Abstract
Background
Salmonella is a major cause of zoonotic illness around the world, arising from direct or indirect contact with a range of animal reservoirs. In the Australian state of New South Wales (NSW), salmonellosis is believed to be primarily foodborne, but the relative contribution of animal reservoirs is unknown.
Methods
The analysis included 4543 serotyped isolates from animal reservoirs and 30,073 serotyped isolates from domestically acquired human cases in NSW between January 2008 and August 2019. We used a Bayesian source attribution methodology to estimate the proportion of foodborne Salmonella infections attributable to broiler chickens, layer chickens, ruminants, pigs, and an unknown or unsampled source. Additional analyses included covariates for four time periods and five levels of rurality.
Results
A single serotype, S. Typhimurium, accounted for 65–75% of included cases during 2008–2014 but < 50% during 2017–2019. Attribution to layer chickens was highest during 2008–2010 (48.7%, 95% CrI 24.2–70.3%) but halved by 2017–2019 (23.1%, 95% CrI 5.7–38.9%) and was lower in the rural and remote populations than in the majority urban population. The proportion of cases attributed to the unsampled source was 11.3% (95% CrI 1.2%–22.1%) overall, but higher in rural and remote populations. The proportion of cases attributed to pork increased from approximately 20% in 2009–2016 to approximately 40% in 2017–2019, coinciding with a rise in cases due to Salmonella ser. 4,5,12:i:-.
Conclusion
Layer chickens were likely the primary reservoir of domestically acquired Salmonella infections in NSW circa 2010, but attribution to the source declined contemporaneously with increased vaccination of layer flocks and tighter food safety regulations for the handling of eggs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-021-06950-7.
Keywords: Salmonella infections, Source attribution, Foodborne disease, Gastroenteritis, Disease reservoir, Zoonosis, Bayesian analysis
Background
Salmonellosis is a common cause of foodborne illness and hospitalisation across Australia. Annual notification rates for salmonellosis in Australia’s most populous state, New South Wales (NSW), ranged between 38.1 and 57.9 per 100,000 population during 2009–2019 [1]. However, for every notified case of salmonellosis, there are an estimated seven cases that go unreported [2]. In 2015, there were an estimated 64 thousand annual episodes of foodborne salmonellosis in Australia that resulted in three thousand hospitalisations and thirteen deaths. The estimated annual societal cost of this illness was AUD 105 million [3–5].
Salmonella can be found in all major livestock and poultry species and a wide array of wild animals; however, the proportion of human Salmonella infections attributable to each of these sources is population- and geography-dependant and often unknown. For instance, in the Netherlands, the primary sources were layers and pigs [6], in Italy and Denmark the primary sources were pigs [7, 8], while in Sweden over 80% of cases were attributed to travel [9]. Source attribution analyses in the Australian states of South Australia and Queensland both identified chickens (broilers and layers) to be the primary sources of illness [10, 11], but also highlighted attribution to nuts or environmental exposures in sub-tropical Queensland [10, 12]. Estimates of the proportion of infections arising from each source can highlight areas needing intervention and inform biosecurity and food safety policy across the entire food supply chain. For instance, a source attribution study in New Zealand in the 2005 demonstrated that > 50% of Campylobacter infections were attributable to poultry, prompting targeted improvements to the food safety standards and practices around poultry. Following these interventions, annual campylobacteriosis notification rates in 2008 declined by 54% compared to 2000–2006 [13] and remained stable over the following decade [14]. Further source attribution analyses demonstrated the decline was due primarily to a 74% reduction in cases attributed to poultry [13]. In NSW, a broad range of interventions were put in place to prevent foodborne salmonellosis following the release of the new NSW Food Safety Strategy 2015–2021. Though salmonellosis notification have declined from approximately 58 per 100,000 in 2014 to 42 per 100,000 in 2018 [1], the proportion of foodborne salmonellosis attributable to different animal reservoirs before and after this period was unknown.
Salmonella enterica subsp. enterica, which accounts for nearly all human cases of salmonellosis, can be classified into over 2500 serotypes [15]. Some serotypes are adapted to specific host species, such as Salmonella ser. Typhi, which has humans as the primary host and Salmonella ser. Gallinarum, which has poultry as the primary host [16]. Other serotypes can be found in one or more animal hosts and can cause human infections e.g. Salmonella ser. Typhimurium. Salmonella can be further differentiated using a range of typing schemes, including phage typing, multi-locus sequence typing (MLST), multiple-locus variable-number tandem repeat analysis (MLVA), whole genome sequencing, and other techniques [15]. Although it has become more common to type Salmonella using MLVA or whole genome sequencing for their superior ability to differentiate types, serotyping remains a standard typing method for Salmonella isolates from animals and humans in Australia.
Several methods have been developed to attribute illness to specific sources [17, 18]. Some rely on linking individual cases or outbreak cases to putative sources through epidemiological investigations, including case–control studies. However, these approaches are labour-intensive and often inconclusive due to small sample sizes. A suitable approach for sporadic cases that does not require food questionnaires is to compare the distribution of Salmonella types in isolates from cases to the distribution of types in putative sources. This approach estimates the overall proportion of cases attributable to each source. Hald et al. adopted a Bayesian source attribution framework [7] and their model and its variants have since been applied widely [10–12, 19–21]. A related approach accounts for genetic relatedness of strains using the asymmetric island model [21, 22]. Recent advances allow these models to be adjusted for covariates to identify trends or differences by subgroup [21]. For instance, recent studies of campylobacteriosis in New Zealand have demonstrated that attribution proportions to different sources vary between rural and urban areas [21]. In Australia, the spatial distribution of salmonellosis varies by serotype (e.g. Salmonella ser. Wangata is associated with proximity to wetlands [23]), suggesting that the proportion of cases attributable to different animal sources may also vary spatially.
We conducted a source attribution analysis for Salmonella infections acquired in New South Wales, Australia between January 2008 and August 2019 using variants of the Hald approach. We adjusted these models to account for trends over time and considered differences by age-group, gender, and rurality.
Methods
Human data
In NSW, treating pathology laboratories are required under public health legislation to report all Salmonella infections to the health department. We used deidentified data for 40,837 human cases notified between January 2008 and August 2019. The available case data included: typing information for the Salmonella isolates, patient gender, 5-year age group, location, and travel history. While nearly all isolates were characterised by serotype, only a minority of these isolates were further characterised with MLVA or phage typing. Location was encoded at the Statistical Area 2 (SA2) level as defined in the Australian Statistical Geography Standard. As we were primarily interested in cases acquired in NSW, we removed all cases recorded as being acquired outside NSW. However, travel history was known only for a small minority of cases, and before May 2010, cases that were known not to have travelled and cases with unknown travel history were recorded identically. Therefore, we also excluded all cases due to serotypes deemed to be travel-associated. A serotype was deemed to be travel-associated if more than half of cases after May 2010 with a known travel history were believed to be acquired outside NSW.
Non-human data
Data for non-human sources were collated from the National Enteric Pathogens Surveillance System (NEPSS) and the New South Wales Food Authority (NSWFA). The two datasets included non-human Salmonella isolates sampled from animals, animal products, farm environments (e.g. animal pens and barns), food, and the natural environment. Isolates considered as from potential animal sources were those derived from the faeces, carcass, or enclosures of an animal or from an animal product. As with human isolates, nearly all isolates were characterised by serotype, but only a minority of these isolates were further characterised with MLVA or phage typing. Isolates that were not serotyped or were from a travel-associated serotype were excluded. We also excluded isolates without a recorded date or with a date before 2008.
Non-human animal isolates were categorised by reservoir animal source where possible from the recorded details. For example, all isolates from pig carcasses; pig faeces; cooked, cured, or raw pig meats; pig offal; pig intestinal contents; and pig farm environments (e.g. bedding) were categorised as porcine. Broiler and layer chickens were treated as separate reservoirs. Chicken meat products for human consumption and environmental samples from broiler farm premises and chicken meat processors were categorised as broiler, while isolates from eggs, egg products (e.g. mayonnaise), and egg farm premises were categorised as layer. Isolates that could not be definitively assigned to an animal reservoir were excluded, e.g. feather meal which might have been derived from broilers or layers and fresh produce which might have been contaminated directly or indirectly by any wild or domestic animal. We excluded isolates from food products with multiple animal origins (e.g. ‘egg and bacon roll’) and isolates with ambiguous descriptions (e.g. ‘meat’). Where possible, putative sources were combined to form categories with at least 100 isolates. Putative sources that could not be reasonably combined into broader categories with at least 100 isolates were excluded.
As source attribution analysis requires the same typing scheme to applied to both source and human data and serotyping was the most consistently and completely applied method, analyses were conducted using serotyped data and isolates without serotyping information were removed from both the human and non-human datasets.
Source attribution modelling framework
We generalised existing Bayesian source attribution methods [20, 21] to estimate changing attribution proportions over time, include covariates for the cases, and adjust for differences between types.
The proportion () of cases in subpopulation during period that were due to type from source was modelled as:
with constraints and and where was the ability of source during period to act as a reservoir of infection for group , was a weight for the relative exposure of humans to contamination from source , was the relative prevalence of type in source during period , and was the relative ability of subtype to lead to human infection. In each group and period the proportion of infections of all types attributed to a source, , was:
while the proportion of cases due to each type, , was:
The estimation of these parameters occurred in two steps. The distribution of types in each source and period () were estimated first, and all other parameters were then estimated repeatedly with draws from the posterior distribution of In the first step, the number of isolates of each type observed in each source () were modelled with multinomial distributions in one of two ways. For sources with many isolates in every period, the relative frequency of types were modelled independently for each period based only on the data collected in that period, i.e. . With a unit Dirichlet prior this resulted in a Dirichlet posterior distribution: For sources with too few samples in each period, the data across the whole study was used for every time period resulting in posterior estimates:
In estimating the remaining parameters, the efficiency of each type () and the exposure weights () were assumed to remain constant over time but source efficiencies () were allowed to vary over time and by subgroup of cases. This was modelled as:
where was a matrix defining a linear predictor based on categorical, ordinal, or continuous covariates for each subgroup (i.e. category of a covariate) of the cases; was a matrix of parameters for each source ; and defined temporal differences in source efficiency by time and source . A reference group was assigned to each covariate, and the associated parameters fixed to 0, while the remaining parameters are given unit normal priors.
The number of human cases in subpopulation during period that were due to pathogen type , were modelled as independent multinomial variables for each period and subgroup , i.e. . The were constrained with a hierarchical log-normal prior:
For predominantly foodborne infections the exposure weights , can be approximated by the relative level of exposure to contaminated food products derived from each source. However, as this is not measured directly, we modelled this as , where was the per capita consumption of food derived from source , and was the prevalence of the pathogen in food derived from source , which was estimated from surveys of animal food products. For each source , we modelled the number of total tests () and positive tests () with an uninformative uniform prior on prevalence, i.e.
Our model framework was extended to include an ‘unsampled source’ by including an additional source with no observed samples, i.e. .
Source attribution models
As a base case we considered a model with no covariates and no temporal variation, which was equivalent to the Modified Hald model applied to the whole study period [20]. As a sensitivity analysis we compared this to a model where all the type efficiency terms were fixed at one, which was equivalent to the Dirichlet model of Liao et al. [21] with the addition of exposure weights. We considered each model with and without an ‘unsampled source'.
We then considered models with combinations of covariates (age, rurality, and gender) with and without temporal variation. In models adjusting for age, we used age categories: 0–4, 5–19, 20–34, 35–64, and 65 and over. The rurality of each case was determined by matching the Australian Bureau of Statistics 2011 Statistical Area 2 (SA2) code provided for the case to one of the five rurality zones (Major Cities, Inner Regional, Outer Regional, Remote, and Very Remote) defined in the 2011 Australian Statistical Geography Remoteness Structure. However, as rurality was defined at the more granular SA1 level and each SA2 is built from SA1s, a few SA2s contained regions with different categories of rurality. Cases associated with these SA2s were assigned the rurality zone closest to the average rurality of the constituent SA1s. In our main analyses, rurality was modelled as an ordinal variable. In a sensitivity analysis we modelled rurality as a nominal categorical variable, combining the five categories down to three categories (‘Major Cities’, ‘Regional’ and ‘Remote’) to increase the number of cases in each category. The small number of cases that had a missing value for gender, age, or location were excluded only from analyses involving the missing covariate.
In models with temporal variation, we considered four three-year periods: 2008–2010, 2011–2013, 2014–2016, and 2017–2019. As there were > 200 isolates per period from broilers and > 450 isolates per period from layers, the relative frequency of serotypes was estimated independently for each period for broilers and layers. As there were relatively few isolates for ruminants and pigs in at least one period (< 30 for pigs and < 20 for ruminants), data across all study years was used to estimate the relative frequency of serotypes in ruminants and pigs in every period.
To improve model convergence, we excluded serotypes that rarely caused disease in humans (i.e. < 10 human cases across the study period) from the analysis.
Exposure by source
Table 1 provides a summary of prevalence assumptions with references, while Table 2 summarises assumptions on food consumption per capita. The datasets used to determine the number of samples positive for each pathogen type did not include data on the number of total tests, so the prevalence of Salmonella in food products was estimated from separate surveys, with an adjustment to reduce the number of positive samples by the fraction of cases from excluded serotypes from each source. As age, sex, location, and time-specific consumption and prevalence data were not available for all food sources, exposure weights and prevalence on animal products were assumed fixed across all time periods and the whole population. We used published national data on Salmonella from the E. coli and Salmonella monitoring (ESAM) program from the early 2000s to inform prevalence in ruminants and pork [24, 25], as publicly available contemporary surveys in NSW had insufficient samples to determine prevalence. The relative exposure of the human population to ruminants, pigs, and broilers were based on the apparent consumption of meats from these sources published by the Australian Bureau of Agricultural and Resource Economics and Sciences (ABARES) [26]. For eggs, we assumed consumption of one egg was equivalent to 200 g of meat (as in our previous work [11]) and used consumption estimates from a 2018 Australian Eggs annual report [27]. In models with an ‘unsampled’ source we adopted the conservative assumption that exposure to the unsampled source was equal to the source or group of sources with the highest consumption.
Table 1.
Salmonella prevalence assumptions by source with references
| Prevalence (95% CI) | Prevalence adjusted for types rare in cases (95% CI) | References | |
|---|---|---|---|
| Chicken meat | 48.4% (42.0–54.8) | 38.6% (32.5–45.0) | NSW-specific data in Table 4 [50] |
| Chicken eggs | 1.76% (0.70–3.59) | 1.44% (0.66–2.72) | Following prior assumptions [11] |
| Pigs | 1.88% (1.57–2.22) | 1.14% (0.90–1.41) | National ESAMa data [24] |
| Ruminants | 0.38% (0.33–0.43) | 0.37% (0.32–0.42) | National ESAMa data [25] |
aE. coli and Salmonella monitoring program
Table 2.
Relative exposure to potential sources of Salmonella, measured by mean consumption of meat and animal products per person per year in Australia
| Relative Exposure (kg/person/year or equivalent) |
References | |
|---|---|---|
| Chicken meat | 47 | ABARESa [26] |
| Chicken eggs | 49b | Australian eggs [27] |
| Pork | 28 | ABARESa [26] |
| Ruminants | 34 | ABARESa [26] |
aAustralian Bureau of Agricultural and Resource Economics and Sciences
bMean consumption was approximately 245 eggs per person per year in the 2017–2018 financial year. The relative exposure was calculated assuming that one egg is equivalent to 200 g of meat
Software
All analyses were conducted in the R programming environment [28]. Bayesian inference was performed using the No U-turn Algorithm with the Stan programming language [29] and the rstan R package [30]. Data cleaning and manipulation was done using the plyr [31], dplyr [32], and tidyr [33] R packages. Data visualisations were made with the ggplot2 R package [34].
Ethical approval and consent to participate
All methods were carried out in accordance with a protocol approved by the Australian National University (ANU) Human Research Ethics Committee (Protocol: 2019/470) and in accordance with all ANU guidelines. The data on human infections were collected under the NSW Public Health Act 2010 and provided to us in a de-identified format by NSW Health. Under the Act, medical practitioners and laboratories were required to notify or report cases of Salmonella infections to NSW public health units.
Results
Data
After excluding isolates from humans with no serotype information (3807 isolates, 9.3%), isolates from cases recorded as having travelled outside NSW (2381 isolates, 5.8%), isolates of Salmonella ser. Paratyphi B bv Java and from serotypes that were commonly travel associated (4089 isolates, 10.0%), and isolates of serotypes that were rare in humans (487 isolates, 1.2%), a total of 30,073 human isolates remained for source attribution analysis (Table 3). Most of the cases associated with these isolates were reported in major cities, with 51% of cases in females. After isolates from some animals were grouped and others were omitted (e.g. wildlife, horses, ducks, quail, and turkeys), the final source groups were broilers (N = 1396), layers (N = 2321), pigs (391), and ruminants (cattle, sheep, and goats; N = 435) (Table 4). The number of non-human isolates in our dataset was lowest in 2017–2019.
Table 3.
Characteristics of Salmonella cases reported between January 2008 and August 2019 in New South Wales after excluding cases without known serotype (N = 3807) travel associated cases (N = 6470) and rare serotypes (N = 487)
| S. Typhimurium (N = 18,802) | Other serotypes (N = 11,271) | All serotypes (N = 30,073) | p value | |
|---|---|---|---|---|
| Gender | 0.010 | |||
| Missing | 47 | 22 | 69 | |
| Female | 9680 (51.6%) | 5633 (50.1%) | 15,313 (51.0%) | |
| Male | 9075 (48.4%) | 5616 (49.9%) | 14,691 (49.0%) | |
| Rurality | < 0.001 | |||
| Missing | 142 | 48 | 190 | |
| Major cities | 14,106 (75.6%) | 7521 (67.0%) | 21,627 (72.4%) | |
| Inner regional | 3434 (18.4%) | 2784 (24.8%) | 6218 (20.8%) | |
| Outer regional | 1070 (5.7%) | 844 (7.5%) | 1914 (6.4%) | |
| Remote | 30 (0.2%) | 48 (0.4%) | 78 (0.3%) | |
| Very remote | 20 (0.1%) | 26 (0.2%) | 46 (0.2%) | |
| Age group | < 0.001 | |||
| Missing | 17 | 6 | 23 | |
| 00–04 | 4191 (22.3%) | 3045 (27.0%) | 7236 (24.1%) | |
| 05–19 | 4186 (22.3%) | 1563 (13.9%) | 5749 (19.1%) | |
| 20–39 | 4986 (26.5%) | 2287 (20.3%) | 7273 (24.2%) | |
| 40–64 | 3434 (18.3%) | 2445 (21.7%) | 5879 (19.6%) | |
| 65 + | 1988 (10.6%) | 1925 (17.1%) | 3913 (13.0%) | |
| Year group | < 0.001 | |||
| 2008–2010 | 4780 (25.4%) | 2059 (18.3%) | 6839 (22.7%) | |
| 2011–2013 | 5441 (28.9%) | 2407 (21.4%) | 7848 (26.1%) | |
| 2014–2016 | 5861 (31.2%) | 3457 (30.7%) | 9318 (31.0%) | |
| 2017–2019 | 2720 (14.5%) | 3348 (29.7%) | 6068 (20.2%) |
Table 4.
The number of serotyped Salmonella isolates from humans and selected non-human sources sampled in NSW between January 2008 and June 2019
| Source | Year | Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | ||
| Human cases | 1759 | 2101 | 2979 | 2864 | 2275 | 2709 | 3416 | 2818 | 3084 | 2489 | 2063 | 1516 | 30,073 |
| Ruminants | 58 | 60 | 49 | 87 | 60 | 33 | 25 | 42 | 10 | 6 | 2 | 3 | 435 |
| Cattle | 50 | 53 | 40 | 70 | 53 | 31 | 22 | 38 | 9 | 6 | 2 | 3 | 377 |
| Ruminants—Othera | 8 | 7 | 9 | 17 | 7 | 2 | 3 | 4 | 1 | – | – | – | 58 |
| Broilersb | 152 | 99 | 98 | 76 | 117 | 37 | 61 | 360 | 185 | 84 | 56 | 71 | 1396 |
| Layersb | 288 | 160 | 155 | 270 | 204 | 251 | 225 | 158 | 109 | 136 | 128 | 237 | 2321 |
| Pigs | 20 | 13 | 42 | 47 | 45 | 79 | 23 | 48 | 51 | 5 | 14 | 4 | 391 |
| Total Included Non-Humanc | 518 | 332 | 344 | 480 | 426 | 400 | 334 | 608 | 355 | 231 | 200 | 315 | 4543 |
| Wildlifed | 12 | 16 | 1 | 3 | 22 | 13 | 5 | 5 | 3 | 3 | – | – | 83 |
| Poultry—Othere | 13 | 7 | 1 | 9 | 1 | 4 | 11 | 1 | 1 | – | – | – | 48 |
| Horses | 2 | 3 | 4 | 7 | 1 | 0 | 4 | 8 | 3 | – | – | – | 32 |
These counts exclude S. Paratyphi B bv Java, types that were rare in humans (less than 10 cases over the period), and travel-associated types (types where more than half of cases with known travel history were acquired outside NSW). A selection of the most common excluded sources are displayed here to illustrate the sparsity of data over time
a Ruminant-other includes sheep and goats
bOther isolates from poultry that could not be linked to either broilers or layers are not included in this table
cThe included sources are ruminants, broilers, layers, and pigs
dWildlife consists of a diverse group of non-captive non-domesticated animalsTyphimurium
ePoultry-other consists of ducks, turkey and quail
Type distribution in sources and humans
S. Typhimurium was the most common serotype in cases and every major source group except for broilers (Additional file 1: Fig. S1, Table S1). In broilers, S. Typhimurium was only the third most common serotype (10.1%, 141/1396) after Salmonella ser. Infantis (30.4%, 425/1396) and Salmonella ser. Sofia (20.6%, 287/1396) (Additional file 1: Fig. S1). The proportion of human cases due to S. Typhimurium was largest in major cities (65.2%), but this proportion declined with increasing rurality (Inner Regional 55.2%; Outer Regional 55.9%; Remote 38.5%; Very Remote 43%) (Table 3). Conversely, cases due to serotypes such as Salmonella ser. Chester and Salmonella ser. Saintpaul were less common in major cities than in regional and remote NSW. The proportion of human cases due to S. Typhimurium declined from 65 to 75% during 2008–2013 to < 50% during 2017–2019. The proportion of cases due to some serotypes rose over the same period, e.g. S. Wangata (2008–2010: 1.0%; 2017–2019: 9.2%), and S. enterica ser. 4,5,12:i:- (2008–2010: 0.0%; 2017–2019: 5.5%). The proportion of layer isolates that were serotyped as S. Typhimurium also declined over the same period (2008–2010: 41.6%, 251/603; 2017–2019: 17.4%, 87/501).
Subtyping
After restricting the analysis to serotyped isolates, only a small minority of non-S. Typhimurium isolates were also typed using phage or MLVA typing (human: 12.6%; reservoirs: 4.5%). While a larger proportion of S. Typhimurium isolates were typed using a least one additional method (human: 95.4%; reservoirs: 73.1%), this additional typing method was not consistent over time or between cases and reservoirs. Nearly all S. Typhimurium isolates from humans were phage typed prior to 2010, but < 7% were phage typed after 2010 (Additional file 1: Table S1). Overall, fewer S. Typhimurium isolates from humans were phage typed (25.1%) than in the major source groups (58.2–84.4%). In contrast, while 93% of S. Typhimurium isolates from cases were MLVA-typed, less than 35% of S. Typhimurium isolates in each major source group were MLVA-typed and coverage varied by year and source. We were therefore unable to use phage typing or MLVA data in our source attribution analyses.
Source attribution without covariates
Table 5 summarises the attribution proportion for models without covariates or temporal variation. All models indicated layers as the leading source of illness. If we assumed all serotypes to be equally efficient at causing disease in humans, very few human cases were attributed to either broilers or pigs: 0.02% and 1.8% respectively. When allowing for differences between serotypes, a greater proportion of cases were attributed to broilers and pigs: 13.1% and 17.8% respectively. Equal- and variable- models with adjustments for unsampled sources attributed 5.8% and 11.3% to the unsampled source respectively and attributed fewer cases to ruminants and pigs than the models without adjustment for an unsampled source. The HaldDP model of Miller et al. [19] failed to converge with our dataset and is not shown.
Table 5.
The percentage of human isolates attributed to ruminants, broilers, layers, pigs, and other unsampled sources with 95% credible intervals for models without covariates or temporal variation
| Ruminants | Broilers | Layers | Pigs | Unsampled | |
|---|---|---|---|---|---|
| Equal- | |||||
| without unsampled source | 41.3 (21.0–58.7) | 0.02 (0.0–0.05) | 56.9 (39.1–74.2) | 1.8 (0.0–8.3) | – |
| with unsampled source | 36.0 (16.9–55.5) | 0.01 (0.0–0.03) | 57.7 (39.7–74.0) | 0.4 (0.0–3.1) | 5.8 (0.0–10.7) |
| Variable- | |||||
| without unsampled source | 24.7 (0.0–55.8) | 13.1 (0.0–44.9) | 44.3 (9.8–76.7) | 17.8 (0.0–39.1) | – |
| with unsampled source | 10.5 (0.0–34.8) | 18.2 (0.0–66.1) | 48.4 (5.4–79.6) | 12.1 (0.0–31.8) | 11.3 (1.2–22.1) |
In the equal- model all Salmonella serotypes are assumed to be equally efficient in their ability to cause infection in humans, while in the variable- model (equivalent to the modified Hald model [20]) serotypes are allowed to differ in their efficiency
Source attribution with covariates
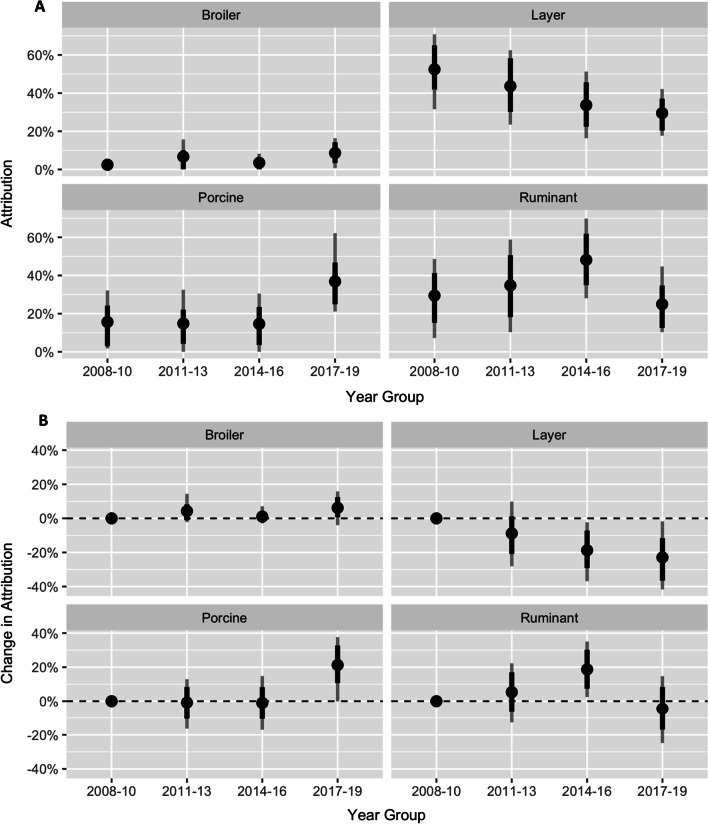
In a temporal model without covariates, the proportion attributed to layers declined significantly from 55% (95% CrI 23–71%) in 2008–2010 to 34% (95% CrI 16–51%) in 2014–2016 and 30% (95% CrI 18–42%) in 2017–2019, with a significant increase in the proportion attributed to pigs in 2017–2019 and a significant increase in attribution to ruminants in 2014–2016 (Fig. 1). When rurality was included in the model, a significantly higher proportion of cases were attributed to layers in major cities, with this proportion declining with increasing rurality (Fig. 2). A similar rural–urban gradient was observed in a non-temporal model, although the proportion attributed to broiler chickens in that model (Additional file 1: Figure S2) was substantially higher than all other models, potentially due to confounding with year (p < 0.001 for association between year and rurality).
Fig. 1.
A Attribution proportion with 95% credible intervals for each of the major source groups for three-year periods and B change over time in the attribution proportion (in percentage points) with 2008–2010 as the reference. Dots indicate posterior mean, while dark and faint lines indicate 80% and 95% credible intervals respectively. The dashed horizontal line indicates no difference
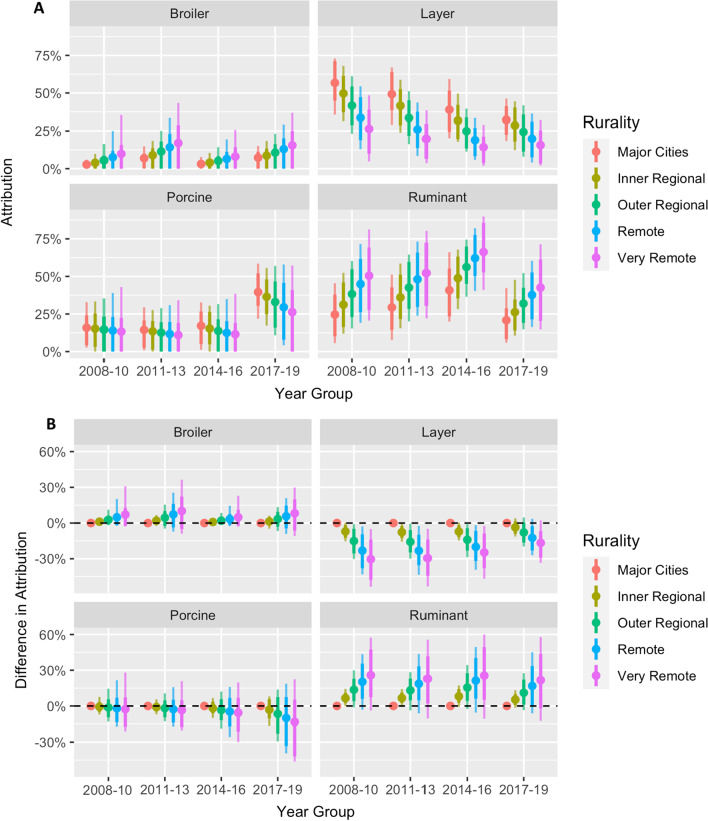
Fig. 2.
A Attribution proportion for each of the major
source groups for cases residing in different rurality zones over time. B The difference in attribution proportion by rurality with residents of major cities as the reference. Dots indicate posterior mean, while dark and faint lines indicate 80% and 95% credible intervals respectively. The dashed horizontal line indicates no difference
There was weaker evidence of a rural–urban gradient in attribution to ruminants, with a higher proportion of infections attributed to ruminants in rural areas (Figs. 2 and Additional file 1: S2), which disappeared in models with an unsampled source (Figures S3 and S4). In a sensitivity analysis where rurality was modelled as categorical variable with three levels (‘Major Cities’, ‘Regional’ and ‘Remote’) all findings remained qualitatively similar (results not shown).
A non-temporal univariable regression analysis found a significant association between attribution proportion and sex, with 5.7% (95% CrI 1.3%-10.1%) fewer male cases attributed to layer chickens. Though a qualitatively similar sex difference was also observed in a model including time and sex, and a model with age and sex, the sex differences were not statistically significant within any time period or age group. There were no consistent associations between age group and attribution proportions.
Estimates of the type-specific parameters, , varied 1000-fold between the most efficient and least efficient serotypes. For instance, in the model including rurality, year-group, and an unsampled source, S. Typhimurium and Salmonella ser. Abortusovis were estimated to be 22.3 (95% CrI 14.0–38.0) and 0.19 (95% CrI 0.07–0.46) times as efficient as S. Infantis, while S. Sofia was estimated about as efficient as S. Infantis ( ratio 0.5; 95% CrI: 0.2–1.7) (Additional file 1: Fig. S5).
Discussion
We estimate that approximately half of all notified cases of non-typhoidal salmonellosis acquired in New South Wales during 2008–2019 were due to direct or indirect transmission from layer chickens. However, the proportion of cases attributed to layers declined from approximately 50% in 2009–2011 to approximately 25% in 2017–2019. The proportion of cases attributed to pork increased from approximately 20% in 2009–2016 to approximately 40% in 2017–2019, coinciding with a rise in cases due to S. enterica ser. 4,5,12:i:-, which has been found to be a persistent coloniser of pigs in Australia [35]. The proportion of cases attributed to layers was lower amongst rural-dwelling than urban-dwelling populations, similar to the rural–urban gradient found in a source attribution analysis of Campylobacter infections in New Zealand [21, 36].
The potential of zoonotic organism to lead to a notified case of foodborne disease depends on many factors including the organism’s ability to survive transport, storage, and food preparation and its virulence in humans. Some source attribution modelling approaches (e.g. those applied to Campylobacter infections [21, 22]) have assumed that all types are equally able to lead to notified human cases. However, this assumption is inappropriate for Salmonella, which is known to have both virulent (e.g. S. Typhimurium) and avirulent (e.g. S. Sofia [37]) serotypes. In our study, S. Typhimurium was more prevalent in cases (63%) than in broilers (10%), layers (32%), pigs (23%), or ruminants (35%), while S. Infantis was less prevalent in cases (3%) than in broilers (30%), layers, (19%) or pigs (7%), indicating that S. Typhimurium is a more efficient foodborne pathogen than S. Infantis. Consequently, models that assumed all serotypes were equally efficient attributed < 0.1% of cases to broilers (in which S. Sofia and S. Infantis were most common). However, this is inconsistent with the common implication of chicken meat in outbreak investigations and case control studies (e.g. [38]). Although models that account for differences between serotypes led to source attribution estimates with much wider confidence intervals, we believe they are more appropriate for modelling source attribution of Salmonella.
The decline in attribution to layers from 2014 onwards may be the result of interventions including vaccination of layers against S. Typhimurium. The NSW Food Safety Strategy 2015–2021, released in 2014, included a specified 30% reduction target for foodborne salmonellosis [39]. Prior to 2014, many reported foodborne outbreaks in NSW were linked to poor handling of eggs and hygiene at retail food service (e.g. [40, 41]). Several measures were implemented to address this, including mandatory education and food safety training on the risks of raw egg use, improved cleaning and sanitising for retail businesses, guidelines for the safe use of eggs which made it an offence in NSW to prepare raw egg foods without adequate processing, and training and guidance for local council Environmental Health Officers to focus on areas of highest food safety risk during inspections. Since 2014, large egg producers across NSW have begun largescale vaccination of layer flocks against S. Typhimurium; estimates based on sales of vaccines indicate approximately 75% of commercial layer flocks were vaccinated in 2017–2019 (Dr. Christopher Morrow, personal communication, April 2021). The introduction of all these measures coincided with a marked decline in S. Typhimurium and total Salmonella incidence in NSW [42].
The models that included an ‘unsampled’ source attempted to quantify the plausible proportion of cases that could be attributed to any animal reservoirs other than domestic chickens, ruminants, and pigs. Since this approach necessarily drew inferences without information about of the relative abundance of different serotypes in the ‘unsampled’ animal reservoirs, these models need to be interpreted cautiously. In our study, the increasing trend in attribution to ‘unsampled’ sources is associated with the increasing incidence of serotypes that were absent or rare in the major food animals included in our analyses, e.g. S. Wangata, Salmonella ser. Birkenhead, Salmonella ser. Waycross, and Salmonella ser. hvittingfoss. S. Wangata was not found in ruminants or pigs in our dataset and was very rare in broilers and layers (< 0.6%) but accounted for 9.0% of cases in 2017–2019 up from only 1.0% in 2008–2010. Similarly, though S. Birkenhead was only represented by a single isolate from layer chickens in 2019, the serotype accounted for 5.7% of cases in 2017–2019, up from 3.3% in 2008–2010. While S. Wangata is associated with contact with a range of wild animals in NSW [23, 43, 44], the animal reservoirs of S. Birkenhead, S. Waycross, and S. Hvittingfoss are unclear. A source attribution analysis of Salmonella infections in the neighbouring state of Queensland linked S. Birkenhead, S. Waycross, and S. Hvittingfoss to nuts (primarily locally grown macadamias) [10] suggesting environmental reservoirs of exposure. In north-eastern NSW, S. Birkenhead infection has been associated with not usually washing or peeling fruit and vegetables before eating raw. S. Hvittingfoss has caused outbreaks linked to contaminated cantaloupes in Australia with closely related isolates identified in bar-tailed godwits in north-western Australia, suggesting a common exposure to an unidentified animal or environmental reservoir [45]. S. Hvittingfoss has also been found in reptiles [46, 47], migratory ducks [48], and feral pigs [49] elsewhere in Australia. Consequently, a substantial part of the attribution to ‘unsampled’ sources in our model may be due to transmission from wild or feral animals through direct contact or indirect contamination of water, food, or the environment.
The present study has a number of strengths. The large dataset of Salmonella cases and a substantial dataset of isolates from major food animals, collected from across the state and over a long time period, have allowed us to identify the leading source of Salmonella infections, attribute separately to egg-laying and meat chickens, and identify temporal and rural–urban gradients in attribution. Furthermore, these key model outcomes were qualitatively robust across multiple model comparisons. In the source attribution framework we adopted attribution is to animal reservoirs rather than routes or vehicles of transmission. For instance, our estimate of the proportion of cases attributed to layers includes infections that may have been acquired through direct contact with layer chickens, consumption of contaminated eggs, contact with manure, or consumption of water or food contaminated by layer chicken faeces at any point between farm and table. Attribution to animal reservoirs is simultaneously a strength—since all possible transmission routes are captured—and a limitation—since the relative contribution of different routes cannot be quantified.
Our study has a number of limitations. No high-resolution molecular subtyping scheme was consistently applied to Salmonella isolates from humans and animals over the whole study period, forcing us to restrict our analyses to the level of serotype. Because the majority of human cases were due to a single serotype, S. Typhimurium, this restriction substantially weakened inferences, leading to wide confidence intervals for many attribution estimates. Though the dataset of reported human cases provided more than adequate sample size, it is estimated that as few as one in eight Salmonella cases are reported [2]. As unreported cases are likely to have milder symptoms than reported cases, more virulent serotypes of Salmonella are likely to be over-represented in our dataset and the attribution proportions are therefore likely indicative of moderate and severe cases. Similarly, rates of healthcare utilisation vary by rurality and other socioeconomic factors that may be associated with exposure to different animal sources, so attribution estimates may be biased towards those sources to which people with high healthcare utilisation rates are most exposed. Travel history was not available for all cases in our dataset, and though we took steps to remove cases not acquired in NSW from the study, we may have removed some locally acquired cases in doing so. The isolates from animal reservoirs were not collected in a single sampling frame. The majority of non-human Salmonella sampling by the NSW Food Authority occurred through investigation of foodborne disease outbreaks and was often sporadic in nature. Some targeted survey work was also undertaken, but this was biased towards testing of layer flocks, or opportunistic Salmonella testing through poultry meat verification work focused on Campylobacter. To date, NSWFA have not conducted comparable ongoing surveys or studies focused on ruminants, pigs, or other potential reservoirs. The remaining isolates from non-human sources collected by the NEPSS included any isolate submitted to Salmonella typing laboratories in Australia and included public, private, and academic sampling efforts. For isolates from non-human sources, we only considered those collected in NSW; however, NSW residents consume food produced across Australia. Though lamb and mutton account for approximately 8% of meat consumption in Australia [26], very few Salmonella isolates from sheep isolates were available, forcing us to combine sheep, cattle, and goats into a single category for source attribution purposes. However, the relatively low consumption of sheep meat and the low prevalence of Salmonella in sheep [25] suggest that sheep are unlikely to be a major source of Salmonella infections in NSW. We only had sufficient samples to estimate the relative frequency of serotypes over time in poultry which may have influenced the estimated trend in source attribution. Finally, we had only limited samples from wild, feral, and companion animals, which may have been especially important for the accurate attribution of rural cases. Though attribution to ‘unsampled’ sources includes these reservoirs, we are unable to estimate the contribution different species or groups of species.
Conclusions
Layer chickens were the primary reservoir for human salmonellosis in NSW during 2009–2019; however, the importance of layers was less in rural populations and declined after 2015, concurrent with changes to food safety regulations and egg industry practices. The apparent increase in attribution to pigs and unsampled sources in 2017–2019 warrants further investigation. Our study highlights the need for a high-resolution typing method to be consistently applied to Salmonella isolates collected from humans, food, animals, and the environment. Using a consistent approach would allow for more precise estimates for source attribution analyses and assist in identifying specific sources in outbreak investigations. Moreover, the mobility of people and food products between Australian states and territories calls for a nationally unified approach to the surveillance of Salmonella. Furthermore, ongoing routine surveillance of Salmonella in food and food animals collected in a consistent sampling frame would improve the sample size and data quality for prevalence and source attribution estimates and allow these estimates to be updated regularly to monitor trends. Source attribution can inform and direct efforts to prevent foodborne disease. Source attribution analyses of data before and after interventions, public health responses, or changes to industry practices can help assess the effects of these changes. Regularly updated source attribution estimates would provide timely guidance for food safety authorities as novel Salmonella types or other epidemiological factors change the risk associated with specific animal reservoirs.
Supplementary Information
Additional file 1: Figure S1. The observed distribution of common1 serotypes in human cases and the major food animal groups in NSW between 2008 and 2019. Each row shows the proportion of isolates due to each serotype in that source (number isolates from the source in parentheses). 1Serotypes within the twenty most common serotypes in one of the sources or in human cases. Figure S2. Source attribution proportions for a model ignoring differences over time, but adjusting for differences by rurality. (A) Attribution proportion for each of the major source groups for cases residing in different rurality zones. (B) The difference in attribution proportion by rurality with residents of major cities as the reference. Dots indicate posterior mean values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. See Figure 2 in main text for model adjusting for rurality and changes over time. Figure S3. Attribution proportion for each of the major source groups for cases residing in different rurality zones over time. Dots and crosses indicate mean and median values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. (Compare Figure 2A). Figure S4. The difference in attribution proportion by rurality with residents of major cities as the reference. Dots and crosses indicate mean and median values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. (Compare Figure 2B). Figure S5. Posterior estimates of the relative efficiency (q) of included serotypes in a model including rurality, year-group, and an unsampled source (see Figures S4 and S5), with S. infantis used as a reference. High relative efficiency indicates a serotype more likely to lead to human disease (e.g. due to high virulence or high survivability) while low relative efficiency indicates serotypes that are common in source animals but rarely cause disease. Dots indicate median values, while thick and thin lines indicate 80% and 95% credible intervals, respectively. Table S1. The percentage of Salmonella isolates of serotype Typhimurium and the percentage of S. typhimurium isolates that were subtyped using phage-typing or MLVA typing. NA indicates that there were no S. typhimurium for the combination of source and year.
Acknowledgements
We would like to thank Health Protection New South Wales, Alison Imlay from the New South Wales Department of Primary Industries (NSWDPI) and the Mathilda Wilmot and Courtney Lane National Enteric Pathogen Surveillance System (NEPSS) hosted at the University of Melbourne (UoM) for providing access to the datasets used in this study. In particular we would like to thank Keira Glasgow and Neil Franklin (Health Protection NSW); Alison Imlay and Themy Saputra (NSWDPI); and Mathilda Wilmot and Courtney Lane (NEPSS/UoM). We would like to acknowledge Declan O’Donovan for assistance with data cleaning, and Dr Thea King from the New South Wales Department of Primary Industries for suggestions around Salmonella enterica ser. 4,5,12:i:- in pigs and potential reservoirs of S. Hvittingfoss and S. Birkenhead.
Abbreviations
- NSW
New South Wales
- CrI
Credible Interval
- AUD
Australian Dollars
- MLST
Multi-locus sequence typing
- MLVA
Multiple-locus variable-number tandem repeat analysis
- SA1
Australian Bureau of Statistics 2011 Statistical Area 1
- SA2
Australian Bureau of Statistics 2011 Statistical Area 2
- NEPSS
National Enteric Pathogens Surveillance System
- NSWFA
New South Wales Food Authority
- ESAM
E. coli And Salmonella Monitoring Program
- ABARES
Australian Bureau of Agricultural and Resource Economics and Sciences
Author contributions
CS, MDK, KG, and conceived the study. AM, CS, and KG designed the study. AM, PMD, and CS collated the data. AM wrote the R code, performed the analysis, and drafted the manuscript. All authors contributed to the interpretation of the results, revised the manuscript, and approved the final manuscript.
Funding
This study was primarily funded by the New South Wales Department of Primary Industries, which also collected a subset of the non-human data and employs C.S. A.M. is supported through an ARC Discovery Project Grant DP180100246 and M.D.K. is supported by an Australian National Health and Medical Research Council Fellowship (Grant No. GNT1145997).
Availability of data and materials
The data used in this study were provided by New South Wales Department of Primary Industries, National Enteric Pathogens Surveillance Scheme, and Health Protection New South Wales; however, restrictions apply to the availability of these data, which were used under agreement for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the relevant third parties.
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with a protocol approved by the Australian National University (ANU) Human Research Ethics Committee (Protocol: 2019/470) and in accordance with all ANU guidelines. The data on human infections were collected under the NSW Public Health Act 2010 and provided to us in a de-identified format by NSW Health. Under the Act, medical practitioners and laboratories were required to notify or report cases of Salmonella infections to NSW public health units.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.National Notifiable Diseases Surveillance System. Australian Government Department of Health. http://www9.health.gov.au/cda/source/cda-index.cfm. Accessed 11 May 2020
- 2.Hall G, Yohannes K, Raupach J, Becker N, Kirk M. Estimating community incidence of Salmonella, Campylobacter, and Shiga Toxin–producing Escherichia coli infections, Australia. Emerg Infect Dis. 2008;14(10):1601–1609. doi: 10.3201/eid1410.071042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk M, Ford L, Glass K, Hall G. Foodborne Illness, Australia, Circa 2000 and Circa 2010. Emerg Infect Dis. 2014;20(11):1857–1864. doi: 10.3201/eid2011.131315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford L, Haywood P, Kirk MD, Lancsar E, Williamson DA, Glass K. Cost of Salmonella infections in Australia, 2015. J Food Prot. 2019;82(9):1607–1614. doi: 10.4315/0362-028X.JFP-19-105. [DOI] [PubMed] [Google Scholar]
- 5.Ford L, Kirk M, Glass K, Hall G. Sequelae of foodborne illness caused by 5 pathogens, Australia, Circa 2010. Emerg Infect Dis. 2014;20(11):1865–1871. doi: 10.3201/eid2011.131316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mughini-Gras L, Enserink R, Friesema I, Heck M, Van Duynhoven Y, Van Pelt W. Risk factors for human salmonellosis originating from pigs, cattle, broiler chickens and egg laying hens: a combined case–control and source attribution analysis. PLoS ONE. 2014;9(2):e87933. doi: 10.1371/journal.pone.0087933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24(1):255–269. doi: 10.1111/j.0272-4332.2004.00427.x. [DOI] [PubMed] [Google Scholar]
- 8.Mughini-Gras L, Barrucci F, Smid JH, Graziani C, Luzzi I, Ricci A, Barco L, Rosmini R, Havelaar AH, Van Pelt W, et al. Attribution of human Salmonella infections to animal and food sources in Italy (2002–2010): adaptations of the Dutch and modified Hald source attribution models. Epidemiol Infect. 2014;142(5):1070–1082. doi: 10.1017/S0950268813001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahlström H, Andersson Y, Plym-Forshell L, Pires SM. Source attribution of human Salmonella cases in Sweden. Epidemiol Infect. 2011;139(8):1246–1253. doi: 10.1017/S0950268810002293. [DOI] [PubMed] [Google Scholar]
- 10.Fearnley EJ, Lal A, Bates J, Stafford R, Kirk MD, Glass K. Salmonella source attribution in a subtropical state of Australia: capturing environmental reservoirs of infection. Epidemiol Infect. 2018;146(15):1903–1908. doi: 10.1017/S0950268818002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass K, Fearnley E, Hocking H, Raupach J, Veitch M, Ford L, Kirk MD. Bayesian source attribution of salmonellosis in South Australia. Risk Anal. 2016;36(3):561–570. doi: 10.1111/risa.12444. [DOI] [PubMed] [Google Scholar]
- 12.Munck N, Smith J, Bates J, Glass K, Hald T, Kirk MD. Source attribution of Salmonella in macadamia nuts to animal and environmental reservoirs in Queensland, Australia. Foodborne Pathog Dis. 2020;17(5):357–364. doi: 10.1089/fpd.2019.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sears A, Baker MG, Wilson N, Marshall J, Muellner P, Campbell DM, Lake RJ, French NP. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg Infect Dis. 2011;17(6):1007–1015. doi: 10.3201/eid1706.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker MG, Grout L, Wilson N. Update on the campylobacter epidemic from chicken meat in New Zealand: the urgent need for an upgraded regulatory response. Epidemiol Infect. 2021;149:1–10. doi: 10.1017/S095026882000299X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Appl Environ Microbiol. 2011;77(22):7877–7885. doi: 10.1128/AEM.05527-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, et al. Comparative genome analysis of Salmonella enteritidis PT4 and Salmonella gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18(10):1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mughini-Gras L, Kooh P, Augustin J-C, David J, Fravalo P, Guillier L, Jourdan-Da-Silva N, Thébault A, Sanaa M, Watier L. Source attribution of foodborne diseases: potentialities, hurdles, and future expectations. Front Microbiol. 1983;2018:9. doi: 10.3389/fmicb.2018.01983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pires SM, Vieira AR, Hald T, Cole D. Source attribution of human salmonellosis: an overview of methods and estimates. Foodborne Pathog Dis. 2014;11(9):667–676. doi: 10.1089/fpd.2014.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller P, Marshall J, French N, Jewell C. sourceR: Classification and source attribution of infectious agents among heterogeneous populations. PLoS Comput Biol. 2017;13(5):e1005564. doi: 10.1371/journal.pcbi.1005564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullner P, Jones G, Noble A, Spencer SEF, Hathaway S, French NP. Source attribution of food-borne zoonoses in New Zealand: a modified hald model. Risk Anal. 2009;29(7):970–984. doi: 10.1111/j.1539-6924.2009.01224.x. [DOI] [PubMed] [Google Scholar]
- 21.Liao S-J, Marshall J, Hazelton ML, French NP. Extending statistical models for source attribution of zoonotic diseases: a study of campylobacteriosis. J R Soc Interface. 2019;16(150):20180534. doi: 10.1098/rsif.2018.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. Tracing the source of Campylobacteriosis. PLoS Genet. 2008;4(9):e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson KMJ, Mor SM, Ward MP, Walsh MG. Divergent geography of Salmonellawangata and Salmonellatyphimurium epidemiology in New South Wales, Australia. One Health. 2019;7:100092. doi: 10.1016/j.onehlt.2019.100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton DR, Smith P, Pointon A: National Salmonella and E. coli Monitoring (ESAM) data from Australian pig carcases from 2000 to 2006. In: Seventh international symposium on the epidemiology and control of foodborne pathogens in pork: 2007; Verona, Italy: Iowa State University Digital Press; 2007: 129–132.
- 25.Jordan D, Morris S. Analysis of ESAM data. Syndey: Meat & Livestock Australia Limited; 2006. [Google Scholar]
- 26.Agricultural commodity statistics. Australian Bureau of Agricultural and Resource Economics and Sciences. https://www.agriculture.gov.au/abares/research-topics/agricultural-outlook/data. Accessed 03 July 2020
- 27.Australian eggs annual report 2017–18: Australian Eggs; 2018.
- 28.R Core Team: R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing; 2020. https://www.R-project.org/
- 29.Stan Development Team: Stan Modeling Language Users Guide and Reference Manual, 2.25. 2020.
- 30.Stan Development Team: RStan: the R interface to Stan. 2020.
- 31.Wickham H. The split-apply-combine strategy for data analysis. J Stat Softw. 2011;40(1):1–29. [Google Scholar]
- 32.Wickham H, François R, Henry L, Müller K: dplyr: A Grammar of Data Manipulation; 2020. https://CRAN.R-project.org/package=dplyr
- 33.Wickham H: tidyr: Tidy Messy Data; 2020. https://CRAN.R-project.org/package=tidyr
- 34.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016. [Google Scholar]
- 35.Weaver T, Valcanis M, Mercoulia K, Sait M, Tuke J, Kiermeier A, Hogg G, Pointon A, Hamilton D, Billman-Jacobe H. Longitudinal study of Salmonella 1,4,[5],12:i:- shedding in five Australian pig herds. Prev Vet Med. 2017;136:19–28. doi: 10.1016/j.prevetmed.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Lake RJ, Campbell DM, Hathaway SC, Ashmore E, Cressey PJ, Horn BJ, Pirikahu S, Sherwood JM, Baker MG, Shoemack P, et al. Source attributed case–control study of campylobacteriosis in New Zealand. Int J Infect Dis. 2020;103:268–277. doi: 10.1016/j.ijid.2020.11.167. [DOI] [PubMed] [Google Scholar]
- 37.Gan E, Baird FJ, Coloe PJ, Smooker PM. Phenotypic and molecular characterization of Salmonella enterica serovar Sofia, an avirulent species in Australian poultry. Microbiology. 2011;157(4):1056–1065. doi: 10.1099/mic.0.047001-0. [DOI] [PubMed] [Google Scholar]
- 38.McPherson ME, Fielding JE, Telfer B, Stephens N, Combs BG, Rice BA, Fitzsimmons GJ, Gregory JE. A multi-jurisdiction outbreak of Salmonella typhimurium phage type 135 associated with purchasing chicken meat from a supermarket chain. Commun Dis Intell. 2006;30(4):449–455. doi: 10.33321/cdi.2006.30.44. [DOI] [PubMed] [Google Scholar]
- 39.New South Wales Food Authority . New South Wales Government food safety strategy 2015–2021: Safe food from paddock to plate. Sydney: New South Wales Food Authority; 2014. [Google Scholar]
- 40.Ward K, Franklin N, Furlong C, Polkinghorne BG, Flint J. New South Wales OzFoodNet Annual Report: 2013. Sydney: Health Protection NSW; 2014. [Google Scholar]
- 41.New South Wales Health Communicable Diseases Branch . New South Wales OzFoodNet Annual Report: 2012. Sydney: Health Protection NSW; 2013. [Google Scholar]
- 42.New South Wales Health Communicable Diseases Branch . New South Wales OzFoodNet Surveillance Report: 2018. Sydney: Health Protection NSW; 2019. [Google Scholar]
- 43.Collins J, Simpson KMJ, Bell G, Durrheim DN, Hill-Cawthorne GA, Hope K, Howard P, Kohlenberg T, Lawrence K, Lilly K, et al. A One Health investigation of Salmonella enterica serovar Wangata in north-eastern New South Wales, Australia, 2016–2017. Epidemiol Infect. 2019;147:1. doi: 10.1017/S0950268819000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson KMJ, Hill-Cawthorne GA, Ward MP, Mor SM. Diversity of Salmonella serotypes from humans, food, domestic animals and wildlife in New South Wales, Australia. BMC Infect Dis. 2018;18(1):1. doi: 10.1186/s12879-018-3563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith HG, Bean DC, Hawkey J, Clarke RH, Loyn R, Larkins J-A, Hassell C, Valcanis M, Pitchers W, Greenhill AR. Salmonella enterica Serovar Hvittingfoss in Bar-Tailed Godwits (Limosa lapponica) from Roebuck Bay, Northwestern Australia. Appl Environ Microbiol. 2020;86(19):1312. doi: 10.1128/AEM.01312-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheelings TF, Lightfoot D, Holz P. Prevalence of Salmonella in Australian reptiles. J Wildl Dis. 2011;47(1):1–11. doi: 10.7589/0090-3558-47.1.1. [DOI] [PubMed] [Google Scholar]
- 47.Iveson JB, Mackay-Scollay EM, Bamford V. Salmonella and Arizona in reptiles and man in Western Australia. J Hygiene. 1969;67(1):135–145. doi: 10.1017/s0022172400041516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoque MA, Burgess GW, Greenhil AR, Hedlefs R, Skerratt LF. Causes of morbidity and mortality of wild aquatic birds at Billabong Sanctuary, Townsville, North Queensland, Australia. Avian Dis. 2012;56(1):249–256. doi: 10.1637/9863-072611-Case.1. [DOI] [PubMed] [Google Scholar]
- 49.Ward MP, Cowled BD, Galea F, Garner MG, Laffan SW, Marsh I, Negus K, Sarre SD, Woolnough AP. Salmonella infection in a remote, isolated wild pig population. Vet Microbiol. 2013;162(2–4):921–929. doi: 10.1016/j.vetmic.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 50.Baseline survey on the prevalence and concentration of Salmonella and Campylobacter in chicken meat on-farm and at primary processing; 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The observed distribution of common1 serotypes in human cases and the major food animal groups in NSW between 2008 and 2019. Each row shows the proportion of isolates due to each serotype in that source (number isolates from the source in parentheses). 1Serotypes within the twenty most common serotypes in one of the sources or in human cases. Figure S2. Source attribution proportions for a model ignoring differences over time, but adjusting for differences by rurality. (A) Attribution proportion for each of the major source groups for cases residing in different rurality zones. (B) The difference in attribution proportion by rurality with residents of major cities as the reference. Dots indicate posterior mean values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. See Figure 2 in main text for model adjusting for rurality and changes over time. Figure S3. Attribution proportion for each of the major source groups for cases residing in different rurality zones over time. Dots and crosses indicate mean and median values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. (Compare Figure 2A). Figure S4. The difference in attribution proportion by rurality with residents of major cities as the reference. Dots and crosses indicate mean and median values, while dark and faint lines indicate 80% and 95% credible intervals, respectively. (Compare Figure 2B). Figure S5. Posterior estimates of the relative efficiency (q) of included serotypes in a model including rurality, year-group, and an unsampled source (see Figures S4 and S5), with S. infantis used as a reference. High relative efficiency indicates a serotype more likely to lead to human disease (e.g. due to high virulence or high survivability) while low relative efficiency indicates serotypes that are common in source animals but rarely cause disease. Dots indicate median values, while thick and thin lines indicate 80% and 95% credible intervals, respectively. Table S1. The percentage of Salmonella isolates of serotype Typhimurium and the percentage of S. typhimurium isolates that were subtyped using phage-typing or MLVA typing. NA indicates that there were no S. typhimurium for the combination of source and year.
Data Availability Statement
The data used in this study were provided by New South Wales Department of Primary Industries, National Enteric Pathogens Surveillance Scheme, and Health Protection New South Wales; however, restrictions apply to the availability of these data, which were used under agreement for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the relevant third parties.