.024 |
| miR-483-5p | |||
| Low vs High | 88/55 | 1.661(1.196–2.307) | 0.010 |
| Multivariate analysis | |||
| Node metastasis | |||
| N0 vs N+ | 65/78 | 1.602(1.192–2.154) | 0.023 |
| Clinical stage | |||
| Ⅲ vs Ⅳ | 87/56 | 2.122(1.476–3.051) | <0.001 |
| Cisplatin | |||
| Sensitive vs Non-sensitive | 75/68 | 0.586(0.426–0.806) | 0.037 |
| MPRL | |||
| Low vs High | 62/81 | 1.763(1.234–2.519) | 0.004 |
| miR-483-5p | |||
| Low vs High | 88/55 | 1.708(1.207–2.416) | 0.014 |
Discussion
The present study shows that MPRL, which is transactivated by E2F1, regulates mitochondrial fission and consequently, cisplatin sensitivity through miR-483–5p. In exploring the mechanism for regulation of miR-483–5p expression, we found that lncRNA MPRL can directly bind to pre-miRNA loop sites and mask its recognition and cleavage by TRBP-DICER complex. Our results reveal a novel regulatory model for mitochondrial fission that affects cisplatin chemosensitivity via lncRNA and miRNA biogenesis in cancer cells (Figure 6E).
LncRNAs have been reported to be involved in a wide range of biological functions, including chromatin modification (17), gene transcription regulation (37), RNA processing (13, 18), and mRNA translation(38). Given the structural similarity of mRNA and lncRNA, the existence of natural miRNA binding sites in lncRNA was not entirely unexpected. The interactions of lncRNAs and miRNAs have been globally mapped by the HITS-CLIP technique (39). Consequently, an increasing number of lncRNAs have been identified as ceRNAs (40). In other words, lncRNAs can act as miRNA sponges (41) and reduce the activity of target miRNAs without essentially altering their biogenesis (21, 32). However, a recent study showed that the lncRNA Uc.283+A prevents pri-miRNA-195 cleavage by Drosha(13). These data extend the RNA-directed regulation of miRNA biogenesis.
Mature miRNAs are generated via two-step processing by DROSHA and DICER. The second processing step is cleavage by DICER in the cytoplasm. DICER cleaves pre-miRNA and generates mature miRNA. Human DICER is a multidomain enzyme that consists of several RNA-binding domains, including the PAZ domain and tandem RNase III domains(14, 42). The PAZ domain recognizes the 2 nt 3’-overhang of the pre-miRNA, while the region between the PAZ and RNase III domains acts as a molecular ruler that defines the mature miRNA size(43). However, DICER requires RNA-binding partners that assist in efficient and accurate pre-miRNA processing, and TRBP has been well demonstrated to be a critical factor in facilitating this procedure(12, 14, 35). By being tightly associated with DICER, TRBP possesses high affinity towards dsRNA with flexible and stretched structure to recruit and position the 3’-end of pre-miRNA to the PAZ domain of DICER, with further verification of the authenticity of the substrate(14, 36) and TRBP also affects product length(35). If DICER is decoupled from TRBP, it is challenged and cannot bind and process pre-miRNA efficiently anymore with RNA abundance in the cell.
In our present study, we found that MPRL with a full length of 2869 nt, which was transactivated by E2F1, directly bound to the loop sites of pre-miR-483 (76 nt) and inhibited its generation. To explore the interaction between these two ncRNAs, we revealed that MPRL masked the pre-miR-483 recognition by TRBP, while a deletion to 1100 nt lost the suppressive function, suggesting truncated MPRL fails to hide the stem region. Moreover, even MPRL(1–1100) incapacitated masking pre-miR-483 stem region, pre-miR-483 was unable to cleaved by DICER since the truncated MPRL(1–1100) binding with pre-miR-483 loop, thereby blocking RNase III domain-dependent cleavage site(44, 45). Although the secondary or tertiary structure of MPRL might affect the contact between pre-miR-483 and TRBP-DICER complex, we reveal for the first time lncRNA size directly influence interaction between RNA and RNA-binding protein without mobilizing other regulatory molecules. Align with our findings, another cytoplasmic lncRNA, NKILA, was found to inhibit IKK-induced IkB phosphorylation by directly masking the phosphorylation sites of IkB from IKK(46). Therefore, to uncover the yashmak of lncRNA, the further study needs to investigate the structure of RNA and the induced functional alteration in cell with large amount of RNA structures.
Emerging data suggest that abnormal mitochondrial morphology may be relevant to various aspects of disease and apoptosis. We first investigated the important function of mitochondrial fission in cisplatin sensitivity. Thus far, it remains unclear whether lncRNA is involved in the regulation of cisplatin chemosensitivity through mitochondrial dynamics. Our present work indicates that MPRL can regulate mitochondrial fission and cisplatin sensitivity through pre-miR-483 processing and miR-483–5p, as well as its direct target FIS1. This work sheds new light on the understanding of mitochondrial fission and chemosensitivity. More importantly, based on our retrospective data, MPRL and miR-483–5p were further identified as biomarkers to predict neoadjuvant chemosensitivity and survival in TSCC patients, which is consistent with the TCGA or GEO database in multiple types of human cancer, suggesting the tumor suppressive role of MPRL. Additionally, patients with low MPRL expression were not sensitive to cisplatin-based chemotherapy and unable to benefit from neoadjuvant chemosensitivity; therefore, the earliest surgery or other strategies should be considered. Finally, our findings have therapeutic implications. Compared to protein, RNA is a malleable evolutionary substrate, and mutation or deletion of the regions outside its functional domains may not interfere with its core functionality(47). Therefore, synthetically engineered MPRLs containing the functional domains that have pre-miR-483 processing functions can be delivered alone or in combination with cisplatin-based chemotherapy to test for their therapeutic effects, which may thus offer a type of RNA therapy.
In summary, numerous studies have reported a range of mechanisms involving RNA-RNA and RNA-protein interactions that regulate miRNA biogenesis, but our study characterizes a regulatory control of miRNA processing based on interactions between two classes of ncRNAs, highlighting lncRNA directly masking recognition and cleavage of pre-miRNA by the TRBP-DICER complex in regulating mitochondrial programs and chemosensitivity. Moreover, detection of MPRL expression is worth taking into account because neoadjuvant chemosensitivity and synthetically engineered MPRLs may be important for overcoming chemoresistance, which needs further exploration.
Supplementary Material
Significance:
These findings uncover a novel mechanism by which lncRNA determines mitochondrial fission and cisplatin chemosensitivity by inhibition of pre-miRNA processing and provide for the first time the rationale for lncRNA and miRNA biogenesis for predicting chemosensitivity and patient clinical prognosis
Translational Relevance.
Long noncoding RNAs (lncRNAs) are emerging new players that control cellular programs and are involved in a network of interactions with other noncoding RNAs (ncRNAs). However, little is known about the mechanism and clinical relevance of lncRNA-miRNA processing. This is the first report of the interaction of lncRNA and pre-miRNA-cropping in chemosensitivity, in which the MPRL level is significantly upregulated in chemosensitive TSCC tissues and TSCC cell lines induced by cisplatin and associated with better patient prognosis. Moreover, MPRL promoted mitochondrial fission and cisplatin chemosensitivity by modulating the miR-483–5p-FIS1 axis by directly binding to pre-miR-483 within the loop region and blocking pre-miR-483 recognition and cleavage by the TRBP-DICER complex. This finding unmasks the tumor suppressive role of MPRL and provides a biomarker panel of MPRL-pre-miR-483-miR-483–5p for accurate prediction of neoadjuvant chemosensitivity and patient survival.
Acknowledgments
We are grateful to Yingjin Lin for critical reading of the manuscript. We appreciate Faya Liang and Sha Fu for their great help in analyzing the tissue slides.
Financial Support: This work was supported by grants from the National Natural Science Foundation of China (81472521, 81402251, 81502350, 81672676, 81772890, 81872194); Guangdong Science and Technology Development Fund (2015A030313181, 2016A030313352, 2017A030311011); Science and Technology Program of Guangzhou (201607010108, 201803010060); Fundamental Research Funds for the Central Universities(16ykpy10); The Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, Sun-Yat-Sen University (KLB09001); Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology (2013163).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Zhong LP, Zhang CP, Ren GX, Guo W, William WN Jr., , Sun J, et al. Randomized phase III trial of induction chemotherapy with docetaxel, cisplatin, and fluorouracil followed by surgery versus up-front surgery in locally advanced resectable oral squamous cell carcinoma. J Clin Oncol. 2013;31:744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson MK, Li Y, Murphy B, Hussain MH, DeConti RC, Ensley J, et al. Randomized phase III evaluation of cisplatin plus fluorouracil versus cisplatin plus paclitaxel in advanced head and neck cancer (E1395): an intergroup trial of the Eastern Cooperative Oncology Group. J Clin Oncol. 2005;23:3562–7. [DOI] [PubMed] [Google Scholar]
- 3.Fan S, Liu B, Sun L, Lv XB, Lin Z, Chen W, et al. Mitochondrial fission determines cisplatin sensitivity in tongue squamous cell carcinoma through the BRCA1-miR-593–5p-MFF axis. Oncotarget. 2015;6:14885–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–8. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017;26:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan S, Chen WX, Lv XB, Tang QL, Sun LJ, Liu BD, et al. miR-483–5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 2015;362:183–91. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–74. [DOI] [PubMed] [Google Scholar]
- 10.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. [DOI] [PubMed] [Google Scholar]
- 11.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. [DOI] [PubMed] [Google Scholar]
- 12.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liz J, Portela A, Soler M, Gomez A, Ling H, Michlewski G, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138–47. [DOI] [PubMed] [Google Scholar]
- 14.Fareh M, Yeom KH, Haagsma AC, Chauhan S, Heo I, Joo C. TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments. Nat Commun. 2016;7:13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. [DOI] [PubMed] [Google Scholar]
- 17.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343–50. [DOI] [PubMed] [Google Scholar]
- 18.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–62. [DOI] [PubMed] [Google Scholar]
- 21.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou LY, et al. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PLoS Genet. 2014;10:e1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Yao Y, Liu B, Lin Z, Lin L, Yang M, et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31:432–45. [DOI] [PubMed] [Google Scholar]
- 24.Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu BH, Chen H, Cai CM, Fang JZ, Wu CC, Huang LY, et al. Epigenetic silencing of JMJD5 promotes the proliferation of hepatocellular carcinoma cells by down-regulating the transcription of CDKN1A 686. Oncotarget. 2016;7:6847–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka A, Youle RJ. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol Cell. 2008;29:409–10. [DOI] [PubMed] [Google Scholar]
- 27.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu F, Megyesi J, Safirstein RL, Price PM. Involvement of the CDK2-E2F1 pathway in cisplatin cytotoxicity in vitro and in vivo. Am J Physiol Renal Physiol. 2007;293:F52–9. [DOI] [PubMed] [Google Scholar]
- 31.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 32.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starega-Roslan J, Galka-Marciniak P, Krzyzosiak WJ. Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer. Nucleic Acids Res. 2015;43:10939–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–6. [DOI] [PubMed] [Google Scholar]
- 38.Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. 2015;34:181–91. [DOI] [PubMed] [Google Scholar]
- 39.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. [DOI] [PubMed] [Google Scholar]
- 41.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. [DOI] [PubMed] [Google Scholar]
- 43.Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, et al. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell. 2014;53:606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. [DOI] [PubMed] [Google Scholar]
- 45.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, et al. Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81. [DOI] [PubMed] [Google Scholar]
- 47.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in the Gene Expression Omnibus (GEO) DataSets (https://www.ncbi.nlm.nih.gov/gds) under the following accession numbers: GSE114929 and GSE115117.
Additional Materials and Methods are detailed in Supplementary Data.