Abstract
Background
Genome-wide association studies (GWAS) have identified multiple common breast cancer susceptibility variants. Many of these variants have differential associations by estrogen receptor (ER) status, but how these variants relate with other tumor features and intrinsic molecular subtypes is unclear.
Methods
Among 106,571 invasive breast cancer cases and 95,762 controls of European ancestry with data on 173 breast cancer variants identified in previous GWAS, we used novel two-stage polytomous logistic regression models to evaluate variants in relation to multiple tumor features (ER, progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and grade) adjusting for each other, and to intrinsic-like subtypes.
Results
Eighty-five of 173 variants were associated with at least one tumor feature (false discovery rate < 5%), most commonly ER and grade, followed by PR and HER2. Models for intrinsic-like subtypes found nearly all of these variants (83 of 85) associated at p < 0.05 with risk for at least one luminal-like subtype, and approximately half (41 of 85) of the variants were associated with risk of at least one non-luminal subtype, including 32 variants associated with triple-negative (TN) disease. Ten variants were associated with risk of all subtypes in different magnitude. Five variants were associated with risk of luminal A-like and TN subtypes in opposite directions.
Conclusion
This report demonstrates a high level of complexity in the etiology heterogeneity of breast cancer susceptibility variants and can inform investigations of subtype-specific risk prediction.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13058-021-01484-x.
Keywords: Breast cancer, Etiologic heterogeneity, Genetic predisposition, Common breast cancer susceptibility variants
Introduction
Breast cancer represents a heterogenous group of diseases with different molecular and clinical features[1]. Clinical assessment of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and histological grade are routinely determined to inform treatment strategies and prognostication[2]. Combined, these tumor features define five intrinsic-like subtypes (i.e., luminal A-like, luminal B–like/HER2-negative, luminal B-like/HER2-positive, HER2-positive/non-luminal, and triple-negative) that are correlated with intrinsic subtypes defined by gene expression panels[2, 3]. Most known breast cancer risk or protective factors are related to luminal or hormone receptor (ER or PR) positive tumors, whereas less is known about the etiology of triple-negative (TN) tumors, an aggressive subtype[4, 5].
Breast cancer genome-wide association studies (GWAS) have identified over 170 common susceptibility variants, most of them single nucleotide polymorphisms (SNPs), of which many are differentially associated with ER-positive than ER-negative disease[6–8]. These include 20 variants that primarily predispose to ER-negative or TN disease[7, 8]. However, few studies have evaluated variant associations with other tumor features, or simultaneously studied multiple, correlated tumor markers to identify source(s) of etiologic heterogeneity[7, 9–13]. We recently developed a two-stage polytomous logistic regression method that efficiently characterizes etiologic heterogeneity while accounting for tumor marker correlations and missing tumor data[14, 15]. This method can help describe complex relationships between susceptibility variants and multiple tumor features, helping to clarify breast cancer subtype etiologies and increasing the power to generate more accurate risk estimates between susceptibility variants and less common subtypes. We recently demonstrated the power of this method in a GWAS to identify novel breast cancer susceptibility accounting for tumor heterogeneity[15].
In this report, we sought to expand our understanding of etiologic heterogeneity across breast cancer subtypes, by applying the two-stage polytomous logistic regression methodology to a large study population from the Breast Cancer Association Consortium (BCAC) for detailed characterization of risk associations with 173 breast cancer risk variants identified by GWAS[6, 7] by tumor subtypes defined by ER, PR, HER2 and tumor grade.
Methods
Study population and genotyping
The study population and genotyping are described in previous publications[6, 7] and in the Additional file 3: Methods. We included invasive cases and controls from 81 BCAC studies with genotyping data from two Illumina genome-wide custom arrays, the iCOGS and OncoArray (106,571 cases (OncoArray: 71,788; iCOGS: 34,783) and 95,762 controls (OncoArray: 58,134; iCOGS: 37,628); Additional file 1: Table S1). All subjects in the study population were female and of European ancestry, with European ancestry determined by ancestry informative GWAS markers as previously described [6]. We evaluated 173 breast cancer risk variants that were identified in or replicated by prior BCAC analyses to be associated with breast cancer risk at a p-value threshold p < 5.0 × 10–8 [6, 7]. Most of these variants (n = 153) were identified because of their association with risk of overall breast cancer, and a small number of variants (n = 20) were identified because of their association specific to ER-negative breast cancer (Additional file 1: Table S2). These 173 variants have not previously been simultaneously investigated for evidence of tumor heterogeneity with multiple tumor markers[6, 7, 15, 16]. Genotypes for the variants marking the 173 susceptibility loci were determined by genotyping with the iCOGS and the OncoArray arrays and imputation to the 1000 Genomes Project (Phase 3) reference panel.
Statistical analysis
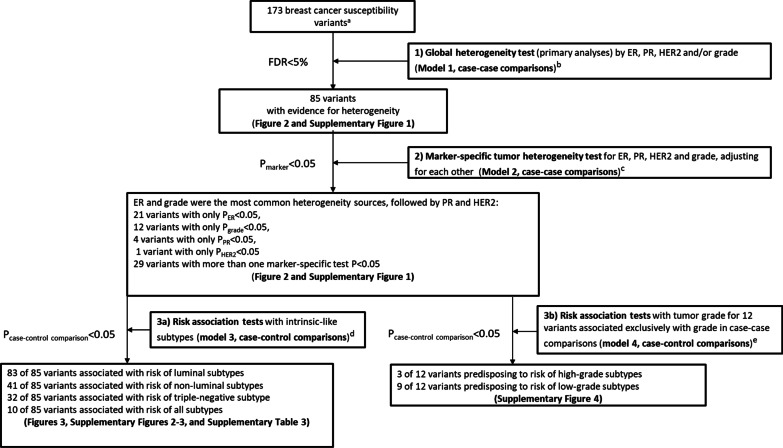
An overview of the analytic strategy is shown in Fig. 1 and a detailed discussion of the statistical methods, including the two-stage polytomous logistic regression, are provided in the Additional file 3: Methods and elsewhere[14, 15]. Briefly, we used two-stage polytomous regression models that allow modelling of genetic association of breast cancer accounting for underlying heterogeneity in associations by combinations of multiple tumor markers using a parsimonious decomposition of subtype-specific case–control odds-ratio parameters in terms of marker-specific case-case odd-ratio parameters[14, 15]. We introduced further parsimony by using the mixed-effect formulation of the model that allows ER-specific case-case parameters to be treated as fixed and similar parameters for other markers (PR, HER2 and grade (as an ordinal variable)) as random. We used an expectation–maximization (EM) algorithm[17] for parameter estimation under this model to account for missing data in tumor characteristics.
Fig. 1.
Overview of the analytic strategy and results from the investigation of 173 known breast cancer susceptibility variants for evidence of heterogeneity of effect according to the estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and grade. aWe evaluated 173 breast cancer risk variants identified in or replicated by prior BCAC GWAS [6, 7], see Methods and Additional file 3: Methods sections for more details. bModel 1 (primary analyses): Mixed-effect two-stage polytomous model (ER as fixed-effect, and PR, HER2 and grade as random-effects) for global heterogeneity tests (i.e. case-case comparisons from stage 2 of the two-stage model) between each individual risk variant and any of the tumor features (separate models were fit for each variant). cModel 2: Fixed-effect two-stage polytomous model for marker-specific tumor heterogeneity tests (i.e. case-case comparisons from stage 2 of the two-stage model) between each individual variant and each of the tumor features (ER, PR, HER2, and grade), mutually adjusted for each other (separate models were fit for each variant). dModel 3: Fixed effect two-stage polytomous model for risk associations with intrinsic-like subtypes (i.e. case–control comparisons from stage 1 of the two-stage model): luminal A-like, luminal B-like/HER2-negative, luminal B-like/HER2-positive, HER2-positive/non-luminal, and triple-negative. eModel 4: Fixed effect two-stage polytomous model for risk associations with tumor grade (i.e. case–control comparisons from stage 1 of the two-stage model) for the 12 variants associated at p < 0.05 only with grade in case-case comparisons (from model 2): grade 1, grade 2, and grade 3
Our primary aim was to identify which of 173 known breast cancer susceptibility variants showed heterogenous risk associations by ER-, PR- and HER2-status and tumor grade. This was tested using a global heterogeneity test by ER, PR, HER2 and/or grade, with a mixed-effect two-stage polytomous model (model 1), fitted separately for each variant. The global null hypothesis was that there was no difference in risk of breast cancer associated with the variant genotype across any of the tumor features being evaluated. We accounted for multiple testing (173 tests, one for each variant) of the global heterogeneity test using a false discovery rate (FDR) < 5% under the Benjamini–Hochberg procedure[18].
For the variants showing evidence of global heterogeneity after FDR adjustment, we further evaluated which of the tumor features contributed to the heterogeneity by fitting a fixed-effects two-stage model (model 2) that simultaneously tested for associations with each tumor feature (this model was fitted for each variant separately). We used a threshold of p < 0.05 for marker-specific tumor heterogeneity tests to describe which specific tumor marker(s) contributed to the observed heterogeneity, adjusting for the other tumor markers in the model. This p-value threshold was used only for descriptive purposes, as the primary hypotheses were tested using the FDR-adjusted global test for heterogeneity described above.
We conducted additional analyses to explore for evidence of heterogeneity. We fitted a fixed-effect two-stage model (model 3) to estimate case–control odd ratios (ORs) and 95% confidence intervals (CI) between the variants and five intrinsic-like subtypes defined by combinations of ER, PR, HER2 and grade: (1) luminal A-like (ER + and/or PR + , HER2-, grade 1 or 2); (2) luminal B-like/HER2-negative (ER + and/or PR + , HER2-, grade 3); (3) luminal B-like/HER2-positive (ER + and/or PR + , HER2 +); (4) HER2-positive/non-luminal (ER- and PR-, HER2 +), and (5) TN (ER-, PR-, HER2-). We also fitted a fixed-effect two-stage model to estimate case–control ORs and 95% confidence intervals (CI) with tumor grade (model 4; defined ordinally as grade 1, grade 2, and grade 3) for the variants associated at p < 0.05 only with grade in case-case comparisons from model 2.
To help describe sources of heterogeneity from different tumor characteristics in models 2 and 3, we performed cluster analyses based on Euclidean distance calculated from the absolute z-statistics that were estimated by the individual marker-specific tumor heterogeneity tests (model 2) and the case–control associations with risk of intrinsic-like subtypes (model 3). The clusters were used only for presentation purposes and were not intended to suggest strictly defined categories, nor are they intended to suggest the variants are associated with tumor markers through similar biological mechanisms. Clustering was performed in R using the function Heatmap as implemented by the package “Complex Heatmap” version 3.1[19]. Additional details for calculating Euclidean distance using absolute z-statistics are provided in Additional file 3: Methods.
We performed sensitivity analyses, in which we estimated the ORs and 95% CI between the variants and the intrinsic-like subtypes by implementing a standard polytomous model that defined the intrinsic-like subtypes using only the available tumor markers data (not using the EM algorithm to account for missing data in tumor markers). We analyzed OncoArray and iCOGS array data separately for all analyses, adjusting for the first ten principal components for ancestry-informative variants, and then meta-analyzed the results.
Results
The mean (SD) ages at diagnosis (cases) and enrollment (controls) were 56.6 (12.2) and 56.4 (12.2) years, respectively. Among cases with information on the corresponding tumor marker, 81% were ER-positive, 68% PR-positive, 83% HER2-negative and 69% grade 1 or 2 (Table 1; see Additional file 1: Table S1 for details by study). Additional file 1: Table S3 shows the correlation between the tumor markers. ER was positively correlated with PR (r = 0.61) and inversely correlated with HER2 (r = -0.16) and grade (r = -0.39). The most common intrinsic-like subtype was luminal A-like (54%), followed by TN (14%), luminal B-like/HER2-negative (13%), Luminal B-like/HER2-positive (13%) and HER2-positive/non-luminal (6%; Table 1). These frequencies varied across BCAC studies because the studies were diverse in both design and country of origin (Additional file 1: Table S1). Notably, there is little population-based data on the frequencies of intrinsic-like subtypes [20, 21]. The overall frequencies in our study population are generally similar to those reported by SEER for non-Hispanic white females and the Scottish cancer registry [20, 21]; however, given the diverse sources of our data, they are not directly comparable to country-specific cancer registries.
Table 1.
Distribution of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and grade and the intrinsic-like subtypes for cases of invasive breast cancer in studies from the Breast Cancer Consortium Association
| Tumor marker | N (%) |
|---|---|
| ER | |
| Negative | 16,900 (19%) |
| Positive | 70,030 (81%) |
| Unknown | 19,641 |
| PR | |
| Negative | 24,283 (32%) |
| Positive | 51,603 (68%) |
| Unknown | 30,685 |
| HER2 | |
| Negative | 47,693 (83%) |
| Positive | 9,529 (17%) |
| Unknown | 49,349 |
| Grade | |
| 1 | 15,583 (20%) |
| 2 | 37,568 (49%) |
| 3 | 24,382 (31%) |
| Unknown | 29,038 |
| Intrinsic-like subtypes | |
| Luminal A-like | 27,510 (54%) |
| Luminal B-like/HER2-negative | 6,804 (13%) |
| Luminal B-like/HER2-positive | 6,511 (13%) |
| HER2-positive/non-luminal | 2,797 (6%) |
| Triple-negative | 7,178 (14%) |
| Unknown | 55,771 |
Luminal A-like (ER + and/or PR + , HER2-, grade 1 & 2); Luminal B-like/HER2-negative (ER + and/or PR + , HER2-, grade 3); Luminal B-like/HER2-positive (ER + and/or PR + , HER2 +); HER2-positive/non-luminal (ER- and PR-, HER2 +), and triple-negative (ER-, PR-, HER2-)
Figure 1 shows an overview of the analytic strategy and results from three main analyses performed separately for each variant: 1) global test for heterogeneity by all tumor markers (model 1; primary hypothesis), 2) marker-specific tumor test for heterogeneity for each marker, adjusting for the others (model 2), and 3) estimation of case–control ORs (95%CIs) by intrinsic-like subtypes (model 3) and by grade (model 4).
Global test for heterogeneity by tumor markers (primary hypothesis)
Mixed-effects two-stage models (model 1) were fitted for each of the 173 variants separately and included terms for ER, PR, HER2 and grade to test for global heterogeneity by any of the tumor features (case-case comparison). This model identified 85 of 173 (49.1%) variants with evidence of heterogeneity by at least one tumor feature (FDR < 5%; Figs. 1, 2; Additional file 1: Fig. S1).
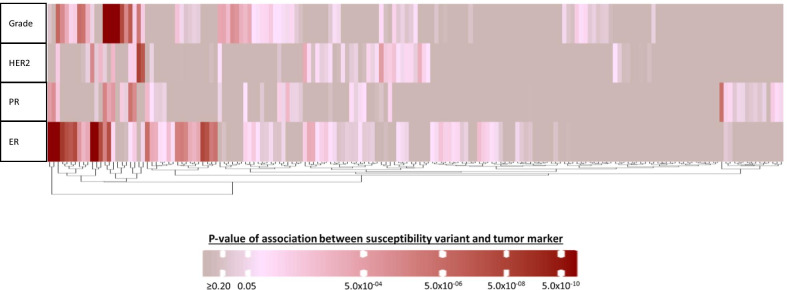
Fig. 2.
Heatmap of the z-values from the fixed-effects two-stage polytomous model for marker-specific heterogeneity tests (case-case comparison from model 2) for the association between each of the 173 breast cancer susceptibility variants and estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) or grade, adjusting for principal components and each tumor marker. Columns represent individual variants. For more detailed information on the context of the figure, see Additional file 1: Fig. S1
Marker-specific tumor test for heterogeneity for each marker, adjusting for other markers
Fixed-effects two-stage models (model 2) were used to test which of the correlated tumor markers was responsible for the observed global heterogeneity (case-case comparison). Figure 2 and Additional file 1: Fig. S1 show results of these analyses clustered by case-case z-values of associations between susceptibility variants and each tumor marker for the 173 variants. For the 85 variants with observed global heterogeneity, these analyses identified ER and grade as the two features that most often contributed to the observed heterogeneity (45 and 33 variants had marker-specific p < 0.05 for ER and grade, respectively), and 29 variants were associated with more than one tumor feature (Figs. 1, 2, Additional file 1: Fig. S1). Eighteen of these 85 variants showed no associations with any individual tumor marker at p < 0.05 (Fig. 2, Additional file 1: Fig. S1). Twenty-one variants were associated at p < 0.05 only with ER, 12 variants only with grade, four variants only with PR and one variant only with HER2 (Fig. 2, Additional file 1: Fig. S1, see footnotes).
Estimation of case–control ORs (95%CIs) by intrinsic-like subtypes (model 3)
Fixed-effects two-stage models for intrinsic-like subtypes (model 3) were fitted for each of the 85 variants with evidence of global heterogeneity to estimate ORs (95% CIs) for risk associations with each subtype (case–control comparisons). Additional file 1: Fig. S2 shows a summary of these analyses for the 85 variants, clustered by case–control z-value of association between susceptibility variants and breast cancer intrinsic-like subtypes, and Additional file 2: Fig. S3 shows forest plots for associations with risk by tumor subtypes. Nearly all (83 of 85) variants were associated with risk (p < 0.05) for at least one luminal-like subtype, and approximately half (41 of 85) of the variants were associated with risk of at least one non-luminal subtype, including 32 variants that were associated with risk of TN disease (Fig. 1, Additional file 1: Fig. S2 footnote ‘h’). Ten variants were associated with risk of all subtypes (Fig. 1, Additional file 1: Fig. S2 footnote ‘j’). Below we describe examples of groups of variants associated with different patterns of associations with intrinsic subtypes (Fig. 3 a-d).
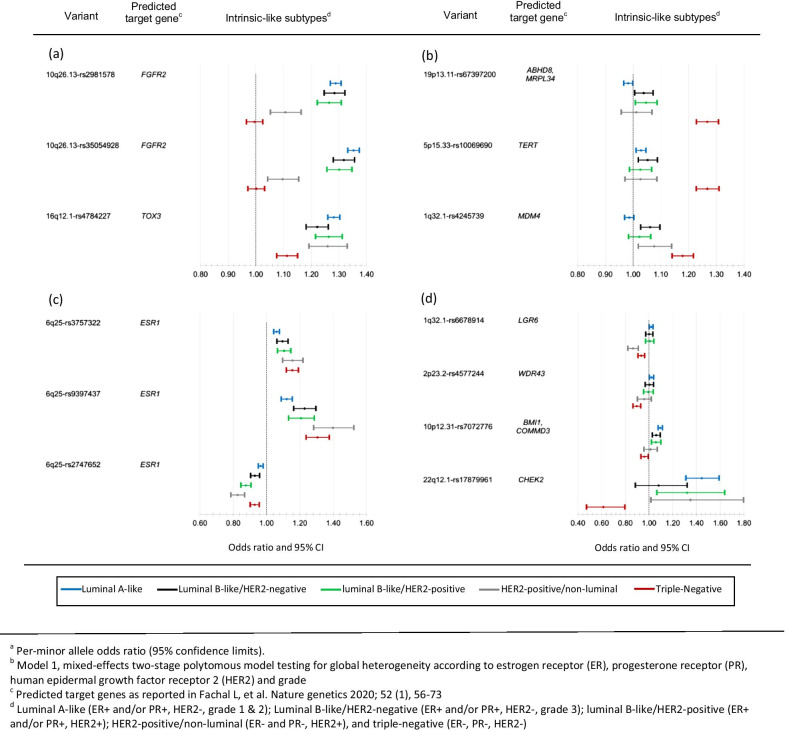
Fig. 3.
Results from fixed-effects two-stage polytomous models for risk associationsa with intrinsic-like subtypes (model 3) for variants with evidence of heterogeneity by tumor markers in the two-stage model (model1)b; panels show examples of variants (a) most strongly associated with luminal-like subtypes, (b) most strongly associated with TN subtypes, (c) associated with all subtypes with varying strengths of association, and (d) associated with luminal A-like and TN subtypes in different directions. See Additional file 1: Fig. S2 for more details
Two variants in linkage disequilibrium (LD, r2 = 0.73) at 10q26.13 (rs2981578 and rs35054928) and 16q12.1-rs4784227 had the strongest evidence of association with risk of luminal-like subtypes (Fig. 3a, Additional file 1: Fig. S2). The two variants at 10q26.13 showed no evidence of associations with TN subtypes, and a weaker association with HER2-positive/non-luminal subtype. In contrast, 16q12.1-rs4784227 was strongly associated with risk of all luminal-like subtypes and, weaker so, with risk of HER2-positive/non-luminal and TN subtypes (Figs. 3a, Additional file 1: Fig. S2).
Three variants 19p13.11-rs67397200, 5p15.33-rs10069690 and 1q32.11-rs4245739 showed the strongest evidence of associations with risk of TN disease. All three of these variants showed weaker or no evidence of associations with risk of the other subtypes (Fig. 3b, Additional file 1: Fig. S2).
Two variants in low LD (r2 = 0.17) at 6q25, rs9397437 and rs3757322, and a third variant in 6q25, rs2747652, which was not in LD (r2 < 0.01) with rs9397437 or rs3757322, showed strong evidence of being associated with risk of all subtypes. rs9397437 and rs3757322 were most strongly associated with risk of TN disease. rs2747652 was most strongly associated with risk of HER2-positive subtypes (Figs. 3c, Additional file 1: Fig. S2).
Five variants were associated with risk of luminal A-like disease in an opposite direction to their association with risk of TN disease. 1q32.1-rs6678914, 2p23.2-rs4577244, and 19p13.11-rs67397200 had weaker evidence of associations with risk of luminal A-like disease compared to associations with risk of TN disease, and 10p12.31-rs7072776 and 22q12.1-rs17879961 (I157T) had stronger evidence of an association with risk of luminal A-like disease compared to their association with risk of TN disease (Fig. 3d, Additional file 1: Fig. S2, for rs67397200 see Fig. 3b).
Estimation of case–control ORs (95%CIs) by tumor grade (model 4)
Case–control associations by tumor grade for the 12 variants that were observed associated at p < 0.05 only with grade in case-case comparisons are shown in Additional file 2: Fig. S4. 13q13.1-rs11571833, 1p22.3-rs17426269 and 11q24.3-rs11820646 showed stronger evidence for predisposing to risk of high-grade subtypes, and the remaining variants showed stronger evidence for predisposing to risk of low-grade subtypes.
When limiting analyses to cases with intrinsic-like subtypes defined only by available tumor marker data, results from case–control analyses were similar, but less precise than results from the two-stage polytomous regression model using the EM algorithm to account for missing tumor marker data (Additional file 1: Table S4).
Discussion
This study demonstrates the extent and complexity of genetic etiologic heterogeneity among 173 breast cancer risk variants by multiple tumor characteristics, using novel methodology in the largest and the most comprehensive investigation conducted to date. We found compelling evidence that about half of the investigated breast cancer susceptibility loci (85 of 173 variants) predispose to tumors with different characteristics. We identified tumor grade, along with confirming ER status, as important determinants of etiologic heterogeneity. Associations with individual tumor features translated into differential associations with the risk of intrinsic-like subtypes defined by their combinations.
Many of the variants with evidence of global heterogeneity predisposed to risk of multiple subtypes, but with different magnitudes. For example, three variants identified in early GWAS for overall breast cancer, FGFR2 (rs35054928 and rs2981578)[22, 23] and 8q24.21 (rs13281615)[22], were associated with luminal-like and HER2-positive/non-luminal subtypes, but not with TN disease. rs4784227 located near TOX3[22, 24] and rs62355902 located in a MAP3K1[22] regulatory element, were associated with risk of all five subtypes. Of the five variants found associated in opposite directions with luminal A-like and TN disease, we previously reported rs6678914 and rs4577244 to have opposite effects between ER-negative and ER-positive tumors[7]. rs17879961 (I157T), a likely causal[16] missense variant located in a CHEK2 functional domain that reduces or abolishes substrate binding[25], was previously reported to have opposite directions of effects on lung adenocarcinoma and lung squamous cell carcinoma and for lung cancer between smokers and non-smokers[26, 27]. Moreover, the risk association of rs17879961 has been reported to vary across tissue locations/cell-types, as this variant has been associated with a higher risk of pancreatic ductal adenocarcinoma [28], chronic lymphocytic leukemia [29], and colorectal cancer [30], and also associated with a lower risk of aerodigestive squamous cell carcinoma [31] and ovarian cancer [32]. To our knowledge, rs67397200 and rs7072776 have not previously been shown to be associated with subtypes in opposite directions. In a prior breast cancer GWAS that applied the two-stage polytomous model for risk variant discovery, we also identified five variants associated with risk of luminal A-like and TN disease in opposite directions [15]. Overall, these findings suggest that the same biological pathway has opposite effects on the susceptibility to different tumor types. This interpretation is supported by functional characterization of rs36115365, a variant on 5p15.33, which was found to have similar cis-regulatory effects on TERT in multiple cancers cell lines from different cancers, but was associated with a higher risk of pancreatic and testicular cancer and a lower risk of lung cancer [33]. Alternatively, a causal variant may differently influence cis-gene regulation and/or alter different biological pathways depending on the cell or tissue of origin [34]. Further studies of these variants are required to clarify the biological mechanisms for these apparent cross-over effects.
In prior ER-negative GWAS, we identified 20 variants that predispose to ER-negative disease, of which five variants were only or most strongly associated with risk of TN disease (rs4245739, rs10069690, rs74911261, rs11374964, and rs67397200)[7, 8]. We confirmed these five variants to be most strongly associated with TN disease. The remaining previously identified 15 variants all showed associations with risk of non-luminal subtypes, especially TN disease, and for all but four variants (rs17350191, rs200648189, rs6569648, and rs322144), evidence of global heterogeneity was observed.
Little is known regarding PR and HER2 as sources of etiologic heterogeneity independent of ER status. Of the four variants that showed evidence of heterogeneity only according to PR, rs10759243[6, 35], rs11199914[36] and rs72749841[6] were previously found primarily associated with risk of ER-positive disease, and rs10816625 was found to be associated with risk of ER-positive/PR-positive tumors, but not other ER/PR combinations[12]. rs10995201 was the only variant found in case-case comparisons to be solely associated with HER2 status, although the evidence was not strong, requiring further confirmation. Previously, rs10995201 showed no evidence of being associated with ER status[37]. Most variants associated with PR or HER2, had not been investigated for PR or HER2 heterogeneity while adjusting for ER[9–13]. We previously reported rs10941679 to be associated with PR-status, independent of ER, and also with grade[10]. We also found suggestive evidence of PR-specific heterogeneity for 16q12-rs3803662[13], which is in high LD (r2 = 0.78) with rs4784227 (TOX3), a variant strongly associated with PR status. Our findings for rs2747652 are also consistent with a prior BCAC fine-mapping analysis across the ESR1 locus, which found rs2747652 to be associated with risk of the HER2-positive/non-luminal subtype and high grade independent of ER[9]. rs2747652 overlaps an enhancer region and is associated with reduced ESR1 and CCDC170 expression[9].
Histologic grade is a composite of multiple tumor characteristics, including mitotic count, nuclear pleomorphism, and degree of tubule or gland formation, therefore susceptibility variants associated with tumor grade could affect multiple biological pathways [38]. Evidence from comparisons of tumor morphology and genomic and molecular alterations suggest that tumor grade is likely a ‘stable’ tumor feature and does not progress from low- to high-grade [39–42], thus the variants associated with grade are likely not associated with grade progression. Among the 12 variants identified with evidence of heterogeneity by grade only, rs17426269, rs11820646, and rs11571833 were most strongly associated with risk of grade 3 disease. rs11571833 lies in the BRCA2 coding region and produces a truncated form of the protein[43] and has been shown to be associated with both risk of TN disease and risk of serous ovarian tumors, both of which tend to be high-grade[44]. To our knowledge, rs17426269 and rs11820646 have not been investigated in relation to grade heterogeneity. The remaining nine variants were all more strongly associated with grade 1 or grade 2 disease. Six of these variants were previously reported to be associated primarily with ER-positive disease[6, 36, 45, 46], highlighting the importance of accounting for multiple tumor characteristics to better illuminate heterogeneity sources.
We identified 18 variants with evidence of global heterogeneity (FDR < 5%), but no significant (marker-specific p < 0.05) associations with any of the individual tumor characteristic(s). This is likely explained by the fact that the test for association with specific tumor markers using fixed-effects models is less powerful than mixed-effects models used to test the primary hypothesis of global heterogeneity by any tumor marker[14].
To help describe and visualize the strength of the evidence for common heterogeneity patterns, we performed clustered analyses of z-values for tumor marker-specific heterogeneity tests and case–control associations with risk of intrinsic-like subtypes. Because they are based on z-values, these clusters reflect differences in sample size and statistical power to detect associations between variants and specific tumor subtypes. Thus, clusters should not be interpreted as strictly defined categories.
A major strength of our study is our large sample size of over 100,000 breast cancer cases with tumor marker information, and a similar number of controls, making this the largest, most comprehensive breast cancer heterogeneity investigation. Our application of the two-stage polytomous logistic regression enabled adjusting for multiple, correlated tumor markers and accounting for missing tumor marker data. This is a more powerful and efficient modeling strategy for identifying heterogeneity sources among highly correlated tumor markers, compared with standard polytomous logistic regression[14, 15]. In simulated and real data analyses, we have demonstrated that in the presence of heterogenous associations across subtypes, the two-stage model is more powerful than polytomous logistic regression for detecting heterogeneity. Moreover, we have demonstrated that in the presence of correlated markers, the two-stage model, incorporating all markers simultaneously, has a much better ability to distinguish the true source(s) of heterogeneity than testing for heterogeneity by analyzing one marker at a time[14, 15]. In prior analyses, we showed that the two-stage polytomous regression is a powerful approach to identify susceptibility variants that display tumor heterogeneity[15]. Notably, in this prior investigation we excluded the genomic regions in which the 173 variants that were investigated in this work are located[15].
Our study also has some limitations. First, many breast cancer cases from studies included in this report had missing information on one or more tumor characteristics. ER tumor status data was available for 81% of cases, but missing data for the other tumor markers ranged from 27 to 46%. To address this limitation, we implemented an EM algorithm that allowed a powerful analysis to incorporate cases with missing tumor characteristics under the assumption that tumor characteristics are missing at random (MAR), i.e., the underlying reason for missing data may depend on observed tumor markers or/and covariate values, but not on the missing values themselves[47]. If this assumption is violated it can lead to an inflated type-one error[14]. However, in the context of genetic association testing, the missingness mechanism would also need to be related to the genetic variants under study, which is unlikely. The 88 variants that did not meet the p-value threshold for significant heterogeneity in the global test, are likely to represent a combination of variants that are associated with risk of all investigated tumor subtypes with similar effects and variants for which we lacked power to detect evidence of global heterogeneity due to weak effect sizes or uncommon allele frequencies. In addition, our study focused on investigating ER, PR, HER2, and grade as heterogeneity sources; future studies with more detailed tumor characterization could reveal additional etiologic heterogeneity sources.
Conclusion
Our findings provide insights into the complex etiologic heterogeneity patterns of common breast cancer susceptibility loci. These findings may inform future studies, such as fine-mapping and functional analyses to identify the underlying causal variants, clarifying biological mechanisms that drive genetic predisposition to breast cancer subtypes. Moreover, these analyses provide precise relative risk estimates for different intrinsic-like subtypes that could improve the discriminatory accuracy of subtype-specific polygenic risk scores [48].
Supplementary Information
Additional file 1. Figures S1 and S2 and Table S1-S4. This file contains supplementary figures 1-2 and supplementary tables 1-4.
Additional file 2. Figures S3 and S4. This file contains supplementary figures S3 and S4.
Additional file 3. Methods. This file contains the supplementary methods.
Additional file 4. Funding and Acknowledgement. This file contains the additional funding not included in the main text, the acknowledgments, and the names of the people in the collaboration groups.
Acknowledgements
A full description of the acknowledgments is provided in the Additional file 4: Funding and Acknowledgement. NBCS Collaborators: greal@rr-research.no. ABCTB Investigators: mythily.sachchithananthan@sydney.edu.au. kConFab/AOCS Investigators: heather.thorne@petermac.org
Abbreviations
- GWAS
Genome-wide association studies
- ER
Estrogen receptor
- PR
Progesterone receptor
- HER2
Human epidermal growth factor receptor 2
- SNPs
Single nucleotide polymorphisms
- FDR
False discovery rate
- TN
Triple-negative
- BCAC
Breast Cancer Association Consortium
- EM
Expectation–maximization
- OR
Odd ratios
- 95% CI
95% Confidence interval
- LD
Linkage disequilibrium
Authors' contributions
Writing group: TUA, HZ, KMI, RLM, FJC, JSi, PKr, DFE, PDPP, MKS, MG-C, NCh; Statistical analysis: HZ, TUA, MG-C, NCh; Provision of DNA samples and/or phenotypic data: KMi, RLM, MKB, JDen, AMD, MLus, QW, ILA, HA-C, VA, KJA, PLA, AAu, AB, HBec, SBe, JBen, MBerm, CBl, SEB, BBon, A-LB-BD, HBra, HBre, AB-W, TB, BBur, SSB, FC, JEC, JC-C, SJC, GC-T, CLC, NBCS, JMC, ACox, SSC, KCz, MBD, PD, TD, MDw, DME, DGE, PAF, JFi, GFl, MG-D, SMG, JAG-S, MMG, GGG, MSG, AG–N, GIG, MGrip, PGu, CAH, PHall, UH, EFH, BAMH-G, BHo, AHol, MJH, RNH, JLHo, AHow, ABCTB, kConFab/AOCS, MJa, AJak, EMJ, MEJ, AJu, RKa, SKaup, RKe, EKh, CMKi, Y-DK, SKou, VNK, UK, KK-S, AWK, KKy, DLa, DGL, ALin, MLin, JLis, AL, W-LL, RJM, AMan, MMan, SMar, MEM, CMc, AMe, UMe, HNe, WGN, JNo, KOf, HO, NO, T-WP-S, AVP, JPet, GPi, DPK, RP, KPu, KPy, PRa, GR, ARo, TRü, ES, SS, DPS, EJS, RKS, MJS, BSch, MES, X-OS, SSm, MCS, JJS, AJS, RMT, WJT, JAT, LRT, MBT, DT, MAT, CMV, CHMVD, EMVV, PWa, CRW, CWe, JWe, RWi, AW, XRY, WZ, FJC, JSi, PKr, DFE, PDPP, MKS, MG-C. All authors read and approved the final version of the manuscript.
Funding
Open Access funding provided by the National Institutes of Health (NIH) This project has been funded in part with Federal funds from the National Cancer Institute Intramural Research Program, National Institutes of Health. Dr. Nilanjan Chatterjee was supported by NHGRI (1R01 HG010480-01). Dr. Haoyu Zhang was supported by National Cancer Institute (1K99 CA256513). OncoArray genotyping was funded by the government of Canada through Genome Canada and the Canadian Institutes of Health Research (GPH-129344), the Ministère de l'Économie, de la Science et de l'Innovation du Québec through Génome Québec, the Quebec Breast Cancer Foundation for the PERSPECTIVE project, the US National Institutes of Health (NIH) (1 U19 CA 148065 for the Discovery, Biology and Risk of Inherited Variants in Breast Cancer (DRIVE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract HHSN268201200008I), Cancer Research UK (C1287/A16563), the Odense University Hospital Research Foundation (Denmark), the National R&D Program for Cancer Control–Ministry of Health and Welfare (Republic of Korea) (1420190), the Italian Association for Cancer Research (AIRC; IG16933), the Breast Cancer Research Foundation, the National Health and Medical Research Council (Australia) and German Cancer Aid (110837). iCOGS genotyping was funded by the European Union (HEALTH-F2-2009–223175), Cancer Research UK (C1287/A10710, C1287/A10118 and C12292/A11174]), NIH grants (CA128978, CA116167 and CA176785) and the Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 (GAME-ON initiative)), an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, the Ministère de l'Économie, Innovation et Exportation du Québec (PSR-SIIRI-701), the Komen Foundation for the Cure, the Breast Cancer Research Foundation and the Ovarian Cancer Research Fund. A full description of the funding is provided in the Additional file 4: Funding and Acknowledgement.
Availability of data and materials
The datasets generated and/or analyzed during the current study are part of the Breast Cancer Association Consortium and would be available with the appropriate permissions, including an application process and appropriate data transfer agreements.
Declarations
Ethics approval and consent to participate
All the studies included in these analyses were approved by local IRBs.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Thomas U. Ahearn, Haoyu Zhang, Montserrat García-Closas, and Nilanjan Chatterjee have contributed equally to this work
Contributor Information
Montserrat García-Closas, Email: montserrat.garcia-closas@nih.gov.
NBCS Collaborators:
Kristine K. Sahlberg, Lars Ottestad, Rolf Kåresen, Ellen Schlichting, Marit Muri Holmen, Toril Sauer, Vilde Haakensen, Olav Engebråten, Bjørn Naume, Alexander Fosså, Cecile E. Kiserud, Kristin V. Reinertsen, Åslaug Helland, Margit Riis, and Jürgen Geisler
ABCTB Investigators:
Christine Clarke, Rosemary Balleine, Robert Baxter, Stephen Braye, Jane Carpenter, Jane Dahlstrom, John Forbes, CSoon Lee, Deborah Marsh, Adrienne Morey, Nirmala Pathmanathan, Rodney Scott, Peter Simpson, Allan Spigelman, Nicholas Wilcken, Desmond Yip, and Nikolajs Zeps
kConFab/AOCS Investigators:
Stephen Fox, Ian Campbell, David Bowtell, Georgia Chenevix-Trench, Amanda Spurdle, Penny Webb, Anna de Fazio, Margaret Tassell, Judy Kirk, Geoff Lindeman, Melanie Price, Melissa Southey, Roger Milne, and Sid Deb
References
- 1.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curigliano G, Burstein HJ, Gnant M, Dubsky P, Loibl S, Colleoni M, Regan MM, Piccart-Gebhart M, Senn HJ et al: De-escalating and escalating treatments for early-stage breast cancer: the St Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol 2017, 28(8):1700–1712. [DOI] [PMC free article] [PubMed]
- 3.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ. Panel m: personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard ME, Boeke CE, Tamimi RM. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim Biophys Acta. 2015;1856(1):73–85. doi: 10.1016/j.bbcan.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103(3):250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindstrom S, Hui S, Lemacon A, Soucy P, Dennis J, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet. 2017;49(12):1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45(4):392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, Healey CS, Kar S, Pooley KA, Lopez-Knowles E, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. 2016;48(4):374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne RL, Goode EL, Garcia-Closas M, Couch FJ, Severi G, Hein R, Fredericksen Z, Malats N, Zamora MP, Arias Perez JI, et al. Confirmation of 5p12 as a susceptibility locus for progesterone-receptor-positive, lower grade breast cancer. Cancer Epidemiol Biomark Prevent. 2011;20(10):2222–2231. doi: 10.1158/1055-9965.EPI-11-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa JD, Garcia-Closas M, Humphreys M, Platte R, Hopper JL, Southey MC, Apicella C, Hammet F, Schmidt MK, Broeks A, et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20(23):4693–4706. doi: 10.1093/hmg/ddr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orr N, Dudbridge F, Dryden N, Maguire S, Novo D, Perrakis E, Johnson N, Ghoussaini M, Hopper JL, Southey MC, et al. Fine-mapping identifies two additional breast cancer susceptibility loci at 9q312. Hum Mol Genet. 2015;24(10):2966–2984. doi: 10.1093/hmg/ddv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20(16):3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Zhao N, Ahearn TU, Wheeler W, García-Closas M, Chatterjee N: A mixed-model approach for powerful testing of genetic associations with cancer risk incorporating tumor characteristics. Biostatistics 2020. [DOI] [PMC free article] [PubMed]
- 15.Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G, Jiang X, O'Mara TA, Zhao N, Bolla MK, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet. 2020;52(6):572–581. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fachal L, Aschard H, Beesley J, Barnes DR, Allen J, Kar S, Pooley KA, Dennis J, Michailidou K, Turman C, et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat Genet. 2020;52(1):56–73. doi: 10.1038/s41588-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via em algorithm. J Roy Stat Soc B Met. 1977;39(1):1–38. [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 19.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 20.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 21.Mesa-Eguiagaray I, Wild SH, Rosenberg PS, Bird SM, Brewster DH, Hall PS, Cameron DA, Morrison D, Figueroa JD. Distinct temporal trends in breast cancer incidence from 1997 to 2016 by molecular subtypes: a population-based study of Scottish cancer registry data. Br J Cancer. 2020;123(5):852–859. doi: 10.1038/s41416-020-0938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9(5):1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 26.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, Caporaso NE, Johansson M, Xiao X, Li Y, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet. 2017;49(7):1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, Broderick P, Zong X, Laplana M, Wei Y, Han Y, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obazee O, Archibugi L, Andriulli A, Soucek P, Malecka-Panas E, Ivanauskas A, Johnson T, Gazouli M, Pausch T, Lawlor RT, et al. Germline BRCA2 K3326X and CHEK2 I157T mutations increase risk for sporadic pancreatic ductal adenocarcinoma. Int J Cancer. 2019;145(3):686–693. doi: 10.1002/ijc.32127. [DOI] [PubMed] [Google Scholar]
- 29.Rudd MF, Sellick GS, Webb EL, Catovsky D, Houlston RS. Variants in the ATM-BRCA2-CHEK2 axis predispose to chronic lymphocytic leukemia. Blood. 2006;108(2):638–644. doi: 10.1182/blood-2005-12-5022. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Wang QS, Wang YJ. The CHEK2 I157T variant and colorectal cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2012;13(5):2051–2055. doi: 10.7314/apjcp.2012.13.5.2051. [DOI] [PubMed] [Google Scholar]
- 31.Lesseur C, Ferreiro-Iglesias A, McKay JD, Bosse Y, Johansson M, Gaborieau V, Landi MT, Christiani DC, Caporaso NC, Bojesen SE et al: Genome-wide association meta-analysis identifies pleiotropic risk loci for aerodigestive squamous cell cancers. PLoS genetics 2021, 17(3):e1009254. [DOI] [PMC free article] [PubMed]
- 32.Ovarian Cancer Association Consortium - Results lookup by region [http://ocac.ccge.medschl.cam.ac.uk/data-projects/results-lookup-by-region/]
- 33.Fang J, Jia J, Makowski M, Xu M, Wang Z, Zhang T, Hoskins JW, Choi J, Han Y, Zhang M, et al. Functional characterization of a multi-cancer risk locus on chr5p1533 reveals regulation of TERT by ZNF148. Nat Commun. 2017;8(1):15034. doi: 10.1038/ncomms15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim-Hellmuth S, Aguet F, Oliva M, Munoz-Aguirre M, Kasela S, Wucher V, Castel SE, Hamel AR, Vinuela A, Roberts AL et al: Cell type-specific genetic regulation of gene expression across human tissues. Science 2020, 369(6509). [DOI] [PMC free article] [PubMed]
- 35.Li X, Zou W, Liu M, Cao W, Jiang Y, An G, Wang Y, Huang S, Zhao X. Association of multiple genetic variants with breast cancer susceptibility in the Han Chinese population. Oncotarget. 2016;7(51):85483–85491. doi: 10.18632/oncotarget.13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darabi H, McCue K, Beesley J, Michailidou K, Nord S, Kar S, Humphreys K, Thompson D, Ghoussaini M, Bolla MK, et al. Polymorphisms in a putative enhancer at the 10q21.2 breast cancer risk locus regulate NRBF2 expression. Am J Hum Genet. 2015;97(1):22–34. doi: 10.1016/j.ajhg.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 39.Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223(2):307–317. doi: 10.1002/path.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schymik B, Buerger H, Kramer A, Voss U, van der Groep P, Meinerz W, van Diest PJ, Korsching E. Is there 'progression through grade' in ductal invasive breast cancer? Breast Cancer Res Treat. 2012;135(3):693–703. doi: 10.1007/s10549-012-2195-1. [DOI] [PubMed] [Google Scholar]
- 41.Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Can Res. 1999;59(7):1433–1436. [PubMed] [Google Scholar]
- 42.Rajakariar R, Walker RA. Pathological and biological features of mammographically detected invasive breast carcinomas. Br J Cancer. 1995;71(1):150–154. doi: 10.1038/bjc.1995.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazoyer S, Dunning AM, Serova O, Dearden J, Puget N, Healey CS, Gayther SA, Mangion J, Stratton MR, Lynch HT, et al. A polymorphic stop codon in BRCA2. Nat Genet. 1996;14(3):253–254. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- 44.Meeks HD, Song H, Michailidou K, Bolla MK, Dennis J, Wang Q, Barrowdale D, Frost D, McGuffog L, Ellis S et al, BRCA2 Polymorphic Stop Codon K3326X and the Risk of Breast, Prostate, and Ovarian Cancers. J Natl Cancer Inst 2016, 108(2). [DOI] [PMC free article] [PubMed]
- 45.Darabi H, Beesley J, Droit A, Kar S, Nord S, Moradi Marjaneh M, Soucy P, Michailidou K, Ghoussaini M, Fues Wahl H, et al. Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene-Environment Study (COGs) Sci Rep. 2016;6:32512. doi: 10.1038/srep32512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Little RJA, Rubin DB: Statistical analysis with missing data. In: Wiley series in probability and statistics. Third edition edn. Hoboken, NJ: Wiley,; 2019: 1 online resource.
- 48.Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet. 2019;104(1):21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figures S1 and S2 and Table S1-S4. This file contains supplementary figures 1-2 and supplementary tables 1-4.
Additional file 2. Figures S3 and S4. This file contains supplementary figures S3 and S4.
Additional file 3. Methods. This file contains the supplementary methods.
Additional file 4. Funding and Acknowledgement. This file contains the additional funding not included in the main text, the acknowledgments, and the names of the people in the collaboration groups.
Data Availability Statement
The datasets generated and/or analyzed during the current study are part of the Breast Cancer Association Consortium and would be available with the appropriate permissions, including an application process and appropriate data transfer agreements.