Abstract
Acetylation is emerging as a posttranslational modification of nuclear proteins that is essential to the regulation of transcription and that modifies transcription factor affinity for binding sites on DNA, stability, and/or nuclear localization. Here, we present both in vitro and in vivo evidence that acetylation increases the affinity of myogenic factor MyoD for acetyltransferases CBP and p300. In myogenic cells, the fraction of endogenous MyoD that is acetylated was found associated with CBP or p300. In vitro, the interaction between MyoD and CBP was more resistant to high salt concentrations and was detected with lower doses of MyoD when MyoD was acetylated. Interestingly, an analysis of CBP mutants revealed that the interaction with acetylated MyoD involves the bromodomain of CBP. In live cells, MyoD mutants that cannot be acetylated did not associate with CBP or p300 and were strongly impaired in their ability to cooperate with CBP for transcriptional activation of a muscle creatine kinase-luciferase construct. Taken together, our data suggest a new mechanism for activation of protein function by acetylation and demonstrate for the first time an acetylation-dependent interaction between the bromodomain of CBP and a nonhistone protein.
Acetylation of histone or nonhistone proteins is emerging as a central process in transcriptional activation (4). Nuclear histone acetyltransferases (HATs) such as GCN5 (6), PCAF (40), and CBP and p300 (22) are transcriptional coactivators (18, 39). Their mode of action is not yet fully understood. They are able to acetylate histones on lysine residues located in the N-terminal histone tails, which protrude from the core nucleosome (38). Histone acetylation is linked to transcriptional activation (33) and participates in the nucleosomal remodeling that accompanies gene activity.
In vitro, HATs are also able to acetylate nonhistone proteins, in particular, general transcription factors such as TFIIE and TFIIF (13) and numerous sequence-specific transcription factors (5, 11, 16, 20, 31), including myogenic factor MyoD (28). MyoD is a transcription factor of the myogenic basic helix-loop-helix family that is central to the process of muscle cell differentiation (7) and that functions by transactivating muscle-specific promoters (36). In vitro, MyoD is acetylated by two HATs, CBP or p300 and PCAF, which are both required for MyoD transactivating activity (25, 42). CBP and p300 are homologous proteins (8; Z. Arany, W. R. Sellers, D. M. Livingston, and R. Eckner, Letter, Cell 77:799–800, 1994) that are found in large multimolecular complexes. They have several highly homologous domains through which they interact with a wide variety of sequence-specific transcription factors and other proteins. In particular, CBP and p300 directly interact with the N-terminal transactivation domain of MyoD (27). CBP and p300 also have in common a domain that displays an intrinsic HAT activity (3, 22). Moreover, CBP and p300 recruit other HATs such as PCAF (40). CBP and p300, PCAF, and other HATs also share a domain of ill-defined function, the bromodomain, which is frequently associated with chromatin-remodeling proteins (12).
Ectopically expressed (28) as well as endogenous (24) MyoD is acetylated in live cells. In vitro, CBP and p300 (24) and PCAF (28) acetylate MyoD on lysines 99 and 102, two lysines located at the boundary of the DNA binding/dimerization domain, the basic helix-loop-helix domain. Acetylation increases MyoD activity on muscle-specific promoters (28). The mechanism of this activation is not yet fully understood but has been reported to involve increased binding to DNA (28). We here show that, in myogenic cells, the fraction of endogenous MyoD that is acetylated is associated with CBP, whereas acetylation of free MyoD is undetectable. This result suggests that acetylated MyoD shows a higher affinity for the HAT than does nonacetylated MyoD. This hypothesis was confirmed by the analysis of nonacetylatable MyoD mutants: these mutants do not form detectable complexes with CBP or p300 in cells. The increased interaction of CBP with acetylated MyoD was confirmed by in vitro experiments. Acetylation of recombinant wild-type MyoD strongly increased its affinity for CBP and p300, as indicated (i) by increased resistance of the complex to high salt concentrations and (ii) by the fact that complex formation required lower doses of acetylated MyoD than nonacetylated MyoD. Acetylation had no effect on complex formation by MyoD point mutants in which key acetylatable lysines (99 and 102) were replaced by arginines. Analysis of CBP deletion mutants revealed that the interaction with acetylated MyoD is mediated by the bromodomain of CBP. The functional relevance of the interaction between acetylated MyoD and CBP was assessed by transient transfection experiments. A nonacetylatable MyoD mutant did not cooperate as efficiently with CBP to activate a muscle creatine kinase (MCK)-luciferase reporter construct as the wild-type MyoD molecule.
Taken together, our results lead us to propose a model in which acetylation of MyoD lysines 99 and 102 triggers the recognition of the acetylated lysines by the bromodomain of CBP, resulting in a strengthened interaction between the HAT and MyoD. In our model, this results in a more efficient recruitment of HAT complexes to muscle promoters, where these enzymes then acetylate other substrates. In support of this model, results of chromatin immunoprecipitation experiments indicated that myogenic terminal differentiation triggers histone H4 acetylation. In summary, our data suggest a new mechanism for protein activation by acetylation and demonstrate an acetylation-dependent interaction between the CBP bromodomain and a nonhistone protein.
MATERIALS AND METHODS
Cell cultures.
Myoblastic C2C12 cells and embryonic C3H 10T1/2 cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 15 and 10% fetal calf serum (FCS; Dominique Deutcher), respectively. To induce terminal differentiation, the cells were placed in differentiation medium (DMEM, 1% FCS). Phenotypic differentiation of C2C12 cells was maximal after 72 h.
Preparation of recombinant proteins and acetylation in vitro.
The bacterial expression vectors containing wild-type MyoD cDNA and all mutated species of MyoD were kind gifts from K. Breitschopf, A. Ciechanover, and S. Leibovich. BL21 (DE3)/pLysS Escherichia coli cells were used for bacterial expression of MyoD. Following induction with IPTG (isopropyl-β-d-thiogalactopyranoside), cells were lysed and MyoD was precipitated by 0.6 M ammonium sulfate as described previously (30). CBP mutants (glutathione S-transferase [GST]–CBP 1-1891 and 1-1696) were prepared as previously described (1, 2). The GST-CBP construct with deleted bromodomain (deletion of amino acids [aa] 1099 to 1220) was derived from GST-CBP 1-1696 by means of XbaI-BsrgI restriction, fill-in with Klenow polymerase, and ligation. The GST-bromodomain construct was obtained by cloning the PCR fragment corresponding to aa 1068 to 1476 of CBP into the pGEMT-Easy vector (Promega), followed by BamHI-EcoRI restriction and cloning into the pGEX vector (Promega). The GST-bromodomain construct with substitutions of alanine for V1115 and Y1125 was obtained from the GST-bromodomain construct using the Quick Change mutagenesis kit (Stratagene). The GST-C/H3 construct was obtained by cloning the PCR fragment corresponding to aa 1619 to 1877 of CBP into the pGEX vector. p300 and PCAF were produced in insect cells using a baculovirus-driven expression system and standard procedures. The His-tagged p300 and p300 with deleted bromodomain were cloned from pBluescript constructs (kind gift from W. Lee Kraus) by NotI- HindIII restriction and ligation into the pCMV eukaryotic expression vector.
Recombinant MyoD acetylation was carried out in vitro by incubating 2 to 4 μg of protein with 0.1 to 2 μg of recombinant HATs for 1 h at 30°C in a buffer containing 50 mM Tris, pH 7.5, 2 mM EDTA, inhibitors of proteases (Complete; Boehringer Mannheim) and of deacetylases (10 mM sodium butyrate; Sigma), and 1 mM acetyl coenzyme A (CoA) per reaction.
Microinjection and myogenic conversion.
C3H 10T1/2 mouse embryonic fibroblasts were seeded on coverslips placed into 35-mm-diameter tissue culture dishes (Nunc) at 7 × 104 cells per dish. Twenty-four hours later, pEMSV-MyoD was microinjected into cells, together with increasing amounts of pCMV-p300 or pCMV-p300 del Br and a mixture of 40,000- and 10,000-molecular-weight dextran-rhodamine, fixable (1% final concentration; Molecular Probes), in a buffer containing 10 mM Tris, pH 7.5, and 100 mM KCl in a final volume of 10 μl. Six hours after microinjection, cells were placed in differentiation medium. After 48 h, cells were fixed with 2% paraformaldehyde, permeabilized with 0.5% Triton X-100, stained with a monoclonal antimyogenin antibody (F.5D; kind gift from W. E. Wright) followed by antimouse fluorescein isothiocyanate (Sigma), and analyzed by fluorescence microscopy.
Semiquantitative GST pull-down.
Increasing amounts of MyoD (2 to 100 ng/point) were incubated for 1 h at 30°C with 2 to 4 μg of GST-CBP or GST immobilized on agarose beads in Tris-EDTA (TE) buffer containing protease inhibitors with or without acetyl-CoA (1 mM; Sigma). TE buffer was chosen as the optimal buffer for in vitro acetylation reactions. Note that nonspecific binding on GST-coated beads was low in this buffer. Beads were washed three times with TE supplemented with the concentrations of KCl indicated in Fig. 2 and 3, and proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with anti-MyoD antibodies (C-20; Santa Cruz Biotechnology). In some experiments, the amount of GST protein was verified using anti-GST antibodies (Z-5; Santa Cruz Biotechnology) and, for the same protein, did not show any variation between samples (data not shown).
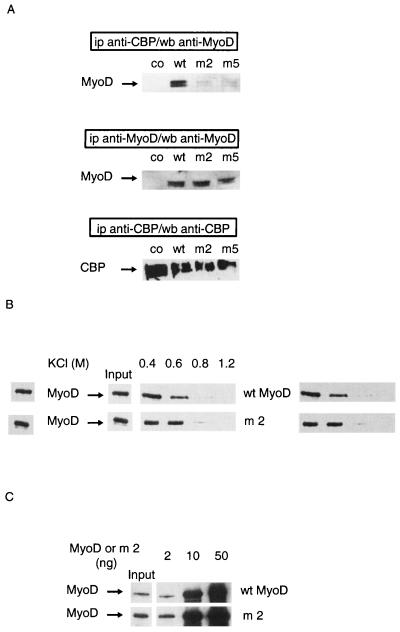
FIG. 2.
Nonacetylatable MyoD mutants do not associate with CBP in cells. Mutants used were m2, in which lysines 99, 102, and 104 were replaced by arginines, and m5, in which lysines 99, 102, 104, 112, 124, and 133 were replaced by arginines. (A) C3H 10T1/2 cells were transfected with expression vectors for wild-type (wt) or point-mutated (m2 or m5) MyoD together with an expression vector for wt CBP 1-2441. A MyoD empty vector vehicle was used in negative controls (co). Extracts from transfected cells were immunoprecipitated (ip) using an anti-CBP antibody or an anti-MyoD antibody as indicated. MyoD and CBP were detected by Western blotting (wb), as indicated. (B) Nonacetylated wt or mutant MyoD (m2) was incubated with GST-CBP-coated beads for 1 h and washed with TE buffer containing the indicated concentrations of KCl. Beads were analyzed by Western blotting using anti-MyoD antibodies. (C) Indicated doses of nonacetylated MyoD (wt or mutated) were incubated in TE buffer with GST-CBP-coated beads. Beads were analyzed by Western blotting using anti-MyoD antibodies. Input, the amount of each MyoD protein, estimated as 2 ng (1/25 of maximal amount used in this experiment).
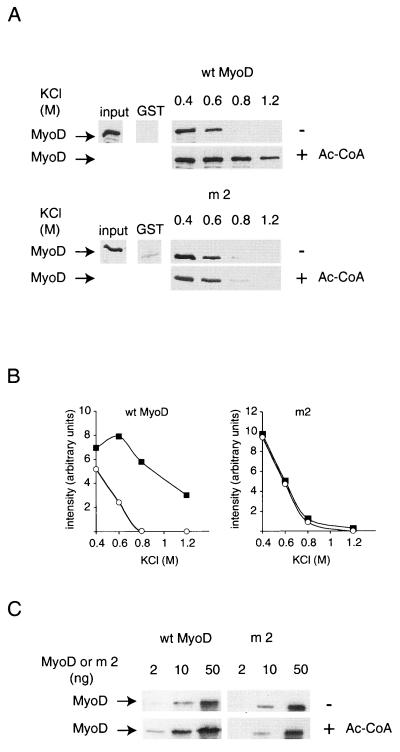
FIG. 3.
Acetylation increases MyoD affinity for CBP in vitro. (A) Equivalent amounts of wild-type (wt) and point-mutated (m2) MyoD were incubated with GST-CBP 1-2441-coated beads for 1 h, with or without acetyl-CoA (1 mM), and washed with TE buffer containing the indicated concentrations of KCl. GST-coated beads were used as a negative control. (B) Autoradiograms of the blots shown in panel A were analyzed by densitometry. Open circle, nonacetylated MyoD; solid square, acetylated MyoD. Increasing doses (2, 10, and 50 ng) of recombinant MyoD were incubated with GST-CBP absorbed onto glutathione-coated beads in the presence or absence of acetyl-CoA. The presence of MyoD in association with CBP was detected by Western blotting as described for Fig. 2.
For experiments illustrated in Fig. 4C to E, MyoD (5 μg) was acetylated by incubation with GST-CBP immobilized on agarose beads in the presence of 1 mM acetyl-CoA (or was not acetylated, for the negative controls) and then separated from the enzyme by incubation in 2 M KCl (15 min at 30°C) followed by centrifugation. The supernatant, which contained free MyoD but no detectable amounts of GST-CBP (as assessed by Western blotting), was diluted and incubated for 15 min with beads coated with GST or with various GST-CBP mutants and pretreated with 0.1% bovine serum albumin. Beads were washed with 1 M KCl followed by PBS; then the proteins were resolved by SDS-PAGE and analyzed by Western blotting with anti-MyoD antibodies. The presence of equal amounts of GST proteins in the lanes with acetylated and nonacetylated MyoD was verified by Western blotting with anti-GST antibodies (Z-5; Santa Cruz Biotechnology; data not shown).
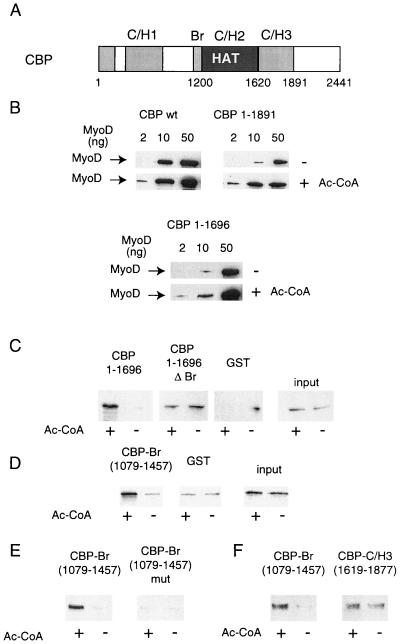
FIG. 4.
CBP bromodomain is involved in the interaction with acetylated MyoD. (A) Diagram of the CBP molecule, showing the C/H3 domain, bromodomain, and HAT domain. (B) Increasing doses (2, 10, and 50 ng) of recombinant MyoD were incubated with GST-CBP absorbed onto glutathione-coated beads in the presence or absence of acetyl-CoA; the presence of MyoD in association with CBP was detected by Western blotting. wt, wild type. (C to F) MyoD was incubated with wt CBP in the presence of acetyl-CoA (Ac-CoA) (or in the absence of Ac-CoA for the controls, as indicated), isolated from the enzyme, and used in a GST pull-down assay with beads coated with the indicated CBP mutants; the presence of MyoD in association with CBP was detected by Western blotting.
Immunoprecipitation and Western blotting.
Immunoprecipitation and Western blotting were performed using standard procedures. Immunoprecipitated proteins were collected on protein G-agarose (Sigma). Beads were washed three times with phosphate-buffered saline (PBS). The samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with PBS containing 10% dry milk, incubated with antibodies followed by peroxidase-conjugated secondary antibodies (Sigma), and developed using the Boehringer Mannheim LumiLight kit as recommended by the manufacturer.
Acetylation of MyoD in CBP complexes was assessed by immunoprecipitation of total cell extracts of differentiating (24 h) C2C12 myoblasts with anti-p300/CBP (NM11; Pharmingen). “Free” MyoD was immunoprecipitated from the supernatant using anti-MyoD antibodies (5.8A; Novocastra). Total MyoD was obtained by direct immunoprecipitation of MyoD from total extracts. All fractions were analyzed by Western blotting using antiacetyllysine antibodies (Upstate Biotechnology) and an anti-MyoD antibody (C-20; Santa Cruz Biotechnology).
Coimmunoprecipitation experiments were performed on extracts from transfected cells. For expression in mammalian cells, MyoD cDNAs from pT7-7 bacterial expression vectors (Sa) were subcloned into pGEMT vector (Promega) and cloned into the EcoRI site of pEMSV-scribe (9, 32). CBP expression was driven by pCMV2N3T CBP (26). Transfections were performed using Lipofectamine (Life Technologies) or Polyfect (Qiagen), as described by the manufacturer, in 10-cm-diameter dishes. Cells received expression vectors (p2N3T CBP [20 μg], pCMV-p300, pCMV-p300 del Br, pEMSV MyoD, or pEMSV MyoD mut2 [10 μg]) and 1 μg of pRSV-βgal as an internal standard for transfection efficiency and were incubated overnight. Extracts were prepared 24 h later, normalized by assaying β-galactosidase activity using a kit from Tropix, immunoprecipitated using anti-CBP antibodies (A-22; Santa Cruz Biotechnology) or anti-p300 antibodies (N-15; Santa Cruz Biotechnology), and analyzed by Western blotting using anti-MyoD antibodies (5.8 A; Novocastra) and either anti-CBP/p300 antibodies (NM11; PharMingen) or an anti-His antibody (H-3; Santa Cruz Biotechnology).
Transient transfection assays.
The C3H 10T1/2 cells were seeded out on 12-well dishes (3 × 104 cells per well) and transfected 24 h later by the calcium phosphate precipitation method. Cells received expression vectors for wild-type MyoD or mutant m2 (100 ng/well), together with 200 ng of MCK-luciferase reporter vector/well and increasing doses of pCMV-CBP, and were incubated overnight at 37°C. Extracts were prepared 36 h later, the luciferase activity was measured using the Luciferase Assay System from Promega, and the expression of wild-type and mutant MyoD was controlled by Western blotting using an anti-MyoD antibody (C-20; Santa Cruz Biotechnology).
Chromatin immunoprecipitation.
C2C12 cells, either continuously growing or after 24 h of differentiation, were fixed using 1% formaldehyde in tissue culture medium for 7 min at 37°C. Chromatin was prepared using a kit from Upstate Biotechnology according to the recommendations of the manufacturer, with eight 10-s sonication pulses at 10-s intervals, which yielded chromatin fragments of an apparent size of 800 bp (as monitored on agarose gels; data not shown). Equivalent amounts of chromatin, as assessed by visualizing serial dilutions on agarose gels (data not shown), were immunoprecipitated using an anti-acetylated histone H4 antibody (Upstate Biotechnology) or irrelevant immunoglobulins (Sigma) for the control. Formaldehyde-induced cross-linking was reversed (4 h at 65°C), and a sequence of the MCK promoter or of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene (as an internal control) was detected by quantitative PCR. In this assay, a fixed amount of immunoprecipitated DNA was amplified together with increasing amounts of a competitor DNA amplified by the same primers.
The primers used were as follows: for MCK, 5′-GGATGAGAGCAGCCACTATG (forward) and 5′-ACCATGGCAGAATTGACAGG (reverse), yielding a 325-bp product corresponding to region −1265 to −945 of the promoter; for GAPDH, 5′-CCAATGTGTCCGTCGTGGATCT-3′ (forward) and GTTGAAGTCGCAGGAGACAACC-3′ (reverse), yielding a 190-bp fragment.
The competitive heterologous DNA fragments were created by amplification of irrelevant plasmids using composite primers. Amplification products were analyzed on agarose gels.
RESULTS
Preferential association between acetylated MyoD and CBP in live cells.
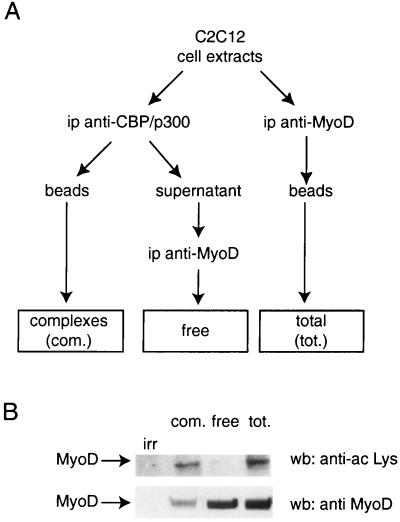
Endogenous MyoD is constitutively acetylated in myogenic cells (24). A comparison of the fraction of MyoD that is complexed to CBP or p300 and the fraction containing free MyoD revealed a nonrandom distribution (Fig. 1). Extracts from C2C12 (a myogenic cell line) cells, sampled after 24 h in differentiation medium, were immunoprecipitated with anti-CBP/p300 antibodies (Fig. 1A, complexes), and the supernatant (in which CBP and p300 could not be detected; data not shown) was immunoprecipitated with anti-MyoD (Fig. 1A, free). In parallel, extracts were directly immunoprecipitated with anti-MyoD antibodies (Fig. 1A, total). All samples were analyzed by Western blotting for the presence of MyoD using anti-MyoD antibodies and for the presence of acetylated proteins using anti-acetylated lysine antibodies (Fig. 1B). Acetylated species were detected only in the fraction of MyoD that was associated with CBP. Although the free fraction contained the majority of cellular MyoD, acetylation could not be detected in this fraction (Fig. 1B). This result suggests that there is a preferential association between acetylated MyoD and CBP and that MyoD acetylation, by CBP itself or by other acetyltransferases, increases MyoD's affinity for CBP.
FIG. 1.
Preferential association between acetylated MyoD and CBP and p300. Myogenic cell extracts were immunoprecipitated (ip) as shown in panel A and analyzed by Western blotting (wb) (B) using anti-acetylated lysine or anti-MyoD as indicated. irr, immunoprecipitation using an irrelevant antibody as a negative control.
Acetylated lysines of MyoD are required for interaction with CBP in cells.
To confirm the hypothesis that acetylated lysines of MyoD are required for interaction with CBP, the interaction between CBP and MyoD point mutants which have lost lysines 99, 102, and 104, and which thus cannot be acetylated (24, 28), was analyzed (Fig. 2). In these experiments, wild-type or mutant MyoD was ectopically expressed in C3H 10T1/2 (nonmuscle) cells together with CBP and extracts were analyzed by immunoprecipitation using anti-CBP antibodies, followed by Western blotting using an anti-MyoD antibody (Fig. 2A). In contrast to the wild-type molecule, the mutants were barely detectable in CBP complexes. This impaired interaction might result from the lack of acetylation of the mutants. Alternatively, the mutated lysines could be directly or indirectly involved in the interaction with CBP. To rule out this possibility, the interaction between mutant MyoD and CBP was analyzed in vitro and compared to that for the nonacetylated wild-type protein. We first analyzed the resistance of the CBP-MyoD complex to high salt concentrations. An excess of recombinant MyoD was incubated with GST-CBP-coated beads, and beads were washed with increasing concentrations of KCl. Under these conditions, both wild-type and mutated MyoD bound to CBP. In addition, the salt concentrations at which the complex was disrupted (above 0.6 M) for the wild-type and mutated forms of MyoD were similar (Fig. 2B). This result was confirmed using a semiquantitative GST pull-down assay in which increasing amounts of MyoD were incubated with GST-CBP-coated beads. After being washed, beads were analyzed by Western blotting using an anti-MyoD antibody. The amounts of MyoD required to detect the protein in the complex were similar for mutated and wild-type MyoD (Fig. 2C), suggesting that the mutant displays an affinity for CBP that is similar to that displayed by the nonacetylated wild-type molecule. These results strongly suggest that MyoD's acetylatable lysines are not themselves directly involved in the interaction with CBP. Thus, when nonacetylated, the two forms of MyoD have similar affinities for CBP, and the observed difference between the mutant and the wild-type proteins in live cells is most likely due to the absence of acetylation of the mutant protein in vivo.
Acetylation increases MyoD affinity for CBP and p300 in vitro.
To further demonstrate that acetylation increases MyoD affinity for CBP, the affinities of acetylated and nonacetylated MyoD were directly compared. First, the resistance of CBP-MyoD complexes to stringent salt conditions was assessed. Whereas the interaction between nonacetylated MyoD and CBP was disrupted above 0.6 M KCl (Fig. 3A and B), the interaction between CBP and acetylated MyoD was readily detectable at 1.2 M KCl. This effect could be due to acetylation of MyoD or, alternatively, to the autoacetylation of CBP. The latter hypothesis was ruled out by experiments using a nonacetylatable MyoD mutant (Fig. 3A and B). This mutant did not show any increased resistance to high salt concentration upon similar treatment.
Second, we used a semiquantitative GST pull-down assay, in which increasing amounts of MyoD were incubated with GST-CBP-coated beads, in the presence or (for the nonacetylated samples) absence of acetyl-CoA. For the wild-type molecule, about fivefold less acetylated MyoD (Fig. 3B) than nonacetylated MyoD was needed to detect an association with CBP. No such effect was observed with a nonacetylatable MyoD mutant. Note that the anti-MyoD antibody detects acetylated and nonacetylated MyoD with equivalent efficiencies (24). An analysis of point mutants indicated that the two acetylatable lysines of MyoD, lysine 99 and lysine 102 (24), were involved in the formation of the complex with CBP, although mutation of lysine 99 alone decreased significantly the strength of the interaction (data not shown). Taken together, these in vitro results indicate that MyoD acetylation strengthens its interaction with the HAT CBP.
The CBP bromodomain is involved in the interaction with acetylated MyoD.
To determine the domains of CBP involved in the interaction with acetylated MyoD, several fragments of CBP were analyzed in the semiquantitative GST pull-down assay. Note that this assay was designed only to compare the binding of acetylated and nonacetylated MyoD and not to compare the affinities of distinct CBP mutants for MyoD. The C-terminal part of CBP was found to be dispensable for the selective interaction with acetylated MyoD (Fig. 4B): CBP 1-1891 interacted more strongly with acetylated than with nonacetylated MyoD. Likewise, deletion of CBP's major domain of interaction with MyoD, the C/H3 domain, did not change the behavior of CBP: this mutant also interacted more strongly with acetylated MyoD than with nonacetylated MyoD (Fig. 4B). This indicates that selective recognition of the acetylated form of MyoD does not require the CBP C/H3 domain.
We next tested the hypothesis of CBP bromodomain involvement in the interaction with acetylated MyoD. For that purpose, we used an indirect assay, in which the detection of protein-protein interactions was uncoupled from the acetylation step, hence allowing the use of HAT-negative fragments of CBP. In this assay, MyoD was incubated with beads coated with wild-type CBP in the presence of acetyl-CoA (or in its absence for the negative controls). MyoD was then separated from the beads by incubation in 2 M KCl and subsequently used in a regular GST pull-down with beads coated with various fragments of CBP (see Materials and Methods). CBP 1-1696 retained acetylated MyoD, whereas association of nonacetylated MyoD was undetectable under the conditions used (Fig. 4C), confirming the difference of affinity between the two forms of MyoD. Deletion of the bromodomain abrogated the difference between acetylated and nonacetylated MyoD (compare CBP 1-1696 and CBP 1-1696ΔBr; note that more of the CBP 1-1696ΔBr protein was used in this assay [data not shown], resulting in a higher background binding of nonacetylated MyoD to this mutant). This result suggests that the bromodomain is involved in the recognition of acetylated MyoD. To further test this hypothesis, a fragment of CBP which includes only the bromodomain (CBP 1079-1457) was tested in the same assay. The bromodomain selectively retained acetylated MyoD but not the nonacetylated species (Fig. 4D). Mutations of highly conserved residues in the CBP bromodomain abrogated the selective interaction with acetylated MyoD (Fig. 4E). In contrast, the C/H3 domain recognized acetylated and nonacetylated MyoD with the same efficiency (Fig. 4F). Taken together, these data indicate that the bromodomain is necessary and sufficient for interaction with acetylated MyoD.
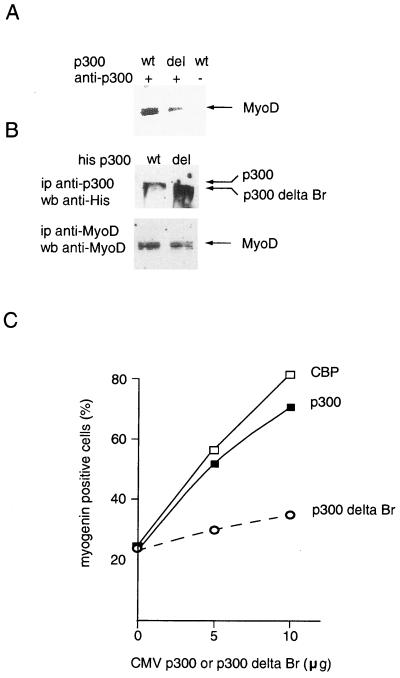
The bromodomains of CBP and p300 are involved in physical and functional interaction with MyoD.
Taken together, our results suggest that, in live cells, the interaction between CBP or p300 and MyoD occurs only when MyoD is acetylated, and involves the bromodomain. Deletion mutants were used to test this hypothesis. For these experiments, we had to use a p300 deletion mutant instead of a CBP deletion mutant due to the impaired HAT activity of the corresponding CBP construct's product. Note that CBP and p300 belong to the same family of HATs and are highly homologous (Arany et al., letter). In addition, ex vivo, they cannot be distinguished from a functional point of view. This is specifically true for myogenic differentiation, for which, to our knowledge, no functional differences have ever been demonstrated. In particular, both CBP and p300 acetylate MyoD with identical functional consequences (24). Coimmunoprecipitation experiments performed on extracts from cells transfected with expression vectors for wild-type MyoD and p300 indicated that deletion of the p300 bromodomain strongly decreased p300's ability to physically interact with MyoD (Fig. 5A), although the mutant was expressed at higher levels than the wild-type molecule (Fig. 5B). This decreased interaction was accompanied by a decreased ability to cooperate with MyoD in a myogenic conversion assay. Nonmuscle cells (C3H 10T1/2) were microinjected with expression vectors for MyoD and CBP or p 300, either wild type or with the bromodomain deleted. Expression of myogenin, a marker of muscle cell differentiation, was assayed 48 h later by immunofluorescence (data not shown). In this assay, p300 and CBP had identical effects on myogenin expression (Fig. 5C): both proteins increased myogenin induction by MyoD. In contrast, the deletion mutant had hardly any effect (Fig. 5C). These data indicate that the bromodomain of CBP is involved in the cooperation between CBP and MyoD. Although we cannot rule out the hypothesis that the bromodomain is also involved through its ability to recognize the nucleosomal histones (10, 17), taken together these data on the physical (A) and functional (C) interaction of CBP and MyoD clearly indicate that the bromodomain participates in the interaction between the two proteins in live cells.
FIG. 5.
CBP bromodomain is involved in physical and functional interaction with MyoD. (A) Nonmuscle cells (C3H 10T1/2) were transfected with expression vectors for MyoD and p300, either wild type (wt) or with the bromodomain deleted. Extracts were immunoprecipitated (ip) with an anti-p300 antibody and analyzed by Western blotting (wb) using an anti-MyoD antibody. (B) Expression of transfected proteins. Extracts of transfected cells were immunoprecipitated and analyzed by Western blotting using the indicated antibodies. (C) C3H 10T1/2 cells were microinjected with expression vectors for MyoD and for p300, either wt or with the bromodomain deleted, together with a rhodamine-coupled injection marker. After 40 h in differentiation medium, cells were fixed and immunostained with an antimyogenin antibody. Shown is the fraction of injected cells (rhodamine positive) which express myogenin; ▪, wt CBP; □, wt p300; ○, p300 delta Br.
Nonacetylatable MyoD mutants show an impaired cooperation with CBP.
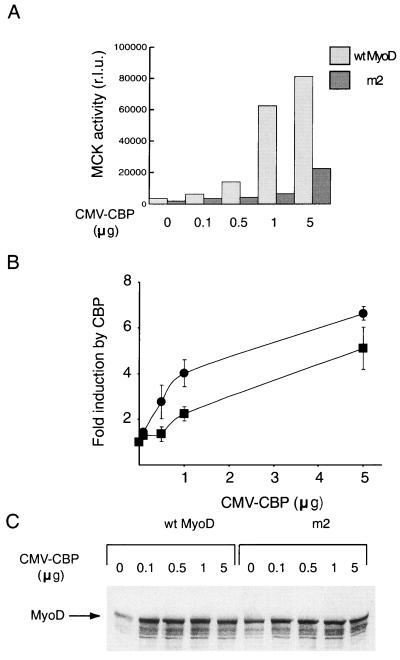
MyoD acetylation has been reported to increase MyoD binding to DNA (28) and/or seems to modify MyoD's affinity for CBP (this study). To demonstrate that the latter effect is central to MyoD activity, we examined whether a MyoD mutant that cannot be acetylated is able to cooperate with CBP. Cells were transfected with a reporter construct in which the luciferase gene is under the control of muscle-specific promoter MCK together with expression vectors for wild-type or mutant (m2) MyoD and increasing doses of an expression vector for CBP (Fig. 6). In the absence of CBP, the m2 mutant was not as efficient as the wild-type molecule in activating MCK, as previously reported (24, 28). The activity of the wild-type molecule was increased in a dose-dependent manner by the expression of CBP, which in the absence of MyoD did not have any effect on promoter activity (data not shown). This cooperation, striking at the highest dose tested, was also seen at lower doses of the CBP expression vector (e.g., 0.5 μg; Fig. 6A and B). In contrast, and although the levels of expression of the mutant and the wild-type molecule were similar (Fig. 6C), no cooperation between the mutant and CBP at low doses of the CBP expression vector was evident (Fig. 6A and B). A significant activation of the MCK reporter by the mutant, however, was observed at high doses. Three- to fivefold more CBP was required to achieve equivalent levels of activation with the mutant form of MyoD than was necessary with the wild-type molecule. Thus, the mutant is deficient in its ability to cooperate with CBP, but cooperation can be rescued, at least partially, by high doses of the HAT. This result suggests, not that the binding to DNA is the limiting step for the mutant's activity, but rather that lack of MyoD acetylation is detrimental to its cooperation with CBP, most likely due to a low affinity for the HAT itself.
FIG. 6.
Impaired cooperation between nonacetylatable MyoD and CBP mutants. Nonmuscle cells (C3H 10T1/2) were transfected with an MCK-luciferase reporter construct, together with a pEMSV MyoD expression vector (100 ng) and indicated doses of CMV-CBP. Results of a representative experiment are shown. (A) Luciferase activity was measured in duplicate. r.l.u., relative light units; wt wild type. (B) Means of three independent experiments ± standard deviations are shown as the fold inductions by CBP (ratio between r.l.u. obtained in the presence of CBP and r.l.u. obtained in its absence). ●, wt MyoD; ▪, m2. (C) Western blot analysis of MyoD expression.
MyoD is not the only target of HATs for endogenous muscle promoter activation.
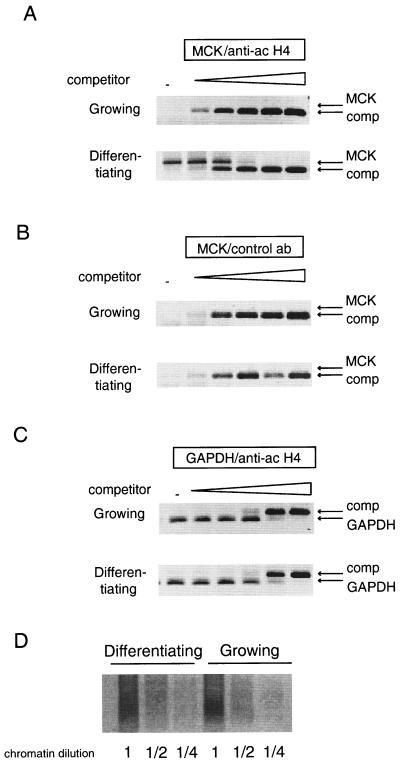
A likely interpretation of our results is that acetylated MyoD acts as a strong bridge between the HAT enzymes and muscle-specific promoters and helps to establish a stable recruitment of the enzymes to the promoters. The functional consequence of HAT's stable recruitment was expected to be the acetylation of other proteins bound to these promoters such as the histones. In order to test this hypothesis, histone acetylation on the MCK promoter was monitored using a chromatin immunoprecipitation assay. In these experiments, equivalent amounts of chromatin (Fig. 7D) were prepared from C2C12 myoblasts, either continuously growing or after 24 h in differentiation medium, and immunoprecipitated using either an antibody directed against acetylated histone H4 (lysine 5, 8, 12, and 14) or an irrelevant antibody as a control. A quantitative PCR assay was used to detect the MCK promoter in the immunoprecipitates. Results (Fig. 7) indicate a significant increase in the amount of MCK promoter retained by the anti-acetylated H4 antibody in extracts from differentiated cells over the amount retained in extracts from growing cells. The MCK promoter was not retained by an irrelevant antibody. In addition, the GAPDH promoter, used as an internal control, did not demonstrate any modulation in histone H4 acetylation in differentiating cells compared with that in growing myoblasts. These results indicate that histone H4 tails associated with the MCK promoter are specifically acetylated during myoblast differentiation and thus that a HAT activity is recruited to this promoter in differentiating cells.
FIG. 7.
MCK-associated histone H4 is acetylated on terminal differentiation. Chromatin was extracted from C2C12 muscle cells, either continuously growing or else differentiating, as indicated. Equivalent amounts of chromatin (as assessed by gel analysis of dilutions; data not shown) were immunoprecipitated using an anti-acetylated (ac) histone H4 (A and C) or an irrelevant antibody (ab) as a control (B). The MCK promoter (A and B) and the GAPDH promoter as an internal control (C) were detected by quantitative PCR analysis using increasing doses of a competitor DNA (no DNA [−]; 100 ag; 1, 10, and 100 fg; and 1 pg) amplified by the same primers. Arrows, bands corresponding to the MCK or GAPDH amplification products, as well as the bands corresponding to the competitor (comp) amplification products. (D) Nondenaturing gel analysis of serial dilutions of chromatin used for immunoprecipitation stained by ethidium bromide.
DISCUSSION
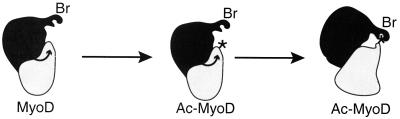
MyoD, like other sequence-specific transcription factors, seems to be regulated by acetylation. The mechanism through which acetylation of MyoD increases its activity is not yet fully understood. MyoD acetylation by PCAF increases its affinity for DNA, at least for MyoD-MyoD homodimers (28). However, homodimers are not the active species in muscle cells (19, 21), and thus the significance of this observation is not clear. Our results support an alternative hypothesis in which the acetylation of MyoD increases the stability of the complex formed between MyoD and the HAT CBP. This is concluded essentially from the analysis of acetylated MyoD in live cells and from results of in vitro testing. Such an effect of acetylation has been recently suggested for another transcription factor (29). Concerning the mechanism of this stabilization, several alternative hypotheses must be considered. The acetylated lysines might be part of MyoD's domain of interaction with CBP, and the acetylation might facilitate this interaction, for example, through neutralization of repulsive charges. However, the region of MyoD in which the acetylated lysines are located has not been reported as being involved in the interaction with CBP or p300, for which a 100-aa N-terminal fragment seems to be sufficient (27). Moreover, point mutation of the lysines did not impair the apparent affinity of nonacetylated MyoD for CBP in vitro (Fig. 2). In addition, the interaction between CBP and acetylated MyoD did not require the C/H3 domain, the major domain of CBP for interaction with the N-terminal region of MyoD (Fig. 4). As an alternative hypothesis, lysine acetylation could result in a conformational change of the MyoD molecule that would positively affect its interaction with CBP. Finally, the acetylated lysines themselves could provide a recognition motif for CBP (Fig. 8). The last hypothesis is supported by the fact that the CBP bromodomain is involved in the interaction. In this regard it is significant that X-ray analysis of the PCAF bromodomain demonstrated that this domain selectively interacts with acetylated lysines (10). A likely hypothesis is thus that the interaction between the CBP bromodomain and acetylated lysines in MyoD provides additional links that strengthen the interaction between the two proteins (Fig. 8). In this model, the bromodomain would be a domain of protein-protein interaction that selectively recognizes amino acid sequences in which lysines are acetylated, as previously suggested (37), much as the SH2 domains are domains that recognize amino acid sequences in which a tyrosine is phosphorylated. Our data provide the first experimental evidence for such a selective interaction between a nonhistone protein and the bromodomain: we show that the bromodomain of CBP, in the absence of the rest of the molecule, discriminates between acetylated and nonacetylated MyoD and selectively binds the acetylated but not the nonacetylated form. This result demonstrates that such a mechanism is not restricted to histones but rather may be more general.
FIG. 8.
A model for the association between acetylated MyoD and CBP (see Discussion). Br, bromodomain. Curved arrows, substrate acetylation by the HAT; asterisks, acetylated lysines on MyoD.
The interaction between recombinant MyoD and recombinant PCAF was weaker than that between MyoD and CBP and, in the absence of CBP, was not significantly strengthened by MyoD acetylation (27; A. Polesskaya and A. Harel-Bellan, unpublished observations). However, it is likely that, in myogenic cells, PCAF is strongly associated with acetylated MyoD through CBP. Indeed, CBP-PCAF complexes are easily detected in muscle cells (A. Polesskaya and A. Harel-Bellan, unpublished observation), suggesting that significant proportions of the two molecules are associated in these cells. Moreover, the binding of MyoD and that of PCAF to CBP are not mutually exclusive, and a trimolecular complex could potentially exist (27). The stabilization of the CBP-MyoD interaction thus likely facilitates the formation of a multimolecular complex that includes PCAF.
Mutation of acetylation sites in MyoD strongly decreased the ability of the molecule to functionally cooperate with CBP (Fig. 6). The mutant, however, was at least partly rescued by a large excess of CBP, suggesting that its defect resides in the lack of a strong interaction with CBP and the HAT complexes. A strong interaction between MyoD and the HATs might have several consequences. Given that sequence-specific transcription factors have been shown to recruit HAT complexes to target promoters (34), a strong interaction between CBP and acetylated MyoD is likely to result in a more efficient recruitment of the HATs to muscle-specific promoters. In that regard, it should be noted that CBP and p300, which are central to the activity of a wide variety of transcription factors, are thought to be present in limiting amounts in cells. Competition of transcription factors for these pivotal coactivators could affect gene regulation (15, 35). In support of this hypothesis is the observation that cbp is subjected to major gene dosage effects in mice (41) and in humans (23). A strong interaction between MyoD and the HATs could thus help in maintaining an adequate fraction of the HATs complexed to MyoD in muscle cells. In this regard, it should be noted that the muscle differentiation program is irreversible, and a strong interaction between MyoD and the HATs could play a role in this phenomenon. The HAT-MyoD complex is likely to be strongly bound to muscle-specific promoters such as MCK, where a significant increase in the acetylation of histone H4 is observed during terminal differentiation of myoblasts (Fig. 7). Acetylation of core histones has been reported to have a very short half-life (14), and permanent reacetylation of histones is likely to be necessary to maintain an “open” structure on the promoter. A strong association between acetylated MyoD and CBP or p300 might thus be crucial to retaining the HAT complex on muscle promoters and maintaining, locally, a state of hyperacetylation.
In summary, our results lead us to propose a model in which MyoD acetylation allows the sequestration of HAT complexes to muscle-specific promoters on which they acetylate other substrates such as histones. These data reveal a new mechanism for transcription factor activation by acetylation. In addition, they provide the first experimental evidence for a selective interaction between an acetylated nonhistone protein and a bromodomain, leading to the generalization of a hypothesis previously formulated for the function of these domains.
ACKNOWLEDGMENTS
This work was supported by grants from the Association pour la Recherche sur le Cancer and from the Association Française contre les Myopathies to A.H.-B. and from the European 5th PCRDT (grant QLRT 1999-00866) to A.H.-B. A.P. was awarded a fellowship from the Federation of European Biochemical Societies (FEBS).
We thank L. Cabanie for the preparation of recombinant proteins, V. Ogryzko, K. Breitschopf, A. Ciechanover, W. Lee Kraus, and S. A. Leibovitch for the kind gift of reagents, and L. L. Pritchard for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali S, Ramirez S, Robin P, Trouche D, Harel-Bellan A. A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res. 1998;26:3869–3870. doi: 10.1093/nar/26.16.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Bayle J H, Crabtree G R. Protein acetylation: more than chromatin modification to regulate transcription. Chem Biol. 1997;4:885–888. doi: 10.1016/s1074-5521(97)90296-9. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 5a.Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–148. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 8.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;265:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 9.Davis R L, Weintraub H, Lassar A B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 10.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 12.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 14.Jackson V, Shires A, Chalkley R, Granner D K. Studies on highly metabolically active acetylation and phosphorylation of histones. J Biol Chem. 1975;250:4856–4863. [PubMed] [Google Scholar]
- 15.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-K, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 16.Kiernan R E, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang K T, Benkirane M, van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraus W L, Manning E T, Kadonaga J T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 19.Lassar A B, Davis R L, Wright W E, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerisation with E12–E47 like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Balbas M A, Bauer U M, Nielsen S J, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 22.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 23.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 24.Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem. 2000;275:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- 25.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez S, Ait-Si-Ali S, Robin P, Trouche D, Harel-Bellan A. The CREB-binding protein (CBP) cooperates with the serum response factor for transactivation of the c-fos serum response element. J Biol Chem. 1997;272:31016–31021. doi: 10.1074/jbc.272.49.31016. [DOI] [PubMed] [Google Scholar]
- 27.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J Y, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 29.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulated transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 30.Thayer M J, Weintraub H. A cellular factor stimulates the DNA-binding activity of MyoD and E47. Proc Natl Acad Sci USA. 1993;90:6483–6487. doi: 10.1073/pnas.90.14.6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita A, Towatari M, Tsuzuki S, Hayakawa F, Kosugi H, Tamai K, Miyazaki T, Kinoshita T, Saito H. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene. 2000;19:444–451. doi: 10.1038/sj.onc.1203329. [DOI] [PubMed] [Google Scholar]
- 32.Trouche D, Grigoriev M, Lenormand J L, Robin P, Leibovitch S A, Sassone-Corsi P, Harel-Bellan A. Repression of c-fos promoter by MyoD on muscle cell differentiation. Nature. 1993;363:79–82. doi: 10.1038/363079a0. [DOI] [PubMed] [Google Scholar]
- 33.Turner B M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 35.Webster G A, Perkins N D. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weintraub H, Dwarki V J, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott S J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5:1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 37.Winston F, Allis C D. The bromodomain: a chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe A P. Transcriptional activation. Switched-on chromatin. Curr Biol. 1994;4:525–528. doi: 10.1016/s0960-9822(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 40.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 41.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 42.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]